Abstract
Objective
Infection with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) can lead to severe pneumonia, but also thrombotic complications and non‐pulmonary organ failure. Recent studies suggest intravascular neutrophil activation and subsequent immune cell–triggered immunothrombosis as a central pathomechanism linking the heterogenous clinical picture of coronavirus disease 2019 (COVID‐19). We sought to study whether immunothrombosis is a pathognomonic factor in COVID‐19 or a general feature of (viral) pneumonia, as well as to better understand its upstream regulation.
Approach and results
By comparing histopathological specimens of SARS‐CoV‐2 with influenza‐affected lungs, we show that vascular neutrophil recruitment, NETosis, and subsequent immunothrombosis are typical features of severe COVID‐19, but less prominent in influenza pneumonia. Activated neutrophils were typically found in physical association with monocytes. To explore this further, we combined clinical data of COVID‐19 cases with comprehensive immune cell phenotyping and bronchoalveolar lavage fluid scRNA‐seq data. We show that a HLADRlow CD9low monocyte population expands in severe COVID‐19, which releases neutrophil chemokines in the lungs, and might in turn explain neutrophil expansion and pulmonary recruitment in the late stages of severe COVID‐19.
Conclusions
Our data underline an innate immune cell axis causing vascular inflammation and immunothrombosis in severe SARS‐CoV‐2 infection.
Keywords: COVID‐19, immunopathology, immunothrombosis, monocytes, neutrophils, SARS‐CoV‐2
Essentials
-
•
It remains unclear if COVID‐19–related immunothrombosis is a general feature of (viral) pneumonia, and how it is regulated in severe SARS‐CoV‐2 infection.
-
•
We compared histopathological specimens of influenza and COVID‐19 lungs and utilized comprehensive immune cell phenotyping, functional in vitro assays, and bronchoalveolar lavage fluid scRNA‐seq data to investigate this further.
-
•
We identified vascular neutrophil recruitment, NETosis, and immunothrombosis to be key features of COVID‐19 as compared to a less prominent role in influenza.
-
•
CD9low HLA‐DRlow monocytes are a prominent feature of severe COVID‐19 and release neutrophil‐attracting chemokines in the lungs, causing pulmonary neutrophil recruitment.
Alt-text: Unlabelled Box
Highlights
-
•
Histopathological comparison of COVID‐19 lungs with influenza specimens shows immunothrombotic vascular occlusions.
-
•
A peripheral blood HLA‐DRlow monocyte population expands in severe disease and releases neutrophil chemokines in the lungs.
-
•
This innate immune cell axis might explain neutrophil expansion and pulmonary recruitment in the late stages of severe COVID‐19.
Alt-text: Unlabelled Box
1. INTRODUCTION
Since its animal–human transmission in late 2019, a novel coronavirus termed severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has spread globally, infecting millions within months.1., 2., 3. Coronavirus disease 2019 (COVID‐19) is characterized by respiratory failure in severe cases, but is also associated with non‐pulmonary organ failure and a systemic prothrombotic state.4 We and others have linked intravascular neutrophil and platelet activation, neutrophil extracellular trap formation (NETosis), and subsequent activation of the coagulation cascade, a process termed “immunothrombosis,” to COVID‐19 progression, providing a possible explanation for multi‐organ involvement.5., 6., 7. However, neutrophil activation and NETosis have been identified as a common effector function in a range of inflammatory disorders, calling into question the specificity of immunothrombosis in contributing to SARS‐CoV‐2–associated acute respiratory distress syndrome (ARDS).8., 9., 10. In addition, upstream regulation of immunothrombosis, which might be amenable to pharmacological treatment, remains poorly understood.11
2. METHODS
Detailed methodology is provided in supporting information.
2.1. Cohort
Details of the analyzed cohorts are stated in Tables S1 and S2 in supporting information. COVID‐19 patients are part of the COVID‐19 Registry of LMU University Hospital Munich (CORKUM, WHO trial ID DRKS00021225) or DeutschesRegister von COVID‐19 Obduzierten Fällen (DeRegCOVID). The study was approved by local ethics committees of Munich and Aachen (No: 20‐245 and 19‐274, EK 092‐20 & EK 119‐20). Data from the 27 patients listed in Table S1 were used for all analyses except histopathology, for which a total of 13 patients detailed in Table S2 were used. For flow cytometry and longitudinal analysis of clinical data patients with pre‐existing diseases like severe kidney or liver failure, immunosuppressive therapy, autoimmune disease, chronic inflammation, or acute extracorporeal membrane oxygenation therapy were excluded. COVID‐19 patients consisted of CoV‐Sev patients requiring intensive care treatment and hospitalized CoV_int patients treated on the ward. We included age‐matched control patients without infectious disease and control non‐COVID pneumonia cases (Table S1).
2.2. Histopathology
Immunohistochemistry stainings were performed on formalin‐fixed paraffinized lung specimens as previously described.12 Specimens were from six COVID‐19 and H1N1 (n = 4) or seasonal influenza (n = 3) patients obtained via the DeRegCOVID register.
2.3. Flow cytometry
Preparation and analysis of patient blood were performed as previously described5 (Table S3 in supporting information). Measurements were performed on BD LSRFortessa Flow Cytometer and analyzed using FlowJo Software (BD). Downsample v3 was used for down‐sampling. The data was concatenated and the t‐SNE FlowJo plugin was used for distributed stochastic neighbor embedding (t‐SNE) of equal and representative numbers of cells for each group. A phenograph algorithm13 was used for unsupervised population detection, and then gated in a supervised manner to assign final subpopulations. Heatmap analysis and visualization were performed with ClustVis.
2.4. Analysis of scRNA seq data
Count matrices for the scRNA‐seq dataset were downloaded from GEO (Accession GSE145926)14 and analyzed using Seurat. Gene‐set enrichment analyses were conducted on Biological Process subset of GeneOntology (GO‐BP).15
2.5. Statistical analysis
Values of individual patients are represented as dots in graphs, and data in main text and figures is mean ± standard error of the mean unless otherwise noted. Unpaired, two tailed t‐tests, or in case of significant F‐test, Mann‐Whitney U tests were used. For regression analysis, the black line represents best‐fit line, shaded area with dashed lines is 95% confidence interval, r 2 and P value (slope non‐zero) are shown in plots. Excel (Microsoft) and Prism (GraphPad) software were used for data analysis, Illustrator (Adobe Inc) for visualization.
3. RESULTS
To better understand the immunopathology of severe SARS‐CoV‐2 infection, we phenotyped peripheral blood leukocytes in controls (Ctrl) and non‐COVID‐19 pneumonia patients (Ctrl_pneu) compared to COVID‐19 patients on normal wards (CoV_int) and requiring intensive care treatment (ARDS cohort, CoV_sev; Figure S1A‐C in supporting information).
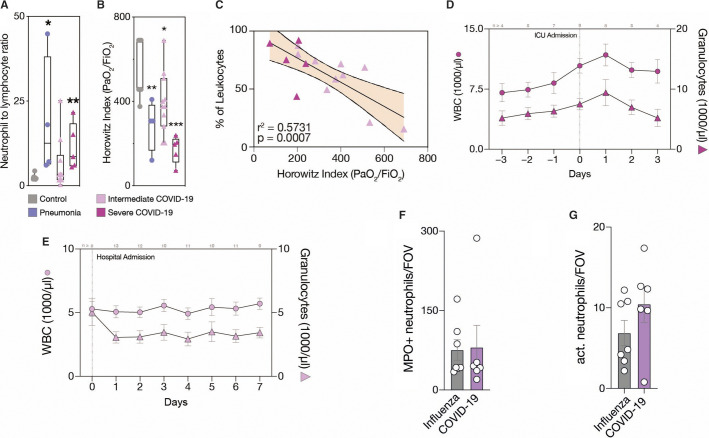
While CoV_int patients showed comparable leukocyte counts to healthy controls, a hallmark of CoV_sev was an expansion of the neutrophil granulocyte compartment, translating into elevated leukocyte counts and an increased neutrophil–lymphocyte ratio (NLR; Figure 1A and Figure S1A). The NLR also correlated with disease severity, as measured by oxygenation index (PaO2/FiO2, Horowitz index; Figure 1B, Figure S1C). In fact, neutrophil counts were the only quantitative immune cell parameter correlating positively with disease severity (Figure 1C, Figure S1D). Longitudinal sampling in the CoV_sev cohort revealed a rise in peripheral neutrophil counts just before manifestation of ARDS requiring mechanical ventilation (Figure 1D). In contrast, CoV_int patients not requiring mechanical ventilation showed stable neutrophil counts throughout the disease course (Figure 1E). This implicates an important role of neutrophils in disease progression.
Figure 1.
COVID‐19 presents dynamic adaptive and innate immunity changes in the peripheral blood. A, Neutrophil‐to‐lymphocyte ratio (NLR) and (B), Horowitz index (PaO2/FiO2) for each patient group. Box‐and‐whiskers plot, two‐tailed unpaired t‐test comparing groups to control (Ctrl). n = 7 Ctrl_healthy, n = 4 Ctrl_pneu, n = 11 CoV_int, n = 5 CoV_sev patients. C, Linear regression of neutrophils with Horowitz index. n = 11 CoV_int, n = 5 CoV_sev. Shaded area is 95% confidence interval. D, E, Time course of daily white blood cell count (WBC) and granulocyte count for CoV_int and CoV_sev normed on hospital or intensive care unit admission. Data is mean ± standard error of the mean (SEM), n per time point is shown above graphs in gray. n = 14 CoV_sev, n = 15 CoV_int. F, Number of neutrophils per field of view in lung sections (FOV). G, Number of activated (citH3+) neutrophils per FOV. F–G, Mean of five high power fields was taken for each sample. n = 6 COVID‐19, n = 7 Influenza. Error bars are SEM. * P ≤ .05, ** P ≤ .01, *** P ≤ .001
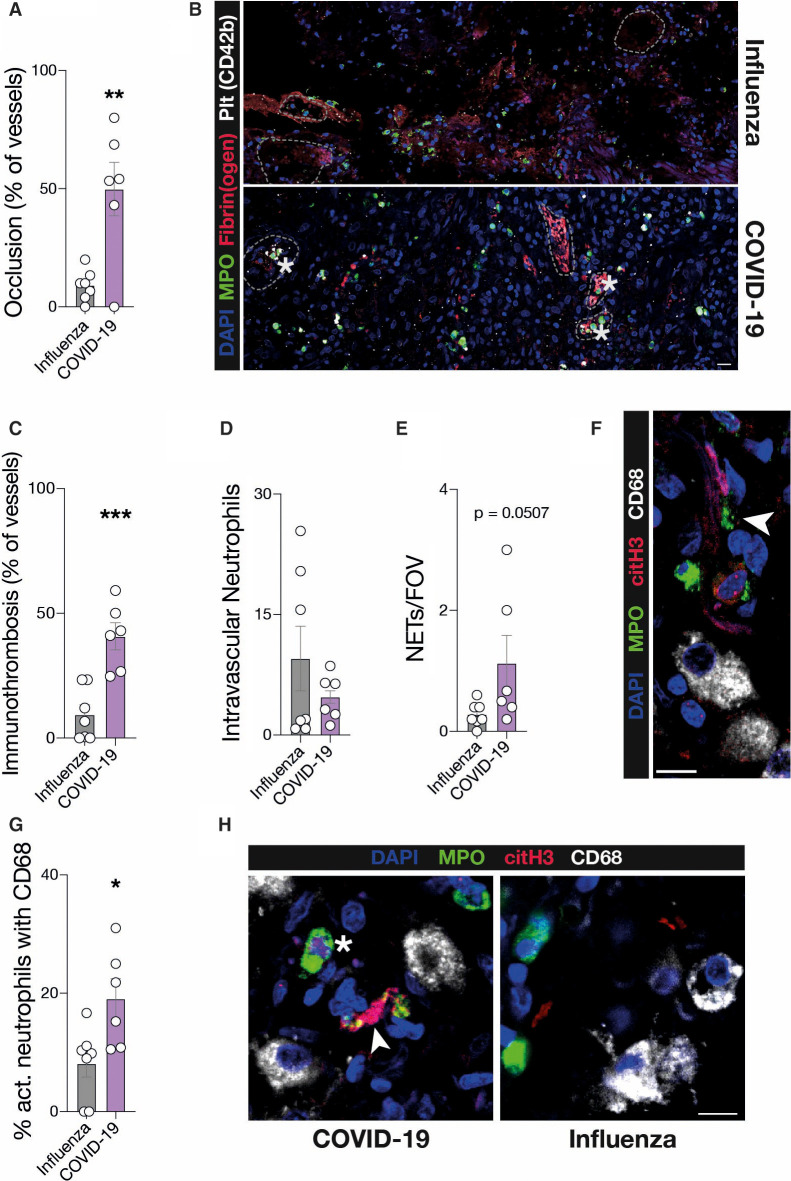
To further examine this link, we histopathologically compared lung specimens of deceased COVID‐19 (n = 6) and influenza (H1N1 or seasonal) pneumonia cases (n = 7). Total neutrophil recruitment as well as activation did not differ between SARS‐CoV‐2 and influenza pneumonia (Figure 1F‐G). However, we detected a significant difference in immunothrombotic occlusion of microvessels in COVID‐19, with 40.8% ± 5.4 of vessels affected versus 9.4% ± 4.0 in influenza (Figure 2A‐C ). Although the overall numbers of intravascular neutrophils were not increased in COVID‐19 lungs (Figure 2D), neutrophil extracellular trap (NET) formation was elevated in COVID‐19 lungs (Figure 2E‐F). In line with these findings, von Willebrand factor (vWF) antigen levels, which are a marker of endothelial injury and are also associated with NETosis, were elevated in severe COVID‐19 cases16., 17. (Figure S1E). These data underline that severe COVID‐19 is also a vascular disease, and show that pulmonary immunopathology in SARS‐CoV‐2 infection might differ from other viral infections, requiring novel treatment strategies.18., 19. On the other hand, the identified correlation with disease severity and involvement in vascular inflammation suggests neutrophils to be causative in the development of organ damage and mortality in COVID‐19.11 In kidney and heart specimens we could indeed see similar trends in immunothrombotic occlusions (Figure S1G, H). Therefore, it is of paramount importance to understand neutrophil recruitment and mobilization, especially to the lungs.
Figure 2.
Vascular neutrophil recruitment, NETosis, and immunothrombosis are defining factors of severe COVID‐19 compared to influenza. A, Percentage of vessels occluded in the lung. Unpaired, two‐tailed t‐test. B, Representative micrographs of vessels in influenza and COVID‐19 lungs. Stars indicate immunothrombosis (see Methods). Dashed lines show vessel borders. Scale bar: 20 µm. C, Percentage of vessels with immunothrombosis as shown in (B). Unpaired, two‐tailed t‐test. D, Number of intravascular neutrophils per field of view (FOV). Mann‐Whitney U test. E, Number of neutrophil extracellular traps (NETs) per FOV. Mann‐Whitney U test. F, Representative micrograph of a NET associated with CD68 + macrophages in a COVID‐19 lung. Arrow indicates NET. Scale bar: 10 µm. G, Percentage of activated (citH3+) neutrophils associated with CD68 + macrophages in the lung. Unpaired, two‐tailed t‐test. H, Representative micrographs of neutrophil associated with macrophages in COVID‐19 and influenza lungs. Arrow indicates NET, star indicates activated neutrophil. Scale bar: 10 µm. C–E, G, Mean of five high power fields was taken for each sample. n = 6 COVID‐19, n = 7 Influenza. Error bars are standard error of the mean. * P ≤ .05, ** P ≤ .01, *** P ≤ .001
As neutrophils are mobilized and recruited to the lungs at advanced stages of COVID‐19, we hypothesized that cellular effectors recruited at earlier disease stages might orchestrate neutrophil influx. When staining lung sections for monocytes and macrophages, we discovered a strong association of monocytic cells with activated and NET‐ting neutrophils, which was increased in COVID‐19 compared to influenza specimens (Figure 2G‐H), although there was no difference in absolute monocyte/macrophage numbers (Figure S1F).
Monocytes are potent phagocytic cells and known attractors of neutrophils in the pulmonary vasculature.20., 21., 22. To better understand their role in advanced COVID‐19, we phenotyped peripheral blood monocytes using a multidimensional flow cytometry‐based panel defining 10 monocyte states (MS1–10; Figure S2A‐D in supporting information).
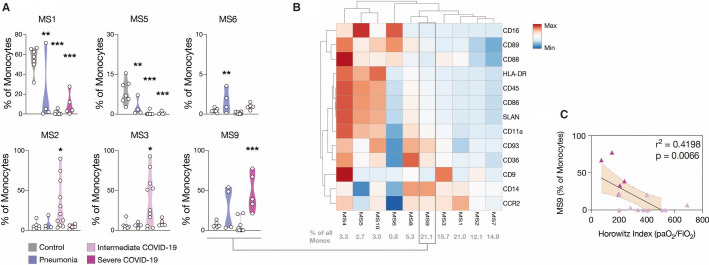
This revealed a striking effect of SARS‐CoV‐2 infection on the monocyte compartment: non‐activated CCR2hi classical monocytes (MS1) and non‐classical monocytes (MS5), together comprising 64% ± 5 of circulating monocytes in non‐infected controls, virtually disappeared in COVID‐19 (Figure 3A , Figure S2F). In contrast, we identified a classical monocyte subpopulation, MS9, which was robustly upregulated in severe COVID‐19. In line with recent scRNA‐seq data, this population showed particularly low CD9 expression levels with upregulation of complement C1q‐receptor (CD93), scavenger‐receptor CD36, and pattern‐recognition receptor CD14, indicating enhanced phagocytic potential (Figure 3B and Figure S2C‐G).23., 24., 25. Indeed, this CD14hiCD9low MS9 cluster correlated significantly with disease severity and represented one of the largest subpopulations comprising approximately 20% of all monocytes in severe COVID‐19 (Figure 3C).
Figure 3.
A HLADRlow CD9low monocyte population expands in severe COVID‐19 and correlates with disease severity. A, Violin plots of the percentages of total monocytes of each patient within each subcluster. One‐way analysis of variance with post‐hoc Dunnett’s multiple comparisons test. n = 7 control (Ctrl), n = 4 Ctrl_pneu, n = 11 CoV_int, n = 5 CoV_sev. B, Heatmap of relative mean fluorescence intensities (MFIs) of subclusters. Percent of cells in each subcluster shown in gray below. C, Linear regression of the MS9 subcluster with Horowitz index. n = 11 CoV_int, n = 5 CoV_sev
To examine the role of this monocyte subpopulation in more detail we performed in vitro assays of sorted monocyte subsets from healthy individuals (see supporting information). Indeed, supernatant of stimulated CD14hiCD16lo monocytes prompted increased migration by neutrophils in a Boyden chamber migration assay (Figure S2H). Furthermore, neutrophils incubated with supernatant specifically from the CD9loHLA‐DRlo monocyte subset showed increased expression of neutrophil activation marker CD163, compared to supernatant from CD16loCD9int/hi classical monocytes (Figure S2I).
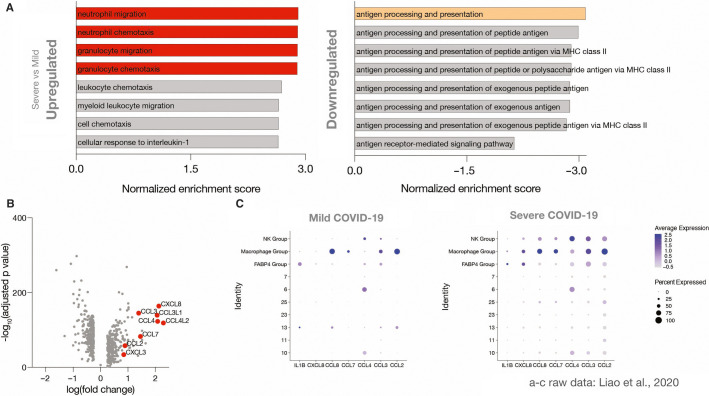
To gain mechanistic insight into monocytes/monocytic macrophages recruited to the lungs in COVID‐19, we re‐analyzed publicly available scRNA‐seq data of bronchoalveolar lavage fluid (BALF) from COVID‐19 patients with mild and severe disease (Figure S3A in supporting information). First, we analyzed pathways that were either up‐ or downregulated when comparing severe and mild COVID‐19 using gene ontology biological process (GO‐BP) analyses.26 Indeed, processes involving neutrophil chemotaxis were robustly upregulated in FABP4‐ pulmonary monocytes/macrophages in severe COVID‐19, specifically CXCL8 (IL8), CCL2, CCL3, CCL4, CCL7, CXCL3, CCL3L1, and CCL4L2 (Figure 4A‐B , Figure S3B).
Figure 4.
The HLADRlow CD9low monocyte population in severe COVID‐19 releases neutrophil chemokines in the lung. In silico reanalysis of publicly available single cell RNA sequencing data published by Liao et al.14 A, Top eight up‐ and downregulated GO‐BP pathways of severe versus mild cases for monocytic macrophages in bronchoalveolar lavage. Fold enrichment is relative to mild cases. B, Volcano plot of relative fold change of severe versus mild cases for monocytic macrophages. CXCL8, CCL2, CCL3, CCL4, CCL7, CXCL3, CCL4L2, and CCL3L1, when significantly increased, are marked in red and annotated. C, Dot plots of average and percentage expression for different cell groups
By analyzing chemokine transcriptomes across all discovered cell types in BALF, we were able to confirm that recruited monocytic macrophages were the key source of neutrophil‐attracting chemokines in the failing lungs (Figure 4C). In line with the proinflammatory CD9low peripheral blood monocyte subset MS9 identified in CoV_sev patients, CD9 expression by monocytic macrophages in the BALF dropped in the severe group. This suggests that blood‐monocyte subset MS9 may give rise to a pulmonary monocytic‐macrophage subset, which in turn contributes to neutrophil activation (Figure S3C).
4. DISCUSSION
Clinically, COVID‐19 presents heterogeneously with lung involvement, but also central nervous system and gastrointestinal symptoms.4 In severe disease, however, this seems to converge into severe immunopathology, i.e., host damage by a dysregulated immune response.11 The mechanisms and regulation of immunopathology in SARS‐CoV‐2 infection are so far incompletely understood. Our data, in agreement with prior studies, underlines that severe COVID‐19 is also a vascular disease with immunothrombotic, neutrophil‐containing vessel occlusions present in the lungs.5., 27. Neutrophils seem to be the key immune cell subset associated with clinical deterioration as neutrophil–lymphocyte ratio and neutrophil counts correlated with disease severity and longitudinal analysis revealed a spike in neutrophil counts preceding respiratory failure. Neutrophil activation and NETosis have been implicated in a wide range of diseases ranging from atherosclerosis to cancer.9., 28. To better understand the involvement of these cells in SARS‐CoV‐2 infection we compared histopathological specimens of COVID‐19 lungs with lethal viral pneumonia caused by H1N1 or seasonal influenza virus. Our data underline neutrophil‐driven immunothrombosis as a key element of severe COVID‐19 as immunothrombotic vessel occlusion and NETosis were strongly elevated compared to influenza pneumonia. While NETosis and innate immunity have also been implicated in influenza our data point to a substantially increased contribution to immunopathology in SARS‐CoV‐2 infection.29., 30. In addition, elevated vWF activity in severe COVID‐19 points to increased endothelial activation and has also been implicated as a marker of NETosis.16 This data might therefore link endotheliitis reported by Varga et al with immunothrombosis in SARS‐CoV‐2 infection.19
So how are neutrophils recruited and activated? Our study points to an innate immune cell axis consisting of CD9low monocytes that release proinflammatory, neutrophil‐attracting chemokines in the failing lungs of severe COVID‐19 patients. Interestingly, this HLA‐DRlow population of monocytes has also been identified in other COVID‐19 studies using single cell RNA‐sequencing of peripheral blood mononuclear cells, highlighting this subset as a general feature of this disease.31., 32.
Principal limitations of the presented study are the limited number of patients and histological specimens analyzed. However, our findings are in line with work from several other groups addressing vascular inflammation in COVID‐19.24., 31.
In summary, our data provide evidence for an innate immune cell axis in vascular inflammation and immunothrombosis observed in severe SARS‐CoV‐2 infection. Targeting neutrophil–monocyte partnership might be a valuable therapeutic approach to dampen disease progression in COVID‐19.
5. CODE AVAILABILITY
All employed code for scRNA‐seq analysis is available from GitHub. (https://github.com/mjoppich/CovidImmune, https://github.com/mjoppich/scrnaseq_celltype_prediction).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Initiation: LN, AL; conceptualization: LN, KP, and KS; methodology: LN, KP, AL, SB, RK MJ; investigation: LN, KP, AL, SB, RK, MH, CG, VP, AE, TW, MZ, BZ, MJ; resources: SM, KS, MM, CS, JCH, RDB, SS, PB; formal analysis: LN, KP, AL, SB, RK, MJ, MM, AE, VP; writing—original draft: LN, KP; writing—editing: all authors; visualization: LN, KP, AL, MJ; supervision: LN, KP, KS; project administration: LN, KP, and KS; funding acquisition: LN, KP, SM, and KS.
DATA AVAILABILITY STATEMENT
Data is available upon reasonable request from authors.
ACKNOWLEDGMENTS
We thank Anna Titova for technical assistance. We would like to thank all CORKUM investigators and staff. The authors thank the patients and their families for their participation in the CORKUM registry. The study was supported by the German Registry of COVID‐19 Autopsies (DeRegCOVID). Open access funding enabled and organized by Projekt DEAL.
H2020 European Research Council
Deutsches Zentrum für Herz‐Kreislaufforschung
Deutsche Herzstiftung
Deutsche Forschungsgemeinschaft
Footnotes
Leo Nicolai, Alexander Leunig, Kami Pekayvaz, and Konstantin Stark contributed equally to this work.
Manuscript handled by: Andreas Greinacher
Final decision: Andreas Greinacher, 10 November 2020
Funding informationThis study was supported by the Deutsche Herzstiftung e.V., Frankfurt a.M. (LN), Deutsche Forschungsgemeinschaft (DFG) SFB 914/3 (SM [B02 and Z01], KS [B02]), the DFG SFB 1123/2 (SM [B06], M.J + RZ [Z02]), the DFG FOR 2033 (S.M.), the German Centre for Cardiovascular Research (DZHK; Clinician Scientist Programme [LN], MHA 1.4VD [SM]), FP7 program (project 260 309, PRESTIGE [SM]), European Research Council (ERC‐2018‐ADG "IMMUNOTHROMBOSIS" [SM], grant aggrement no. 833440), FöFoLe project 1015/1009 (LN), and the clinician scientist programme PRIME by the DFG ‐ 413 635 475 (KP, RK). This work was supported by the German Registry of COVID‐19 Autopsies (DeRegCOVID, www.DeRegCOVID.ukaachen.de), funded by the Federal Ministry of Health (ZMVI1‐2520COR201; PB), and the Federal Ministry of Education and Research in the framework of the Network University Medicine (DEFEAT PANDEMIcs, 01KX2021 and STOP‐FSGS‐01GM1901A; both to PB).
Supporting Information
Supplementary Material
REFERENCES
- 1.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Guan X., Wu P., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou F., Yu T., Du R., et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolai L., Leunig A., Brambs S., et al. Immunothrombotic dysregulation in COVID‐19 pneumonia is associated with respiratory failure and coagulopathy. Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.120.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuo Y., Yalavarthi S., Shi H., et al. Neutrophil extracellular traps (NETs) as markers of disease severity in COVID‐19. medRxiv. 2004;2020(2020):2009. [Google Scholar]
- 7.Middleton E.A., He X.‐.Y., Denorme F., et al. Neutrophil extracellular traps (NETs) contribute to immunothrombosis in COVID‐19 acute respiratory distress syndrome. Blood. 2020 doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caudrillier A., Kessenbrock K., Gilliss B.M., et al. Platelets induce neutrophil extracellular traps in transfusion‐related acute lung injury. J Clin Invest. 2012;122(7):2661–2671. doi: 10.1172/JCI61303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warnatsch A., Ioannou M., Wang Q., Papayannopoulos V. Neutrophil extracellular traps license macrophages for cytokine production in atherosclerosis. Science. 2015;349(6245):316–320. doi: 10.1126/science.aaa8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lood C., Blanco L.P., Purmalek M.M., et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus‐like disease. Nat Med. 2016;22(2):146. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao X. COVID‐19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020:1–2. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marx C., Novotny J., Salbeck D., et al. Eosinophil‐platelet interactions promote atherosclerosis and stabilize thrombosis with eosinophil extracellular traps. Blood. 2019;134(21):1859–1872. doi: 10.1182/blood.2019000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine J.H., Simonds E.F., Bendall S.C., et al. Data‐driven phenotypic dissection of AML reveals progenitor‐like cells that correlate with prognosis. Cell. 2015;162(1):184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao M., Liu Y., Yuan J., et al. Single‐cell landscape of bronchoalveolar immune cells in patients with COVID‐19. Nat Med. 2020 doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 15.Ashburner M., Ball C.A., Blake J.A., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perdomo J., Leung H.H.L., Ahmadi Z., et al. Neutrophil activation and NETosis are the major drivers of thrombosis in heparin‐induced thrombocytopenia. Nat Commun. 2019;10(1):1–14. doi: 10.1038/s41467-019-09160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubin D.B., Wiener‐Kronish J.P., Murray J.F., et al. Elevated von Willebrand factor antigen is an early plasma predictor of acute lung injury in nonpulmonary sepsis syndrome. J Clin Invest. 1990;86(2):474–480. doi: 10.1172/JCI114733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvelli J., Demaria O., Vély F., et al. Association of COVID‐19 inflammation with activation of the C5a–C5aR1 axis. Nature. 2020 doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varga Z., Flammer A.J., Steiger P., et al. Endothelial cell infection and endotheliitis in COVID‐19. Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreisel D., Nava R.G., Li W., et al. In vivo two‐photon imaging reveals monocyte‐dependent neutrophil extravasation during pulmonary inflammation. Proc Natl Acad Sci. 2010;107(42):18073. doi: 10.1073/pnas.1008737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maus U.A., Waelsch K., Kuziel W.A., et al. Monocytes are potent facilitators of alveolar neutrophil emigration during lung inflammation: role of the CCL2‐CCR2 axis. J Immunol. 2003;170(6):3273–3278. doi: 10.4049/jimmunol.170.6.3273. [DOI] [PubMed] [Google Scholar]
- 22.Dhaliwal K., Scholefield E., Ferenbach D., et al. Monocytes control second‐phase neutrophil emigration in established lipopolysaccharide‐induced murine lung injury. Am J Respir Critical Care Med. 2012;186(6):514–524. doi: 10.1164/rccm.201112-2132OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapellos T.S., Bonaguro L., Gemünd I., et al. Human monocyte subsets and phenotypes in major chronic inflammatory diseases. Front Immunol. 2019;10(2035) doi: 10.3389/fimmu.2019.02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silvin A., Chapuis N., Dunsmore G., et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID‐19. Cell. 2020 doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulte‐Schrepping J., Reusch N., Paclik D., et al. Severe COVID‐19 is marked by a dysregulated myeloid cell compartment. Cell. 2020 doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Ackermann M., Verleden S.E., Kuehnel M., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid‐19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teijeira Á., Garasa S., Gato M., et al. Cxcr1 and cxcr2 chemokine receptor agonists produced by tumors induce neutrophil extracellular traps that interfere with immune cytotoxicity. Immunity. 2020 doi: 10.1016/j.immuni.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Koupenova M, Corkrey HA, Vitseva O, et al. The role of platelets in mediating a response to human influenza infection. 2019;10(1):1‐18. [DOI] [PMC free article] [PubMed]
- 30.Tang B.M., Shojaei M., Teoh S., et al. Neutrophils‐related host factors associated with severe disease and fatality in patients with influenza infection. Nat Commun. 2019;10(1):1–13. doi: 10.1038/s41467-019-11249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilk A.J., Rustagi A., Zhao N.Q., et al. A single‐cell atlas of the peripheral immune response in patients with severe COVID‐19. Nat Med. 2020;1–7 doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giamarellos‐Bourboulis E.J., Netea M.G., Rovina N., et al. Complex immune dysregulation in COVID‐19 patients with severe respiratory failure. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Data is available upon reasonable request from authors.