Viruses remain a significant public health concern worldwide, and the threat is associated with the ability of the viruses to mutate frequently and adapt to different hosts. A point or stand‐off biosensor that can detect and classify viruses from indoor and outdoor environments would be suitable for viral surveillance. This review critically evaluates the prospects and challenges of LIF‐LiDAR technology for real‐time stand‐off detection and classification of potentially pathogenic viruses in various environments.
Summary
Viruses remain a significant public health concern worldwide. Recently, humanity has faced deadly viral infections, including Zika, Ebola and the current severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). The threat is associated with the ability of the viruses to mutate frequently and adapt to different hosts. Thus, there is the need for robust detection and classification of emerging virus strains to ensure that humanity is prepared in terms of vaccine and drug developments. A point or stand‐off biosensor that can detect and classify viruses from indoor and outdoor environments would be suited for viral surveillance. Light detection and ranging (LiDAR) is a facile and versatile tool that has been explored for stand‐off detection in different environments including atmospheric, oceans and forest sensing. Notably, laser‐induced fluorescence‐light detection and ranging (LIF‐LiDAR) has been used to identify MS2 bacteriophage on artificially contaminated surgical equipment or released amidst other primary biological aerosol particles in laboratory‐like close chamber. It has also been shown to distinguish between different picornaviruses. Currently, the potentials of the LIF‐LiDAR technology for real‐time stand‐off surveillance of pathogenic viruses in indoor and outdoor environments have not been assessed. Considering the increasing applications of LIF‐LiDAR for potential microbial pathogens detection and classification, and the need for more robust tools for viral surveillance at safe distance, we critically evaluate the prospects and challenges of LIF‐LiDAR technology for real‐time stand‐off detection and classification of potentially pathogenic viruses in various environments.
Introduction
Microbial pathogens such as bacteria, viruses, fungi and parasites are a major public health concern. Of these, viruses have proven to be a great threat in recent times. Humanity has been faced with deadly viral infections including Zika virus, Ebolavirus and the ongoing severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) pandemic. The viral threat is attributed to their ability to mutate frequently and adapt to different hosts. To outcompete these viruses, there is the need for robust detection and surveillance of emerging strains. This will ensure that humanity is prepared in terms of vaccine and drug development. A promising approach in this respect is a point or stand‐off biosensor that can detect and classify viruses phylogenetically or based on their pathogenicity from indoor and outdoor environments. Stand‐off detection and identification systems are defined by their ability to detect and classify chemical, biological and explosive (CBE) hazards without contact with the hazardous material(s). The stand‐off detection range may be considered short (also referred to as non‐contact), medium or long, depending on the distance between the stand‐off detector and the material being targeted for detection. The stand‐off detection range has been variably defined. Even though some studies have presented < 10 m and > 10 m as short and long stand‐off range, respectively (Bogue, 2018), this review adopts the detection distances < 20 m, 20–100 m and > 100 m as short, medium and long stand‐off detection range, respectively, in consonance with many other reports (Sedlacek et al., 2002; Huestis et al., 2010; Babichenko et al., 2018; Fellner et al., 2020). Stand‐off detection methods are suitable for operations in high risk and harsh environments, as they could provide information about CBE hazards in real‐time from safe distances of several centimetres to up to a kilometre (Jonsson et al., 2009; Babichenko et al., 2018; Fellner et al., 2020). Based on these advantages, stand‐off detection will be an ideal approach for viruses, which can be highly contagious and dangerous.
Light detection and ranging (LiDAR) is a group of techniques that are based on the principle that when a pulsed laser (light amplification by stimulated emission of radiation) impinges on particles, the particles absorb and/or emit signals that can be characterized using a suitable detector (Buteau et al., 2008). The emitted signals could be a backward scattered or polarized light, or fluorescence (Yang et al., 2016). LiDAR is a facile and versatile tool that has been applied in remote environmental monitoring, including atmospheric, oceans and forest sensing (Eitel et al., 2016; Almeida et al., 2019). LiDAR has also become a powerful air‐ or space‐borne altimetric tool (Flood, 2001; Forfinski‐Sarkozi and Parrish, 2016). In addition, the LiDAR sensor is fast becoming a key component of autonomous vehicles, to provide information about the surroundings of the vehicles with high resolution (Royo and Ballesta‐Garcia, 2019; Tang et al., 2020).
Based on the nature of the scattered light, four basic types of LiDAR, including Rayleigh, Mie, Raman and fluorescence have been described (Veerabuthiran, 2003; Zhang et al., 2011). The laser‐induced optical behaviours have been exploited for the detection and characterization of biogenic materials both in laboratory and field conditions using LiDAR platforms. Raman scattering, for example, has been applied spectroscopically to detect and identify pathogenic bacteria in clinical samples (Ho et al., 2019) and determine antibiotics susceptibility and transcriptomic profile that reflect antibiotics resistance in bacteria (Germond et al., 2018). A combination of Rayleigh and Raman scattering effectively characterized extracellular vesicles (EVs) and lipoproteins, including differentiating tumour‐derived EVs from normal blood cells (Enciso‐Martinez et al., 2020a,b). Similarly, laser‐induced fluorescence (LIF) has been used for detection and/or identification of microorganisms and pollens (Pan et al., 2010, 2011; Babichenko et al., 2018; Swanson and Huffman, 2018).
Of the different types of LiDAR, fluorescence LiDAR, particularly the LIF‐LiDAR, has demonstrated high potentials for stand‐off detection and characterization of primary biological aerosol particles (PBAPs) (Buteau et al., 2008; Li et al., 2019). LIF‐LiDAR has been used to identify MS2 bacteriophage on artificially contaminated surgical equipment (Babichenko et al., 2018). LIF‐LiDAR has also been used to identify MS2 bacteriophage amidst other PBAPs in laboratory‐like close chamber and open field settings (Sivaprakasam et al., 2004; Baxter et al., 2007; Jonsson et al., 2009; Farsund et al., 2010) and thus highlighting the potentials of the technique for viral detection indoors or outdoors. In addition, LIF‐LiDAR was used to both detect and identify each virus from a pool of pure isolates of seven viruses of the Picornavirus family (Gabbarini et al., 2019). However, the potentials of the LIF‐LiDAR technology have not been assessed for real‐time stand‐off surveillance of pathogenic viruses in the environment. In view of the increasing applications of LIF‐LiDAR for potential pathogens detection, and the need for more robust tools for non‐contact viral surveillance at long range, this review critically evaluates the prospect and challenges of LIF‐LiDAR technology for stand‐off surveillance of potentially harmful viruses in in‐built or outdoor environments.
In this review, we critically provide an overview of the types of LiDAR, LIF‐LiDAR working principles, suitability and prospects of LIF‐LiDAR for virus surveillance, challenges of LIF‐LiDAR approach in virus surveillance and a perspective on overcoming the potential challenges.
Types of LiDAR
Based on the nature of the scattered light, many types of LiDAR, including Rayleigh, Mie, Raman and resonance fluorescence, have been described (Veerabuthiran, 2003; Zhang et al., 2011). Both Rayleigh and Mie scattering are elastic, meaning both incidence and scattered light have the same wavelength, and no energy is lost as the incidence light impinges on the particles (Veerabuthiran, 2003). For Rayleigh, the incident light has a much longer wavelength than the size of the impinged particle, while Mie scattering occurs when the particle has much larger size than the wavelength of the impinging light (Veerabuthiran, 2003). Unlike Rayleigh and Mie scattering, Raman scattering is inelastic and involves energy (or wavelength) shift, based on the molecular nature of the particles impinged by the light; the Raman energy shift is unique to different molecules and could be used to characterize different particles (Turner et al., 2016). Resonance fluorescence scattering is also inelastic and occurs when the wavelength of the incidence light corresponds to the absorption line of the atoms, ions or molecules, such that the atoms are excited to a higher energy level by the incoming light and re‐emit photons of the same or longer wavelength than the incident light (Abo, 2005; Gardner and Collins, 2015). Although elastic scatterings could be used to map clouds and detect PBAPs such as pollens, bacterial and fungal cells, or spores at low concentrations and long range, those types of LiDAR have poor resolution for particles of different composition but similar sizes. A detailed characterization of biological particles at long range will therefore require inelastic scattering (Simard et al., 2004; Buteau et al., 2006). Figure 1 summarizes the differences between Rayleigh, Mie and Raman scattering.
Fig. 1.
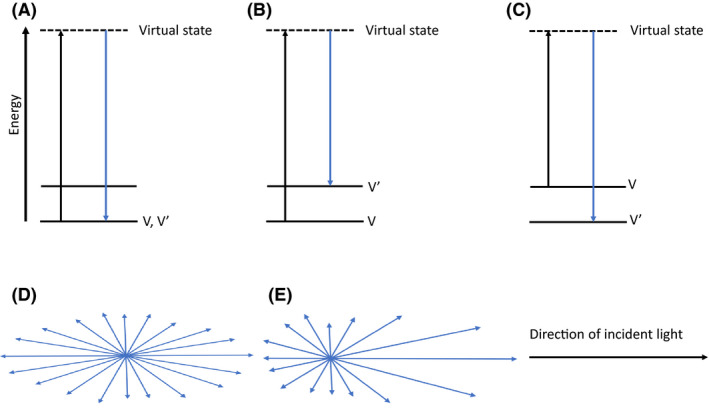
Schematic representations of Rayleigh, Mie and Raman scattering.
A. Energy diagram of Rayleigh scattering. There is no change in frequency of the incident and scattered light (V = V’).
B. Energy diagram of Stokes Raman scattering. There is a shift in frequency of the scattered light such that (V> V’).
C. Energy diagram of anti‐Stokes Raman scattering. There is a shift in the frequency of the scattered light such that (V < V’).
D. Rayleigh scattering. This occurs when the wavelength of the incident light is greater than the diameter of the impinged particle (e) Mie scattering. This occurs when the wavelength of the incident light is less than the diameter of the impinged particle. V = frequency of incident light. V’ = frequency of scattered light.
LIF‐LiDAR: Working principles and its suitability for viral surveillance
The detailed working principle of LIF‐LiDAR has been well described in various reports (Joshi et al., 2013; Wojtanowski et al., 2015; Yang et al., 2016). Briefly, a LIF‐LiDAR has a source which emits laser of desired excitation wavelength to be directed to a target such as bioaerosols, and a receiving system which collects the echoed signals (such as a scattered light and fluorescence) from the laser‐interrogated target. Typically, a receiving system is comprised of a telescope which collects the scattered light, a dichroic mirror which separates the light into spectra based on frequencies, an array of photomultiplier tubes which receive high frequency light for signal amplification, and a spectrograph which receives and directs the lower frequency (longer wavelength) light to gated intensified charge coupled device (ICCD) camera (Joshi et al., 2013). In addition, a LIF‐LiDAR system has a data acquisition platform, which displays the amplified signals in a visible/readable format.
Microorganisms contain biomolecules in various subcellular structures and the biomolecules possess intrinsic fluorescence ability (Yang et al., 2016). Hence, when microorganisms are impinged with light of an appropriate wavelength, they emit fluorescence of maximal intensity (Yang et al., 2016). The fluorescence excitation and emission spectra are a function of the nature of fluorophores (the fluorogenic biomolecules) in the biomolecular architecture of the microorganisms and are discriminatory enough for identification and classification of the microorganisms (Buteau et al., 2006; Joshi et al., 2013). Viruses vary in their structures, even though they are basically comprised of nucleic acid (DNA or RNA) encased in a protein capsid (Louten, 2016) and some viruses, in addition to this basic structure, also possess enzymes and lipid‐derived envelopes (Mesters et al., 2006). Altogether, viruses possess different fluorophores that can be interrogated by light/laser to identify and/or classify them.
Prospects of LIF‐LiDAR for viral detection and classification
The fluorogenic ability of viruses has been well‐established. Sivaprakasam and colleagues reported fluorescence intensities of 16 bioaerosols, including MS2 bacteriophage, at 350, 450 and 550 nm spectra bands when the bioaerosols were excited with 266 and 355 nm laser pulses at 400 nanosecond intervals (Sivaprakasam et al., 2004). Notably, the highest fluorescence intensity was obtained for the bacteriophage at the 350 spectra band and 266 nm excitation wavelength. At the two excitation wavelengths, the spectral signature of MS2 bacteriophage was discriminatory against 15 aerosol particles, including vegetative bacteria, spores and proteins. In contrast, a small scale LIF‐LiDAR platform built to remotely detect biological weapons using a single excitation wavelength of 266 nm showed a significant overlap (14.8%) between MS2 bacteriophage and dirty diesel exhaust fluorescence spectra (Baxter et al., 2007). This discrepancy highlights the potentials of multiwavelength excitation to increase the discriminatory power of LIF‐LiDAR for virus detection and identification in aerosols.
Field trials of two LiDAR systems, namely from the Norwegian Defense Research Establishment (FFI) and Swedish Defense Research Agency (FOI), have been reported for simulated detection of different bacterial species, ovalbumin and MS2 bacteriophage, in a plume of expected atmospheric interferents, such as pollens, oil fog, smoke and diesel fumes have been reported (Jonsson et al., 2009). Using 355 nm excitation wavelength pulsed at 5 ns and 10 Hz pulse repetition frequency (PRF), fluorescence spectra with peak within the 450 nm band was acquired for the MS2 bacteriophage released at a concentration of 3 × 109 – 1.5 × 1011 plaque‐forming units per millilitre (PFU/ml), 230 m – 1 km away from the LiDAR systems and 100–200 m upwind. A comparison of 355 and 294 nm excitation wavelengths for stand‐off detection of bioaerosols containing bacterium, MS2 bacteriophage and ovalbumin released in a semi‐closed chamber that was 210 m away from the LIF‐LiDAR platform indicated a better spectral resolution with the 294 than 355 nm (Farsund et al., 2012). Farsund et al. also observed highest fluorescence intensity for MS2 bacteriophage within the 350 nm spectral band when the virus was excited with the 294 nm wavelength. In another study, a LiDAR platform named BC‐Sense device obtained a unique spectral signature of 430 nm maximal fluorescence for MS2 bacteriophage when the virus was excited with a 248 nm laser of PRF of 10 Hz (Babichenko et al., 2018). The spectral profile of BC‐Sense device was discriminatory enough to identify MS2 bacteriophage on several surfaces that were co‐contaminated with bacteria and fungi and placed about 20 m away from the LIF‐LiDAR with a limit of detection of 9.5 × 104 PFU/cm2. However, this limit of detection is high and may not be suitable for stand‐off virus surveillance.
Autofluorescence of F2 bacteriophage has been observed to depend almost entirely on amino acid contents (particularly tryptophan and tyrosine) of its nucleocapsid rather than its RNA genome; when F2 bacteriophage was excited with a UV light of 265–295 nm, it emitted fluorescence spectra within 320 nm band (Kitchell et al., 1977). A latter study using two bacteriophages, Ø6 and Ø12, and their bacterial hosts showed that the fluorescence signature of tryptophan varies with different protein environments and hence tryptophan fluorescence profile of the two phages differs from their Pseudomonad hosts (Alimova et al., 2004). These findings suggest that, with appropriate laser excitation wavelengths, viruses could be discriminated from bacteria and other biogenic materials.
A study which investigated spectrally resolved fluorescence cross sections of bioaerosols at 266, 273, 280, 365 and 405 nm excitation wavelengths reported excitation‐emission spectra of Venezuelan equine encephalitis virus (VEEV), as well as MS2 bacteriophage and several vegetative bacteria and spores. VEEV and MS2 bacteriophage strongly emitted fluorescence within 280–400 nm spectra band, with peak fluorescence occurring at 310–320 nm, when the viruses were interrogated with 266, 273, and 280 nm wavelengths (Pan et al., 2014). Interestingly, Pan and colleagues also observed dissimilar excitation‐emission signatures between VEEV and MS2 bacteriophage, with the former showing strong fluorescence only within the 280–400 nm band, while the latter had strong fluorescence within 280–400 nm and 400–600 nm bands. VEEV and MS2 bacteriophage belong to two different families of viruses, Togaviridae and Leviviridae, respectively, and suggest that viruses belonging to different families may have different excitation‐emission spectra signature that could be explored for LIF‐based viral classification.
A LIF platform was used to identify seven viruses that belong to the Picornavirus family. When the viral samples were interrogated with a 266 nm laser of 10 kHz PRF in the LiDAR set up, the viruses emitted fluorescence spectra between 350 and 700 nm. These spectra signatures were unique to the viruses except for two serotypes—Coxsackie A7 and A9, which had poor separation based on principal component analysis (PCA) (Gabbarini et al., 2019). However, further analysis showed that support vector machine (SVM), a supervised machine learning algorithm developed for classification and regression analysis, especially using complex data (Bhavsar and Panchal, 2012; Pisner and Schnyer, 2020) and neural network algorithm, perfectly classified the seven viruses investigated. Further, the limit of detection of LIF platform was 2 × 104 – 2 × 105 tissue culture 50% infectious dose (TCID50)/ml for hepatitis A, although the limit of detection was not determined for the other six viruses.
Collectively, the studies highlighted above indicate that viruses have excitation–emission spectral profile that could be explored to differentiate them from one another, from other microbial agents and from environmental interferents such as pollens and hydrocarbon effluents. However, despite these promising findings, the suitability of LIF‐LIDAR for stand‐off viral surveillance in real‐life to warrant field deployment of the technique has not been established.
Challenges to LIF‐LiDAR approach in viral surveillance
First, most studies reporting on viral fluorescence spectra used purified viral isolates and hence the results obtained may not reflect a real‐life environment where viruses exist amidst plethora of chemical and biological interferents (Pan, 2015). Even though environmental interference was simulated in some of the studies in the previous section, the spectrum of the interfering simulants used was too narrow to capture the varieties of PBAPs and other aerosol components in the atmosphere.
Second, the concentration of viruses in the atmosphere, indoor air or surfaces, may be less than those detectable by the LIF platforms tested. Although data on viral concentration in indoor and outdoor air are scant, total virus‐like particles (VLPs) of 1 × 105/m3 and 2.6 × 105/m3 have been reported for indoor and outdoor air, respectively (Prussin et al., 2015). In addition, a metagenomics‐based study in Korea indicated that the VLPs concentration of near surface atmosphere (about 1 m above the ground level) varies seasonally from 1.7 × 106 to 4.0 × 107 VLP/m3 with winter and spring having the highest and lowest VLP concentrations, respectively (Whon et al., 2012). The total VLPs in outdoor environments that were reported by Whon et al. and Prussin et al. were lower than the viral concentration that was measured in the Jonsson et al., 2009 study (3 × 109 – 1.5 × 1011 PFU/ml). Although the reported total VLP concentrations are comparable to the 104 PFU/cm2 and 2 × 104 – 2 × 105 TCID50/ml limits of viral detection of the LIF platforms used by Babichenko et al. and Gabbarini et al., respectively (Babichenko et al., 2018; Gabbarini et al., 2019), the limits of detection of the LIF platforms are likely less than the atmospheric viral concentrations when individual viruses are considered rather than total VLP.
Third, bioaerosol surveillance using LIF‐LiDAR requires the use of high‐energy pulsed excitation laser (Baxter et al., 2007; Farsund et al., 2012) and this raises genuine biosafety concerns, especially that a high‐energy laser could cause irreversible damage to the eyes (Sayed, 2014; Birtel et al., 2017). Although some researchers have tried to reduce the high‐energy laser to eye‐safe energy level by enlarging the beam radius of the laser to 10 mm (Duschek et al., 2017), biosafety is still not guaranteed.
Fourth, a key advantage of the LIF‐LiDAR is the potential for a long‐range detection of harmful bioaerosols, especially those classified as biological weapons. However, choosing the right excitation wavelength for stand‐off interrogation of viruses in the atmosphere faces some difficulties. For instance, whereas studies have indicated that shorter excitation wavelengths (248–294 nm) elicit more intense and distinct fluorescence spectra for viruses than longer excitation wavelengths such as 355 nm (Sivaprakasam et al., 2004; Farsund et al., 2012; Babichenko et al., 2018), the shorter wavelengths are more easily attenuated in the atmosphere than the longer wavelengths. Hence, choosing the appropriate excitation wavelength may be difficult.
The fifth hurdle is about the cost of LIF‐LiDAR platforms, which is due to various factors; it is expected that the cost will be directly proportional to the stand‐off distance, and the area to be monitored among other factors. The average cost of UV‐LIF bioaerosol sensors runs into hundreds of thousands of US Dollars (Swanson and Huffman, 2018), which is prohibitive for their commercialization and routine application, especially in poor‐resourced countries, which coincidentally have higher burden of infectious pathogens, including viruses (Fenollar and Mediannikov, 2018).
LIF‐LiDAR for viral surveillance: What will it take?
Excitation‐emission spectra of viruses are needed
Although extensive characterization of laser excitation‐emission spectra of bacteria, fungi and pollens have been undertaken (O’Connor et al., 2011; Pöhlker et al., 2011; Dartnell et al., 2013), similar studies on viruses are lacking. Hence, to adapt LIF‐LiDAR for stand‐off viral surveillance, studies are needed to determine the optimum excitation–emission spectra of viruses that are potentially highly pathogenic to humans. This baseline data are required for building databases that are needed for real‐time detection, classification and identification of viruses. Such studies should consider irradiating different classes of virus with a range of UV‐laser excitation wavelengths to determine laser specifications that would yield maximum intensity and most distinct signature spectra for individual viruses and classes of viruses.
A multiwavelength excitation approach is required
A key requirement for viral surveillance using LIF‐LiDAR is the ability to discriminate viruses from other microbes or materials. Viruses differ from microorganisms, in terms of biomolecular composition, in many respects. Notably, reduced nicotinamide adenine dinucleotide (NADH), a redox carrier and co‐enzyme in energy metabolic pathways in viable cells, and other co‐enzymes (e.g. riboflavin–vitamin B2) and vitamins which are fluorogenic, are absent in viruses. Interestingly, NADH is the most dominant fluorogenic co‐enzyme in microbial cells, with well‐characterized fluorescence excitation–emission spectra (Pöhlker et al., 2011). Direct excitation wavelength of NADH ranges from 340 to 370 nm while its emission wavelength is 440–470 nm (Kaye et al., 2005; Pöhlker et al., 2011; Babichenko et al., 2018; Fennelly et al., 2018). Reported excitation and emission wavelengths for viruses are mostly in the range of 248–295 nm and 310–450 nm, respectively (Sivaprakasam et al., 2004; Farsund et al., 2012; Babichenko et al., 2018; Gabbarini et al., 2019). Hence, probing bioaerosol clouds with excitations of 340–370 nm and 248–295 nm wavelengths may yield spectra features to discriminate viruses from viable bacteria and fungi. Similarly, non‐viable bacterial and fungal cells, and spores and pollens, which may lack NADH, equally possess fluorogenic biopolymers that could discriminate them from viruses (Pöhlker et al., 2011). Bacteria, fungi and pollens possess cellulose and sporopollenin, respectively, and in their cell walls, and fungi also have chitin as one of their cell wall components. These biopolymers are all absent in viruses. Interrogating a bioaerosol particle with carefully selected multiple wavelengths could discriminate viral from non‐viral particles based on their fluorescence spectra signatures.
Size differences could enhance viral discrimination
Viruses could also be discriminated from other PBAPs based on their differences in size. Apart from members of the Poxviridae which are up to 300 nm in diameter, human pathogenic viruses are generally small, often less than 200 nm in diameter (Gelderblom, 1996). Bacteria and fungi, on the other hand, are generally larger than viruses. With the size differences, an excitation wavelength, which is smaller than microbial cells but larger than viral particles, will elicit Rayleigh and Mie scattering upon impinging a virus and bacterium or a fungus, respectively. It also suggests that Rayleigh scattering could be used to locate viruses among bacteria and fungi using < 300 nm excitation wavelengths.
Discrimination of viruses by Rayleigh‐LIF‐LiDAR hyphenation and time‐resolved fluorescence
The differential capacity of Rayleigh scattering is enhanced in a system combining Rayleigh with an inelastic scattering (Enciso‐Martinez et al., 2020a,b). Hence, a LiDAR system for viral detection and identification in real‐life environment will potentially use a multiwavelength excitation approach to probe bioaerosol clouds to acquire Rayleigh scattering information which will help to locate the viruses and LIF spectra data for virus classification/identification. In addition, the specificity of the Rayleigh‐LIF‐LiDAR platform could be enhanced by using multiple channels of resolution for acquiring the LIF spectra (Kaye et al., 2005; Ruske et al., 2017). The specificity of the hyphenated LiDAR platform could further be enhanced by acquiring data on fluorescence lifetimes (time‐resolved LIF data) of the viruses and other aerosol particles, as was recently demonstrated for detection, classification and identification of some CBE agents (Fellner et al., 2020).
The time‐resolved LIF is based on observations that the fluorescence decay time depends on the immediate environment of the fluorophores. Some of the variables that may influence fluorescence lifetimes include the concentration of ions in the environment and the presence of neighbouring fluorophores (Meier et al., 2010). Since viruses differ from one another and from other PBAPs by amino acid composition, and lack co‐enzymes, such as NADH and riboflavin which are present in viable cells, the microenvironment, and by extension, the fluorescence decay time of fluorophores is expected to differ significantly among viruses and from other fluorogenic materials. This suggests that a combination of Rayleigh, LIF and time‐resolved LIF‐LiDAR will be a potentially robust platform for detection and specific identification of viruses in real‐life environments where interferents which possess overlapping fluorescence spectra may be present. However, for this to be achieved, studies are needed to generate reference data regarding the fluorescence lifetimes of viruses, bacteria, fungi, spores, pollens and other potential interferents in the atmosphere.
Machine learning algorithms are required
Considering that a feasible LiDAR platform for viral surveillance will likely acquire both elastic and inelastic radiation data that involves multiple spectra features, analysis of the acquired data may be very challenging. The primary focus of the data analysis will be to determine whether an interrogated PBAP is a virus, what virus it is, and if it poses a threat or not. Machine learning algorithms (MLA) with high predictive power and trained with a robust database of viral spectra features and fluorescence lifetimes will be required to build an online virus classifying platform for real‐time viral surveillance. A decision tree (implemented in the online virus classifying platform) could be grown from the size, spectra, fluorescence lifetimes datasets and lists of potentially harmful viral agents to indicate if a virus identified poses a threat (positive alarm) or not (negative alarm), following a model that has previously been described (Fischbach et al., 2015).
Several MLAs, such as support vector machine, principal component analysis, decision tree, neural network and wavelet transform, have been used for extraction of relevant spectra features and reduction of high‐dimensional spectra data to lower dimensions and thereby reducing computational complexity and increasing classification accuracy (Sobolev and Babichenko, 2013; Gabbarini et al., 2019). Genetic algorithm (an evolutionary algorithm), which has higher classification accuracy than SVM, principal component analysis, Fisher’s linear discriminant and forward feature selection, has also been reported (Nyhavn et al., 2011). Future studies on viral surveillance using LiDAR could explore the appropriate algorithms, as well as consider other algorithms such as the t‐Distributed Stochastic Neighbour embedding, which is suitable for both linear and non‐linear data, for high‐dimensional spectra data reduction.
Cost reduction and biosafety concern
There is a high potential for miniaturization and significant reduction of the cost of LIF‐LiDAR platforms for bioaerosol agents’ surveillance. Although early LIF platforms that have been built for bioaerosol surveillance require high end instrumentation and therefore make them quite expensive for widespread use, recent LIF platforms have used innovative instrumentation such as the replacement of Q‐switch solid‐state lasers with UV light‐emitting laser diodes (LED) as an effective and inexpensive laser excitation source (Zhang et al., 2013; Swanson and Huffman, 2018).
Biosafety concern should be thoroughly addressed before considering LIF‐LiDAR for viral surveillance, especially in residential or other places where people are likely going to be exposed to the high‐energy laser. Although the laser safety concern cannot be completely addressed presently, keeping the excitation wavelength below 400 nm, which is considered ‘eye‐safe’ (Franks, 1991) should be considered. In addition, standard safety precautions for use of laser, including laser‐safe eye goggle should be mandatory (Smalley, 2011).
Conclusion
Viral infections have caused unprecedented hardship to humanity, partly due to the ability of viruses to mutate fast and adapt to new hosts. To manage and avert viral infections that evolve into outbreaks like the current COVID‐19 pandemic, robust viral surveillance and monitoring of emerging viruses is imperative. Here, we have assessed the potential of LIF‐LiDAR approach for detection of viruses before, during and beyond pandemics, by looking at the prospects, challenges and perspectives of the approach that may be associated with stand‐off viral surveillance. Taking together all the factors that have been addressed in this review, we conclude that substantial effort is still required to achieve a long‐distance viral surveillance in the atmospheric environment using the LIF‐LiDAR approach. However, the platform may have successful short stand‐off range applications in food and medical virology. For instance, LIF‐LiDAR is amenable to virus surveillance on fomites such as medical equipment, medical laboratory workbench, theatre tables and conveyor belts in food industries, among others.
Conflict of interest
The authors have declared that no competing interests exist.
Author contributions
OO and COO conceived and prepared the first draft of the manuscript. OQ critically reviewed the draft. All the authors approved the final version of the manuscript.
Acknowledgements
OO and COO were supported by WACCBIP‐ACE PhD fellowship (ACE02‐WACCBIP: Awandare) and a DELTAS Africa grant (DEL‐15‐007: Awandare). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (107755/Z/15/Z to Awandare) and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government.
Microbial Biotechnology (2021) 14(1), 126–135
Funding informationOO and COO were supported by WACCBIP‐ACE PhD fellowship (ACE02‐WACCBIP: Awandare) and a DELTAS Africa grant (DEL‐15‐007: Awandare). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (107755/Z/15/Z to Awandare) and the UK government.
References
- Abo, M. (2005) Resonance Scattering Lidar In Springer Series in Optical Sciences. Weitkamp, C. (ed). New York, NY: Springer, pp. 307–323. [Google Scholar]
- Alimova, A. , Katz, A. , Podder, R. , Minko, G. , Wei, H. , Alfano, R. R. , and Gottlieb, P. (2004) Virus particles monitored by fluorescence spectroscopy: a potential detection assay for macromolecular assembly. Photochem Photobiol 80: 41. [DOI] [PubMed] [Google Scholar]
- Almeida, D. R. A. , Stark, S. C. , Chazdon, R. , Nelson, B. W. , Cesar, R. G. , Meli, P. , et al (2019) The effectiveness of lidar remote sensing for monitoring forest cover attributes and landscape restoration. For Ecol Manage 438: 34–43. [Google Scholar]
- Babichenko, S. , Gala, J. L. , Bentahir, M. , Piette, A. S. , Poryvkina, L. , Rebane, O. , et al (2018) Non‐contact, real‐time laser‐induced fluorescence detection and monitoring of microbial contaminants on solid surfaces before, during and after decontamination. J Biosens Bioelectron 9: 255. [Google Scholar]
- Baxter, K. , Castle, M. , Barrington, S. , Withers, P. , Foot, V. , Pickering, A. , and Felton, N. (2007) UK small scale UVLIF lidar for standoff BW detection In Electro‐Optical Remote Sensing, Detection, and Photonic Technologies and Their Applications. Kamerman, G. W. , Steinvall, O. K. , Lewis, K. L. , Krapels, K. A. , Carrano, J. C. , and Zukauskas, A. (eds). Bellingham, Washington: SPIE, p. 67390Z. [Google Scholar]
- Bhavsar, H. , and Panchal, M. H. (2012) A review on support vector machine for data classification. Int J Adv Res Computer Eng Technol 1: 185–189. [Google Scholar]
- Birtel, J. , Harmening, W. M. , Krohne, T. U. , Holz, F. G. , Issa, P. C. , and Herrmann, P. (2017) Retinal injury following laser pointer exposure – a systematic review and case series. Deutsches Arzteblatt International 114: 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue, R. (2018) Remote chemical sensing: a review of techniques and recent developments. Sensor Review 38: 453–457. [Google Scholar]
- Buteau, S. , Simard, J.‐R. , Déry, B. , Roy, G. , Lahaie, P. , Mathieu, P. , et al (2006) Bioaerosols laser‐induced fluorescence provides specific robust signatures for standoff detection In Chemical and Biological Sensors for Industrial and Environmental Monitoring II. Christesen, S. D. , Sedlacek, A. J. , Gillespie, J. B. , and Ewing, K. J. (eds). Bellingham, Washington: SPIE, p. 637813. [Google Scholar]
- Buteau, S. , Simard, J.‐R. , Lahaie, P. , Roy, G. , Mathieu, P. , Déry, B. , et al (2008) Bioaerosol standoff monitoring using intensified range‐gated laser‐induced fluorescence spectroscopy In Advanced Environmental Monitoring. Kim, Y. J. , and Platt, U. (eds). Dordrecht, the Netherlands: Springer Netherlands, pp. 203–216. [Google Scholar]
- Dartnell, L. R. , Roberts, T. A. , Moore, G. , Ward, J. M. , and Muller, J. P. (2013) Fluorescence characterization of clinically‐important bacteria. PLoS One 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duschek, F. , Fellner, L. , Gebert, F. , Grünewald, K. , Köhntopp, A. , Kraus, M. , et al (2017) Standoff detection and classification of bacteria by multispectral laser‐induced fluorescence. Advanced Optical Technologies 6: 75–83. [Google Scholar]
- Eitel, J. U. H. , Höfle, B. , Vierling, L. A. , Abellán, A. , Asner, G. P. , Deems, J. S. , et al (2016) Beyond 3‐D: the new spectrum of lidar applications for earth and ecological sciences. Remote Sens Environ 186: 372–392. [Google Scholar]
- Enciso‐Martinez, A. , van der Pol, E. , Lenferink, A. T. M. , Terstappen, L. W. M. M. , van Leeuwen, T. G. , and Otto, C. (2020) Synchronized Rayleigh and Raman scattering for the characterization of single optically trapped extracellular vesicles. Nanomed Nanotechnol Biol Med 24: 102109. [DOI] [PubMed] [Google Scholar]
- Enciso‐Martinez, A. , Van Der Pol, E. , Hau, C. M. , Nieuwland, R. , Leeuwen, T. G. V. , Terstappen, L. W. M. M. , and Otto, C. (2020) Label‐free identification and chemical characterisation of single extracellular vesicles and lipoproteins by synchronous Rayleigh and Raman scattering. Journal of Extracellular Vesicles 9: 1730134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsund, Ø. , Rustad, G. , Kåsen, I. , and Haavardsholm, T. V. (2010) Required spectral resolution for bioaerosol detection algorithms using standoff laser‐induced fluorescence measurements. IEEE Sens J 10: 655–661. [Google Scholar]
- Farsund, Ø. , Rustad, G. , and Skogan, G. (2012) Standoff detection of biological agents using laser induced fluorescence—a comparison of 294 nm and 355 nm excitation wavelengths. Biomedical Optics Express 3: 2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellner, L. , Kraus, M. , Gebert, F. , Walter, A. , and Duschek, F. (2020) Multispectral LIF‐based standoff detection system for the classification of CBE hazards by spectral and temporal features. Sensors 20: 2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennelly, M. J. , Sewell, G. , Prentice, M. B. , O’Connor, D. J. , and Sodeau, J. R. (2018) Review: the use of real‐time fluorescence instrumentation to monitor ambient primary biological aerosol particles (PBAP). Atmosphere 9: 1. [Google Scholar]
- Fenollar, F. , and Mediannikov, O. (2018) Emerging infectious diseases in Africa in the 21st century. New Microbes New Infect 26: S10–S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach, T. , Duschek, F. , Hausmann, A. , Pargmann, C. , Aleksejev, V. , Poryvkina, L. , et al (2015) Standoff detection and classification procedure for bioorganic compounds by hyperspectral laser‐induced fluorescence. Chemical, Biological, Radiological, Nuclear, and Explosives (CBRNE) Sensing XVI 9455: 945508. [Google Scholar]
- Flood, M. (2001) Laser altimetry: from science to commercial lidar mapping. Photogramm Eng Remote Sensing 67: 1209–1218. [Google Scholar]
- Forfinski‐Sarkozi, N. , and Parrish, C. (2016) Analysis of MABEL bathymetry in Keweenaw Bay and implications for ICESat‐2 ATLAS. Remote Sens 8: 772. [Google Scholar]
- Franks, J. K. (1991) What is eye safe? In Eyesafe Lasers: Components, Systems, and Applications. Johnson, A. M. (ed). SPIE, pp. 2–8. [Google Scholar]
- Gabbarini, V. , Rossi, R. , Ciparisse, J.‐F. , Malizia, A. , Divizia, A. , De Filippis, P. , et al (2019) Laser‐induced fluorescence (LIF) as a smart method for fast environmental virological analyses: validation on Picornaviruses. Sci Rep 9: 12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, C. S. , and Collins, R. L. (2015) Lidar: Resonance In Encyclopedia of Atmospheric Sciences. North G. R. (ed). Amsterdam, The Netherlands: Elsevier Inc., pp. 305–308. [Google Scholar]
- Gelderblom, H. R. (1996) Structure and classification of viruses In Medical Microbiology. Baron, S. (ed). Galveston, TX, USA: University of Texas Medical Branch at Galveston; Chapter 41. Available from: https://www.ncbi.nlm.nih.gov/books/NBK8174/. [PubMed] [Google Scholar]
- Germond, A. , Ichimura, T. , Horinouchi, T. , Fujita, H. , Furusawa, C. , and Watanabe, T. M. (2018) Raman spectral signature reflects transcriptomic features of antibiotic resistance in Escherichia coli . Communications Biology 1: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, C. S. , Jean, N. , Hogan, C. A. , Blackmon, L. , Jeffrey, S. S. , Holodniy, M. , et al (2019) Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat Commun 10: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huestis, D. L. , Smith, G. P. , and Oser, H. (2010) Laser‐based standoff detection of surface‐bound explosive chemicals In Micro‐ and Nanotechnology Sensors, Systems, and Applications II. International Society for Optics and Photonics, p. 76790G. [Google Scholar]
- Jonsson, P. , Elmqvist, M. , Gustafsson, O. , Kullander, F. , Persson, R. , Olofsson, G. , et al (2009) Evaluation of biological aerosol stand‐off detection at a field trial In Optically Based Biological and Chemical Detection for Defence V. Carrano, J. C. , and Collins, C. J. (eds). Bellingham, Washington: SPIE, p. 74840I. [Google Scholar]
- Joshi, D. , Kumar, D. , Maini, A. K. , and Sharma, R. C. (2013) Detection of biological warfare agents using ultra violet‐laser induced fluorescence LIDAR. Spectrochimica Acta ‐ Part A: Molecular and Biomolecular Spectroscopy 112: 446–456. [DOI] [PubMed] [Google Scholar]
- Kaye, P. H. , Stanley, W. R. , Hirst, E. , Foot, E. V. , Baxter, K. L. , and Barrington, S. J. (2005) Single particle multichannel bio‐aerosol fluorescence sensor. Opt Express 13: 3583. [DOI] [PubMed] [Google Scholar]
- Kitchell, B. B. , Merrill, S. P. , and Henkens, R. W. (1977) Optical and hydrodynamic studies of the structure of bacteriophage f2 II. Fluorescence of the capsid. BBA Section Nucleic Acids and Protein Synthesis 475: 536–547. [DOI] [PubMed] [Google Scholar]
- Li, X. , Huang, S. , and Sun, Z. (2019) Technology and equipment development in laser‐induced fluorescence‐based remote and field detection of biological aerosols. Journal of Biosafety and Biosecurity 1: 113–122. [Google Scholar]
- Louten, J. (2016) Virus structure and classification In Essential Human Virology. Varma V. (ed). Amsterdam, The Netherlands: Elsevier, pp. 19–29. [Google Scholar]
- Meier, J. D. , Xie, H. , Sun, Y. , Sun, Y. , Hatami, N. , Poirier, B. , et al (2010) Time‐resolved laser‐induced fluorescence spectroscopy as a diagnostic instrument in head and neck carcinoma. Otolaryngol Head Neck Surg 142: 838–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesters, J. R. , Tan, J. , and Hilgenfeld, R. (2006) Viral enzymes. Curr Opin Struct Biol 16: 776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyhavn, R. , Moen, H. J. F. , Farsund, Ø. , and Rustad, G. (2011) Optimal classification of standoff bioaerosol measurements using evolutionary algorithms. Chemical, Biological, Radiological, Nuclear, and Explosives (CBRNE) Sensing XII 8018: 801806. [Google Scholar]
- O’Connor, D. J. , Iacopino, D. , Healy, D. A. , O’Sullivan, D. , and Sodeau, J. R. (2011) The intrinsic fluorescence spectra of selected pollen and fungal spores. Atmos Environ 45: 6451–6458. [Google Scholar]
- Pan, Y. Le (2015) Detection and characterization of biological and other organic‐carbon aerosol particles in atmosphere using fluorescence. J Quant Spectrosc Radiat Transfer 150: 12–35. [Google Scholar]
- Pan, Y. Le , Hill, S. C. , Pinnick, R. G. , House, J. M. , Flagan, R. C. , and Chang, R. K. (2011) Dual‐excitation‐wavelength fluorescence spectra and elastic scattering for differentiation of single airborne pollen and fungal particles. Atmos Environ 45: 1555–1563. [Google Scholar]
- Pan, Y.‐L. , Hill, S. C. , Pinnick, R. G. , Huang, H. , Bottiger, J. R. , and Chang, R. K. (2010) Fluorescence spectra of atmospheric aerosol particles measured using one or two excitation wavelengths: Comparison of classification schemes employing different emission and scattering results. Opt Express 18: 12436. [DOI] [PubMed] [Google Scholar]
- Pan, Y.‐L. , Hill, S. C. , Santarpia, J. L. , Brinkley, K. , Sickler, T. , Coleman, M. , et al (2014) Spectrally‐resolved fluorescence cross sections of aerosolized biological live agents and simulants using five excitation wavelengths in a BSL‐3 laboratory. Opt Express 22: 8165. [DOI] [PubMed] [Google Scholar]
- Pisner, D. A. , and Schnyer, D. M. (2020) Support vector machine In Machine Learning. Mechelli A. and Vieira S. (eds). Cambridge, United Kingdom: Elsevier, pp. 101–121. [Google Scholar]
- Pöhlker, C. , Huffman, J. A. , and Pöschl, U. (2011) Autofluorescence of atmospheric bioaerosols – fluorescent biomolecules and potential interferences. Atmospheric Measurement Techniques Discussions 4: 5857–5933. [Google Scholar]
- Prussin, A. J. , Garcia, E. B. , and Marr, L. C. (2015) Total concentrations of virus and bacteria in indoor and outdoor air. Environmental Science and Technology Letters 2: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo, S. , and Ballesta‐Garcia, M. (2019) An overview of lidar imaging systems for autonomous vehicles. Applied Sciences 9: 4093. [Google Scholar]
- Ruske, S. , Topping, D. O. , Foot, V. E. , Kaye, P. H. , Stanley, W. R. , Crawford, I. , et al (2017) Evaluation of machine learning algorithms for classification of primary biological aerosol using a new UV‐LIF spectrometer. Atmospheric Measurement Techniques 10: 695–708. [Google Scholar]
- Sayed, M. S. (2014) Ocular damage secondary to lights and lasers: How to avoid and treat if necessary. World Journal of Ophthalmology 4: 1. [Google Scholar]
- Sedlacek, A. J. III , Ray, M. D. , Higdon, N. S. , and Richter, D. A. (2002) Short‐range noncontact detection of surface contamination using Raman lidar In Vibrational Spectroscopy‐based Sensor Systems. Christesen S. D. and Sedlacek A. J. III (eds). Bellingham, Washington: International Society for Optics and Photonics, pp. 95–104. [Google Scholar]
- Simard, J. R. , Roy, G. , Mathieu, P. , Larochelle, V. , McFee, J. , and Ho, J. (2004) Standoff sensing of bioaerosols using intensified range‐gated spectral analysis of laser‐induced fluorescence. IEEE Trans Geosci Remote Sens 42: 865–874. [Google Scholar]
- Sivaprakasam, V. , Huston, A. L. , Scotto, C. S. , and Eversole, J. D. (2004) Multiple UV wavelength excitation and fluorescence of bioaerosols In Chemical and Biological Point Sensors for Homeland Defense II. Sedlacek, A. J. III , Christesen, S. D. , Vo‐Dinh, T. , and Combs, R. J. (eds). SPIE, p. 71. [DOI] [PubMed] [Google Scholar]
- Smalley, P. J. (2011) Laser safety: risks, hazards, and control measures. Laser Therapy 20: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev, I. , and Babichenko, S. (2013) Application of the wavelet transform for feature extraction in the analysis of hyperspectral laser‐induced fluorescence data. Int J Remote Sens 34: 7218–7235. [Google Scholar]
- Swanson, B. E. , and Huffman, J. A. (2018) Development and characterization of an inexpensive single‐particle fluorescence spectrometer for bioaerosol monitoring. Opt Express 26: 3646. [DOI] [PubMed] [Google Scholar]
- Tang, L. , Shi, Y. , He, Q. , Sadek, A. W. , and Qiao, C. (2020) Performance test of autonomous vehicle lidar sensors under different weather conditions. Transportation Research Record: Journal of the Transportation Research Board 2674: 319–329. [Google Scholar]
- Turner, D. D. , Goldsmith, J. E. M. , and Ferrare, R. A. (2016) Development and applications of the ARM Raman lidar. Meteorological Monographs 57: 18.1–18.15. [Google Scholar]
- Veerabuthiran, S. (2003) Exploring the atmosphere with lidars. Resonance 8: 47–51. [Google Scholar]
- Whon, T. W. , Kim, M.‐S. , Roh, S. W. , Shin, N.‐R. , Lee, H.‐W. , and Bae, J.‐W. (2012) Metagenomic characterization of airborne viral DNA diversity in the near‐surface atmosphere. J Virol 86: 8221–8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtanowski, J. , Zygmunt, M. , Muzal, M. , Knysak, P. , Młodzianko, A. , Gawlikowski, A. , et al (2015) Performance verification of a LIF‐LIDAR technique for stand‐off detection and classification of biological agents. Opt Laser Technol 67: 25–32. [Google Scholar]
- Yang, H. , Sun, Y. F. , Wang, T. D. , Zhao, X. S. , Dong, Y. , and Qing, F. (2016) Study on Bioagents or Bioaerosols standoff detection by lidar. MATEC Web of Conferences 44: 1–5. [Google Scholar]
- Zhang, J. , Tong, Y. , Yang, X. , Gong, J. , and Gong, W. (2011) Detection of atmospheric composition based on lidar. J Phys: Conf Ser 276: 012036. [Google Scholar]
- Zhang, P. , Zhao, Y. , Liao, X. , Yang, W. , Zhu, Y. , and Huang, H. (2013) Development and calibration of a single UV LED based bioaerosol monitor. Opt Express 21: 26303. [DOI] [PubMed] [Google Scholar]


