Abstract
Although the COVID‐19 pandemic peaked in March/April 2020 in France, the prevalence of infection is barely known. Using high‐throughput methods, we assessed herein the serological response against the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) of 1847 participants working in three sites of an institution in Paris conurbation.
In May–July 2020, 11% (95% confidence interval [CI]: 9.7–12.6) of serums were positive for IgG against the SARS‐CoV‐2 N and S proteins, and 9.5% (95% CI: 8.2–11.0) were neutralizer in pseudo‐typed virus assays. The prevalence of seroconversion was 11.6% (95% CI: 10.2–13.2) when considering positivity in at least one assay. In 5% of RT‐qPCR positive individuals, no systemic IgGs were detected. Among immune individuals, 21% had been asymptomatic. Anosmia (loss of smell) and ageusia (loss of taste) occurred in 52% of the IgG‐positive individuals and in 3% of the negative ones. In contrast, 30% of the anosmia–ageusia cases were seronegative, suggesting that the true prevalence of infection may have reached 16.6%. In sera obtained 4–8 weeks after the first sampling, anti‐N and anti‐S IgG titers and neutralization activity in pseudo‐virus assay declined by 31%, 17%, and 53%, resulting thus in half‐life of 35, 87, and 28 days, respectively.
The population studied is representative of active workers in Paris. The short lifespan of the serological systemic responses suggests an underestimation of the true prevalence of infection.
Keywords: bioluminescence, COVID‐19, ELISA, LuLISA, SARS‐CoV‐2
In May 2020, 11% of workers at Institut Curie living in Paris conurbation were seropositive and 9.5% had detectable but short‐lived neutralizing antibodies of SARS‐CoV‐2. Some 21% of these neutralizing sera actually belong to asymptomatic individuals. Only 2% of the PCR‐detected infections had not been followed by humoral immune response.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) causing the coronavirus disease 2019 (COVID‐19) emerged in 2019 in China [1, 2, 3] before being detected in a patient living in the Paris conurbation in December 2019 [4]. From January 2020, the virus spread exponentially leading to a risk of Paris conurbation intensive care units saturation. Accordingly, on March 17, a lockdown was imposed by the French authorities to slow down the virus progression. To date, the exposure of the French population during that period remains poorly documented. In contrast with RT quantitative PCR (RT‐qPCR) assays, which are positive for only 2–3 weeks after infection [5], a more efficient way to monitor virus propagation is a serological study of representative populations since specific and lasting antibodies are generated in the great majority of infected subjects [6, 7]. However, studying the anti‐SARS‐CoV‐2 serological response of large cohorts is challenging and requires robustness, specificities, sensitivities and high‐throughput capabilities of the measurement methods, which are exceeding the performance of currently marketed serological assays.
Here, we developed original bioluminescence‐based serological assays allowing a high‐throughput assessment of the specific antibody responses to the spike (S) and nucleoprotein (N) proteins of SARS‐CoV‐2 and their ability to neutralize the virus fusion with a permissive human cell line.
We monitored individual serology against SARS‐CoV‐2 in a large cohort of workers in three institution sites following the March–April 2020 peak of the COVID‐19 pandemic in Paris (France) and over the next 6 months. More than half of Institut Curie workers (n = 1847), a hospital and research center specialized in oncology, volunteered for this Curie‐O‐SA longitudinal serological study. The participants had only been marginally in contact with COVID‐19 patients and are domiciled in the Paris conurbation and are thus representative of an urban population of healthy active adults living in a big metropolitan area. In the course of this survey, we found a high prevalence of immunization, although endowed with rather short‐lived immune responses.
Results
Cohort description, assay development, and validation
Blood samples were collected from 1847 volunteers at the three sites of the Institut Curie located in three cities of Paris conurbation (Ile‐de‐France): Paris, Saint‐Cloud, and Orsay from April 28 until July 31 for the initial time‐point. None of the individuals showed clinical signs of COVID‐19 or had been subjected to a standard RNA detection of SARS‐CoV‐2, using RT‐qPCR, within 14 days prior to blood sampling. All participants were invited to complete a web‐based questionnaire, which included demographic variables, symptom occurrences, and whether these had led to a sick leave, treatment, and/or hospitalization. The participant cohort had a strong (77.4%) female bias (Table 1); the mean age was 38 and ranged between 19 and 75 years old. The hospital‐working staff represented 72.7% of the volunteers, the rest being researchers and administrative staff.
Table 1.
Serological assay results and working groups
| Institute | Hospital | Research center | ||||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Total | 1847 | 100 | 1342 | 72.7 | 505 | 27.3 |
| Female | 1429 | 77.4 | 1074 | 80.0 | 355 | 70.3 |
| Male | 418 | 22.6 | 268 | 20.0 | 150 | 29.7 |
| Age (mean) | 38 | 40 | 38 | |||
| 0–38 years | 943 | 51.1 | 656 | 48.9 | 287 | 56.8 |
| >38 years | 904 | 48.9 | 686 | 51.1 | 218 | 43.2 |
| RT‐qPCR | 189 | 10.2 | 181 | 13.5 | 8 | 1.2 |
| Positive | 66 | 34.9 | 63 | 34.8 | 3 | 37.5 |
| Serological tests | 1847 | 100.0 | 1342 | 100.0 | 505 | 100.0 |
| IgG/N positive | 183 | 9.9 | 151 | 11.3 | 32 | 6.3 |
| IgG/S positive | 181 | 9.8 | 149 | 11.1 | 32 | 6.3 |
| PTN positive | 176 | 9.5 | 146 | 10.9 | 30 | 5.9 |
| Sero. positive | 215 | 11.6 | 171 | 12.7 | 44 | 8.7 |
| Female | 160 | 11.2 | 134 | 12.5 | 26 | 7.3 |
| Male | 55 | 13.2 | 37 | 13.8 | 18 | 12.0 |
| 0–38 years | 107 | 11.3 | 80 | 12.2 | 27 | 9.4 |
| >38 years | 108 | 11.9 | 91 | 13.3 | 17 | 7.8 |
Three serological assays were carried out on these 1847 sera samples in multiwell plates at the Institut Pasteur. Luciferase‐linked immunosorbent assays (LuLISA) were used to assess specific IgG for SARS‐CoV‐2 nucleoprotein (N) and spike (S) proteins in these serum samples. The LuLISA [10] is expanding the sensitivity, the dynamic range, and the scalability in comparison with the gold standard ELISA [8] as detailed in Supporting Information Figs. S1 and S2. A neutralization activity assay using pseudo‐typed virus, also named pseudo‐neutralization test (PNT), was undertaken [9] to assess in parallel of LuLISA the ability of serum components to neutralize the fusion of a SARS‐CoV‐2 spike pseudo‐typed lentiviral vector encoding a luciferase gene using a permissive human cell line (HEK 293T), which is constitutively expressing the human ACE2 receptors (Supporting Information Fig. S3). The specificity threshold of the three methods were established by using serum samples from 54 COVID‐19 patients (March 2020, Institut Cochin), 234 prepandemic negative healthy donors from a blood bank (2014–2018, EFS/ICAReB), and 75 negative serums from prepandemic breast cancer patients (2012, Institut Curie) (Fig. 1A–C). The positivity thresholds were set to 98% specificity for LuLISA assay allowing the detection of anti‐N IgG (10,400 RLU/s) and anti‐S IgG (8400 RLU/s) and to a confidence level of 99% in the case of PNT assay (28 783 RLU/s) established on prepandemic negative sera.
Figure 1.
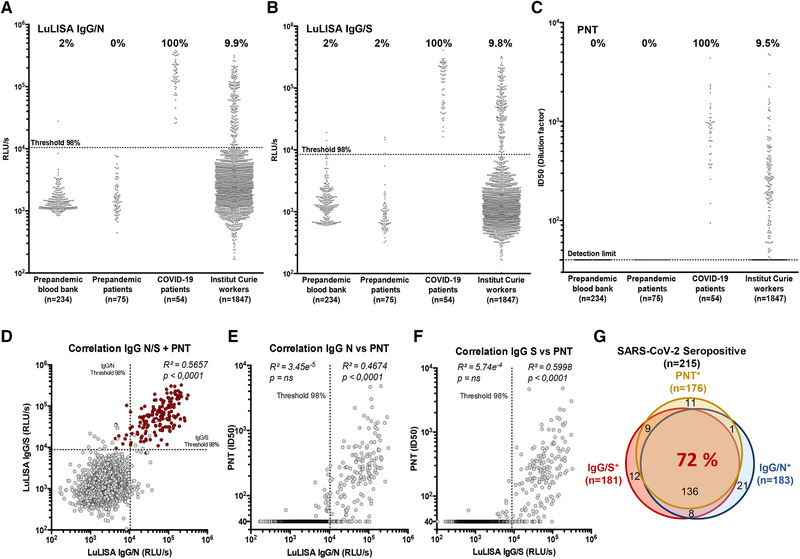
Serological responses to SARS‐CoV‐2 among Institut Curie workers using LuLISA IgG/N, IgG/S, and PNT assays. (A–C) Sera from prepandemic samples from healthy donors (blood bank), prepandemic patients (breast cancer), COVID‐19 patients (RT‐PCR positive), and Institut Curie workers were evaluated in LuLISA IgG/N (A) or IgG/S (B) and PNT (C) assays. For LuLISA, raw values are represented. Sera were considered positive for anti‐N or ‐S IgG if the value was above the 98% threshold (See Supporting Information Fig. S1 for calculation details). For PNT assay, values after ID50 calculation are represented (see Supporting Information Fig. S3 for calculation details and Fig. S7 for raw values). Negative sera are represented with an ID50 below detection limit (40). Percentages of positive are indicated above each series. (D) Correlation plots between LuLISA IgG/N, IgG/S, and PNT (red dots) or between PNT and LuLISA IgG/N (E) or IgG/S (F). Thresholds at confidence index of 98% are shown (dotted lines). Correlation coefficients (R 2) and associated p values from Pearson test (one‐tailed) are indicated above each corresponding area. Numerical values of each combination of assays are summarized with a Venn diagram (G) in overlapping areas. Proportion (%) of triple‐positive individuals is indicated in red.
The robustness of the specificity thresholds and dynamic ranges were assessed using dilution series of COVID‐19 positive sera (Supporting Information Figs. S2 and S3). The specificity for SARS‐CoV‐2 anti‐N IgG was assessed against purified nucleoproteins of SARS‐CoV‐1 as well as seasonal coronaviruses (HCoV) HKU, OC43, NL63, and 229E (Supporting Information Figs. S4 and S5).
High prevalence of anti‐SARS‐CoV‐2 IgG response in the study cohort
For the Institut Curie workers, using a 98% specificity threshold, the seroprevalence of IgG directed against N and S proteins was 9.9% (183/1847, 95% CI: 8.6–11.4) and 9.8% (181/1847, 95% CI: 8.5–11.3), respectively (Fig. 1A and B and Table 1). Among all the serums tested, 9.5% (176/1847, 95% CI: 8.2–11.0) displayed a pseudo‐neutralization activity against the pseudo‐virus (Fig. 1C). Considering each of these assays independently as a marker of specific immune response leads to a 11.6% (215/1847, 95% CI: 10.2–13.2) positivity of immunization.
The correlative plots (Fig. 1D) indicates that the responses against the N and S are linked when both are above their respective threshold (R² = 0.57). Correlation between PNT and LuLISA is mainly detectable when high levels of both IgG against N and S are detected (red dots in Fig. 1D). Moreover, above the 98% specificity threshold, a higher correlation is observed between PNT and LuLISA anti‐spike IgG (IgG/S) (R² = 0.60) (Fig. 1F) than between PNT and LuLISA anti‐nucleoprotein IgG (IgG/N) (R² = 0.47) (Fig. 1E). Remarkably, out of the 215 seropositive samples, 72% are positive for the three assays (Fig. 1G), 9.7% are positive only for IgG/N, 5.6% for IgG/S, and 5.1% for PNT.
Prevalence and kinetic of symptoms and serological responses
Based on the web‐based survey, 54% (1007/1847) participants mentioned at least one symptom (Supporting Information Table S1). Symptomatic workers were more seropositive (16.8%, 170/1007, CI 95%: 14.6–19.3) than asymptomatic workers (5.3%, 45/840, CI 95%: 3.9–7.1) (Table 2 and Fig. 2A). Hence, SARS‐CoV‐2 infection may have been asymptomatic in at least 20.9% (45/215, 95% CI: 16.5–28.2) of the cases (Fig. 2B). The amount of anti‐N IgG was higher in the symptomatic versus asymptomatic patients, while the levels of anti‐S or the neutralization capacity in pseudo‐virus assay did not differ (Supporting Information Fig. S6). This discrepancy suggests that anti‐N IgG may be generated in the course of a mild infection.
Table 2.
Correlation between serological assays and symptoms
| Total (1847) | Seropositive (215) | Seronegative (1632) | |||||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | p (chi‐square) | |
| Any symptom | 1007 | 54 | 170 | 79 | 837 | 51 | 1E‐14 |
| Anosmia/ageusia | 159 | 9 | 111 | 52 | 48 | 3 | 1E‐76 |
| Myalgia | 359 | 19 | 103 | 48 | 256 | 16 | 3E‐29 |
| Fever | 305 | 17 | 86 | 40 | 219 | 13 | 6E‐23 |
| Tiredness | 741 | 40 | 142 | 66 | 599 | 37 | 2E‐16 |
| Cough | 239 | 13 | 61 | 28 | 178 | 11 | 7E‐13 |
| Unusual headache | 416 | 23 | 88 | 41 | 328 | 20 | 6E‐12 |
| Shortness of breath | 663 | 36 | 119 | 55 | 544 | 33 | 2E‐10 |
| Rhinitis | 380 | 21 | 74 | 34 | 306 | 19 | 9E‐08 |
| Intestinal symptoms | 259 | 14 | 50 | 23 | 209 | 13 | 3E‐05 |
| Conjunctivitis | 91 | 5 | 17 | 8 | 74 | 5 | 3E‐02 |
Figure 2.
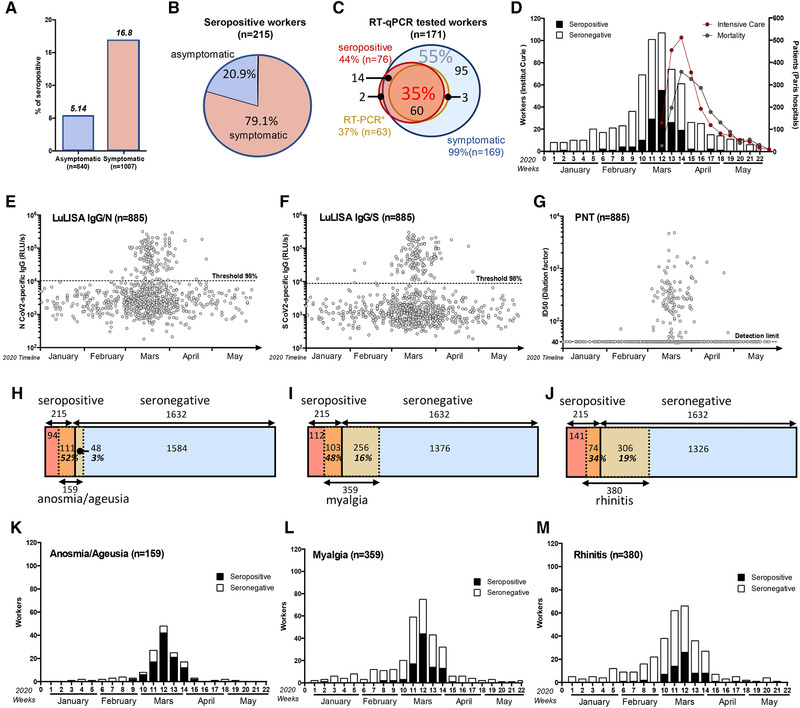
Temporal distribution of symptoms appearance and serology correlates with COVID‐19 outbreak in France. (A) Seroprevalence among asymptomatic and symptomatic workers. (B) Proportion of asymptomatic and symptomatic seropositive workers. (C) Correlation between symptom reporting (blue circle), serological profile (red circle), and RT‐qPCR result (orange circle) among RT‐qPCR tested workers. Proportion (%) of triple‐positive individuals is indicated in red and RT‐qPCR negative, seronegative among symptomatic workers in blue. (D–G) Temporal distribution of serological test results according to the first symptom onset. (D) Number of workers (left y‐axis) reporting at least one symptom but seronegative (white) or seropositive (black). Curves of intensive care admission and mortality in all Paris hospitals are also plotted (right y‐axis). (E–G) Individual test results according to date of symptom onset. (E) LuLISA IgG/N, (F) LuLISA IgG/S, and (G) PNT. (H–J) Prevalence of symptoms according to serology status. Seropositive workers are represented in red and seronegative in blue. Symptomatic workers are represented in orange/yellow. Number of workers for each area is indicated. Percentage represents the proportion of symptomatic in seropositive (orange area) individuals and symptomatic in seronegative ones (yellow area). (K–M) Prevalence of symptoms during pandemic outbreak according to serology status. Plots represent the number of workers reporting symptoms (y‐axis) per week in 2020 (x‐axis). Only the three most representative symptoms from Table 2 are plotted: Anosmia/ageusia as an example of temporally and clinically correlated to COVID‐19 (H and K), Myalgia as clinically only correlated (I and L), and rhinitis as poorly correlated (J and M).
A correlation between serological tests, RT‐qPCR, and symptoms was performed (Fig. 2C). In the 171 individuals tested by RT‐qPCR, 169 (99%) reported symptoms, only 76 (44.4%, CI 95%: 37.9–53.1) were positive in serological assays. Among these 171 RT‐qPCR tested workers, 55% of them were RT‐qPCR negative, seronegative for anti‐N and anti‐S IgG and PNT, but symptomatic, whereas 35% were positive, seropositive, and symptomatic. Moreover, no IgG antibodies were detected in three subjects out of 63 with a positive SARS‐CoV‐2 RT‐qPCR, indicating that a systemic anti‐N or S IgG response may not always be present following a proven SARS‐CoV‐2 infection. However, low levels of anti‐SARS‐CoV‐2 IgM were detected using a commercial lateral flow assay, in one of these three subjects (data not shown). Except for one case, all anosmia/ageusia cases without detectable systemic IgG (n = 48) were associated with other COVID‐19 typical symptoms and occurred in late February, March, or April, suggesting that they represent true SARS‐CoV‐2 infections. Indeed, one of them was associated with a positive SARS‐CoV‐2 RT‐qPCR test, and in seven cases, anti‐SARS‐CoV‐2 IgM was detected using lateral flow assays (data not shown). Thus, in addition to the 215 SARS‐CoV‐2 immune cases detected by our survey, the cohort may feature an additional 48 infection cases devoid of detectable systemic IgG antibodies. Assuming that the incidence (52%) of the anosmia/ageusia symptom is similar in immune and nonimmune individuals, the true prevalence of SARS‐CoV‐2 infection in this population would then be more than 11.6% (215/1847) and as high as 16.6% ((215 + (48/0.52) = 307); 307/1847).
A date for the symptom onset was mentioned in 885 out of 1007 cases. Symptoms were mostly (61%) reported in March 2020 (Fig. 2D), consistent with the reported epidemic development as well as the number of Parisian hospital admissions published daily by Santé Publique France (the French governmental public health agency) [10]. The intensity of immune responses according to the date of symptom occurrence is reported in Fig. 2E–G. The decrease seen in April (14.8%) probably reflects the efficacy of the population lockdown on the disease spread. The March peak of symptom occurrence represented 82% of the seropositive individuals compared to 56% in people devoid of COVID‐19 specific IgG. Although some workers displayed an immune response corresponding to symptoms dated as early as the first week of February 2020, a sharp peak of seropositive individuals corresponded to symptoms declared in March. These results indicate that the virus was circulating in early February in the Paris conurbation then achieving a high prevalence in March.
The frequency of declared symptoms was significantly much higher in seropositive workers (79%) than in those devoid of COVID‐19 specific IgG (51%) (Table 2). If fever (66%, 142/215) was the most frequent symptoms in the seropositive population, it was also noted in individuals lacking antibodies (37%, 599/1632) suggesting a low correlation with a COVID‐19 infection (chi‐square scores 2E–16) (Table 2). In contrast, anosmia/ageusia and myalgia symptoms were highly prevalent (52%, 111/215 and 48%, 103/215, respectively) in the seropositive group but were rare in the seronegative group 3% (48/1632) and 15.7% (256/1632), respectively (Fig. 2H–J), resulting in a high correlation with COVID‐19 (chi‐square scores 5E–76 and 3E–29) (Table 2). Only anosmia/ageusia symptoms were temporally correlated with the epidemic peak in March, whereas other symptoms such as myalgia and rhinitis (Fig. 2K–M) were declared by seronegative workers mainly before but also after this peak, suggesting the consequences of other circulating infections.
Decrease of antibody titer and neutralization activity with time
To follow the antibody titers and neutralizing activity over time, a second blood sample (t 1) was obtained 4–8 weeks after the first one (t 0) from more than 1000 individuals. For the 120 samples of individuals previously found positive, the results are reported in Fig. 3A, D, and G according to the time interval between symptom onset and sampling. A clear decrease in the antibody titers and neutralization activity in pseudo‐virus assay was observed. The half‐lives of the antibody titers were 35, 87, and 28 days for anti‐N, anti‐S IgG, and neutralization activity in pseudo‐virus assay, respectively. A paired analysis showed a systematic decreased response (p < 0.0005) (Fig. 3B, E, and H). The titers of antibodies decreased by 31% and 17% for anti‐N and anti‐S IgG, respectively, for a majority of workers (>75%) and this correlated with a major decrease in the neutralization activity in pseudo‐virus assay (53%) (Fig. 3C, F, and I). Interestingly, some workers sera became negative in our assays: 15% (16/107) for LuLISA IgG/N (Fig. 3C), 14% (10/71) for PNT (Fig. 3I), and 5% (4/84) for LuLISA IgG/S (Fig. 3F). Thus, past a few months, a serological‐based survey of SARS‐CoV‐2 may run a risk of underestimating the number of formerly infected individuals.
Figure 3.
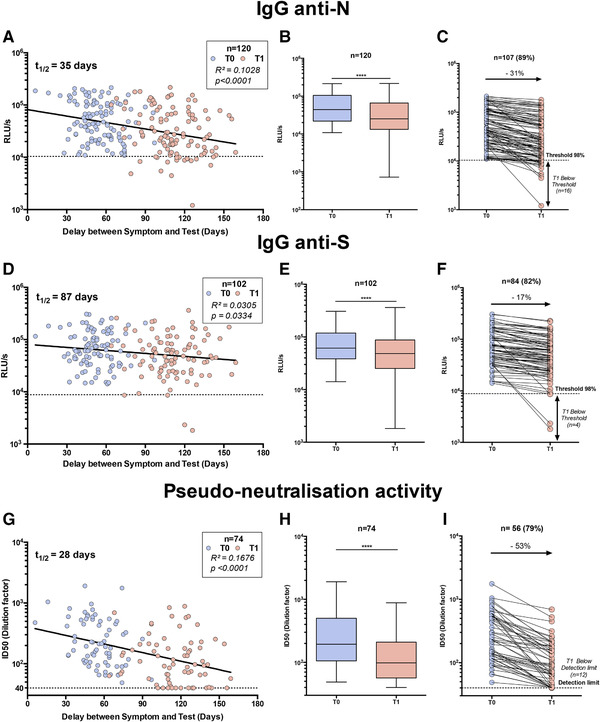
Serological profile follow‐up overtime. Workers whose serum was positive for IgG anti‐N (A–C), anti‐S (D–F), and pseudo‐neutralization activity (G–I) at the first blood sampling (t 0) were reassessed together with serum obtained 6–12 weeks later (t1 ). (A–G) Test values according to delay between date of symptom onset and the two serum analyses: t 0 (blue dots) and t 1 (red dots). Linear regression is plotted. Coefficient of determination and associated p value are indicated. (B–H) Whisker‐plots summarizing test value for both tests (t 0 and t 1). Statistical significance was determined using a Wilcoxon test (****p < 0.0001). (C–I) Individual follow‐up of seropositive workers with a decreasing value. Variation of mean (t 0 /t 1) is indicated in %.
Discussion
We report here the longitudinal study Curie‐O‐SA describing the natural immune response against the SARS‐CoV‐2 in a large population of healthy subjects working in the Paris conurbation following the March 2020 peak. Three bioluminescence‐based and sensitive high‐throughput assays including a pseudo‐typed virus neutralization activity assay allowed repeated measurements on a large number of samples. In contrast with other studies focusing on hospitalized patients or based on the occurrence of symptoms [11, 12, 13, 14, 15, 16], this study included more than half of all the employees working at three distant locations of a non especially COVID‐exposed institution with a corresponding web‐based questionnaire filled up by more than 96% of the participants.
This survey evidenced a high prevalence (11.6–16.6%) of previous infection by the SARS‐CoV‐2 in March/April 2020 along with the following points: (1) the levels of IgG/N and IgG/S were highly correlated beyond twice their positive thresholds the viral pseudo‐neutralization capacity beyond three times its positive threshold; (2) 21% of infections had been asymptomatic; (3) at least 5% of the RT‐qPCR confirmed infections and 30% (92 of 307) of the very probable infection cases according to symptoms did not develop any detectable anti‐N or anti‐S IgG antibodies nor a serum neutralization capacity; (4) the systemic IgG/N and IgG/S immunity associated with neutralization activity on pseudo‐typed virus decreased rapidly with a half‐life between 4 and 12 weeks following infection.
Still, there are some limitations in our study. (1) High‐throughput methods for assessing IgM or IgA responses were not ready at the time of the study. Assessing these isotypes may indeed be relevant since a commercial lateral flow assay detected anti‐SARS‐CoV‐2 IgM in seven out of 48 individuals featuring anosmia/ageusia but devoid of IgG response. (2) For regulatory and logistic reasons, the blood sampling started 4–6 weeks after the epidemic peak and was performed over a 2.5‐month period. The interval between the infection and the blood draw varied between 2 and 18 weeks. We may thus have missed the antibody response peak in some cases and, again, underestimated the true prevalence of the infection. (3) For logistic reasons, the blood samplings from research center staff were delayed by an average 2 weeks, possibly leading to a slight underestimation of the true prevalence of SARS‐CoV‐2 infection in the research center. (4) The use of a web‐based auto‐questionnaire leaves some space for inaccurate or selective memories as well as input errors and missing values. Indeed, despite the high response rate, the symptom onset date was missing in 18% of the cases (187/1007). Symptoms were declared by 79% of the immunized individuals but also by 51% of the individuals without antibodies, suggesting either a still higher proportion of infections without detectable systemic antibodies or the limit of self‐reported symptom questionnaire. (5) Although more than half of the employees participated to the study, a selection bias is still possible with a recruitment of those with the most symptoms leading to an overestimation of SARS‐CoV‐2 infection prevalence. (6) Neutralization assays had to be performed on pseudo‐typed virus because it could not be done using SARS‐CoV‐2 active viruses on such a large number of samples within a biosafety level three (BCL3) laboratory.
In accordance with recent studies 38/1847 individuals were RT‐qPCR positive but negative for serological tests [17] and all subject among the 215/1847 did not display a common scheme of coordinated immune response [18], which included all the parameters studied. Indeed, the largest response was the sequential occurrence of anti‐N IgG, followed by the anti‐S IgG and then the pseudo‐typed virus neutralizing activity. A few individuals were endowed with neutralizing sera without any detectable IgG against S, either because their S‐specific IgG had high affinity and a very low concentration or because other Ig than IgG were responsible for the pseudo‐typed virus neutralization activity as recently suggested for S‐specific IgA or IgM [19]. The lower prevalence of the PNT activity could be either related to a lower sensitivity of the assay or to an immune response decrease of the as evidenced in Fig. 3. Indeed, although most of the symptoms occurred from March to early April, the blood samplings were performed between May and July, leading to a variable interval between an eventual infection and the antibody response study.
Our experiments pointed out a clear cross‐recognition of IgG for SARS‐CoV‐1 and SARS‐CoV‐2 nucleoproteins but none with any of the seasonal HCoV, aside from samples displaying a very high IgG/N content (Supporting Information Fig. S4). This may be expected in view of the relatively few short peptide patterns common to all the aligned antigens sequences (Supporting Information Fig. S5). Altogether, since there are no documented cases of SARS‐CoV‐1 observed in France during the 2003 Asian outbreak and afterward, we do not expect any bias of the seroprevalence for SARS‐CoV‐2 with SARS‐CoV‐1 or the seasonal HCoV in Paris from February to June.
The seroprevalence we observed for this population of active workers in Paris conurbation (11.6%) is much higher than the one we found in a nation‐wide representative population in France in May (n = 3592, 4.9%) [20] but much lower than the response we saw in a population living in precarious conditions such as homeless (n = 543, 50.5%) or people not born in France living in workers’ residences (n = 127, 88.7%) in Paris conurbation performed with the same LuLISA and PNT methods in July [21]. Our results are consistent with other large‐scale serological studies (mostly with single time‐point and no symptom reports) in conurbations that have been subjected to the SARS‐CoV‐2 epidemic. Higher seroprevalence was found in New York City (n = 5129, 22.7%, March [22]; n = 72 401, 41.5%, March [23]), Madrid, Spain (n = 2590, 31.6%, April) [24], similar in Saint Petersburg, Russia (n = 1038, 10.8%, May) [25] and lower in Geneva (Swiss, n = 2766, 8.5%, April) [26], Wuhan, China (n = 17 368, 3.8%, March–April) [27] or nine cities of Rio Grande do Sul, Brazil (n = 4500, 0.22%, May) [28]. Seroprevalence in healthcare workers (HCW) highly exposed to COVID is also contrasted. It was high in London (n = 2167, 31.6%, May–June 2020) [12] and Paris (n = 154, 21.1%, March) [29] but lower in Essen, Germany (n = 316, 1.6%, March) [13], Madrid, Spain (n = 578, 9.3%, April) [11], and Milano, Italy (n = 789, 10.8%, March) [15].
Among seropositive individuals, 20% had been asymptomatic in this study, which is less than what has been mentioned in other studies' reports, although this is highly dependent on the recording and reporting methods: 40% in Madrid area [11], 50% in Boston area [30], and up to 80% locally in France on September 2020. This last result is a likely consequence of reinforced mask wearing policies [31] since such efficacy was also observed in Wuhan with 86% of asymptomatics in January 2020 [32], plausibly due to mask‐attenuated infectious load [31].
The pattern of symptoms displayed by the immune subjects are consistent with those reported elsewhere [33, 34]. Our results further emphasize the predictive value and specificity of the anosmia/ageusia symptoms. Aside from fatigue, only 51% of the studied individuals, 77% of the immunized individuals declared some symptoms. The higher prevalence of infection in the hospital workers (12.7%) than in the research center (8.7%) is certainly not related to difference in confinement since most contaminations occurred in March 2020 before or at the time of the lockdown. Because the hospital treated very few COVID patients, it is likely that the observed contaminations in the hospital staff are resulting from public transportation use as well as social encounters rather than work related. The difference of the RT‐qPCR number performed between the hospital and research center is mainly due to the healthcare staff priority access to SARS‐CoV‐2 RT‐qPCR assays in March and April 2020.
In this first report, we studied the immune response of 120 positive individuals 6–12 weeks after the first blood sampling. A majority of these individuals (>75%) displayed a diminishing anti‐SARS Cov2 response. Notably, the anti‐N IgG decreased faster (31%) than anti‐S IgG (17%), suggesting that if anti‐N IgG titration is a reliable marker for prevalence follow‐up during the early stage of the COVID‐19 pandemic, this assay may be less relevant for delayed studies and should be seconded with an anti‐S IgG assay. This observation emphasizes the difficulty to estimate the real seroprevalence in a large population. Interestingly, the very slow drop of anti‐S IgG titer also reported in other studies [23, 35] did not correlate with the major decrease of neutralization activity on pseudo‐typed viruses observed herein (53%). Since our pseudo‐neutralization assay is exclusively associated with anti‐S response, the neutralization activity we observed might be explained by the occurrence of other Ig isotypes, such as IgM or IgA, which would disappear much faster than the IgG from subsequent blood samples [19]. This illustrates again the serological complexity of any long‐lasting immunity.
From an epidemiological perspective, the 11.6–16.6% seroprevalence results may still underestimate the number of individuals who have been infected by the SARS‐CoV‐2 because, as discussed earlier, we also observed a lack of systemic IgG response among the RT‐qPCR positive individuals along with a gradual loss of the virus‐specific IgG titer. In the present epidemic, the rather fast decrease in antibody titers is also hindering any retrospective assessment of its true extent. Since Paris may be considered as representative of the world hard‐hit conurbations, such high prevalence of a SARS‐CoV‐2 previous infection along with a short‐lived immune response are raising the issues of possible reinfection and virus persistence in a high‐density population and may be important parameters to guide future public health policies.
Materials and methods
Participants and web‐based questionnaire
This study was registered and received ethical approval by the Comité de Protection des Personnes Méditerranée III (2020.04.18 bis 20.04.16.49458, 27/4/2020) registered in the clinical trial database (NCT04369066). Following informed consent, 18 years of age or older volunteer participant outside of any SARS‐CoV‐2 acute infectious episode in the last 7 days, working at one of the three Institut Curie locations (Paris, Orsay, or Saint‐Cloud) completed a web‐based questionnaire (Ennov Clinical) detailed in the Supporting Information. A 5 mL blood sample was taken from all participants in dry tubes. After clotting, blood was centrifuged 10 min at 2000 g. Supernatant serum was separated and frozen. Sera to be tested were thawed and distributed into 96‐well plates at the Institut Curie. An aliquot of a pool of positive (or negative) sera was distributed into six wells of each plate in a unique dispatching pattern allowing an unambiguous identification of plates.
These positive and negative wells were used to control for any drift of the measurements. The plates were then assessed at the Institut Pasteur. For the longitudinal analysis (Fig. 3), the t 0 and t 1 samples were analyzed in the same experiment from frozen serum samples.
SARS‐CoV‐2‐specific IgG assays
Development and validation of the LuLISA method are described in the Methods section of the Supporting Information. Briefly, N‐ and S‐specific IgGs were assessed using an ELISA‐based assays on sera incubated in antigen‐coated wells (Supporting Information Figs. S1 and S2) [36, 37, 38]. Antigens have been produced as follows. Full‐length N protein from SARS‐CoV‐2, SARS‐CoV1, HCoV‐HKU, HCoV‐OC43, HCoV‐NL63, and HCoV‐229E (UniProtKB ID: P0DTC9, P59595, Q5MQC6, P33469, Q6Q1R8, P15130, respectively) were produced with a (His)6 tag in the E. coli, purified on Ni‐NTA affinity column, and then size‐exclusion chromatography was performed. The ectodomain (amino acids 1–1211) of the S protein (UniProtKB ID: P0DTC2) was expressed with a trimerization foldon domain from the bacteriophage T4 fibritin and a Twin‐Strep‐tag and purified from a streptavidin‐affinity column and then a size‐exclusion chromatography was performed. White 384‐well plates with flat bottoms (Fluoronunc C384 Maxisorp, Nunc) were coated with either 1 μg/mL of nucleoprotein or spike protein in PBS buffer, 50 μL/well for 3 h at room temperature, or overnight at 4°C. Wells were washed using a plate washer (Zoom, Berthold Technologies, Germany) two cycles of three times with 100 μL of PBS/Tween 20 0.1%. Sera were diluted 200 times in PBS, nonfat milk 3%, and Tween 20 0.1%. Note that 50 μL of serum dilutions were incubated for 1 h at room temperature in their respective wells. Wells were washed two cycles of three times with 100 μL of PBS/Tween 20 0.1%. The Anti‐Fc IgG VHH (Fc1) was derived from an antibody from immunized alpaca (Vicugna pacos) [39] and expressed as a tandem with an optimized catalytic domain nanoKAZ from Oplophorous gracilirostris luciferase [40]. Purified Fc1‐nanoKAZ 1 ng/mL (400 × 106 RLU·s–1·mL–1) in PBS, nonfat milk 3%, and Tween 20 0.1% was loaded (50 μL/well) and incubated for 30 min at room temperature. Wells were washed two cycles of three times with 100 μL of PBS/Tween 20 0.1%. Emptied wells were loaded with 50 μL of the luciferin solution, resulting from hikarazine‐108 hydrolysis (Q‐108) at 13 μM. The plate was orbitaly shaken for 3 s and the collected photons were counted during 0.5 s per well and measured two times in a plate luminometer (Mithras2; Berthold, Wildbad, Germany).
Pseudo‐neutralization assays
Pseudo‐typed vectors were produced and titrated as previously described [41]. Inhibition assay of neutralization of pseudo‐typed virus–cell fusion by serum contents [9] is detailed in the Methods section of the Supporting Information and Fig. S3. Briefly, sera were decomplemented at 56°C during 30 min in a water bath and diluted from 1/40 to 1/40960 by successive fourfold dilutions, mixed, and co‐incubated with 300 TU of pseudo‐typed vector at room temperature during 30 min under agitation. Mix is then plated in tissue culture treated black 96‐well plate clear bottom (Costar) with 20 000 HEK 293T‐hACE2 cells in suspension in culture medium DMEM‐glutamax (Gibco) + 10% FCS (Gibco) + Pen/Strep (Gibco). After 48‐h incubation at 37°C 5% CO2, the medium is completely removed by aspiration and bioluminescence is measured using a Luciferase Assay System (Promega) on an EnSpire plate reader (PerkinElmer).
Statistical analysis
Seropositivity was defined as the presence of detectable anti‐SARS‐CoV‐2 antibodies against N or S. The proportion of seropositive samples was compared by time between onset of symptoms and collection of blood sample using chi‐square test.
LuLISA and pseudo‐neutralization of sera were compared by delay since onset of symptoms using the Kruskall–Wallis nonparametric test. The chi‐square test was used to evaluate the association between investigated factors and neutralization levels.
All analyses were performed using GraphPad Prism 8 (GraphPad Software, LLC). These results and the raw data of the LuLISA IgG/N and IgG/S and PNT are provided in the Supporting Information Table S1.
Author Contributions
PNT design: FA and PC; PNT processing: FA and PS; LuLISA design: SG; LuLISA processing: SG and TR; target design: MG, ZC, and NE; target production: FD, ED, OR‐LG, and SP; substrate synthesis: YJ; study logistics and sampling collection: DL, VG, FC‐B, AS, MDG, and OL; automate and plate handling: OH; data analysis: FA, TR, AIL, and OL; contribution to text and figure editing: FA, SG, AIL, YJ, and TR; writing manuscript: FA, TR, and OL.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202049058.
Conflict of interest
YJ, SG, and TR have patented the proluciferins (hikarazines) synthesis and uses (EP 3395803/WO 2018197727, 2018) and applied for a patent, which includes claims describing the LuLISA. FA and PC have applied for a patent claiming the PNT. The rest of the authors declare no commercial or financial conflict of interest.
Abbreviations
- CI
confidence interval
- COVID‐19
coronavirus disease 2019
- IgG/N
anti‐nucleoprotein IgG
- IgG/S
anti‐spike IgG
- LuLISA
luciferase‐linked immunosorbent assay
- PNT
Pseudo‐neutralization test
- RT‐qPCR
RT quantitative PCR
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
Supporting information
Supporting Information
Acknowledgements
All authors thank all the volunteers from Institut Curie for their participation to this study. The help of the personnel in the Departments of Diagnostic and Theranostic Medicine, of early phase clinical trials, and of the outpatient clinics is gratefully acknowledged. Authors thank Sylvie Arnaud, Anne Blondel, Anne‐Claire Coyne, Aurélie Dos Santos, and Cécile Simondi for their contribution in blood sampling and study logistics at Institut Curie, Pr Frédéric Pène from Institut Cochin for access to COVID‐19 patient samples and Dr. Yves Jacob from Institut Pasteur for the four HCoV genes. The authors thank Sébastien Brulé, Sylviane Hoos, Dr. Bertrand Raynal, and Dr. Patrick England from the Molecular Biophysics Platform for the quality control of protein targets at the Institut Pasteur. The authors thank Dr. Hélène Munier‐Lehmann for access to automate and supply management at the Unit of Chemistry and Biocatalysis, Institut Pasteur. The authors thank Pr J. Di Santo and Dr. D. Duffy for their comments. This work was made possible thanks to the financial support obtained through the "URGENCE nouveau coronavirus" fundraising campaign of Institut Pasteur and the financial support of the Fondation Total. This study was funded in part by a grant from Fondation de France and by Institut Curie Institutional funding. The luciferin synthesis development has been supported by DARRI (ValoExpress 2016–2018). LuLISA development has been supported by IARP Pasteur‐Carnot MI (2019‐2020). We thank Dr. Florence Miller for her technical advices and the technical support of Berthold France and Berthold Technologies Germany.
Contributor Information
Thierry Rose, Email: rose@pasteur.fr.
Olivier Lantz, Email: olivier.lantz@curie.fr.
Data availability statement
All raw data are available in the Supporting Information.
References
- 1. Guan, W. J. , Ni, Z. Y. , Hu, Y. , Liang, W. H. , Ou, C. Q. , He, J. X. , Liu, L. et al., Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020. 382: 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu, F. , Zhao, S. , Yu, B. , Chen, Y. M. , Wang, W. , Song, Z. G. , Hu, Y. et al., A new coronavirus associated with human respiratory disease in China. Nature 2020. 579: 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu, N. , Zhang, D. , Wang, W. , Li, X. , Yang, B. , Song, J. , Zhao, X. , et al., A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020. 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lescure, F. X. , Bouadma, L. , Nguyen, D. , Parisey, M. , Wicky, P. H. , Behillil, S. , Gaymard, A. et al., Clinical and virological data of the first cases of COVID‐19 in Europe: a case series. Lancet Infect. Dis. 2020. 20: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wolfel, R. , Corman, V. M. , Guggemos, W. , Seilmaier, M. , Zange, S. , Muller, M. A. , Niemeyer, D. et al., Virological assessment of hospitalized patients with COVID‐2019. Nature 2020. 581: 465–469. [DOI] [PubMed] [Google Scholar]
- 6. Guo, L. , Ren, L. , Yang, S. , Xiao, M. , Chang, D. , Yang, F. , Dela Cruz, C. S. et al., Profiling early humoral response to diagnose novel coronavirus disease (COVID‐19). Clin. Infect. Dis. 2020. 71: 778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang, A. T. , Garcia‐Carreras, B. , Hitchings, M. D. T. , Yang, B. , Katzelnick, L. C. , Rattigan, S. M. , Borgert, B. A. et al., A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv 2020. 10.1101/2020.1104.1114.20065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. GeurtsvanKessel, C. H. , Okba, N. M. A. , Igloi, Z. , Bogers, S. , Embregts, C. W. E. , Laksono, B. M. , Leijten, L. et al., An evaluation of COVID‐19 serological assays informs future diagnostics and exposure assessment. Nat. Commun. 2020. 11: 3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grzelak, L. , Temmam, S. , Planchais, C. , Demeret, C. , Tondeur, L. , Huon, C. , Guivel‐Benhassine, F. et al., A comparison of four serological assays for detecting anti‐SARS‐CoV‐2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020. 12. 10.1126/scitranslmed.abc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.SPF, Santé Publique France (French National Public Health Agency). 2020.. https://www.santepubliquefrance.fr/dossiers/coronavirus-covid-19.
- 11. Garcia‐Basteiro, A. L. , Moncunill, G. , Tortajada, M. , Vidal, M. , Guinovart, C. , Jimenez, A. , Santano, R. et al., Seroprevalence of antibodies against SARS‐CoV‐2 among health care workers in a large Spanish reference hospital. Nat. Commun. 2020. 11: 3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grant, J. J. , Wilmore, S. M. S. , McCann, N. S. , Donnelly, O. , Lai, R. W. L. , Kinsella, M. J. , Rochford, H. L. et al., Seroprevalence of SARS‐CoV‐2 antibodies in healthcare workers at a London NHS Trust. Infect. Control Hosp. Epidemiol. 2020: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Korth, J. , Wilde, B. , Dolff, S. , Anastasiou, O. E. , Krawczyk, A. , Jahn, M. , Cordes, S. et al., SARS‐CoV‐2‐specific antibody detection in healthcare workers in Germany with direct contact to COVID‐19 patients. J. Clin. Virol. 2020. 128: 104437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Plebani, M. , Padoan, A. , Fedeli, U. , Schievano, E. , Vecchiato, E. , Lippi, G. , Lo Cascio, G. et al., SARS‐CoV‐2 serosurvey in health care workers of the Veneto Region. Clin. Chem. Lab. Med. 2020. 10.1515/cclm-2020-1236 [DOI] [PubMed] [Google Scholar]
- 15. Valenti, L. , Bergna, A. , Pelusi, S. , Facciotti, F. , Lai, A. , Tarkowski, M. , Berzuini, A. et al., SARS‐CoV‐2 seroprevalence trends in healthy blood donors during the COVID‐19 Milan Outbreak. MedRxiv 2020. 10.1101/2020.1105.1111.20098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou, P. , Yang, X. L. , Wang, X. G. , Hu, B. , Zhang, L. , Zhang, W. , Si, H. R. et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020. 579: 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gudbjartsson, D. F. , Norddahl, G. L. , Melsted, P. , Gunnarsdottir, K. , Holm, H. , Eythorsson, E. , Arnthorsson, A. O. et al., Humoral immune response to SARS‐CoV‐2 in Iceland. N. Engl. J. Med. 2020. 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rosado, J. , Pelleau, S. , Cockram, C. , Merkling, S. H. , Nekkab, N. , Demeret, C. , Meola, A. et al., Serological signatures of SARS‐CoV‐2 infection: implications for antibody‐based diagnostics. MedRxiv 2020. 10.1101/2020.1105.1107.20093963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sterlin, D. , Mathian, A. , Miyara, M. , Mohr, A. , Anna, F. , Claer, L. , Quentric, P. et al., IgA dominates the early neutralizing antibody response to SARS‐CoV‐2. MedRxiv 2020. 10.1101/2020.1106.1110.20126532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Vu, S. , Jones, G. , Anna, F. , Rose, T. , Richard, J.‐B. , Bernard‐Stoecklin, S. , Goyard, S. et al., Prevalence of SARS‐CoV‐2 antibodies in France: results from nationwide serological surveillance. MedRxiv 2020. 10.1101/2020.1110.1120.20213116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roederer, T. , Mollo, B. , Vincent, C. , Nikolay, B. , Llosa, A. , Nesbitt, N. , Vanhomwegen, J. et al., High seroprevalence of SARS‐CoV‐2 antibodies among people living in precarious situations in Ile de France. MedRxiv 2020. 10.1101/2020.1110.1107.20207795 [DOI] [Google Scholar]
- 22. Rosenberg, E. S. , Tesoriero, J. M. , Rosenthal, E. M. , Chung, R. , Barranco, M. A. , Styer, L. M. , Parker, M. M. et al., Cumulative incidence and diagnosis of SARS‐CoV‐2 infection in New York. Ann. Epidemiol. 2020. 48: 23–29,e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wajnberg, A. , Amanat, F. , Firpo, A. , Altman, D. R. , Bailey, M. J. , Mansour, M. , McMahon, M. et al., Robust neutralizing antibodies to SARS‐CoV‐2 infection persist for months. Science 2020. 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Galan, I. , Velasco, M. , Casas, M. L. , Goyanes, M. J. , Rodriguez‐Caravaca, G. , Losa, J. E. , Noguera, C. et al., SARS‐CoV‐2 seroprevalence among all workers in a teaching hospital in Spain: unmasking the risk. MedRxiv 2020. 10.1101/2020.1105.1129.20116731. [DOI] [Google Scholar]
- 25. Barchuk, A. , Skougarevskiy, D. , Titaev, K. , Shirokov, D. , Raskina, Y. , Novkunkskaya, A. , Talantov, P. et al., Seroprevalence of SARS‐CoV‐2 antibodies in Saint Petersburg, Russia: a population‐based study. MedRxiv 2020. 10.1101/2020.1111.1102.20221309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stringhini, S. , Wisniak, A. , Piumatti, G. , Azman, A. S. , Lauer, S. A. , Baysson, H. , De Ridder, D. et al., Seroprevalence of anti‐SARS‐CoV‐2 IgG antibodies in Geneva, Switzerland (SEROCoV‐POP): a population‐based study. Lancet 2020. 396: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu, X. , Sun, J. , Nie, S. , Li, H. , Kong, Y. , Liang, M. , Hou, J. et al., Seroprevalence of immunoglobulin M and G antibodies against SARS‐CoV‐2 in China. Nat. Med. 2020. 26: 1193–1195. [DOI] [PubMed] [Google Scholar]
- 28. Silveira, M. F. , Barros, A. J. D. , Horta, B. L. , Pellanda, L. C. , Victora, G. D. , Dellagostin, O. A. , Struchiner, C. J. et al., Population‐based surveys of antibodies against SARS‐CoV‐2 in Southern Brazil. Nat. Med. 2020. 26: 1196–1199. [DOI] [PubMed] [Google Scholar]
- 29. Tubiana, S. , Burdet, C. , Houhou, N. , Thy, M. , Manchon, P. , Blanquart, F. , Charpentier, C. et al., High‐risk exposure without personal protective equipment and infection with SARS‐CoV‐2 in healthcare workers: results of the CoV‐CONTACT prospective cohort. MedRxiv 2020. 10.1101/2020.1109.1117.20194860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Naranbhai, V. , Chang, C. C. , Beltran, W. F. G. , Miller, T. E. , Astudillo, M. G. , Villalba, J. A. , Yang, D. et al., High seroprevalence of anti‐SARS‐CoV‐2 antibodies in Chelsea, Massachusetts. J. Infect. Dis. 2020. 10.1093/infdis/jiaa1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gandhi, M. and Rutherford, G. W. , Facial masking for covid‐19 ‐ potential for “variolation” as we await a vaccine. N. Engl. J. Med. 2020. 10.1056/NEJMp2026913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li, R. , Pei, S. , Chen, B. , Song, Y. , Zhang, T. , Yang, W. and Shaman, J. , Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS‐CoV‐2). Science 2020. 368: 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Havers, F. P. , Reed, C. , Lim, T. , Montgomery, J. M. , Klena, J. D. , Hall, A. J. , Fry, A. M. et al., Seroprevalence of Antibodies to SARS‐CoV‐2 in 10 Sites in the United States, March 23‐May 12, 2020. JAMA Intern. Med. 2020. 10.1001/jamainternmed.2020.4130. [DOI] [PubMed] [Google Scholar]
- 34. Perez‐Saez, J. , Lauer, S. A. , Kaiser, L. , Regard, S. , Delaporte, E. , Guessous, I. , Stringhini, S. and Azman, A. S. , Serology‐informed estimates of SARS‐CoV‐2 infection fatality risk in Geneva, Switzerland. Lancet Infect. Dis. 2020. 10.1016/S1473-3099(1020)30584-30583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Figueiredo‐Campos, P. , Blankenhaus, B. , Mota, C. , Gomes, A. , Serrano, M. , Ariotti, S. , Costa, C. et al., Seroprevalence of anti‐SARS‐CoV‐2 antibodies in COVID‐19 patients and healthy volunteers up to 6 months post disease onset. Eur. J. Immunol. 2020. 10.1002/eji.202048970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coutant, E. P. , Gagnot, G. , Hervin, V. , Baatallah, R. , Goyard, S. , Jacob, Y. , Rose, T. et al., Bioluminescence profiling of NanoKAZ/NanoLuc luciferase using a chemical library of coelenterazine analogues. Chemistry 2020. 26: 948–958. [DOI] [PubMed] [Google Scholar]
- 37. Coutant, E. P. , Goyard, S. , Hervin, V. , Gagnot, G. , Baatallah, R. , Jacob, Y. , Rose, T. et al., Gram‐scale synthesis of luciferins derived from coelenterazine and original insights into their bioluminescence properties. Org. Biomol. Chem. 2019. 17: 3709–3713. [DOI] [PubMed] [Google Scholar]
- 38. Goyard, S. , Balbino, B. , Chinthrajah, R. S. , Lyu, S. C. , Janin, Y. L. , Bruhns, P. , Poncet, P. et al., A highly sensitive bioluminescent method for measuring allergen‐specific IgE in microliter samples. Allergy 2020. 10.1111/all.14365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hermans, W. J. J. , Bezemer, S. and Mijnsbergen, Y. M. , Single‐domain antigen‐binding proteins that bind mammalian IgG. In B.V., B. I. (Ed.), C07K 16/42 (2006.01). 2009. [Google Scholar]
- 40. Inouye, S. , Sato, J. , Sahara‐Miura, Y. , Yoshida, S. and Hosoya, T. , Luminescence enhancement of the catalytic 19 kDa protein (KAZ) of Oplophorus luciferase by three amino acid substitutions. Biochem. Biophys. Res. Commun. 2014. 445: 157–162. [DOI] [PubMed] [Google Scholar]
- 41. Iglesias, M. C. , Mollier, K. , Beignon, A. S. , Souque, P. , Adotevi, O. , Lemonnier, F. and Charneau, P. , Lentiviral vectors encoding HIV‐1 polyepitopes induce broad CTL responses in vivo. Mol. Ther. 2007. 15: 1203–1210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
All raw data are available in the Supporting Information.


