Abstract
To investigate the relationship between Bacille Calmette‐Guérin (BCG) vaccination and SARS‐CoV‐2 by a bioinformatics approach, two datasets for the SARS‐CoV‐2 infection group and BCG‐vaccinated group were downloaded. Differentially Expressed Genes were identified. Gene ontology and pathways were functionally enriched, and networking was constructed in NetworkAnalyst. Lastly, the correlation between post‐BCG vaccination and COVID‐19 transcriptome signatures was established. A total of 161 DEGs (113 upregulated DEGs and 48 downregulated genes) were identified in the SARS‐CoV‐2 group. In the pathway enrichment analysis, a cross‐reference of upregulated Kyoto Encyclopedia of Genes and Genomes pathways in SARS‐CoV‐2 with downregulated counterparts in the BCG‐vaccinated group, resulted in the intersection of 45 common pathways, accounting for 86.5% of SARS‐CoV‐2 upregulated pathways. Of these intersecting pathways, a vast majority were immune and inflammatory pathways with top significance in interleukin‐17, tumor necrosis factor, NOD‐like receptors, and nuclear factor‐κB signaling pathways. Given the inverse relationship of the specific differentially expressed gene pathways highlighted in our results, the BCG‐vaccine may play a protective role against COVID‐19 by mounting a nonspecific immunological response and further investigation of this relationship is warranted.
Keywords: BCG vaccine, COVID‐19, differentially expressed genes, in silico analysis, networking, pathways
1. INTRODUCTION
The coronavirus pandemic now affects over 2 million people with nearly 130,000 deaths reported thus far, resulting in unprecedented ramifications in global health, social infrastructure, and economic trade. Since its emergence in Wuhan, China in 2019, SARS‐coronavirus‐2 (SARS‐CoV‐2), the causative agent of the disease, has spread rapidly across the globe to over 185 countries. Curiously, the degree of intensity has varied markedly between countries—even those with similar climates, geography, and/or healthcare infrastructure. The transmission pattern does not appear to follow previous SARS‐CoV virus transmission 1 or climatic zones based on human transmission. 2 , 3 , 4 , 5 The incidence of the COVID‐19 pandemic varies widely with the highest number of cases in the United States, Italy, China, and the lowest in regions of the world historically vulnerable to infectious diseases such as sub‐Saharan Africa. The disproportionately high morbidity and mortality of COVID‐19 infection in some countries have been linked to several factors including, but not limited to, differences in efforts to mitigate the disease spread (i.e., social distancing, travel restrictions), circulating SARS‐CoV strains, and vaccination policies.
To confront COVID‐19, it is critical to characterize the virus' biological and immunological pathways. Recent studies have uncovered the mechanism of viral entry of the SARS‐CoV‐2 relies on human angiotensin‐converting enzyme 2 (hACE2) acting as the host cell receptor. hACE2 interacts with the glycoprotein spike (S) of SARS‐CoV‐2 to form a complex structure, a receptor‐binding motif similar to some biologically related viruses, including the pathogen responsible for the Severe Acute Respiratory Syndrome (SARS). 6 , 7 , 8 , 9 , 10 , 11 Another study, which analyzed the transcriptional changes in the immune genes of three COVID patients, reported increased inflammatory responses to the virus, revealing increased T cell activation and cytokine expression, indicating proinflammatory pathways may be prognostic markers and/or serve as potential targets in COVID‐19 disease. 12 Increased interleukin (IL)‐6 expression and tumor necrosis factor (TNF‐α) have also been reported, furthering the notion that cytokines are playing a key role in COVID‐induced pneumonia. 13 Given the virus' respiratory pathology, in addition to its unique geographical distribution, some have hypothesized that protection by the Bacille Calmette‐Guérin (BCG) vaccination, a vaccine originally developed to protect against tuberculosis (TB) disease, maybe playing a critical role against COVID‐19 spread. 14 , 15 , 16 , 17 , 18 , 19
BCG is a century‐old vaccine that is given as an attenuated live strain of Mycobacterium bovis used to confer immunity against some strains of TB. Various studies have indicated that BCG vaccination has a role extending far beyond TB treatment alone, eliciting nonspecific effects (NSEs) within the innate immune system alongside the adaptive immune response. 20 BCG vaccination has been identified for its protective role specifically against respiratory viral infections, including influenza A and respiratory syncytial virus (RSV), 21 and is the standard therapy for certain types of bladder cancer. Although the exact mechanism remains elusive, BCG‐protection is thought to be conferred by epigenetic and immunological moderation of the immune response through the release of proinflammatory cytokines (TNF‐α, IL‐6, and interferon γ [IFN‐γ]) and the role of vitamin D. 22 , 23 , 24 More specifically, one proposed mechanism dictates that during the antimicrobial response (such as in M. tb infection), toll‐like receptors (TLRs) upregulate the expression of vitamin D receptor (VDR) on immune cells and pulmonary epithelial cells. VDR then binds to calcitriol, the active form of vitamin D, and together regulates the transcriptional activity of several antimicrobial peptides such as cathelicidin and β‐defensin. 25 , 26 , 27 , 28 Additionally, vitamin D itself alters T‐cell activation and IFN‐γ stimulation, increasing the expression of several proinflammatory cytokines. Overall, the NSEs following BCG vaccination are conferred by epigenetic and transcriptional modulation of the innate immune system, as evidenced by its far‐reaching role in several viral infections beyond TB alone.
Furthermore, it has been hypothesized that countries with nationalized BCG vaccination policies show decreased morbidity and mortality to COVID‐19 when compared to those where no such uniform policy exists (such as the United States or Italy); however, these preliminary data are limited given it is yet to be peer‐reviewed and fails to account for several confounding factors such as age, testing rates, 29 , 30 and the accuracy of the BGC World Atlas. 31 Nonetheless, given the safety of BCG vaccination and the well‐characterized role of NSEs, it is theorized that BCG can serve as a temporary and safe solution until a targeted vaccination becomes available. Currently, four clinical trials are already underway in Australia, the Netherlands, and the United States involving BCG versus placebo‐controlled trials in healthcare workers involved in Covid‐19 patient care (ClinicalTrial.gov; NCT04347876, NCT04327206, NCT04328441, and NCT04348370). Furthermore, there is an additional observational study in Egypt for tuberculin positivity in COVID‐19 patients versus COVID‐19 negative, BCG‐vaccinated parallel cohort (NCT04350931).
Overall, the far‐reaching effects of BCG vaccination testify to its dynamic role: Eliciting NSEs, curbing inflammation in cancer models, and reducing viremia in several distinct pathogens, including RSV, influenza A, yellow fever, and herpes simplex virus. 21 , 32 Altogether, this study aims to elucidate the relationship between BCG vaccination and SARS‐CoV‐2 through bioinformatic analysis of the biological and immunological pathways underlying both. Defining this relationship is the first step to understanding the role BCG vaccination may play in putative immunity against COVID‐19.
2. MATERIALS AND METHODS
2.1. Data source
The microarray data analyzed in this study were obtained from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE147507, published March 25, 2020. This data set analyzed the gene expression profile of a normal human bronchial epithelial (NHBE) cell line, derived from a 79‐year‐old Caucasian female, after SARS‐CoV‐2 viral infection. The following samples were analyzed; SARS‐CoV‐2 infected NHBE cells (GSM4432381, GSM4432382, and GSM4432383) compared to mock‐treated NHBE cells (GSM4432378, GSM4432379, and GSM4432380).
To compare the transcriptomic alterations in SARS‐CoV‐2 viral infection with host immune response following administration of BCG vaccine, the RNA sequencing data (GSE87186) were retrieved from the SRA database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87186) in the Biojupies Analysis Notebook (https://amp.pharm.mssm.edu/biojupies/analyze) and processed by ARCHS4 (all RNA‐seq and ChIP‐seq sample and signature search) pipeline (https://amp.pharm.mssm.edu/archs4). 33 In this data set, the immunogenicity of a recombinant BCG vaccine in healthy BCG‐naïve adults and those with no prior exposure to M. tb was evaluated. We analyzed 40 samples of eight adult vaccine recipients at five different timing postvaccination; representing Days 0, 14, 28, 56, and 84 (GSM2324141–GSM2324180). A separate analysis comparing each time point to Day 0 was first performed followed by a combined analysis for all datasets (Days 14, 28, 56, and 84) compared to Day 0.
2.2. Data preprocessing and differential expression analysis
After background correction of raw expression data, mapped multiple probes to the same genes were summarized to gene levels by using the median. Genes were filtered out based on two criteria: If variance percentile rank lower than the threshold (set at 15%) or low relative abundance (average expression signal) below 5%. To ensure similar expression distributions of each sample across the entire experiment, normalization by the log2 transformation method was performed. The quality of the normalized data set was checked with the box plot and density plot, and the Benjamini and Hochberg method was selected for the multiple testing correction. Using the Limma R package available on Bioconductor (http://bioconductor.org/packages/release/bioc/html/limma.html), differentially expressed genes (DEGs) were identified in the microarray data set. Genes with a false discovery rate (FDR) < 0.05 and log2‐fold change (FC) ≥ 1.0 were considered as significantly differentially expressed and subjected to further analysis. For the RNA seq data of the BCG vaccine, gene expression signature was generated by comparing gene expression levels of the postvaccination groups with the control group on Day 0. Altered patterns of gene expression were defined through each experimental time (FC > 1, p < .05). To visually identify the similarities and differences between different cell line samples, principal component analyses (PCA) were plotted to project high‐dimensional data into lower dimensions using a linear transformation.
2.3. Functional enrichment analysis
Gene Ontology (GO) database for functional annotation of genes was used to analyze the DEGs at the functional level in terms of biological processes, molecular function, and cellular component domains. 34 Pathway functional analysis was performed on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, Wikipathway, and Reactome pathways. 35 The significantly overrepresented GO and pathways of the upregulated and downregulated DEGs were identified by The Search Tool for the Retrieval of Interacting Genes (STRING) database version 11.0 (https://string-db.org/) and validated in Enrichr website (http://amp.pharm.mssm.edu/Enrichr/) 36 with p‐value < .05 as the cut‐off criterion.
2.4. Protein‐protein interaction (PPI) network construction
The STRING database was used to construct protein interaction pairs of the screened DEGs based on their function and scores. The setting was adjusted with high confidence (combined score) > 0.9 and the existence of experimental evidence. The network was visualized in Network Analyst (https://www.networkanalyst.ca/), a visual analytics platform for comprehensive gene expression profiling with the following threshold; degree distribution of 20 and betweenness at 10. The hub nodes in the PPI network were then identified based on the connectivity degree in the network statistics (number of neighbors).
2.5. Gene regulatory networks
The gene–miRNA interactome was constructed from experimentally validated miRNA–gene interaction data collected from TarBase v7.0 (http://diana.imis.athena-innovation.gr/) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) databases and plotted in Network Analyst. Potential transcription factors (TFs) among DEGs were first screened in the Panther (http://pantherdb.org/), JASPAR TF binding site profile database (http://jaspar.genereg.net/), and the ENCODE ChIP‐seq data (https://www.encodeproject.org/chip-seq/transcription_factor/). Additionally, upstream regulatory networks for the DEGs were built and inferred networks combining transcription factor enrichment analysis and PPI network expansion with kinase enrichment analysis were constructed in Expression2Kinase web application (http://amp.pharm.mssm.edu/X2K/).
2.6. Correlations between postvaccination host immune response and COVID transcriptome signatures
Through KEGG pathway enrichment analysis, downregulated KEGG signaling pathways following BCG vaccination were intersected with upregulated pathways in SARS‐CoV‐2 infection using Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/). Deregulated genes in each pathway were explored and the direction of expression was compared between SARS‐CoV‐2 and BCG‐vaccinated experiments in KEGG mapper (https://www.genome.jp/kegg/mapper.html).
3. RESULTS
3.1. Transcriptomic changes in SARS‐CoV‐2 infection
3.1.1. Data exploration
Raw data for the reads are provided in Table S1. After normalization, the quality was checked in density and box plots (Figure S1A,B). Mock‐treated and infected samples showed a clear discrimination pattern in PCA (Figure S1C).
3.1.2. DEG screening
Of 23,710 genes screened in the experiment, 7107 genes with constant values were removed. A total of 161 annotated genes were identified to be differentially expressed, which included 113 upregulated DEGs and 48 downregulated genes (Figure S1D). The top variable genes between the SARS‐CoV‐2 and mock‐treated samples are shown in Figure 1A. Using the most variable genes, heatmap (Figure 1B) showed activation of genes in the SARS‐CoV‐2 group and global downregulation in mock‐treated one. The detailed information on DEGs is listed in Table S2.
Figure 1.
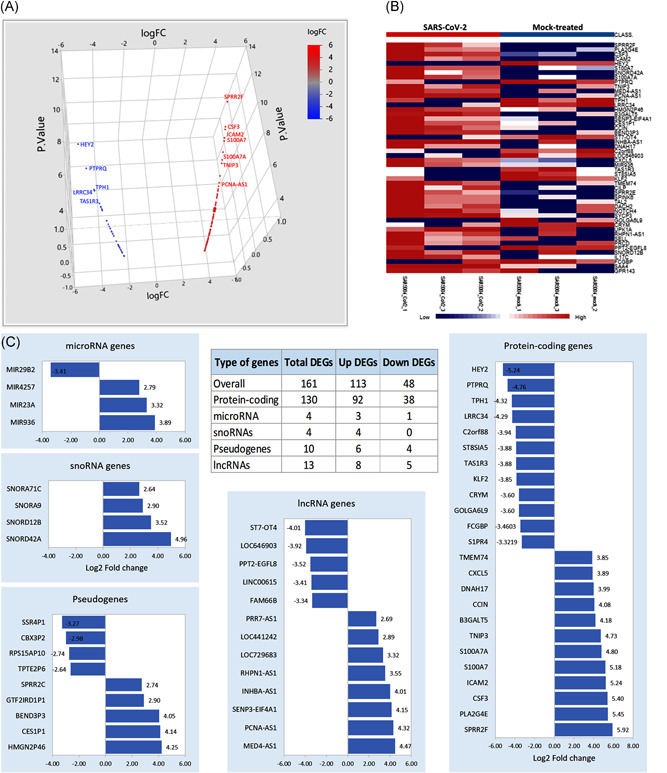
Functional annotations of differentially expressed genes (DEGs). (A) Three‐dimensional scatter plot for the upregulated (red) and downregulated (blue) DEGs. (B) Gene set enrichment analysis showing the top 100 variable genes between the six samples. (C) The numbers of upregulated and downregulated DEGs. The relative expression level of DEGs is stratified by the type of genes
The upregulated genes with the highest expression level include: (a) Small proline‐rich protein 2F (SPRR2F), which is an important structural protein that provides a protective barrier in stratified squamous epithelium and is associated with antimicrobial response; (b) Granulocyte‐colony stimulating factor (CSF3), a proinflammatory cytokine, has been shown to impair CD8+ T cell functionality and act as a modulator of T cell and dendritic cell functions, (c) intercellular adhesion molecule 2 (ICAM2) is expressed on bronchial epithelial, mediates adhesive interactions important for antigen‐specific immune response, NK‐cell mediated clearance, and lymphocyte recirculation, (d) S100A7A, immunogenic‐related calcium‐binding protein, regulated by TLR4, and associated with psoriasis, (e) TNFAIP3‐interacting protein 3 (TNIP3), which binds to zinc finger protein and inhibits nuclear factor‐κB (NF‐κB) activation induced by TNF‐α, TLR4, and (f) PCNA‐AS1, the antisense and regulator for proliferating cell nuclear antigen gene (Table S2).
Among the most significantly downregulated genes, the following are noted: (a) Hes Related Family BHLH Transcription Factor With YRPW Motif 2 (HEY2) is a nuclear transcription factor that represses DNA and negatively regulates miR‐146a, IL‐6, IL‐1β, and TNF‐α expression, (b) protein tyrosine phosphatase receptor type Q (PTPRQ) phosphatase which catalyzes the dephosphorylation of phosphotyrosine and phosphatidylinositol and plays roles in cellular proliferation and differentiation and is required for auditory function, and (c) taste 1 receptor member 3 (TAS1R3) encoding for G‐protein‐coupled receptor involved in taste responses (Table S2).
3.1.3. Functional annotations of genes
To analyze the aberrant gene expression pattern in SARS‐CoV‐2 infection, further functional analysis, and annotation for DEGs were performed (Figure 1C). Thirteen long noncoding RNAs (lncRNAs) were deregulated following the SARS‐CoV‐2 treatment. Annotation analysis revealed that PPT2‐EGFL8 readthrough (PPT2‐EGFL8, FC = −3.5169, p = 5.86E−06) was previously associated with circulating phospho‐ and sphingolipid concentrations, and others were associated with gastric, breast, and prostate cancer such as INHBA‐antisense RNA 1 (INHBA‐AS1, FC = 4.0, p = 2.41E−07), RHPN1 antisense RNA 1 (RHPN1‐AS1, FC = 3.54, p = 4.85E−06), and ST7 overlapping transcript 4 (ST7‐OT, FC = 4–4.01, p = 2.35E−07).
Genes for four miRNAs were deregulated; miR‐936 (FC = 3.88, p = 5.4E−07), miR‐23a (FC = 3.32, p = 1.8E−05), and miR‐4257 (FC = 2.79, p = 3.1E−04) were upregulated, and miR‐29b‐2 was downregulated (FC = −3.4, p = 1.12E−05). Four small nucleolar RNA (snoRNAs) genes were upregulated; including two snoRNAs, C/D box: SNORD42A (FC = 4.95, p = 1.69E−10) and SNORD12B (FC = 3.51, p = 5.86E−06), and two snoRNAs, H/ACA box: SNORA9 (FC = 2.90, p = 1.8E−04) and SNORA71C (FC = 2.63, p = 6.8E−04). In addition, 10 pseudogenes (six upregulated and four downregulated DEGs) were identified to be deregulated. Despite being nonfunctional DNA segments that resemble functional genes, some might contain inherited or acquired promoter elements and exert beneficial regulatory function (www.GeneCards.org).
As depicted in Figure 1C, DEGs included 130 protein‐coding genes (92 up and 38 down). Annotation of these genes was revealed to enclose seven transcription factors; three were activated; (a) TAL bHLH transcription factor 2 (TAL2), a basic helix‐loop‐helix transcription factor (FC = 3.74, p = 4.7E−04), (b) dachshund family transcription factor 2 (DACH2), a winged helix/forkhead transcription factor (FC = 3.68, p = .0006), and (c) AF4/FMR2 family member 2 (AFF2), a DNA‐binding transcription factor (FC = 3.40, p = .0023), and four were downregulated, (a) HEY2, basic helix‐loop‐helix transcription factor (FC = −5.24, p = 3.44E−08), and three C2H2 zinc finger transcription factor, (b) Kruppel like factor 2 (KLF2, FC = −3.85, p = 2.6E−04), (c) early growth response 2 (EGR2, FC = −2.83, p = .024), and (d) zinc finger and BTB domain containing 16 (ZBTB16, FC = −2.74, p = .0327). Furthermore, three genes for extracellular matrix structural protein were under‐expressed; (a) IgGFc‐binding protein (FCGBP, FD = −3.46, p = 8.26E−06), (b) collagen α‐3(VI) chain (COL6A3, FC = −2.93, p = .00015), and (c) complement component C1q receptor (CD93, FC = −2.74, p = .0004). Additionally, phospholipid‐transporting ATPase IM (ATP8B4), an active transporter (FC = −2.88, p = .0002) and retroviral‐like aspartic protease 1 (ASPRV1), a viral or transposable element protein, (FC = −2.83,p = .0002) were also downregulated. Two serine protease inhibitors were also downregulated; insulin‐like growth factor‐binding protein 5 (IGFBP5, FC = −2.65, p = .0006) and Serpin B10 (SERPINB10, FC = −2.81, p = .00028).
In contrast, multiple proteases were upregulated in SARS‐CoV‐2 infection compared to mock‐treated samples. Of these, SENP3‐EIF4A1 readthrough gene (SENP3‐EIF4A1, FC = 4.15, p=8.93E‐08), chymotrypsin‐like protease (CTRL‐1, FC = 2.88, p = .00019), T‐cell differentiation antigen CD6 (FC = 2.88, p = .00021), HTRA4 (FC = 2.92, p = .00016), ABHD1 (FC = 2.83, p = .0002), and cytosolic carboxypeptidase 3 AGBL3 (FC = 2.85, p = .00024). Moreover, of the upregulated DEGs, the active transporter, aquaporin (AQP7, FC = 3.07, p = .0095), the gap junction protein gamma 2 (GJC2, FC = 3.32, p = 1.87E‐05), multiple membrane traffic proteins as MX dynamin‐like GTPase 1 (MX1, FC = 2.71, p = .0004) and synaptotagmin 5 (SYT5, FC = 2.88, p = .0002), and ion channels as γ‐aminobutyric acid type A receptor subunit rho2 (GABRR2, FC = 2.74, p = .0004) and bestrophin 4 (BEST4, FC = 2.91, p = .00017) were identified (Figure 1C and Table S2).
3.1.4. Functional enrichment analysis of DEGs
To explore the functions of the DEGs, the 113 upregulated and 48 downregulated genes were subjected to GO and KEGG pathway enrichment analysis. As shown in Figure 2A–C, the significantly enriched GO terms for upregulated genes were mainly related to the acute inflammatory response (GO:0002526, FDR = 6.46E−08), response to virus (GO:0009615, FDR = 7.27E−08), and stress responses (GO:0006950, FDR = 8.37E−14). Among regulatory pathways, the regulation of immune response (GO:0050776, FDR = 2.23E−08) and viral genome replication (GO:0045071, FDR = 1.60E−07) were the most enriched. Lastly, cytokine activity (GO:0005125, FDR = 2.66E−09), chemokine activity (GO:0008009, FDR = 0.0018), and chemotaxis (GO:0006935, FDR = 5.07E−07) were further significantly enriched in upregulated genes (Table S3).
Figure 2.
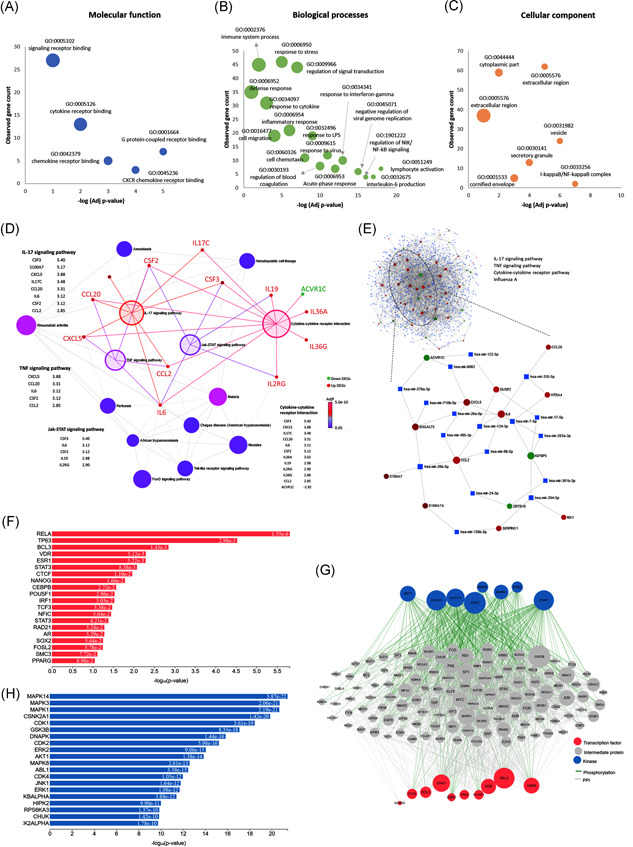
Functional enrichment analysis and gene regulatory networks. (A) Gene ontology analysis for molecular function, (B) for biological processes, (C) for cellular components, (D) The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. Nodes represented pathways, with color based on its significance. Four enlarged highlighted nodes represented the most significant pathways. Bipartite gene networks with these nodes showed the association with upregulated (red) and downregulated (green) genes. (E) Gene–microRNA interaction. The data source for interaction pairs: TarBase and miRTarBase. Red and green circles represented upregulated and downregulated differentially expressed genes, respectively. Blue squares represented the microRNAs. The full network is shown with extracted nodes (ellipse) for four significant KEGG pathways. (F) Transcription factor enrichment analysis showing putative transcription factors that most likely to regulate the differences in gene expression. (G) Upstream regulatory network that connects the enriched transcription factors to kinases through known protein–protein interactions. (H) Kinase enrichment analysis. Candidate enriched protein kinases that most likely regulate the formation of the identified transcriptional complexes. They are ranked based on the overlap between known kinase‐substrate phosphorylation interactions and the proteins in the protein–protein interaction subnetwork created in (G)
KEGG pathway analysis (Figure 2D) showed that upregulated genes were mainly enriched in the IL‐17 signaling pathway (KEGG ID: hsa04657, FDR = 1.39E−15), TNF signaling pathway (hsa04668, FDR = 5.34E−15), NOD‐like receptor (NLR) signaling pathway (hsa04621, FDR = 4.07E−10), cytokine–cytokine receptor interaction (hsa04060, FDR = 2.08E−07), Jak‐STAT signaling pathway (hsa04630, FDR = 5.38E−06), NF‐κB signaling pathway (hsa04064, FDR = 1.33E−07), and influenza A (hsa05164, FDR = 1.10E−07). Other viral pathways included Kaposi's sarcoma‐associated herpesvirus infection (hsa05167), herpes simplex infection (hsa05168), measles (hsa05162), hepatitis C (hsa05160), and Epstein‐Barr virus infection (hsa05169) (Table S4). In contrast, no significant GO terms or pathways were significant for the downregulated genes.
As depicted in Figure 2D, upregulated pathways are highly connected with inflammatory cytokines exhibiting significant crosstalk in particular. IL‐6 (also known as B‐stimulatory factor‐2) is involved in the final differentiation of B cells into immunoglobulin‐secreting cells and the induction of acute‐phase reactants. IL‐6 has sequence similarity with granulocyte colony‐stimulating factors 2 and 3 (CSF2 and CSF3), which are also highly expressed following SARS‐CoV‐2 infection. Furthermore, several interleukins (IL17C, IL19, IL36A, and IL36G) were noted to serve as proinflammatory cytokines for the regulation of dendritic cells and T cells, as well as interleukin‐2 receptor subunit gamma (IL2RG), which encodes a common gamma chain essential for IL‐receptor function. Overexpression of two members of the CC chemokine family was observed: C–C Motif chemokine ligand 2 (CCL2) and C–C motif chemokine ligand 2 (CCL20), whose function was to induce the migration of monocytes and lymphocytes, respectively (www.GeneCards.org).
3.1.5. Gene regulatory networks
The gene–miRNA interactions network was constructed (Figure 2E). It consisted of 77 seeds (significant genes), 1136 nodes, and 1776 edges mapped to the corresponding molecular interaction databases. KEGG pathway enrichment analysis of the network was significant for four pathways; namely, IL‐17 signaling pathway (p = 7E−05, Hits = 5/93), TNF signaling pathway (p = .0017, Hits = 4/110), cytokine–cytokine receptor pathway (p = .012, Hits = 5/294), and influenza A (p = .044, Hits = 3/167). After extraction of nodes relevant to these immune‐related pathways, the densely connected microRNAs in the cluster module included: miR‐26b‐5p, miR‐26a‐5p, miR‐124‐3p, miR‐7‐5p, miR‐17‐5p, miR‐335‐5p, mirR‐24‐3p, miR‐203a‐3p, and miR‐122‐5p.
Upstream regulatory cell signaling layers responsible for the observed pattern in gene expression following SARS‐CoV‐2 infection is depicted in Figure 2F–H. The network included integrated transcription factors and kinases with PPI. The top enriched transcription factors were RELA proto‐oncogene (p = 1.55E−06), BCL3 (p = .0014), TP63 (p = 2.9E−05), and VDR (p = .0051). RELA, an NF‐κB subunit, has a key role in mediating inflammation, differentiation, cell growth, tumorigenesis, and apoptosis (www.GeneCards.org). It is associated with 12 overlapping targets in the DEGs list (IER3, STAT1, NFKB2, ICAM1, DRAM1, NEDD9, IL32, NFKBIA, TNFAIP3, NFKBIZ, and TNIP1). Another NF‐κB regulator, BCL3, has a paradoxical effect depending on its subcellular localization: In the nucleus, it regulates transcriptional activation of NF‐κB target gene; however, in the cytoplasm, it inhibits the nuclear translocation of the NF‐κB p50 subunit (www.GeneCards.org). From the DEGs defined following SARS‐CoV‐2 infection, six genes (IER3, STAT1, NFKB2, NFKBIA, IRF9, and BIRC3) were targeted by BCL3. Lastly, the nuclear receptor for calcitriol, VDR, was significantly associated with the upregulated DEGs (NFKB2, NFKBIA, and IRAK2).
3.1.6. TB pathway
Gene set enrichment analysis (GSEA) showed an over‐representation of SARS‐CoV‐2 genes in the TB pathway (ID: hsa05152, p = 6.09E−04). In this pathway, 116 DEGs (of 179 genes) were upregulated (Figure S2). IL6, TLR9, and TNF were at the top of that list. VDR and CEPB are further noted for their critical regulation of inflammatory and immune responses, including SARS‐CoV‐2 infected cells.
3.2. Transcriptomic changes following BCG vaccine administration
3.2.1. DEGs
After preprocessing of the expression data from the four different stages postvaccination (Figure S3), 696, 66, 49, and 80 DEGs were detected at 14, 28, 56, and 84 days, respectively. As depicted in Figure 3A, THE principal component analysis showed the clustering of samples with discrete demarcation from the initial stage at Day 0 (controls). A heatmap of the most variable genes demonstrated an unclear difference in the transcriptomic pattern at different time points (Figure 3B). As shown in Figure 3C, the volcano plot illustrated deregulated genes. A total of 696, 66, 49, and 80 DEGs, respectively, were identified (Figure 3D). The altered expression patterns across four different time points showed host immune responses to be constantly changing post‐BCG vaccination. Stratified analysis according to the direction of DEGs showed similar results with no overlapping between up and down DEGs at each stage (Figure 3E,F).
Figure 3.

Differentially expressed genes (DEGs) and functional enrichment analysis following Bacille Calmette‐Guérin (BCG) vaccination. (A) Principal component analysis after normalization, showing a cluster of samples postvaccination on Days 14 through 84. (B) Clustergram showing the hierarchical clustering of the top 2500 variable genes among the five groups. (C) Volcano plot representing log2‐fold change and −log10 (adjusted p‐value). (D) Number of DEGs at each stage postvaccination (Days 14 vs. 0, 28 vs. 0, 56 vs. 0, and 84 vs. 0) (E, F) Venn diagram showing the intersection between upregulated and downregulated DEGs at a different stage. The numbers of upregulated and downregulated DEGs are demonstrated in blue and red boxes, respectively. (G) Protein–protein interaction (PPI) network for DEGs following BCG vaccination. String interactome for PPI showing upregulated (red nodes) and downregulated (green nodes) genes. (H) A cluster of inhibited Kyoto Encyclopedia of Genes and Genomes pathways. Circled in blue the gene list of the selected pathways. (I) Top significant pathways inhibited following BCG vaccination (enriched in the cluster)
3.2.2. Functional enrichment analysis
Activated and inhibited GO and pathway terms for each stage are listed in detail in Tables S5–S12. GO enrichment analysis was carried out in three categories, including biological processes (BP), molecular functions (MF), and cellular components (CC). The most significant terms for BP, MF, and CC were the translation (GO:0006412, FDR = 8.59E−10), RNA binding (GO:0003723, FDR = 6.98E−08), and cytosolic ribosome (GO:0022626, FDR = 1.30E−08) at Day 14; short‐chain fatty acid catabolic process (GO:0019626, FDR = 0.0008), azole transmembrane transporter activity (GO:1901474, FDR = 0.0088), and nucleolar ribonuclease P complex (GO:0005655, FDR = 0.1409) at Day 28; protein import into peroxisome matrix (GO:0016558, FDR = 0.0029), β‐1,3‐galactosyltransferase activity (GO:0048531, FDR = 0.0046), and manchette (GO:0002177, FDR = 0.0121) at Day 56; and positive regulation of T‐helper 2 cell differentiation (GO:0045630, FDR = 0.0088), primary lysosome (GO:0005766, FDR = 0.0246), and alanine transmembrane transporter activity (GO:0022858, FDR = 0.0121) at Day 84 postvaccination.
Pathway enrichment analysis showed multiple activated pathways at different stages. The most significant upregulated pathways at an early stage (Day‐14 and Day‐28 groups) were (a) ribosome (hsa03010, FDR = 3.83E−12) including multiple mitochondrial ribosomal proteins (as MRPL17, MRPS21, RPL23A, MRPL24, MRPL34, and MRPL36), and ribosomal proteins for small subunits (RPS14, RPS15, RPS16, RPS19, and RPS28), and large subunits (RPL18A, RPL36, RPLP2, RPL13, and RPL15) and (b) propionyl‐CoA catabolism (R‐HSA‐71032, FDR = 0.0001) via modulating methyl malonyl CoA epimerase (MCEE), methylmalonic aciduria type A (MMAA), and propionyl‐CoA carboxylase (PCCB) genes. However, at later stages, the top upregulated pathways were aminoacyl‐tRNA biosynthesis (hsa00970, FDR = 0.0246), scavenging of heme from plasma (R‐HSA‐2168880, FDR = 0.0056), and heme biosynthesis (WP561, FDR = 0.020).
In contrast, of the inhibited pathways after 2 weeks of vaccination were cytokine–cytokine receptor interaction (hsa04060, FDR = 2.63E−04), JAK‐STAT signaling pathway (hsa04630, FDR = 0.047), and extracellular matrix–receptor interaction (hsa04512, FDR = 0.05456). Cell adhesion molecules (hsa04514, FDR = 0.0055) and cytokines and inflammatory response (WP530, FDR = 0.137) were downregulated in the same cohorts 1 month after vaccination. However, delayed host immune response at Day 56 showed downregulation of NLR signaling pathway (hsa04621, FDR = 0.0029), PI3K‐Akt signaling pathway (hsa04151, FDR = 0.0057), and signaling by interleukins (R‐HSA‐449147, FDR = 0.0048), whereas inhibited IFN signaling (R‐HSA‐913531, FDR = 0.0014) and IFN‐α/β signaling (R‐HSA‐909733, FDR = 0.0015) were inactivated in patients at Day 84 postvaccination.
3.2.3. PPI network
On comparing combined postvaccination samples (Days 14, 28, 56, and 84) versus Day 0, 76 DEGs (41 up and 35 down) were defined. A PPI network was constructed from the combined DEGs. The clusters with densely connected nodes in the PPI network were detected (Figure 3G). KEGG enrichment analysis revealed 121 significant downregulated pathways. Eleven pathways were extracted in a separate subnetwork module; namely, influenza A, IL‐17 signaling pathway, TNF signaling pathway, chemokine signaling pathway, NLR signaling pathway, PI3K‐Akt signaling pathway, NF‐κB signaling pathway, JAK‐STAT signaling pathway, MAPK signaling pathway, Gap junction, and Leukocyte transendothelial migration (Figure 3H,I). The number of nodes was 2076, whereas the corresponding edges counted for 6123. To identify hub genes involved in the host immune response following BCG vaccination, a further GSEA of the cluster was performed. Downregulated genes with high strength of enrichment as NFKB1 (degree of connectivity with other genes = 91), RELA (degree = 95), IKBKB (degree = 60) genes were enriched in eight (out of 11) pathways, followed by PIK3CA (degree = 119), PIK3CD (degree = 59), PRKCB (degree = 26), RAF1 (degree = 69), AKT1 (degree = 221), and AKT2 (degree = 48) were involved in six (out of 11) pathways.
3.3. Comparison between transcriptomic signature in SARS‐CoV‐2 infection and BCG vaccination
3.3.1. Intersecting common pathways
A total of 52 enriched KEGG pathways were upregulated in COVID‐19 infection. On cross‐reference of these upregulated pathways with downregulated KEGG pathways in the BCG vaccination experiment, 45 common pathways were intersected accounting for 86.5% of SARS‐CoV‐2 upregulated pathways (Figure 4A–C). These pathways were categorized into the following groups: (1) Cellular processes, (2) organismal systems, (3) environmental information, and (4) human diseases. The top significant ones in the SARS‐CoV‐2 experiment were the IL‐17 signaling pathway (FDR = 1.39E−15, Hits = 14/92; overlapping genes in the pathway compared to the total gene set in the pathway), the TNF signaling pathway (FDR = 5.34E−15, Hits = 14/108 genes), and the NLR signaling pathway (FDR = 4.07E−10, Hits = 12/166 genes). Other immune‐related pathways were cytokine–cytokine receptor interaction (FDR = 2.08E−07, Hits = 11/263 genes), NF‐κB signaling pathway (FDR = 1.33E−07, Hits = 8/93 genes), and JAK‐STAT signaling pathway (FDR = 5.38E−06, Hits = 8/160 genes). In the same 45 pathways, viral infectious agents included influenza A (FDR = 1.10E−07, Hits = 10/168 genes), measles (FDR = 1.33E−07, Hits = 9/133 genes), and herpes simplex virus 1 infection (FDR = 1.33E−07, Hits = 10/181 genes), whereas bacterial infections included Legionellosis (FDR = 0.0014, Hits = 7/54 genes) and TB (FDR = 0.0039, Hits = 5/172 genes). Overlapping genes within each pathway that were significantly upregulated in SARS‐CoV‐2 infection and downregulated following BCG vaccination at the four different time points (14, 28, 56, and 84 days) are demonstrated in Table S13. Although different genes were modified at each time point, the pathways remained constantly downregulated post‐BCG vaccination.
Figure 4.

Common pathways between SARS‐CoV‐2 and BCG vaccination experiments. (A) The intersection between KEGG pathways of SARS‐CoV‐2 infected cells and BCG vaccination. (B) The expression level of some hub genes following BCG vaccination showing downregulation postvaccination as an example of the reversed direction of expression. (C) Displays 45 upregulated KEGG signaling pathways in the SARS‐CoV‐2 cell line which are downregulated following BCG vaccination. Bars represent the observed gene count for each pathway. The degree of color represents the degree of significance (the more intensity, the higher significance). KEGG signaling pathways are categorized according to the functional hierarchical classification system. (D) The expression intensity of genes in the tuberculosis KEGG pathway. Colored by the log fold change of DEGs in (1) SARS‐CoV‐2 infection compared to mock‐treated cells and (2) following BCG vaccination. BCG, Bacille Calmette‐Guérin, KEGG, Kyoto Encyclopedia of Genes and Genomes
3.3.2. Comparative enrichment analysis in the TB pathway
On comparing 116 DEGs in SARS‐CoV‐2 infected cells which were over‐represented in the 179 gene set of TB pathways with their expression pattern after BCG vaccination, the top upregulated inflammatory markers (IL6/IL12, TLR1/2/9, TNF, C3, IL1A/B, NFKB1, TGFB2, AKT1, and HLA‐DPA1/B1), and transcription factors (NFKB1 and STAT1) showed reversed direction post‐BCG vaccination. Additionally, the downregulated DEGs in SARS‐CoV‐2, including several nonreceptor serine/threonine‐protein kinase (AKT3, CAMK2D, and MAPK10), a monocyte differentiation antigen (CD14), a membrane trafficking regulatory protein (LAMP2), growth factor (TGFB3), an apoptotic regulator (BCL2), and a proinflammatory cytokine (IL18) switched their direction following BCG vaccine administration (Figure 4D). Similar paradox directions of expression level in some DEGs for SARS‐CoV‐2 cells and BCG vaccine samples were also noted in Figure S4–S10.
4. DISCUSSION
4.1. Overview
To gain insight into the engendering mechanism of the SARS‐CoV‐2 infection, the gene expression profiles of the virus were systematically analyzed through bioinformatics techniques. In this study, a total of 161 DEGs, including 113 upregulated and 48 downregulated, were screened. The biological functions of these DEGs were explored based on GO function and pathway enrichment data. Further analysis indicated significant upregulation in 45 distinct KEGG pathways in SARS‐CoV‐2 infection overlapped with pathways downregulated following the BCG vaccination. In addition to the pathway analysis, this study describes potential kinases, transcription factors, miRNAs, and lncRNAs differentially expressed in Sars‐CoV‐2 infection (Figure 2C). Several candidate DEGs, including IL‐6, CCL20, CSF2, ICAM, and CXCL1/2 were highlighted for their key roles in respiratory viral infection, TB, and overall immune function. To elucidate the relationship between COVID‐19 infection and BCG‐vaccination, the common pathways were categorized into the following groups: Inflammatory and immunoregulatory, signaling, and infectious pathways.
4.2. Common inflammatory and immunoregulatory pathways
Of SARS‐CoV‐2 infected cells, analysis implicates several upregulated genes enriched in inflammatory pathways, most significantly in the IL‐17 and NLR pathways. Conversely, these same pathways were downregulated in the BCG‐vaccinated group, suggesting BCG vaccination may compensate for aspects of pathway dysregulation in SARS‐CoV‐2 infection. In IL‐17 signaling, the family of IL‐17 mediates protective innate immunity against external pathogens and plays a central role in the self‐clearance of intracellular pathogens. 37 , 38 Additionally, T helper cells (Th17), which themselves produce Il‐17, are known to be key players in the pathogenesis of chronic inflammatory diseases and autoimmune tissue destruction. 39 Elevated Th17 responses and IL‐17 pathways are seen in COVID‐19 patients and have been linked to “cytokine storms,” a surge of proinflammatory molecules that are associated with the acute respiratory distress syndrome (ARDS) typically seen in these patients. 40 An antagonistic effect is noted in BCG vaccination, which has been shown to decrease Th17 cell maturation, suppress Th17 response, and prevent the production of IL‐17 in the Rhesus macaque model, thereby conferring nonspecific immunity. 23 , 41 Furthermore, the IL‐17 gene family eliminates self‐reactive T cells through the production of granulocyte colony‐stimulating factor (G‐CSF) and several chemokines upregulated in Sars‐CoV‐2 infected cells, including CXCL1, CXCL2, CCL20, and IL‐17 39 , 42 (Figure 2D).
The other major upregulated inflammatory pathway, the NLR‐signaling pathway, detects various pathogens and stimulates innate immune responses against them, driving the subsequent activation of cytokine, NF‐κB, and MAPK pathways. Upregulation of NLR signaling without appropriate negative feedback regulation can contribute to pathological tissue damage. 43 Innate and adaptive immune response to an invading pathogen relies on the ability of the body to recognize foreign elements and is stimulated by inflammasomes, which are intracellular multiprotein complexes such as NLR. 44 NLRC5 (short for NLR family CARD domain containing 5) regulates major histocompatibility complex class I expression during a viral infection. In accordance with this, a study reported knockdown of NLRC5 resulted in decreased levels of CD8+ cells in influenza A virus. 45 Like IL‐17 signaling, the upregulation of NLR signaling may further be contributing to the cytokine storms evident in COVID‐19 patients. These same pathways are markedly downregulated post BCG vaccination, suggesting BCG vaccination could potentially attenuate these pathways.
4.3. Common signal transduction pathways
Several signal transduction pathways were upregulated in Sars‐CoV‐2 and inversely downregulated in the BCG‐vaccinated group with TNF, NF‐κB, MAPK, and JAK/STAT signaling pathways conferring the most statistical significance. TNF signaling upregulation is heavily involved in the complex regulation of immune cells. Following trimerization with either TNFR1 (expressed nearly ubiquitously) or TNFR2 (expressed mostly in immune cells), the TNF signaling pathway can activate NF‐κB and MAPK signaling pathways downstream of it. Upon viral infection, NF‐κB proteins regulate the transcription of many genes, including antimicrobial peptides, cytokines, chemokines, stress‐response proteins, and antiapoptotic proteins. 46 In a study of Middle Eastern Respiratory Syndrome, a viral infection caused by a member of the coronavirus family, researchers found the virus downregulated antiviral cytokines (TNF‐α) as it induced proinflammatory cytokines (IL‐1β, IL‐6, and IL‐8) during initial infection. 47 In a study of SARS, yet another member of the coronavirus family, TNF‐α was similarly downregulated and induced NF‐κB activation. 46 In the SARS pathogen, ACE2 was determined to be the functional receptor of the virus through the regulation of TNF‐α, which ultimately permitted viral entry and promoted respiratory pathogenesis. Altogether, the literature on other members of the coronavirus family indicates disruption of the TNF‐signaling pathway, which is in accordance with the marked upregulation of TNF pathway signaling noted in our findings. In BCG vaccinated models in vitro, exposure of human peripheral blood mononuclear cells (PBMCs) to BCG treatment boosted IL‐6 and TNF‐α expression in response to lipopolysaccharide stimulation. 22 Similarly, BCG‐immunized adults produced high TNF‐α and IL‐1β expression 3‐month postvaccination. 23 In addition to sustained cytokine response at 3 months, the production of heterologous Th1 and Th17 remained elevated 1‐year post BCG vaccination, concluding the vaccine created long‐term immune responses to pathogens besides M. tb alone. 23 Again, we note the same TNF pathway upregulated in the SARS‐CoV‐2 virus is downregulated in BCG vaccination. Downstream of TNF, the NF‐κB signaling pathway similarly affects a broad range of biological processes, including adaptive immune, inflammatory, and stress responses. Upon viral infection, NF‐κB proteins regulate the transcription of many genes, including antimicrobial peptides, cytokines, chemokines, stress‐response proteins, and antiapoptotic proteins. 46 Among the SARS‐CoV‐2 upregulated genes in the NF‐κB pathway were CSF‐2 and IL‐6 are of particular note given their role as innate immune system mediators. 48 , 49 The upregulation of CSF‐2, produced by endothelial and immune cells, is similarly upregulated in other respiratory diseases, including mycoplasma pneumoniae and M. tb, by promoting neutrophil and macrophage inflammatory response. The upregulation of IL‐6 has been noted in other members of the coronavirus family, including the pathogenic agent responsible for SARS. In murine models, SARS‐coronavirus spike protein, which is evolutionarily conserved in Sars‐CoV‐2, induced the upregulation of IL‐6 through the NF‐κB pathway. Higher levels of proinflammatory cytokines, including IL‐6, TNF, CSF‐2, IL‐1b, and IL‐8 are noted in COVID‐19 patients with increasing levels as a predictor of the severity of pneumonia. 50
Additionally, MAPK signaling was upregulated in the Sars‐CoV‐2 infected group but inversely downregulated in the BCG‐vaccinated group. MAPK regulates cellular processes of proliferation, stress responses, as well as immune defense. 44 , 51 , 52 The MAPK signaling cascade is activated in response to external stress signals of the three MAP kinases (ERK, JNK, and p38 isoforms) with subsequent stimulation of proinflammatory cytokines by JNK and p38 signaling. 53 , 54 , 55 In the nucleus, activated JNK and p38 promote multiple effector proteins, including NF‐κB, c‐Jun, STAT1. 56 These effector proteins have been implicated in viral infections such as influenza A and HSV‐1, and their dysregulation by pathogens is associated with an impaired antiviral response by the host. 56 , 57 The downregulation of the MAPK pathway in the BCG‐vaccinated group (Figure 3I) may be able to partially antagonize the upregulation induced by the SARS‐CoV‐2 virus. Additionally, our results indicated the top three upregulated kinases in the SARS‐CoV‐2 group were MAKP14, MAPK3, and MAPK1, further corroborating the central role MAPK plays in viral pathogenesis of SARS‐CoV‐2 infection (Figure S6).
Lastly, JAK‐STAT signaling, which is a major pathway involved in cytokine signaling and subsequent inflammation, was upregulated in SARS‐CoV‐2 infection as well. IFNs induce the JAK/STAT signaling pathway, which in turn activates transcription of widespread immune protection. For this reason, several viruses have evolved to target the JAK/STAT pathway themselves. 58 Several studies have recently advocated for the potential benefit of commercially available Jak 1 and 2 inhibitors as an anti‐inflammatory treatment in COVID‐19 cases 58 ; however, there is concern from the scientific community that JAK inhibitors may promote the evolution of SARS‐CoV‐2 virus as this has been reported in herpes viruses. BCG vaccination may be key in attenuating the JAK/STAT pathway without entirely impairing IFN‐mediated response. In vitro experiments show that BCG vaccination induces two members of the suppressor of cytokine signaling (SOCS) family, SOCS1/3, eliciting a negative feedback regulator of the JAK/STAT signaling cascade via IFN‐γ regulation. 59
Overall, although TNF, NF‐κB, MAPK, and JAK/STAT signaling were upregulated in SARS‐CoV‐2 infection, they were markedly downregulated in BCG, suggesting BCG vaccination may be targeting these same pathways. The exact mechanism remains elusive, but identification of these pathways and their targets is key to mitigating pathogenesis and merits further investigation.
4.4. Infectious and human disease‐signaling pathways
BCG vaccination downregulated several pathways related to viral and bacterial inflammatory disease pathways as well, including influenza A, measles, herpes simplex virus 1, legionellosis, and TB, all of which were upregulated in SARS‐CoV‐2 infection. Prior studies have demonstrated nonspecific effects of BCG against viral infection 21 and bacterial infections. 32 , 60 In one such study, mice inoculated with BCG displayed overall increased resistance to encephalomyocarditis, murine hepatitis, type 1 and 2 herpes simplex, foot‐and‐mouth disease, and A0 and A2 influenza viruses. 61 Intercellular adhesion molecules (ICAM) included in the upregulated DEGs (Table S1), are receptors exploited by viruses for entry into host cells, intercellular signaling, and continued survival. In the human rhinovirus and influenza virus, ICAM‐1 is involved in viral protein uncoating, delivering the viral RNA genome into the cytoplasm of host cells across the lipid bilayer. 62 , 63 Other studies corroborate this, showing that most viruses interact with and induce ICAM‐1 expression, including HIV, human parainfluenza virus, and rhinovirus infection. 64 , 65 , 66 The presence of cytokines such as IFN‐γ, TGF‐β, and TNF‐α induces the expression of ICAM on human TB infected macrophages, further activating T‐Cells. 67 Intercellular adhesion molecules (or CD54), included in the upregulated DEGs, are receptors utilized for pathogen entry into host cells that promote intercellular signaling and survival of the virus in the host cell. In human rhinovirus infection and influenza virus, ICAM‐1 is involved in viral protein uncoating, delivering the viral RNA genome into the cytoplasm of host cells across a lipid bilayer. 62 , 63 Other studies corroborate this showing that most viruses interact with and induce ICAM‐1 expression, including HIV, human parainfluenza virus, and rhinovirus infection. 64 , 65 , 66 The presence of cytokines such as IFN‐γ, TGF‐β, and TNF‐α induces expression of ICAM on macrophages for activation of T cells, this was observed in human TB infected macrophages. 67 BCG immunotherapy of bladder tumor cells showed mycobacteria infection‐induced expression of ICAM‐1 molecules, thereby eliciting immune response through improved antigen presentation to T lymphocytes. 68 BCG is thought to inhibit inflammasome activation via zinc metalloproteases to improve immunogenicity 60 , 69 and remains the current standard therapy for non‐muscle‐invasive bladder cancer.
4.5. The role of VDR and other enriched transcription factors
Compared to mock‐treated cells, SARS‐CoV‐2 infected cells exhibited significant enrichment of VDR transcription factor, one of the major downstream signaling transduction nuclear receptor in TB KEGG pathway, and was significantly associated with the upregulated DEGs, namely, NFKB2, NFKBIA, and IRAK2, which are related to NF‐κB and Toll‐like receptor signaling pathways. Normally, VDR binds to calcitriol, the active form of vitamin D. The VDR‐calcitriol complex then interacts with retinoid‐X‐receptor to form a heterodimer capable of regulating transcriptional activity, leading to several pleiotropic transcriptional effects. Similar to its upregulation in SARS‐CoV‐2, VDR gene expression is continuously upregulated during HIV 70 and influenza virus infection. 71 Previously reported, VDR signaling may repress cytokine gene expression in activated T‐cells, consequently reducing inflammatory response. 28 , 72 Part of the mechanism of action of the BCG vaccine against TB is through increased production of vitamin D, which binds with its nuclear receptor VDR, resulting in the generation of antimicrobial peptides (cathelicidin and β‐defensin) and death of intracellular M. tb. 44 In short, vitamin D plays an important role in modulating the innate and adaptive immune response alongside its classically characterized role in bone health. Long before the scientific basis was understood, vitamin D was unknowingly and empirically being used to help TB patients who were sent to sanatoriums for sun‐light exposure and prescribed cod‐liver‐remedies rich in vitamin D. 73 Since then, vitamin D has been well characterized for its immunomodulatory properties, including the regulation of IFN‐γ, a key activator of macrophage response, 74 and promoting the production of regulatory T cells. 75 In addition to its role in M. tb, 76 cathelicidin is a highly conserved protein that has been shown to direct antiviral activity in several respiratory viral infections including respiratory syncytial virus, human rhinovirus (HRV), and influenza A. 77 , 78 , 79 , 80 , 81 , 82 In vitro, vitamin D has been shown to counteract the M. tb‐induced downregulation of cathelicidin by actually recovering cathelicidin levels and promoting Th1 cell differentiation. Following BCG vaccination, vitamin D levels remain elevated, indicating vitamin D is being upregulated long after the initial inoculation. It has been postulated that increased calcitriol recruits dendritic cells from the site of inoculation to the lymph nodes, accounting for the sustained response. 24 This serves as a possible mechanism of protective effects of BCG vaccination in relation to consistently elevated plasma vitamin D concentration present for as long as 9 months as reported by the authors. 24 Vitamin D deficiency has been shown to contribute to the pathogenesis of ARDS. 83 In children, the mutation in the VDR gene single‐nucleotide polymorphism reported in children was associated with a viral infection. 82 Given these findings, the transcriptional upregulation of VDR in SARS‐CoV‐2 may be contributing to the dysregulation of immunological response by upregulating cytokine response, interrupting microbial peptides, and promoting immune invasion in the lungs; therefore, the well‐characterized role of VDR in BCG vaccination could serve as a potential biomarker against COVID‐19 disease.
In addition to VDR, our bioinformatic analysis showed the deregulation of multiple transcription factors (TFs) in SARS‐CoV‐2 infection. Specifically, TAL2, DACH2, and AFF2 have upregulated; however, the functional role in the viral immune response is unclear. Conversely, transcription factors HEY2, KLF2, EGR2, and ZBTB16 were under‐expressed and were previously reported to have an eminent role in the fine‐tuning immune response. Among these, HEY2 was the most downregulated TF gene. HEY2 is a known transcription repressor, modulating cardiovascular development, neurogenesis, and oncogenesis. 84 A recent study demonstrated the role of HEY2 in inflammation, namely, chronic periodontitis, via negative regulation of IL‐6, IL‐1β, and TNF‐α expressions. 85 Similarly, KLF2 is a key player in T cell differentiation, trafficking, quiescence, and survival, in addition to the regulation of endothelial function. Mice with KLF2 deficiency experienced an activated T cell phenotype with severely reduced T cells in the periphery and increased susceptibility to HIV‐1 infection. 86 , 87 Next, the EGR2 transcription factor can control adaptive innate immunity and lead to functional impairment of T cells. 88 After infection with influenza virus, EGR2 knockout mice exhibited prolonged viral shedding, impaired CD4+ T‐cell response, and infiltration of memory precursor type CD8+ T cells into the lung with decreased IFN‐γ, TNF‐α, and granzyme B. 89 Lastly, the ZBTB16 transcription factor can mediate the ubiquitination and subsequent proteasomal degradation of target proteins. 90 Transcription factor enrichment analysis unravels multiple transcription factors modulating the deregulated genes in SARS‐CoV‐2 infected cells. RELA, a well‐known antiviral transcription factor, was the top enriched transcription factor for DEGs following SARS‐CoV‐2 infection. It is previously reported to promote the growth of cytopathic RNA viruses by extending the lifespan of infected cells and serve as the replicative niche of intracellular pathogens. 91 TP63 was also shown to be a cellular regulator of the human papillomavirus life cycle. 92
4.6. MicroRNAs, snoRNAs, and pseudogenes
In the present study, we observed some deregulated miRNAs and highlighted the relationship between DEGs and miRNA networking which may account for the transcriptomic changes evident in SARS‐CoV‐2 expression. Of particular note, mir‐26a‐5p, miR‐26b‐5p, and miR‐124‐3p, which have all been well characterized for their involvement in viral and bacterial inflammatory pathways, particularly influenza A, RSV, and M. tb, were noted here for their putative respiratory pathogenesis. In M. tb infection, miR‐26a‐5p can modulate macrophage IFN‐γ responsiveness. 93 In another study, this same microRNA was shown to be downregulated in influenza A virus in humans, contributing to viral pathogenesis. 94 The other family member, miR‐26b‐5p, can inhibit viral replication in the vesicular stomatitis virus and Sendai virus by inducing type‐1 IFN expression. 95 RSV acts to upregulate miR‐26b, which in turn inhibits Toll‐like receptor TLR4, a key sentinel in adaptive immune response. 96 A similar effect on TLRs is seen in the pathogenesis of M. tb in alveolar macrophages wherein microRNA‐124 negatively regulates TLR signaling. SARS‐CoV‐2 infected cells exhibited overexpression of miR‐23a, which was previously shown to promote the replication of human herpes simplex virus type 1 via downregulating interferon regulatory factor 1 (IRF1), an innate antiviral molecule. 97 Altogether, the highlighted cluster of microRNAs is likely regulating viral pathways relevant to SARS‐CoV‐2 infection. Posttranscriptional regulation of some genes altered in SARS‐CoV‐2 infected cells are, therefore, worthy of experimental validation.
snoRNAs and pseudogenes were found to be deregulated in SARS‐CoV‐2 cells. SnoRNAs can serve as a source of short regulatory RNA species involved in the control of processing and translation of various messenger RNAs. Despite their exact role in COVID‐19 and TB being unclear and complicated, these ncRNAs were found to be upregulated in virus‐infected human cells. They can guide chemical modifications of structural RNAs and have a role in cell–cell communication. 98 SnoRNAs were reported to have a paradox effect; they can act as mediators of host antiviral response, and on the contrary, are utilized by viruses to evade innate immunity and complete their life cycle. 98 Furthermore, different groups of pseudogenes were activated in human cells upon triggering inflammation with known cellular protein, including virus particles and chunks of bacterial cell walls, highlighting the unique functions of pseudogenes in host immune response. 99
5. LIMITATIONS
To the best of our knowledge, our study is the first to inquire into the regulatory mechanisms underlying the host immune response against SARS‐CoV‐2 infection and to provide a putative protective role of BCG vaccination. Altogether, several DEGs with a high connectivity degree in the PPI network were identified in this study, which showed the strong inverse correlation in SARS‐CoV‐2 infection and BCG vaccination with upregulation in the former and downregulation in the latter. This inverse relationship suggests BCG vaccination may mediate key pathways in SARS‐CoV‐2 infection through NSEs. Nevertheless, it is essential to recognize some limitations of administering BCG vaccines, including, the heterogeneity of BCG response due to pharmacogenomic variation of patients, contraindications of BCG‐vaccination for immunosuppressed patients, and BCG response variations in age groups. 100 Although the exact mechanisms with which BCG elicits protective effects are not entirely understood, its efficacy in infectious, oncogenic, and inflammatory diseases are well documented and may confer similar effects in COVID‐19. More research is warranted to determine whether the pathways upregulated genetically in SARS‐CoV‐2 infected cells function the same in vivo.
6. CONCLUSION
In summary, the bioinformatics approach identified key DEGs involved in the pathogenesis of COVID‐19. Significant nodes in the PPI network, including high degrees in IL‐17 signaling, TNF signaling pathway, NLR, and NF‐κB give novel insights into the inflammatory and signal transduction pathways mediating SARS‐CoV‐2 viral infection. The 45 common pathways upregulated in SARS‐CoV‐19 and downregulated in BCG vaccination suggest BCG vaccination may help to mitigate this pathway dysregulation. Although further experimental validation is warranted to verify our discoveries, the results give insights into potential biomarkers and targeted therapy to address the ongoing COVID‐19 pandemic. We maintain BCG vaccination may be a safe and cost‐effective alternative in incurring partial protection against the COVID‐19 pandemic until more targeted measures can be produced and implemented.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Eman A. Toraih and Emad Kandil designed the study. Eman A. Toraih and Mohammad H. Hussein analyzed the data. Eman A. Toraih and Emad Kandil interpreted the data. Eman A. Toraih, Jessica A. Sedhom, and Titilope M. Dokunmu searched the literature and wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Supporting information
Supporting information.
Supporting information.
Toraih EA, Sedhom JA, Dokunmu TM, et al. Hidden in plain sight: The effects of BCG vaccination in the COVID‐19 pandemic. J Med Virol. 2021;93:1950–1966. 10.1002/jmv.26707
Contributor Information
Eman A. Toraih, Email: etoraih@tulane.edu.
Emad Kandil, Email: ekandil@tulane.edu.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Kissler SM, Tedijanto C, Goldstein E, Grad Y, Lipsitch M. Projecting the transmission dynamics of SARS‐CoV‐2 through the postpandemic period. Science. 2020;5793:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chinazzi M, Davis JT, Ajelli M, et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID‐19) outbreak. Science. 2020;368:395‐400. 10.1126/science.aba9757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kucharski AJ, Russell TW, Diamond C, et al. Early dynamics of transmission and control of COVID‐19: a mathematical modelling study. Lancet Infect Dis. 2020;20:553‐558. 10.1016/S1473-3099(20)30144-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harbert R, Cunningham SW, Tessler M. Spatial modeling could not differentiate early SARS‐CoV‐2 cases from the distribution of humans on the basis of climate in the United States. PeerJ. (2020);8:e10140. 10.7717/peerj.10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deshwal VK. COVID 19: A comparative study of Asian, European, American continent. Int J Sci Res Eng Dev. 2020;3:436‐440. [Google Scholar]
- 6. Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS‐CoV‐2 and other human coronaviruses. Trends Immunol. 2020;41:1‐5. 10.1016/j.it.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;180:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li W, Moore MJ, Vasilieva N, et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265‐269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS‐CoV‐2 entry by using human ACE2. Cell. 2020;181:894‐904. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ong EZ, Chan YFZ, Leong WY, et al. A dynamic immune response shapes COVID‐19 progression. Cell Press. 2020;27:879‐882. 10.1016/j.chom.2020.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang C, Wu Z, Li J, Zhao H, Wang G. The cytokine release syndrome (CRS) of severe COVID‐19 and interleukin‐6 receptor (IL‐6R) antagonist tocilizumab may be the key to reduce the mortality. Int J Antimicrob Agents. 2020;55(5):105954. 10.1016/j.ijantimicag.2020.105954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hegarty PK, Service NH, Kamat AM, Dinardo A. BCG vaccination may be protective against Covid‐19. medRxiv. 2020. 10.13140/RG.2.2.35948.10880 [DOI] [Google Scholar]
- 15. Gursel M, Gursel I. Is global BCG vaccination coverage relevant to the progression of SARS‐CoV‐2 pandemic? [published online ahead of print April 5, 2020]. Med Hypotheses. 10.1016/j.mehy.2020.109707 [DOI] [Google Scholar]
- 16. Dayal D, Gupta S. Connecting BCG vaccination and COVID‐19: additional data. medRxiv. 2020:2755657. 10.1101/2020.04.07.20053272 [DOI] [Google Scholar]
- 17. Miller A, Reandelar MJ, Fasciglione K, Roumenova V, Li Y, Otazu GH. Correlation between universal BCG vaccination policy and reduced morbidity and mortality for COVID‐19: an epidemiological study. J Chem Inform Model. 2012;53:1689‐1699. [Google Scholar]
- 18. Shet A, Ray D, Malavige N, Santosham M, Bar‐Zeev N. Differential COVID‐19‐attributable mortality and BCG vaccine use in countries. medRxiv. 2020. 10.1101/.04.01.20049478 [DOI] [Google Scholar]
- 19. Berg MK, Yu Q, Salvador CE, Melani I, Kitayama S. Mandated Bacillus Calmette‐Guérin (BCG) vaccination predicts flattened curves for the spread of COVID‐19. Sci Adv. 2020;6(32):eabc1463. 10.1126/sciadv.abc1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shann F. Nonspecific effects of vaccines and the reduction of mortality in children. Clinical Ther. 2013;35:109‐114. 10.1016/j.clinthera.2013.01.007 [DOI] [PubMed] [Google Scholar]
- 21. Moorlag SJ, Arts RJ, van Crevel R, Netea MG. Non‐specific effects of BCG vaccine on viral infections. Clin Microbiol Infect. 2019;25:1473‐1478. [DOI] [PubMed] [Google Scholar]
- 22. Netea MG, Schlitzer A, Placek K, Joosten LA, Schultze JL. Innate and adaptive immune memory: an evolutionary continuum in the host's response to pathogens. Cell Host Microbe. 2019;25:13‐26. [DOI] [PubMed] [Google Scholar]
- 23. Kleinnijenhuis J, Quintin J, Preijers F, et al. Long‐lasting effects of BCG vaccination on both heterologous Th1/Th17 responses and innate trained immunity. J Innate Immun. 2014;6:152‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lalor MK, Ben‐Smith A, Gorak‐Stolinska P, et al. Population differences in immune responses to Bacille Calmette‐Guérin vaccination in infancy. J Infect Dis. 2009;199:795‐800. 10.1086/597069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McMahon L, Schwartz K, Yilmaz O, Brown E, Ryan LK, Diamond G. Vitamin D‐mediated induction of innate immunity in gingival epithelial cells. Infect Immun. 2011;79:2250‐2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lagishetty V, Liu NQ, Hewison M. Vitamin D metabolism and innate immunity. Mol Cell Endocrinol. 2011;347:97‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prietl B, Treiber G, Pieber TR, Amrein K. Vitamin D and immune function. Nutrients. 2013;5:2502‐2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin R. Crosstalk between vitamin D metabolism, VDR signalling, and innate immunity. BioMed Res Int. 2016;2016:73–77. 10.1155/2016/1375858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hensel J, Mcandrews KM, Mcgrail DJ, Dowlatshahi DP. Exercising caution in correlating COVID‐19 incidence and mortality rates with BCG vaccination policies due to variable rates of SARS CoV‐2 testing. medRxiv. 2020. 10.1101/2020.04.08.20056051 [DOI] [Google Scholar]
- 30. Anthony SJ, Johnson CK, Greig DJ, et al. Global patterns in coronavirus diversity. Virus Evol. 2017;3:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kirov S. Association between BCG policy is significantly confounded by age and is unlikely to alter infection or mortality rates. medRxiv. 2020. 10.1101/2020.04.06.20055616 [DOI] [Google Scholar]
- 32. Arts RJW, Moorlag SJCFM, Novakovic B, et al. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe. 2018;23:89‐100 [DOI] [PubMed] [Google Scholar]
- 33. Lachmann A, Torre D, Keenan AB, et al. Massive mining of publicly available RNA‐seq data from human and mouse. Nature Comm. 2018;9:1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27e30‐30e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;128:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cua DJ, Tato CM. Innate IL‐17‐producing cells: the sentinels of the immune system. Nat Rev Immunol. 2010;10:479‐488. [DOI] [PubMed] [Google Scholar]
- 38. Rudner XL, Happel KI, Young EA, Shellito JE. Interleukin‐23 (IL‐23)‐IL‐17 cytokine axis in murine Pneumocystis carinii infection. Infect Immun. 2007;75:3055‐3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuwabara T, Ishikawa F, Kondo M, Kakiuchi T. The role of IL‐17 and related cytokines in inflammatory autoimmune diseases. Mediators Inflamm. 2017;2017:1‐11. 10.1155/2017/3908061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu D, Yang X. TH17 responses in cytokine storm of COVID‐19: An emerging target of JAK2 inhibitor Fedratinib. J Microb Immun Infect. 2020;53:368‐370. 10.1016/j.jmii.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dijkman K, Sombroek CC, Vervenne RAW, et al. Prevention of tuberculosis infection and disease by local BCG in repeatedly exposed rhesus macaques. Nat Med. 2019;25:255‐262. [DOI] [PubMed] [Google Scholar]
- 42. Ye P, Rodriguez FH, Kanaly S, et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony‐stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519‐527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhong Z, Umemura A, Sanchez‐Lopez E, et al. NF‐κB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164:896‐910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu T, Zhang L, Joo D, Sun SC. NF‐κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lupfer CR, Stokes KL, Kuriakose T, Kanneganti T. Deficiency of the NOD‐Like receptor NLRC5 results in decreased CD8+ T cell function and impaired viral clearance. J Virol. 2017;91:e00377‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang W, Ye L, Ye L, et al. Up‐regulation of IL‐6 and TNF‐α induced by SARS‐coronavirus spike protein in murine macrophages via NF‐κB pathway. Virus Res. 2007;128:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kindrachuk J, Ork B, Hart BJ, et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East Respiratory Syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob Agents Chemother. 2015;59:1088‐1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Begum NA, Ishii K, Kurita‐Taniguchi M, et al. Mycobacterium bovis BCG cell wall‐specific differentially expressed genes identified by differential display and cDNA subtraction in human macrophages. Infect Immun. 2004;72:937‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Verreck FA, de Boer T, Langenberg DM, Zanden L, Ottenhoff TH. Phenotypic and functional profiling of human proinflammatory type‐1 and anti‐inflammatory type‐2 macrophages in response to microbial antigens and IFN‐gamma‐ and CD40L‐mediated costimulation. J Leukoc Biol. 2006;79:285‐293. [DOI] [PubMed] [Google Scholar]
- 50. Chen, G. , et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020:1‐10. 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Ann Rev Immunol. 2002;20:55‐72. [DOI] [PubMed] [Google Scholar]
- 52. Arthur JSC, Ley SC. Mitogen‐activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679‐692. [DOI] [PubMed] [Google Scholar]
- 53. Kaminska B. MAPK signalling pathways as molecular targets for anti‐inflammatory therapy—from molecular mechanisms to therapeutic benefits. Biochim Biophys Acta. 2005;1754:253‐262. [DOI] [PubMed] [Google Scholar]
- 54. Zhang Y, Dong C. Regulatory mechanisms of mitogen‐activated kinase signaling. Cell Mol Life Sci. 2007;64:2771‐2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Soares‐Silva M, Diniz FF, Gomes GN, Bahia D. The mitogen‐activated protein kinase (MAPK) pathway: role in immune evasion by trypanosomatids. Frontiers Microbiol. 2016;7:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kumar R, Khandelwal N, Thachamvally R, et al. Role of MAPK/MNK1 signaling in virus replication. Virus Res. 2020;253:48‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hirasawa K, Kim A, Han HS, Han J, Jun HS, Yoon JW. Effect of p38 mitogen‐activated protein kinase on the replication of encephalomyocarditis virus. J Virol. 2003;77:5649‐5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Richardson P, Griffin I, Tucker C, et al. Baricitinib as potential treatment for 2019‐nCoV acute respiratory disease. Lancet. 2020;395:e30‐e31. 10.1016/S0140-6736(20)30304-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Imai K, Kurita‐Ochiai T, Ochiai K. Mycobacterium bovis bacillus Calmette‐Gueérin infection promotes SOCS induction and inhibits IFN‐γ‐stimulated JAK/STAT signaling in J774 macrophages, FEMS Immunol . Med Microb. 2003;39:173‐180. [DOI] [PubMed] [Google Scholar]
- 60. Iqbal NT, Hussain R. Non‐specific immunity of BCG vaccine: a perspective of BCG immunotherapy. Trials Vaccinol. 2014;3:143‐149. [Google Scholar]
- 61. Floc'h F, Werner GH. Increased resistance to virus infections of mice inoculated with BCG (Bacillus Calmette‐Guérin). Ann Immunol. 1976;127:173‐186. [PubMed] [Google Scholar]
- 62. Othumpangat S, Noti JD, McMillen CM, Beezhold DH. ICAM‐1 regulates the survival of influenza virus in lung epithelial cells during the early stages of infection. Virol. 2016;487:85‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bella J, Rossmann MG. ICAM‐1 receptors and cold viruses. Pharmacochem Lib. 2000;31:291‐297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Christensen JP, Johansen J, Marker O, Thomsen AR. Circulating intercellular adhesion molecule‐1 (ICAM‐1) as an early and sensitive marker for virus‐induced T cell activation. Clin Exp Immunol. 1995;102:268‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gao J, Choudhary S, Banerjee AK, De BP. Human parainfluenza virus type 3 upregulates ICAM‐1 (CD54) expression in a cytokine‐independent manner. Gene Expr. 2000;9:115‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scheglovitova O, Scanio V, Fais S, et al. Antibody to ICAM‐1 mediates enhancement of HIV‐1 infection of human endothelial cells. Arch Virol. 1995;140:951‐958. [DOI] [PubMed] [Google Scholar]
- 67. DesJardin LE, Kaufman TM, Potts B, Kutzbach B, Yi H, Schlesinger LS. Mycobacterium tuberculosis‐infected human macrophages exhibit enhanced cellular adhesion with increased expression of LFA‐1 and ICAM‐1 and reduced expression and/or function of complement receptors, FcgammaRII and the mannose receptor. Microbiol. 2002;148:3161‐3171. [DOI] [PubMed] [Google Scholar]
- 68. Jackson AM, Alexandroff AB, McIntyre M, Esuvaranathan K, James K, Chisholm GD. Induction of ICAM 1 expression on bladder tumours by BCG immunotherapy. J Clin Pathol. 1994;47:309‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Master SS, Rampini SK, Davis AS, et al. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe. 2008;3:224‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nevado J, Tenbaum SP, Castillo AI, Sánchez‐Pacheco A, Aranda A. Activation of the human immunodeficiency virus type I long terminal repeat by 1 alpha, 25‐dihydroxyvitamin D3. J Mol Endocrinol. 2007;38:587‐601. [DOI] [PubMed] [Google Scholar]
- 71. Rieder FJJ, Gröschel C, Kastner MT, et al. Human cytomegalovirus infection downregulates vitamin‐D receptor in mammalian cells. J Steroid Biochem Mol Biol. 2017;165:356‐362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. White JH. Vitamin D signaling, infectious diseases, and regulation of innate immunity. Infect Immun. 2008;76:3837‐3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jarrett P, Scragg R. A short history of phototherapy, vitamin D and skin disease. Photochem Photobiol Sci. 2017;16:283‐290. [DOI] [PubMed] [Google Scholar]
- 74. Lemire JM, Archer DC, Beck L, Spiegelberg HL. Immunosuppressive actions of 1,25‐dihydroxyvitamin D3: preferential inhibition of Th1 functions. J Nutr. 1995;125:1704S‐1708S. [DOI] [PubMed] [Google Scholar]
- 75. Griffin MD, Xing N, Kumar R. Vitamin D and its analogs as regulators of immune activation and antigen presentation. Ann Rev Nutr. 2003;23:117‐145. [DOI] [PubMed] [Google Scholar]
- 76. Sonawane A, Santos JC, Mishra BB, et al. Cathelicidin is involved in the intracellular killing of mycobacteria in macrophages. Cell Microbiol. 2011;13:1601‐1617. [DOI] [PubMed] [Google Scholar]
- 77. Sousa FH, Casanova V, Findlay F, et al. Cathelicidins display conserved direct antiviral activity towards rhinovirus. Peptides. 2017;95:76‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Currie SM, Findlay EG, McHugh BJ, et al. The human cathelicidin LL‐37 has antiviral activity against respiratory syncytial virus. PLOS One. 2013;8:e73659. 10.1371/journal.pone.0073659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gunville CF, Mourani PM, Ginde AA. The role of vitamin D in prevention and treatment of infection. Inflamm Allergy Drug Targets. 2013;12:239‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liu PT. Toll‐like receptor triggering of a vitamin D‐mediated human antimicrobial response. Science. 2006;311:1770‐1773. 10.1126/science.1123933 [DOI] [PubMed] [Google Scholar]
- 81. Roth DE, Soto G, Arenas F, et al. Association between vitamin D receptor gene polymorphisms and response to treatment of pulmonary tuberculosis. J Infect Dis. 2004;190:920‐927. [DOI] [PubMed] [Google Scholar]
- 82. Kresfelder TL, Janssen R, Bont L, Venter M. Confirmation of an association between single nucleotide polymorphisms in the VDR gene with respiratory syncytial virus‐related disease in South African children. J Med Virol. 2011;83:1834‐1840. [DOI] [PubMed] [Google Scholar]
- 83. Dancer RCA, Parekh D, Lax S, et al. Vitamin D deficiency contributes directly to the acute respiratory distress syndrome (ARDS). Thorax. 2015;70:617‐624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fischer A, Gessler M. Hey genes in cardiovascular development. Trends Cardiovasc Med. 2003;13:221‐226. [DOI] [PubMed] [Google Scholar]
- 85. Lina S, Lihong Q, Di Y, Bo Y, Xiaolin L, Jing M. microRNA‐146a and Hey2 form a mutual negative feedback loop to regulate the inflammatory response in chronic apical periodontitis. J Cell Biochem. 2018;120:645‐657. [DOI] [PubMed] [Google Scholar]
- 86. Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Krüppel‐like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996‐2001. [DOI] [PubMed] [Google Scholar]
- 87. Carlson CM, Endrizzi BT, Wu J, et al. Kruppel‐like factor 2 regulates thymocyte and T‐cell migration. Nature. 2006;442:299‐302. [DOI] [PubMed] [Google Scholar]
- 88. Miao T, Symonds ALJ, Singh R, et al. Egr2 and 3 control adaptive immune responses by temporally uncoupling expansion from T cell differentiation. J Exp Med. 2017;214:1787‐1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Du N, Kwon H, Li P, et al. Role for EGR2 in the T‐cell response to influenza. Proc Natl Acad Sci. 2014;111:16484‐16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB‐Cullin 3‐Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001‐1007. [DOI] [PubMed] [Google Scholar]
- 91. Bais SS, Ratra Y, Khan NA, et al. Chandipura virus utilizes the prosurvival function of RelA NF‐κB for its propagation. J Virol. 2019;93:e00081‐19. 10.1128/JVI.00081-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mighty KK, Laimins LA. p63 is necessary for the activation of human papillomavirus late viral functions upon epithelial differentiation. J Virol. 2011;85:8863‐8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ni B, Rajaram MV, Lafuse WP, Landes MB, Schlesinger LS. Mycobacterium tuberculosis decreases human macrophage IFN‐γ responsiveness through miR‐132 and miR‐26a. J Immunol. 2014;193:4537‐4547. [DOI] [PubMed] [Google Scholar]
- 94. Tambyah PA, Sepramaniam S, Mohamed Ali J, et al. microRNAs in circulation are altered in response to influenza A virus infection in humans. PLOS One. 2013;8:e76811. 10.1371/journal.pone.0076811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liu C, Zhang L, Xu R, Zheng H. MiR‐26b inhibits virus replication through positively regulating interferon signaling. Viral Immunol. 2018;31:676‐682. [DOI] [PubMed] [Google Scholar]
- 96. Liu S, Gao L, Wang X, Xing Y. Respiratory syncytial virus infection inhibits TLR4 signaling via up‐regulation of miR‐26b. Cell Biol Int. 2015;39:1376‐1383. [DOI] [PubMed] [Google Scholar]
- 97. Ru J, Sun H, Fan H, et al. MiR‐23a facilitates the replication of HSV‐1 through the suppression of interferon regulatory factor 1. PLOS One. 2014;9:e114021. 10.1371/journal.pone.0114021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Stepanov GA, Filippova JA, Komissarov AB, Kuligina EV, Richter VA, Semenov DV. Regulatory role of small nucleolar RNAs in human diseases. BioMed Res Int. 2015;2015:206849‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti‐inflammatory therapeutics. eLife. 2013;2:e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rowland R, McShane H. Tuberculosis vaccines in clinical trials. Expert Rev Vaccines. 2011;10:645‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


