Abstract
This study assesses the clinical performance of three anti‐SARS‐CoV‐2 assays, namely EUROIMMUN anti‐SARS‐CoV‐2 nucleocapsid (IgG) ELISA, Elecsys anti‐SARS‐CoV‐2 nucleocapsid (total antibodies) assay, and LIAISON anti‐SARS‐CoV‐2 spike proteins S1 and S2 (IgG) assay. One hundred and thirty‐seven coronavirus disease 2019 (COVID‐19) samples from 96 reverse‐transcription polymerase chain reaction confirmed patients were chosen to perform the sensitivity analysis. Non‐SARS‐CoV‐2 sera (n = 141) with a potential cross‐reaction to SARS‐CoV‐2 immunoassays were included in the specificity analysis. None of these tests demonstrated a sufficiently high clinical sensitivity to diagnose acute infection. Fourteen days since symptom onset, we did not find any significant difference between the three techniques in terms of sensitivities. However, Elecsys performed better in terms of specificity. All three anti‐SARS‐CoV‐2 assays had equivalent sensitivities 14 days from symptom onset to diagnose past‐COVID‐19 infection. We also confirmed that anti‐SARS‐CoV‐2 determination before Day 14 is of less clinical interest.
Keywords: COVID‐19, cut‐off, SARS‐CoV‐2, serology, symptom onset
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), the causative agent of coronavirus disease 2019 (COVID‐19), has led to significant morbidity and mortality. 1 The number of confirmed cases exceeds 7.8 million and the number of deaths worldwide stands at 431,541. 2
The considered standard method of reference for the diagnosis of SARS‐CoV‐2 infection is (real‐time) reverse‐transcription polymerase chain reaction (RT‐PCR) in respiratory samples. 3 However, the accuracy of the method depends on several factors including pre‐analytical variables like sample type, collection, transport, and storage. 4 The time since infection and the viral load are other factors affecting the sensitivity of the RT‐PCR. 3 In addition, RT‐PCR is not able to detect past infection 5 and the throughput of RT‐PCR is also limited because it requires a high workload, skillful operators, expensive instrumentation, and crucial biosafety measures. 6 Access to RT‐PCR tests remains limited in many countries worldwide while the virus is present in 188 countries. 7
The detection of anti‐SARS‐CoV‐2 antibodies represents an additional method for the diagnosis of COVID‐19, especially in patients who present late, with a low viral load. 8 Detection of anti‐SARS‐CoV‐2 antibodies is also useful to identify convalescent plasma donors and to screen the population to determine seroprevalence. 9 , 10
A wide range of serology immunoassays has therefore been developed to complement the RT‐PCR, with different SARS‐CoV‐2 antigen targets and formats. 3 More than 100 manufacturers have notified that they are offering or plan to offer serological testing. 11 Due to the widespread dissemination of these methods and the limited experience with these new assays, it is essential for laboratories to independently validate these methods to assure they are in line with the expected analytical and clinical performance. 3 , 5 , 8 , 12 , 13 , 14 , 15 This is also the reason why some national authorities are planning broad validation campaigns to ensure they will offer the population approved and controlled immunoassays which are the cornerstone to fight this pandemic.
The aim of the present study is to assess and compare the clinical performance of three fully automated anti‐SARS‐CoV‐2 immunoassays, namely: EUROIMMUN anti‐SARS‐CoV‐2‐nucleocapsid (IgG) ELISA, Elecsys anti‐SARS‐CoV‐2‐nucleocapsid (total antibodies) assay, and LIAISON anti‐SARS‐CoV‐2 spike proteins S1 and S2 (IgG) assay.
2. MATERIALS AND METHODS
2.1. Study design
This retrospective study was conducted from May 6 to 25, 2020 at the clinical biology laboratory of the Clinique Saint‐Luc Bouge (SLBO, Namur, Belgium). A total of 137 serum samples were obtained from 96 COVID‐19 patients confirmed positive for SARS‐CoV‐2 by RT‐PCR. Antibody kinetics since the onset of symptoms was evaluated in the full cohort of patients. Non‐SARS‐CoV‐2 sera (n = 141) with a potential cross‐reaction to SARS‐CoV‐2 immunoassays were included in the specificity analysis. Clinical performance were evaluated on three different platforms. Analytical performance are only reported for the newly available EUROIMMUN nucleocapsid assay as the analytical performance of the Elecsys anti‐SARS‐CoV‐2‐nucleocapsid (total antibodies) assay, and LIAISON anti‐SARS‐CoV‐2 spike proteins S1 and S2 (IgG) assay have already been reported elsewhere. 15 , 16 , 17 , 18
2.2. Sample collection
Blood samples were collected from patients into serum‐gel tubes (BD Vacutainer® 8.5 ml tubes; Becton Dickinson) or in lithium‐heparin plasma tubes (BD Vacutainer® 4.0 ml tubes) according to standardized operating procedures. The manufacturer recommendations authorize the use of these two matrices. Samples were centrifuged for 10 min at 1885g (ACU Modular® Pre Analytics, Roche Diagnostics®). One hundred thirty‐seven sera from 96 COVID‐19 patients were collected from March 21 to May 25, 2020.
The study population displayed the following characteristics: There were 45 females and 51 males aged 24 to 93 years (mean age = 63 years). Information on the days since the onset of symptoms was retrieved from medical records. Symptoms included fever, cough, fatigue, muscle aches, chest pain or pressure, difficulty breathing or shortness of breath, headache, sore throat, diarrhea, loss of taste, and loss of smell. Fever was the most frequent symptom (68.1%), followed by cough (60.4%), fatigue (58.2%), difficulty breathing (45.1%), and muscle aches (31.9%).
Non‐SARS‐CoV‐2 sera with a potential cross‐reaction to the SARS‐CoV‐2 immunoassay were collected before December 2019. Thirty‐seven samples were kindly provided by the Department of Laboratory Medicine of Iris Hospitals South in Brussels. Samples were stored in the laboratory serum biobank at −20°C. Frozen samples were thawed one hour at room temperature on the day of the analysis. Re‐thawed samples were vortexed before the analysis.
2.3. Analytical procedures
Three anti‐SARS‐CoV‐2 immunoassays were evaluated.
The anti‐SARS‐CoV‐2‐nucleocapsid ELISA (EUROIMMUN Medizinische Labordiagnostika AG) is used for the in vitro semiquantitative detection of IgG (also IgA and IgM, according to the insert kit of the manufacturer) to SARS‐CoV‐2 in human serum and plasma. 19 All measurements were performed on the EUROIMMUN Analyzer I‐2P®. The result of a sample is given in the form of a ratio (extinction of patient sample/extinction of calibrator). According to the manufacturer, a ratio < 0.80 is considered negative, a ratio ≥ 0.80 to < 1.10 considered borderline, and a ratio ≥ 1.10 considered positive. 19
The Elecsys anti‐SARS‐CoV‐2 nucleocapsid electrochemiluminescent immunoassay (ECLIA) (Roche Diagnostics) is used for the in vitro qualitative detection of total antibodies (including IgG) to SARS‐CoV‐2 in human serum and plasma. All measurements were performed on the Cobas® e801 module. The test result is given as a cut‐off index (COI). According to the manufacturer, a result <1.00 is considered negative while a result ≥1.00 is considered positive. 5
The LIAISON SARS‐CoV‐2 spike proteins S1/S2 assay (DiaSorin) is used for the in vitro quantitative detection of IgG to SARS‐CoV‐2 in human serum and plasma. All measurements were performed on the LIAISON‐XL analyzer. The test result is given as arbitrary units per ml (AU/ml). According to the manufacturer, a result < 12.0 is considered negative, a result ≥12.0 to <15.0 considered borderline, and a result ≥5.0 considered positive. 15
Only one calibration curve was done, and one batch of reagent was used for each of these platforms.
The RT‐PCR for SARS‐CoV‐2 determination in respiratory samples (nasopharyngeal swab samples) was performed on the LightCycler® 480 Instrument II (Roche Diagnostics®) using the LightMix® Modular SARS‐CoV E‐gene set.
2.4. Assessment of analytical performance (EUROIMMUN assay)
2.4.1. Precision
Precision was evaluated by using two pools of human and two internal quality controls provided by the manufacturer. Precision estimations were obtained by means of triplicate measurements of aliquots for a total of five consecutive days. Aliquots were stored at −20°C between analyses. The calculation was performed according to the Clinical and Laboratory Standards Institute (CLSI) EP15‐A3 protocol. 20
2.4.2. Limit of blank, detection, and quantification
The diluent provided by the manufacturer (diluent universal) was used as a blank sample to determine the limit of blank (LOB), detection (LOD), and quantification (LOQ). The LOB has been determined by running the blank sample on three separate occasions to verify that the results are well < 0.80. The LOD and LOQ have been determined by running 30 analyses of the blank sample using the following equations according to the SH GTA 04 document—revision 1 of the COFRAC. 21
-
‐
LOD = mean of the 30 measurements + 3 × standard deviation
-
‐
LOQ = mean of the 30 measurements + 10 × standard deviation.
2.4.3. Linearity
Linearity was evaluated according to CLSI EP‐06. A sample with high total antibody levels (i.e., 9.52) was analyzed and diluted by a factor of 2 on 5 consecutive dilutions. The manufacturer's diluent was used for the dilution. Observed values were compared to the expected ones and polynomial regression was calculated.
2.4.4. Carry‐over evaluation
A sample with a high IgG value (i.e., 8.92) was run in triplicate (A1, A2, A3) and followed by a negative sample (i.e., 0.10) also run in triplicate (B1, B2, B3). The carry‐over formula used is (B1‐B3)/(A3‐B3) × 100. A carry‐over below 1% is considered negligible.
2.4.5. Assessment of the clinical specificity
One‐hundred forty‐one non‐SARS‐CoV‐2 sera were analyzed for determining the cross‐reactivity and establishing specificity. Thirty‐eight sera from COVID‐19 negative healthy subjects and 103 sera from patients with a potential cross‐reaction to the SARS‐CoV‐2 immunoassay were included in this study. Potential cross‐reactive samples included positive antinuclear antibodies (n = 5), anti‐treponema pallidum antibodies (n = 3), anti‐thyroid peroxidase antibodies (n = 3), antibodies RAI + (search for irregular agglutinins) (n = 5), chikungunya antibody (n = 1), direct coombs (n = 1), hepatitis B antigen (n = 7), hepatitis C antibodies (n = 7), hepatitis E antibodies (n = 4), human immunodeficiency virus antibodies (n = 2), IgA Chlamydia pneumoniae (n = 1), IgM Borrelia + IgA Helicobacter pylori (n = 1), IgM C. pneumoniae (n = 1), IgG Chlamydia trachomatis (n = 1), IgG Coxiella burneti (n = 2), IgM C. burneti (n = 1), IgM cytomegalovirus (n = 13), IgM Epstein‐Barr virus viral capsid (n = 5), IgM Mycoplasma pneumoniae (n = 6), IgM parvovirus B19 (n = 8), IgM Toxoplasma gondii (n = 11), influenza A antibodies (n = 4), influenza A and B (n = 1), high level of total IgG (17.40 g/L) (normal range, 7.00–16.00 g/L) (n = 1), both high levels of total IgM (5.26 g/L; normal range, 0.4–2.3 g/L) and total IgG (28.67 g/L) (n = 1), rheumatoid factor (n = 6), urinary tract infection with Escherichia coli (n = 1), urinary tract infection with Klebsiella oxytoca (n = 1). All these samples were collected before the COVID‐19 pandemic and were stored at −20°C. The calculation of the specificity was stratified by excluding these cross‐reactive samples from the pool of healthy subjects and by combining the two cohorts.
2.4.6. Assessment of the clinical sensitivity
One hundred and thirty‐seven sera obtained from 94 COVID‐19 patients were analyzed to calculate the clinical sensitivity. Samples were subdivided according to the following different categories since symptom onset: 0–6 days: 23 sera; 7–13 days: 27 sera; 14–20 days: 24 sera; 21–27 days: 23 sera; 28 days or more: 40 sera. Clinical sensitivity for SARS‐Cov‐2 serological test depending on the onset of COVID‐19 symptoms was carried out with the manufacturer's cut‐off and with ROC curve adapted cut‐offs.
2.5. Statistical analysis
Descriptive statistics were used to analyze the data. Sensitivity was defined as the proportion of correctly identified COVID‐19‐positive patients since symptom onset. Specificity was defined as the proportion of naïve patients or healthy volunteers classified as negative. The ROC area under the curve (AUC) was calculated as the fraction of positive and negative determined according to the manufacturer's cut‐off values for positive results. Samples included for ROC curves analyses were sera obtained from at least 2 weeks after symptoms onset (n = 87), sera selected to assess cross‐reactivity (n = 103), and sera from healthy volunteers (n = 38). Data analysis was performed using GraphPad Prism® software (version 8.2.1) and MedCalc® software (version 14.8.1). p < .05 was used as a significance level. Our study fulfilled the Ethical principles of the Declaration of Helsinki.
3. RESULTS
3.1. Assessment of analytical performance (EUROIMMUN assay)
Repeatability and reproducibility results are summarized in Supporting Information data 1. Coefficients of variation (CV) are equal or lower to 7.6%. The limit of blank, detection, and quantification was 0.033 ± 0.013, 0.072, and 0.164, respectively. For the linearity assessment, the regression equation was: Y = 3.3 + 1.7x − 0.12x 2 with a correlation coefficient (R 2) of 0.99. Regarding the carry‐over, the following ratios have been obtained for the different samples and the different runs: A1 = 8.92, A2 = 8.90, A3 = 9.19, B1 = 0.10, B2 = 0.11 and B3 = 0.10. The calculated carry‐over was 0.0%.
3.2. Assessment of specificity
3.2.1. EUROIMMUN anti‐SARS‐CoV‐2‐nucleocapsid (IgG) ELISA
The calculated specificity was 96.5% (136 of 141) (95% CI, 91.9%–98.8%) by using the manufacturer's cut‐off (i.e., ratio ≥ 0.80) and considering borderline results as false positive. Five false‐positive results were observed with two IgM CMV, one HIV antibody, one hepatitis B Ag, and one in a healthy volunteer (respective ratios of 1.11, 1.35, 1.81, 0.82, and 0.96) using the manufacturer's cut‐off. If considering borderline results as negative (n = 2), the specificity increased to 97.9% (138 of 141) (95% CI, 93.9%–99.6%). Using an optimized cut‐off (i.e. ratio > 0.40 COI), specificity was 94.3% (133 of 141) (95% CI, 89.1%–97.5%). The calculated specificity was 97.4% (95% CI, 86.2%–99.9%) and 96.1% (95% CI, 90.4%–98.9%) for healthy volunteers and cross‐reactive samples, respectively, by using the manufacturer's cut‐off (Table 1).
Table 1.
Clinical performance of three anti‐SARS‐CoV‐2 immunoassays since symptom onset with the manufacturer's cut‐off and with optimized cut‐offs
| 0–6 d | 7–13 d | 14–20 d | 21–27 d | ≥28 d | Specificity (%) and 95% CI | Specificity (%) and 95% CI | Specificity (%) and 95% CI | |||
|---|---|---|---|---|---|---|---|---|---|---|
| n | 23 | 27 | 24 | 23 | 40 | (combined, n = 141) | (HVs, n = 38) | (cross‐reactivity, n = 103) | ||
| EUROIMMUN, IgGa | True positive | 4 | 19 | 23 | 20 | 36 | 96.5 (91.9 | 97.4% (86.2%–99.9%) | 96.1% (90.4%–98.9%) | |
| Ratio ≥ 0.80 | False negative | 19 | 8 | 1 | 3 | 4 | ||||
| Sensitivity (%) and 95% CI | 17.4 (5.0 | 70.4 (49.8 | 95.8 (78.9 | 87.0 (66.4 | 90.0 (76.3 | |||||
| True positive | 4 | 20 | 23 | 21 | 39 | 94.3 (89.1 | NA | NA | ||
| Ratio > 0.40 | False negative | 19 | 7 | 1 | 2 | 1 | ||||
| Sensitivity (%) and 95% CI | 17.4 (5.0 | 74.1 (53.7 | 95.8 (78.9 | 91.3 (72.0 | 97.5 (86.8 | |||||
| LIAISON, IgG | True positive | 3 | 9 | 20 | 20 | 37 | ||||
| AU/ml ≥ 12.0 | False negative | 20 | 18 | 4 | 3 | 3 | 97.9 (93.9%–99.6%) | 100% (90.8‐100%) | 97.1% (91.7%–99.4%) | |
| Sensitivity (%) and 95% CI | 13.0 (2.8%–33.6%) | 33.3 (16.5%–54.0%) | 83.3 (62.6%–95.3%) | 87.0 (66.4%–97.2%) | 92.5 (79.6%–98.4%) | |||||
| AU/ml > 3.94 | True positive | 6 | 15 | 23 | 22 | 39 | ||||
| False negative | 17 | 12 | 1 | 1 | 1 | 91.5 (85.6%–95.5%) | NA | NA | ||
| Sensitivity (%) and 95% CI | 26.1 (10.2%–48.4%) | 55.6 (35.3%–74.5%) | 95.8 (78.9%–99.9%) | 95.7 (78.1%–99.9%) | 97.5 (86.8%–99.9%) | |||||
| Elecsys, total antibodies | True positive | 4 | 19 | 21 | 20 | 39 | ||||
| COI ≥ 1.00 | False negative | 19 | 8 | 3 | 3 | 1 | 100 (97.4%–100%) | 100% (90.8‐100%) | 100% (96.5‐100%) | |
| Sensitivity (%) and 95% CI | 17.4 (5.0%–38.8%) | 70.4 (49.8%–86.3%) | 87.5 (67.6%–97.3%) | 87.0 (66.4%–97.2%) | 97.5 (86.8%–99.9%) | |||||
| True positive | 6 | 23 | 22 | 21 | 40 | |||||
| COI > 0.165 | False negative | 17 | 4 | 2 | 2 | 0 | 100 (97.4%–100%) | NA | NA | |
| Sensitivity (%) and 95% CI | 26.1 (10.2%–48.4%) | 85.2 (66.3%–95.8%) | 91.7 (73.0%–99.0%) | 91.3 (72.0%–98.9%) | 100 (91.2%–100%) |
Abbreviations: CI, confidence interval; HV, healthy volunteer.
The EUROIMMUN IgG assay is also sensitive to IgA and IgM, according to the insert kit of the manufacturer.
3.2.2. Elecsys anti‐SARS‐CoV‐2‐nucleocapsid (total antibodies) assay
The calculated specificity was 100% (141 of 141) (95% CI, 97.4%–100%) by using the manufacturer's cut‐off (i.e., ≥1.00). Using an optimized cut‐off (i.e., >0.165 COI) did not alter the specificity. The calculated specificity was 100% (95% CI, 90.8%–100%) and 100% (95% CI, 96.5%–100%) for healthy volunteers and cross‐reactive samples, respectively, by using the manufacturer's cut‐off (Table 1).
3.2.3. LIAISON anti‐SARS‐CoV‐2 spike proteins S1 and S2 (IgG) assay
The calculated specificity was 97.9% (138 of 141) (95% CI, 93.9%–99.6%) by using the manufacturer's cut‐off (i.e., ≥12.0 AU/ml) and considering borderline results as false positive. Three false‐positive results were observed with one IgM T. gondii, one IgM CMV, and one with a high level of total IgM (5.26 g/L) and high level of total IgG (28.67 g/L), with respective values of 32.0, 18.6, and 14.4 AU/ml using the manufacturer's cut‐off. If considering borderline results as negative (n = 1), the specificity increased to 98.6% (139 of 141) (95% CI, 95.0%–99.8%). Using optimized cut‐off (ratio > 3.94 AU/ml), specificity was 91.5% (129 of 141) (95% CI, 85.6%–95.5%). The calculated specificity was 100% (95% CI, 90.8%–100%) and 97.1% (95% CI, 91.7%–99.4%) for healthy volunteers and cross‐reactive samples, respectively, by using the manufacturer's cut‐off (Table 1).
3.2.4. Assessment of sensitivity
The calculated sensitivities classified according to different time categories since symptom onset are represented in Table 1. Fourteen days before symptom onset, the sensitivities (ranging from 70.4% to 85.2%) were not high enough to be reliably used in clinical practice, especially considering the LIAISON IgG assay.
3.2.5. EUROIMMUN anti‐SARS‐CoV‐2‐nucleocapsid (IgG) ELISA
After 2 weeks since symptom onset, the sensitivity was 90.8% (79 of 87) (95% CI, 82.7%–96.0%) by using the cut‐off provided by the manufacturer and considering borderline results (n = 1) as positive. Using the optimized cut‐off, the sensitivity (i.e., ratio > 0.40) was 95.4% (83 of 87) (95% CI, 86.6%–98.7%) (Figure 1). The sensitivity increased to 97.5% (39 of 40) (95% CI, 86.8%–99.9%) from 28 days since symptom onset (Table 1).
Figure 1.
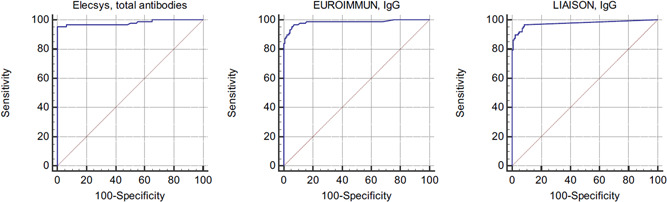
ROC curve analysis of three anti‐SARS‐CoV‐2 immunoassays at more than 2 weeks after the symptom onset (n = 87)
3.2.6. Elecsys anti‐SARS‐CoV‐2‐nucleocapsid (total antibodies) assay
After 2 weeks since symptom onset, the sensitivity was 92.0% (80 of 87) (95% CI, 84.1%–96.7%) by using the cut‐off provided by the manufacturer. Using the optimized cut‐off, the sensitivity (i.e., >0.165 COI) was 95.4% (83 of 87) (95% CI, 88.6%–98.7%) (Figure 1). The sensitivity increased to 100% (40 of 40) (95% CI, 91.2%–100%) from 28 days since symptom onset (Table 1).
3.2.7. LIAISON anti‐SARS‐CoV‐2 spike proteins S1 and S2 (IgG) assay
After 2 weeks since symptom onset, the sensitivity was 88.5% (77 of 87) (95% CI, 79.9%–94.4%) by using the cut‐off provided by the manufacturer and considering borderline results (n = 1) as positive. Using the optimized cut‐off, the sensitivity (i.e., >3.94 AU/ml) was 96.6% (84 of 87) (95% CI, 90.3%–99.3%) (Figure 1). The sensitivity increased to 97.5% (39 of 40) (95% CI, 86.8%–99.9%) from 28 days since symptom onset (Table 1).
4. DISCUSSION
Serological testing is a useful strategy for the diagnosis, characterization of the course of the disease, for identifying convalescent plasma donors as well as for epidemiological study, lockdown exit programs, and COVID‐19 vaccine development. 5 , 6 , 8 , 14 , 22 To date, peer‐reviewed data concerning the performance of SARS‐CoV‐2 immunoassays remains limited, but it is crucial for society to be confident in the results of these assays. Therefore, independent validations of these methods before broad introduction into routine clinical practice is mandatory, given the limited experience of the scientific community with these new assays. 6 , 10 , 14 , 23 , 24 , 25 , 26 , 27 We report here the external validation of the EUROIMMUN anti‐SARS‐CoV‐2‐nucleocapsid (IgG) ELISA. Our results show satisfactory analytical performance. Repeatability and reproducibility studies determined on two different pools of sera from patients and two internal quality controls were ≤7.3% and ≤7.6%, respectively. The carry‐over was negligible, and we found a LOQ of 0.164, which is lower than the optimized cut‐off of 0.40 we found. Satisfactory analytical performance have also recently been reported for the Elecsys and LIAISON anti‐SARS‐CoV‐2 assays and were not reassessed in this study. 15 , 16 , 17 , 18 , 28
4.1. Specificity of the three automated assays
The Elecsys assay had a perfect specificity, considering both the manufacturer and the ROC curve adapted cut‐off. The LIAISON assay had up to three false‐positive results and the EUROIMMUN assay had up to five false‐positive results. Tang et al. 17 found a specificity of 98.7% on the Elecsys assay using 153 presumed negative specimens. There were two false‐positive results from two patients with negative RT‐PCR results but with symptoms. Given that approximately 20% of the RT‐PCR results might be falsely negative in COVID‐19 patients, 14 , 29 the fact that Tang et al. considered these two patient results as false positive is questionable. For instance, Zhao et al. 9 found that combining RT‐PCR and antibody detection significantly improved the sensitivity of pathogenic diagnosis for COVID‐19. In our study, only samples collected before the COVID‐19 pandemic were included, excluding any confusion. Using a higher patient cohort of blood donors and ICU patients collected before the COVID‐19 outbreak (n = 456), Egger et al. 18 only observed one false‐positive result on the Elecsys assay. Considering the LIAISON assay, Tré‐Hardy et al. 15 found a specificity of 100% and 99% using the manufacturer's cut‐off or an adapted cut‐off (i.e., >6.1 AU/ml), respectively. We found lower specificities of 97.9% and 91.5% using the manufacturer's cut‐off or our adapted cut‐off (i.e., >3.94 AU/ml). Plebani et al. found similar specificities of 96.8% and 88.9% using the manufacturer's cut‐off or an adapted cut‐off (i.e., >6.2 AU/ml).
The higher specificity observed in the study of Tré‐Hardy et al. 15 is probably due to the lower number of samples included (n = 81) for the specificity calculation compared to our study (n = 141) and the one of Plebani et al. (n = 191). 30 Interestingly, adapted cut‐offs proposed on the LIAISON assay were all lower (>3.94, >6.1 AU/ml, 15 >6.2 AU/ml 30 ) than the manufacturer's cut‐off (i.e., ≥12.0 AU/ml) using three independent cohorts of patients. The performance of these optimized cut‐offs are not considered clinically different as there is an overlap between 95% confidence intervals.
4.2. Sensitivity of the three automated assays
Current data suggest that seroconversion occurs approximately 7–14 days after symptom onset. 6 , 14 , 31 , 32 Although the Elecsys and the EUROIMMUN assays detected more positive results earlier after onset of symptoms than the LIAISON assay, none of the assays demonstrated a high enough clinical sensitivity to diagnose acute infection (i.e., <14 days). From 14 days since symptom onset, sensitivities increased for all assays, especially using optimized cut‐offs. Using manufacturer's cutoffs resulted in 8, 10, and 7 false‐negative specimens for the EUROIMMUN, LIAISON and Elecsys assays, respectively. Optimized cut‐offs gave less false‐negative results (4, 3, and 4, respectively). Due to the overlapping of confidence intervals at 95% between assays, we cannot conclude that one assay had a significantly higher true positivity rate.
Two studies having included less patients with symptoms since at least 14 days evaluated the performance of the Elecsys assay. 17 , 18 Tang et al. found a sensitivity of 89.4% (n = 47) 17 and Egger et al. a sensitivity of 100% (n = 18). By using the manufacturer's cut‐off, we found a somewhat similar sensitivity compared to Tang et al. (i.e., 92.0%). However, they did not determine an optimized cut‐off to increase the performance of the test. Fourteen days after RT‐PCR positivity, Tré‐Hardy et al. 15 found a sensitivity of 91% and 100%, using the manufacturer's and an optimized cut‐off on the LIAISON assay. It is important to note that with the confidence interval around 100% they found (92%–100%) was consistent with our results (i.e., 96.6% sensitivity; 95% CI, 90.3%–99.3%). Plebani et al. 30 published results in agreement with our finding with a sensitivity of 97.1% for the LIAISON assay.
5. CONCLUSION
All three anti‐SARS‐CoV‐2 assays had equivalent sensitivities 14 days from symptom onset to diagnose past‐COVID‐19 infection. We also confirmed that anti‐SARS‐CoV‐2 determination before Day 14 is of less clinical interest. However, the Elecsys assay had a higher specificity compared to the EUROIMMUN and the LIAISON assays. Further studies specifically designed to evaluate long‐term evolution of antibody response are also needed.
AUTHOR CONTRIBUTIONS
Julien Favresse, Julie Cadrobbi, Christine Eucher, Marc Elsen, Kim Laffineur, Jean‐Michel Dogné, and Jonathan Douxfils performed the research; Julien Favresse, Julie Cadrobbi, Christine Eucher, Marc Elsen, Kim Laffineur, Jean‐Michel Dogné, and Jonathan Douxfils designed the research study, Roche Diagnostics and EUROIMMUN contributed essential reagents or tools; Julien Favresse, Julie Cadrobbi, Christine Eucher, Marc Elsen, Kim Laffineur, Jean‐Michel Dogné, and Jonathan Douxfils analyzed the data; Julien Favresse wrote the paper.
DISCLOSURES
Among the authors, Jonathan Douxfils is chief executive officer and founder of QUALIblood SA and reports personal fees from Diagnostica Stago, Roche, Roche Diagnostics, Daiichi‐Sankyo, and Portola, outside the submitted work. Roche Diagnostics generously provided the kits for the validation.
Supporting information
Supporting information.
ACKNOWLEDGMENT
We wish to thank the personnel of the Saint‐Luc Bouge laboratory for their technical assistance.
Favresse J, Cadrobbi J, Eucher C, et al. Clinical performance of three fully automated anti‐SARS‐CoV‐2 immunoassays targeting the nucleocapsid or spike proteins. J Med Virol. 2021;93:2262–2269. 10.1002/jmv.26669
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the main manuscript and supplementary material of this article. The detailed clinical/biological/radiological data of COVID‐19 confirmed patients are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Fauci AS, Lane HC, Redfield RR. COVID‐19—navigating the uncharted. N Engl J Med. 2020;382:1268–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization . Coronavirus disease 2019 (COVID‐19) Situation Report – 129.
- 3. Vashist SK. In vitro diagnostic assays for COVID‐19: recent advances and emerging trends. Diagnostics. 2020;10(4):202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID‐19). Clin Chem Lab Med. 2020;58:1070–1076. [DOI] [PubMed] [Google Scholar]
- 5. Winter AK, Hegde ST. The important role of serology for COVID‐19 control. Lancet Infect Dis. 2020;20:758–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS‐CoV‐2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020;58:1081–1088. [DOI] [PubMed] [Google Scholar]
- 7. Coronavirus Resource Center . https://coronavirus.jhu.edu/map.html. Accessed May 16, 2020.
- 8. Farnsworth CW, Anderson NW. SARS‐CoV‐2 serology: much hype, little data. Clin Chem. 2020;66:875–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019 [published online ahead of print March 28, 2020]. Clin Infect Dis. 2020. 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tré‐Hardy M, Blairon L, Wilmet A, et al. The role of serology for COVID‐19 control: population, kinetics and test performance do matter. J Infect. 2020;81:e91–e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. FDA . https://www.fda.gov/medical-devices/emergencysituations-medical-devices/emergency-use-authorizations
- 12. Kirkcaldy RD, King BA, Brooks JT. COVID‐19 and postinfection immunity: limited evidence, many remaining questions. JAMA. 2020;323:2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diamandis P, Prassas I, Diamandis EP. Antibody tests for COVID‐19: drawing attention to the importance of analytical specificity. Clin Chem Lab Med. 2020;58:1144–1145. [DOI] [PubMed] [Google Scholar]
- 14. Bohn MK, Lippi G, Horvath A, et al. Molecular, serological, and biochemical diagnosis and monitoring of COVID‐19: IFCC taskforce evaluation of the latest evidence. Clin Chem Lab Med. 2020;58:1037–1052. [DOI] [PubMed] [Google Scholar]
- 15. Tre‐Hardy M, Wilmet A, Beukinga I, Dogne JM, Douxfils J, Blairon L. Validation of a chemiluminescent assay for specific SARS‐CoV‐2 antibody. Clin Chem Lab Med. 2020;58:1357–1364. [DOI] [PubMed] [Google Scholar]
- 16. Favresse J, Eucher C, Elsen M, Marie TH, Dogne JM, Douxfils J. Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS‐CoV‐2 total antibodies. Clin Chem. 2020;66:1104–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang MS, Hock KG, Logsdon NM, et al. Clinical performance of the Roche SARS‐CoV‐2 serologic assay. Clin Chem. 2020;66:1107–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Egger M, Bundschuh C, Wiesinger K, et al. Comparison of the Elecsys(R) Anti‐SARS‐CoV‐2 immunoassay with the EDI enzyme linked immunosorbent assays for the detection of SARS‐CoV‐2 antibodies in human plasma. Clin Chim Acta. 2020;509:18–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.EUROIMMUN insert kit, anti‐SARS‐CoV‐2 (IgG) nucleocapsid assay. 2020.
- 20. CLSI . User Verification of Precision and Estimation of Bias. Approved guideline. CLSI Document EP15‐A3. 3rd ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- 21. Cofrac. Guide Technique d′Accréditation de Vérification (Portée A)/Validation (Portée B) des Méthodes en Biologie Médicale –Document SH GTA 04 (révision 01). 2015.
- 22. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med. 2020;26:845–848. [DOI] [PubMed] [Google Scholar]
- 23. Tang MS, Hock KG, Logsdon NM, et al. Clinical performance of two SARS‐CoV‐2 serologic assays. Clin Chem. 2020.66(8):1055–1062. 10.1093/clinchem/hvaa120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Montesinos I, Gruson D, Kabamba B, et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti‐SARS‐CoV‐2 antibodies. J Clin Virol. 2020;128:104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lippi G, Salvagno GL, Pegoraro M, et al. Assessment of immune response to SARS‐CoV‐2 with fully automated MAGLUMI 2019‐nCoV IgG and IgM chemiluminescence immunoassays. Clin Chem Lab Med. 2020;58:1156–1159. [DOI] [PubMed] [Google Scholar]
- 26. Tré‐Hardy M, Wilmet A, Beukinga I, et al. Analytical and clinical validation of an ELISA for specific SARS‐CoV‐2 IgG, IgA, and IgM antibodies [published online ahead of print July 15, 2020]. J Med Virol. 2020:jmv.26303. 10.1002/jmv.26303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mairesse A, Favresse J, Eucher C, et al. High clinical performance and quantitative assessment of antibody kinetics using a dual recognition assay for the detection of SARS‐CoV‐2 IgM and IgG antibodies [published online ahead of print August 25, 2020]. Clin Biochem. 2020. 10.1016/j.clinbiochem.2020.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lau C, Hoo S, Yew S, et al. Evaluation of the Roche Elecsys antio‐SARS‐CoV‐2 Assay. MedRxiv. 2020. 10.1101/2020.06.28.20142232 [DOI]
- 29. Stowell S, Guarner J. Role of serology in the COVID‐19 pandemic [published online ahead of print May 1, 2020]. Clin Infect Dis. 2020.ciaa510. 10.1093/cid/ciaa510 [DOI] [Google Scholar]
- 30. Plebani M, Padoan A, Negrini D, Carpinteri B, Sciacovelli L. Diagnostic performances and thresholds: the key to harmonization in serological SARS‐CoV‐2 assays? Clin Chim Acta. 2020;509:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Favresse J, Eucher C, Elsen M, et al. Unexpected kinetics of anti‐SARS‐CoV‐2 total antibodies in two patients with chronic lymphocytic leukemia. Br J Haematol. 2020;190(4):e187–e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Favresse J, Eucher C, Elsen M, Laffineur K, Dogne JM, Douxfils J. Response of anti‐SARS‐CoV‐2 total antibodies to nucleocapsid antigen in COVID‐19 patients: a longitudinal study. Clin Chem Lab Med. 2020;58:e193–e196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that supports the findings of this study are available in the main manuscript and supplementary material of this article. The detailed clinical/biological/radiological data of COVID‐19 confirmed patients are not publicly available due to privacy or ethical restrictions.


