Abstract
Circular RNAs (circRNAs) are a recently discovered class of noncoding RNAs found in many species across the eukaryotic kingdom. These intriguing RNA species are formed through a unique mechanism that is known as back splicing in which the 5′ and 3′ termini are covalently joined. Recent research has revealed that viruses also encode a repertoire of circRNAs. Some of these viral circRNAs are abundantly expressed and are reported to play a role in disease pathogenesis. A growing number of studies also indicate that host circRNAs are involved in immune responses against virus infections with either an antiviral or proviral role. In this review, we briefly introduce circRNA, its biogenesis, and mechanism of action. We go on to summarize the latest research on the expression, regulation, and functions of viral and host‐encoded circRNAs during the host–virus interaction, with the aim of highlighting the potential of viral and host circRNAs as a suitable target for diagnostic biomarker development and therapeutic treatment of viral‐associated diseases. We conclude by discussing the current limitations in knowledge and significance of elucidating the roles of circRNAs in host–virus interactions, as well as future directions for this emerging field.
Keywords: circRNA, circular RNA, miRNA sponge, noncoding RNA, virus
Circular RNA (circRNA) is a novel noncoding RNA that is formed through backsplicing. Upon virus infection, virus‐induced host circRNAs and/or viral‐derived circRNAs promote proviral or antiviral host immune responses. Here, we provide a summary of the key current evidence of host and viral circRNA expression, regulation, and functions during host–virus interactions. The diagnostic and therapeutic potential of circRNAs is also discussed.
Abbreviations
- 2‐5A
2′,5′‐linked oligoadenylates
- BARTs
BamHI A rightward transcripts
- BSJ
backsplice junctions
- ceRNA
competing endogenous RNA
- circRNA
circular RNA
- circRNP
NF90/NF110‐circRNA protein
- DHX9
DExH‐Box helicase 9
- dsRNA
double‐stranded RNA
- EBV
Epstein–Barr virus
- EMCV
encephalomyocarditis virus
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HDV
hepatitis delta virus
- HPV
human papillomavirus
- IAV
influenza virus
- KSHV
Kaposi sarcoma herpesvirus
- LANA‐1
latency‐associated nuclear antigen
- m1ψ
N1‐methylpseudouridine
- m6A
N6‐methyladenosine
- MAPK
mitogen‐activated protein kinase
- MDA5
melanoma differentiation‐associated gene 5
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- miRNA
microRNA
- ncRNA
non‐coding RNA
- NMD
nonsense‐mediated decay
- OAS1
5′ oligoadenylate synthase 1
- OASL
OAS‐like protein
- PAN
polyadenylated nuclear
- PEDV
porcine endemic diarrhea virus
- pgRNA
pregenomic RNA
- PKR
protein kinase R
- PPRs
pattern recognition receptors
- RBP
RNA‐binding protein
- RIG‐I
retinoic acid‐inducible gene‐I
- RTA
replication and transcription activator
- SV40
simian virus 40
- TGEV
transmissible gastroenteritis virus
- TLR
Toll‐like receptor
- TRIM59
tripartite motif‑containing protein 59
- vIRF4
viral interferon regulatory factor 4
Introduction
Viruses are obligate intracellular parasites that fully rely on their host’s biochemical machinery for growth and reproduction, but are inert when present outside the host. A subset of pathogenic viruses known as oncogenic viruses can cause cancer upon viral infection. To reproduce inside the host, viruses utilize many cellular factors and pathways for efficient replication. Meanwhile, the infected host cells respond by expressing and/or activating factors (ranging from RNA and proteins to lipids) and pathways to restrict virus replication.
Innate immunity presents the first line of defense against viral pathogens and is deployed rapidly. A repertoire of sensors recognize specific pathogen‐derived patterns that culminate in the mobilization of a range of innate immune effectors and regulators such as interferon release and induction of an antiviral state [1]. At the same time, the viruses also deploy various mechanisms to evade the host immune response fueling the evolution of mechanisms to sense and censor viruses and virus‐derived RNA. It is the net effect of these virulence factors of the pathogen and the host immune defense that likely determines the outcome of the virus infection. However, our present understanding of the role of non‐coding RNAs (ncRNAs) during host–virus interaction is comparatively lacking.
With the improvement of biochemical methods and the advent of high‐throughput sequencing, an intriguing class of ncRNA, namely circular RNA (circRNA), has been discovered [2, 3, 4, 5, 6, 7]. circRNAs were previously thought to be the byproduct of mis‐splicing during mRNA processing in eukaryotic cells, but have now become one of the most attention‐grabbing molecular species in research focus. circRNA expression is ubiquitous throughout the eukaryotic kingdoms. circRNAs are primarily generated through a unique mechanism known as backsplicing, a process that differs from canonical splicing whereby the upstream 3′ splice acceptor is covalently joined to the downstream 5′ splice acceptor. To date, three hypothetical models for circRNA biogenesis mechanisms have been widely accepted (Fig. 1), including RNA‐binding protein (RBP)‐mediated [6, 8, 9], intron pairing‐driven [10, 11], and lariat‐driven circularization [12, 13].
Fig. 1.
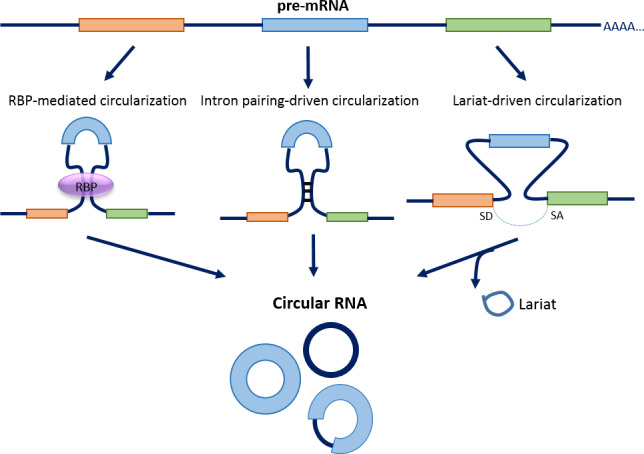
Biogenesis of circRNA. Three hypothetical models for circRNA biogenesis mechanisms have been widely accepted, including (1) RBP‐mediated circularization [6, 8, 9]. RBP (e.g., QKI, MBL) is a trans‐factor that regulates circularization through the binding of specific sequence motifs on introns flanking exon(s) to be circularized on a linear pre‐mRNA, and dimerize to facilitate backsplicing; (2) intron pairing‐driven circularization [10, 11]. This model utilizes the presence of inverted complementary sequences in flanking intronic regions (e.g., Alu repeats) that may promote alternative circularization through intron pairing; and (3) Lariat‐driven circularization which starts with canonical splicing, resulting in the production of linear mRNA with skipped exon(s) within a long intron lariat. Subsequently, internal splicing facilitates the removal of flanking intronic sequence, allowing the generation of circRNA [12, 13]. SA, splicing acceptor; SD, splicing donor.
circRNAs formed through backsplicing lack free termini; thus, these can easily escape from hydrolysis by the numerous cellular exonucleases such as RNase R. Reduction in exonuclease susceptibility allows circRNAs to have significant longer half‐lives than linear RNA [3]. The abundance of most circRNAs is low at a hundred to a thousand times lower than that of the cognate linear RNAs. circRNAs can be derived from either exons or introns or from both, and with a great diversity in length, from less than a hundred to thousands of nucleotides. Thousands of circRNAs are reported to be highly expressed in tissue and developmental stage‐specific manners [14] with more than 20 % of expressed genes in examined cells and tissues able to produce circRNA transcripts [15]. circRNAs are abundant, evolutionarily conserved across species, and predominantly reside in the cytoplasm. Nuclear‐retained circRNAs are predicted to regulate transcription, whereas circRNAs that are predominantly found in the cytoplasm are more likely to be involved in post‐transcriptional gene regulation. To date, several biological functions have been proposed for circRNAs (Fig. 2), including microRNA (miRNA) sponge [16, 17, 18, 19, 20], RBP sponge [9, 21], transcriptional regulator [12, 22], mRNA trap [9], protein regulator [23, 24], and as templates for translation into protein [25, 26, 27]. Comprehensive review of circRNA biogenesis, detection, and function would be beyond the scope of this work and has been recently reviewed elsewhere [28, 29, 30, 31].
Fig. 2.
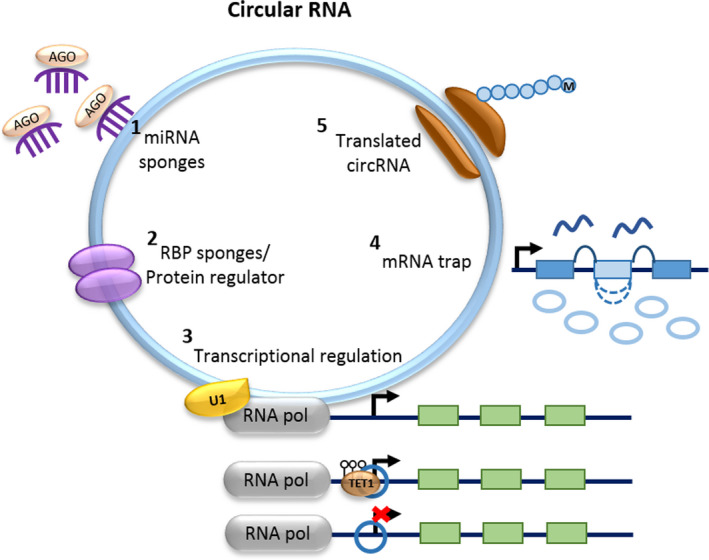
Biological functions of circRNAs. circRNA (ciRS‐7, cir‐ITCH) acts as a (1) miRNA sponge that may compete with other classes of RNA for miRNA‐binding sites and perturb the complex network of interaction and gene regulation [16, 17, 18, 19, 20]; (2) RBP sponge [9, 21] and protein regulator [23, 24]. circRNAs (e.g., circFoxo3) are able to bind, store, sort, and sequester proteins to particular subcellular locations, and act as dynamic scaffolding molecules that modulate protein–protein interactions; (3) regulates transcription of parental gene in cis‐ or trans‐manners through interacting with U1 snRNP and promote transcription of their parental genes (e.g., circEIF3J and circPAIP2); recruiting TET1 demethylase to induce DNA demethylation of promoter or interacting with DNMT1 promoter that results in DNMT1 silencing (e.g., circFECR1) [12, 22]; (4) mRNA trap (e.g., circMbl) whereby the formation of circRNA competes with linear mRNA production [9]; and (5) template for circRNA translation (e.g., circβ‐catenin) as it contains an open reading frame (ORF) [25, 26, 27].
Recent studies have revealed that viruses also encode a repertoire of circRNAs. Some viral circRNAs are abundantly expressed and are reported to play a role in disease pathogenesis. Evidence of how host circRNAs changes in response to virus infections is also slowly emerging. In this review, we provide a summary of key current evidence of host and viral‐derived circRNA expression, their regulation, and their biological functions during host–virus interaction. In addition, the medical potential of host and viral circRNAs as diagnostic and prognostic markers and as therapeutic targets for viral‐associated diseases will be discussed.
Viral‐derived circRNAs
circRNAs were first identified in RNA viruses as viroids via electron microscopy [32]. Years later, endogenous circRNA was first discovered in the humans deleted in colon cancer (DCC) gene [33]. With the functional characterization of eukaryotic circRNAs, viruses are being revisited for the possibility in encoding bona fide circRNAs backspliced from viral genes. Recent circRNA analyses have discovered and validated the presence of viral circRNAs in DNA viruses such as herpesviruses, papillomavirus, and others (see more below). These viral circRNAs are different from viroids that also have single‐stranded circular RNA structure as the genetic content ranging from 220 to 457 bp in size and lacking protein‐coding capability [34]. Although viral circRNAs and viroids have parallels, viroids are pathogenic self‐replicating infectious agents of higher plants with no effects in humans and animals, whereas viral circRNAs are nonreplicating, noninfectious RNA transcripts derived via backsplicing of viral genes. Importantly, the circularity of plant viroid RNA genomes has functional relevance to their replication cycle via the rolling circle mechanism, which is different from viral circRNAs formed via backsplicing [35]. The landscape of viral‐derived circRNAs across different viruses and their characterization including biological functions are summarized in Fig. 3 and discussed below.
Fig. 3.
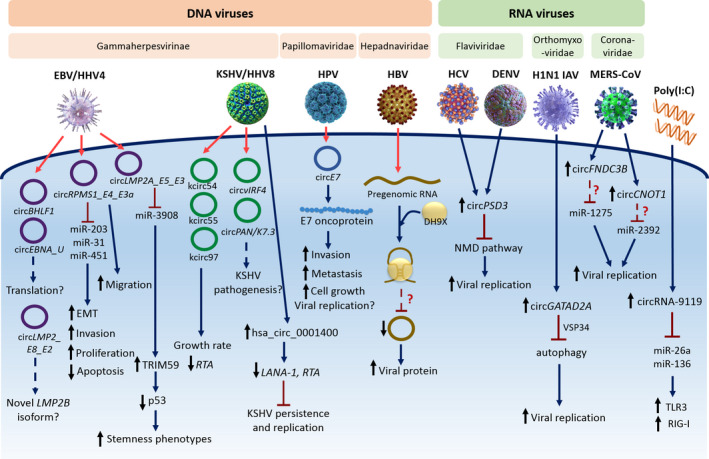
Mechanism of actions of viral and host circRNAs during viral infection. DNA viruses (EBV [38, 39, 42, 43], KSHV [39, 40, 47], HPV [49, 50], and HBV [52, 53])—transcribed viral circRNAs contribute to disease pathogenesis by acting as miRNA sponge, oncoprotein producer, and affecting hallmarks of cancer. RNA viruses (HCV [70], DENV [70], H1N1 IAV [69], and MERS‐CoV [71]) regulate host circRNA expression to promote viral replication. Poly(I:C) treatment upregulates host circRNA to trigger TLR3 and RIG‐I signaling pathway through miRNA sponging.
EBV circRNAs
Epstein–Barr virus (EBV) is one of the most common human viruses, infecting approximately 90% of the world’s population [36]. Primary EBV infection during early childhood is usually asymptomatic and delayed onsets can be associated with infectious mononucleosis. EBV infection also accounts for various malignancies in B lymphocytes and epithelial cells including Burkitt’s lymphoma (BL), Hodgkin lymphoma (HL), extranodal nasal‐type natural killer/T‐cell lymphoma (NKTL), nasopharyngeal carcinoma (NPC), and EBV‐associated gastric carcinoma (EBVaGC). Other than protein‐coding gene expression, EBV synthesizes many essential viral noncoding RNAs that regulate diverse biological processes including viral replication, host immune evasion, and cellular transformation [37].
circRNAs encoded by EBV and a closely related tumorigenic herpesvirus, Kaposi sarcoma herpesvirus (KSHV), were first described in a series papers by three different groups [38, 39, 40]. Erik Flemington’s group showed that EBV expresses a spectrum of viral circRNAs between latent and lytic cycles across various cell lines with different latency status [38]. Notably, some viral circRNAs have been found to be expressed at levels comparable or higher than host circRNA levels such as circRPMS1_E4_E3a and circBHLF1, which suggests potential biological implications. Some of these circRNAs such as circRPMS1_E4_E3a and circEBNA_U are also expressed broadly across different EBV latencies. Whereas a majority of the circRNAs were formed via backsplicing of one or more exons, a few were found to include intronic regions. Intriguingly, most of the EBV circRNAs identified are encoded from latency genes and a significant proportion of expressed EBV circRNA were upregulated upon lytic reactivation. Further subcellular localization study demonstrated that EBV circRNAs can be localized to either cytoplasm, nucleus, or both. Importantly, comparison between EBV and rhesus macaque lymphocryptovirus (rLCV)‐expressed circRNAs showed that circRPMS1_E4_E3a and circEBNA_U are conserved among gammaherpesvirinae, even though there is only 65% nucleotide homology between rLCV and EBV genomes, suggesting conserved functional relevance [41]. On the other hand, Yuan Chang’s group has identified and characterized a number EBV circRNAs, including abundant circRPMS1 isoforms (known as circBARTs in Toptan et al.’s study) in EBV‐positive posttransplant lymphoproliferative disease samples (PTLDs) [39]. Essentially, further characterization by the group showed that other than detection in EBV‐positive cell lines, circRPMS1_E4_E3a was also found in EBV‐positive AIDS‐associated lymphoma, EBV‐positive gastric carcinoma, and NPC xenografts.
To date, limited functions have been ascribed to these EBV‐derived circRNAs with only circRNAs from RPMS1 and LMP2A genes being reported. circRPMS1_E4_E3a is located within the EBV BamHI A rightward transcript (BART) locus that is responsible for the majority of EBV noncoding RNA transcription [37]. Two studies have demonstrated that circRPMS1_E4_E3a (known as ebv‐circRPMS1 in Huang et al.’s study and as circRPMS1 in Liu et al’s work) promotes EBV‐associated epithelial cancer tumorigenesis by acting as a miRNA sponge. Liu et al. [42] showed that circRPMS1_E4_E3a was upregulated in NPC tissues with higher expression in metastatic NPC and was associated with shorter survival time. Moreover, circRPMS1_E4_E3a promotes cell proliferation and invasion ability, as well as suppress apoptosis in EBV‐positive NPC cells through sponging multiple miRNAs and promoting epithelial–mesenchymal transition (EMT). Likewise, Huang et al. [43] showed that ectopic expression of circRPMS1_E4_E3a in EBVaGC cells increased cell migration rate compared with the control group, as well as downregulate 11 of 14 human miRNAs.
Another EBV circRNA, circLMP2A which is formed through the backsplicing of exon 5 to exon 3 of the LMP2A gene, was recently reported to promote cancer stemness properties of EBVaGC cells through a circLMP2A/miR‐3908/TRIM59/p53 axis [44]. circLMP2A contains three predictive binding sites of miR‐3908, with sites 1 and 3 crucial for its sponging ability. Sponging of miR‐3908 by circLMP2A freed the miRNA target, tripartite motif‑containing protein 59 (TRIM59), an E3 ligase, to potentially ubiquitinate and degrade the tumor suppressor p53. Gong et al.’s finding is consistent with the loss of p53 in gastric cancers promoting tumorigenesis via conferring cell stemness and inducing EMT [45]. Importantly, high expression of circLMP2A correlates with an enhanced metastasis rate and the poor prognosis of EBVaGC patients, suggesting its potential use in EBVaGC diagnosis and treatment.
KSHV circRNAs
Kaposi’s sarcoma (KS)‐associated herpesvirus (KSHV) is the causative agent for Kaposi’s sarcoma, the most common cancer in untreated HIV‐infected individuals, organ transplant recipients, and immunocompromised individuals [46]. KSHV was recently confirmed to generate circRNAs from several KSHV genes. KSHV circRNAs are ubiquitously but differentially expressed depending on the virus life cycle. The most abundant KSHV circRNAs are derived from viral interferon regulatory factor 4 (vIRF4) and PAN (polyadenylated nuclear) region [39]. However, according to Toptan et al., analysis of circRNA from the KSHV PAN region is challenging as the region includes both sense (PAN) and antisense (K7.3) transcripts which have yet to be found spliced and there is no obvious sequence feature accounting for hypervariable and bidirectional generation of circPAN/K7. Intriguingly, the two abundant KSHV circRNAs, circvIRF4 and circPAN/K7.3, have a different expression pattern in lytic replication. The expression of circvIRF4 remained unchanged compared with the expression of linear vIRF4, while circPAN/K7.3 expression was induced upon lytic reactivation, mirroring the expression of its linear counterpart. No change in circvIRF4 expression after lytic cycle induction suggests the transcription activity of the vIRF4 operon does not directly regulate its biogenesis [47]. It is possible that a nonlytic cycle‐regulated splicing mechanism that recognizes alternative splice donor and acceptor sites is involved in the circularization of vIRF4 [39].
Curiously, circvIRF4 and circPAN/K7.3 were found packaged into nuclease‐resistant viral particles. This is similar to the packaging of KSHV linear transcripts including PAN, suggesting a function for viral circRNAs at the initial steps of primary infection prior to the onset of the viral transcription program [47]. However, further investigations are required to determine whether these circRNAs exhibit a similar function to those of other transcripts and proteins incorporated into the KSHV virions to regulate cellular immune response for successful establishment of KSHV infection.
Importantly, abundant KSHV circRNAs can be detected in KS patient samples [47]. RT–PCR revealed a higher detection rate of circvIRF4 and circPAN/K7.3 than of linear LANA‐1 (latency‐associated nuclear antigen) transcripts in KS tissue samples. Basescope in situ hybridization showed that a majority of the valid archival formalin‐fixed, paraffin‐embedded (FFPE) KS tissue biopsy specimens were also positive for circvIRF4 and circPAN/K7.3. In addition, KSHV circRNAs can be detected in freshly prepared parallel plasma and serum samples and to a certain extent, the old sera samples.
Tagawa and team also reported the presence of another list of KSHV circRNAs in KSHV‐infected cell lines and fresh lymph node biopsies from KSHV‐positive patient samples including circvIRF4 and circPAN/K7.3. They showed that KSHV circRNA expression correlates with the increased expression of replication and transcription activator (RTA) in the patient samples. Furthermore, differential growth rate can be observed in KSHV‐infected and noninfected cells that stably express viral circRNAs (kcirc54, kcirc55, and kcirc97) compared with control cells. Interestingly, transient but not stable expression of the same viral circRNAs caused mild repression of KSHV RTA but not LANA‐1 [40]. Further investigations are needed to determine the reason behind the different regulation pattern of viral genes by KSHV circRNAs in transient and stably expressing cells.
HPV circRNAs
Human papillomavirus (HPV) is a double‐stranded DNA oncovirus that contributes to the development of oropharyngeal cancer and other female reproductive system‐related cancers [48]. HPV vaccination provides protection against several virulent HPV subtypes and reduces the risk of HPV‐related cancers. Based on the RNA‐seq data from HPV‐infected tissues, Zhao et al. predicted a list of HPV circRNA candidates. circE7 that is abundant and contains the entire open reading frame of the E7 oncogene was selected for further characterization as the E7 oncogene is strongly related with HPV‐associated oncogenesis [49]. circE7 can be detected from multiple high‐risk HPVs including HPV16 and HPV18, with HPV16 being the most abundant species. Importantly, the study showed that rather than its linear E7 mRNA, N6‐methyladenosine (m6A)‐modified circE7 and cytoplasmic‐enriched circE7 serve as the main template for E7 oncoprotein translation through a cap‐independent mechanism and the translation can be enhanced in a heat‐shock regulated manner. Moreover, E7 oncoprotein from circE7 is essential for HPV16’s ability to transform CaSki cervical carcinoma cells and for its transformed behavior in vitro and in tumor xenografts. The authors also suggest other potential functions for circE7 such as regulating HPV replication and host cell transformation, but further experiments are needed for validation.
Based on the study by Chamseddin and colleagues, circE7 was also detected in anal squamous cell carcinoma (ASCC), which is a rare, potentially fatal malignancy primarily linked to HPV. A higher level of circE7 as assessed by RNA‐seq was significantly associated with improved survival rate in ASCC, and similar trend was also found in the analysis of RNA‐seq from HPV‐positive head and neck cancer and cervical cancers from The Cancer Genome Atlas (TCGA) data. These phenomena can be explained by the higher level of circE7 that allows for increased expression of E7 oncoprotein, which in turn activates a stronger immune response against HPV‐related cancers [50]. However, this hypothesis remains to be clarified. Thus, further studies are necessary to support such hypothesis and to confirm the role of circE7 in different HPV‐related cancers.
HBV circRNAs
Hepatitis B virus (HBV) infection is a major health concern worldwide as it increases the risk of development of liver cirrhosis and hepatocellular carcinoma [51]. RNA‐seq data revealed that HBV encodes a ~ 2.5 kb circRNA derived from intronless pregenomic RNA (pgRNA), which is predominantly found in the cytoplasm, with some in the nucleus [52]. Interestingly, circularization of this circRNA requires mechanisms similar to the pre‐mRNA splicing and RNA homologous recombination at two different junction sites of pgRNA. The latter junction involves a 113 bp repetitive sequence at the 3′ and 5′ end of pgRNA. However, further study is required to identify the specific mechanism. In addition, Sekiba and team also reported a circRNA derived from HBV pgRNA that interacts with DExH‐Box helicase 9 (DHX9), a RNA helicase that is involved in innate immune responses against DNA viruses [53]. Knockdown of DHX9 leads to increased production of HBV circRNA but decreased level of mRNAs that are essential for viral protein translation, hence a decrease in the level of viral surface proteins. DHX9 is known to suppress circRNA formation by preventing the binding of complementary sequences [54]. The authors suggest that DHX9 can bind to the inverted repeat sequences at the 5′ and 3′ ends at the HBV pgRNA to inhibit the circRNA biogenesis. This regulation of HBV circRNA by DHX9 is thought to potentially be a crucial mechanism in the regulation of viral circRNA and viral protein levels during HBV infection [53]. Additional efforts are needed to further investigate and prove the involvement of DHX9 in pgRNA circularization.
Viral circRNA database
The first viral circRNA database, VirusCircBase was introduced in 2020 [55]. Utilizing a de novo method for viral circRNA prediction, 1592 high‐confidence circRNAs from DNA and RNA viruses with lengths ranging from 200 bp to 1 kb and at least 5 reads detected on the backsplice junctions (BSJs) were reported. Notably, up to 99% of viral circRNAs were virus‐specific and had no homologs in other viral species. In addition to the predicted viral circRNAs, the interaction between viral circRNAs with host miRNAs was also analyzed. Based on the competing endogenous RNA (ceRNA) network concept, circRNAs could sponge miRNAs and indirectly modulate expression of miRNA‐targeted mRNAs. Gene Ontology (GO) term enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of circRNAs were performed based on the affected miRNA‐targeted host genes that were indirectly targeted by the viral circRNAs. Consistently, KEGG pathway analysis shows them to be significantly associated with cancer, such as viral carcinogenesis and multiple cancers suggesting important roles for viral circRNAs in cancer. The availability of the VirusCircBase can facilitate further discovery and investigation of viral circRNAs in the context of disease development. The functional enrichment analysis of the predicted viral circRNAs will help to narrow down the potential functions of viral circRNAs; thus, in vivo and in vitro experiments can be planned based on the suggested roles.
Biogenesis of viral circRNAs
Although evidence for viral circRNAs is emerging, the mechanisms for biogenesis of most viral circRNAs remain unknown. Ungerleider et al. in their review reported that inverted repeats such as Alu repeats were not found for the flanking introns of the gammaherpesvirus BSJs identified to date, though short inverted repeats may be a possible mechanism to promote RNA circularization [56]. Interestingly, based on the circRNA analysis by Cai and team, it was reported that viruses might utilize the intron pairing‐driven circularization model for circRNA biogenesis. Their analysis indicated that more than 90% of the viral circRNAs are flanked by short repeat or reverse complementary sequences that are essential for the RNA circularization [55]. Huang et al. also reported that exon 3a to exon 4 of the EBV RPMS1 gene is flanked by long introns containing inverted complementary sequence on each side. This may ease the circularization of RNA and contribute to its abundance across various EBV‐infected cell lines and patient samples [43]. It is worth noting that in the gammaherpesviruses, many viral circRNAs are differentially expressed in latent and lytic states, suggesting specific cis regulation. However, more research is required for further understanding of viral circRNA biogenesis and the regulatory factors involved in circularization.
Host‐derived circRNAs in antiviral immunity
Host innate immune surveillance mechanisms such as the pattern recognition receptors (PRRs) are responsible to react against viral infection to maintain internal homeostasis. Upon detection of viruses, innate immune system is activated and antiviral cytokines are released to protect the cells against viral infection. Notably, findings from several recent studies have begun to shed light on the potential role of host circRNAs in the regulation of antiviral immune responses.
Effects of exogenous circRNAs on host immune response
Chen et al. reported that intracellular introduction of purified exogenous circRNAs, although lacking 5′triphosphate, potently stimulate innate immune genes, including retinoic acid‐inducible gene‐I (RIG‐I), melanoma differentiation‐associated gene 5 (MDA5), 5′ oligoadenylate synthase 1 (OAS1), OAS‐like protein (OASL), and protein kinase R (PKR) with RIG‐I being the most upregulated. Interestingly, it was found that RIG‐I is not only able to recognize circRNAs but also able to differentiate endogenous from exogenous circRNA based on the self versus nonself intronic regions that are essential for RNA circularization [57]. Specifically, circRNAs spliced by endogenous spliceosomes are associated with a diverse and distinctive set of RBPs, whereas exogenous circRNAs lack the binding of host RBPs and thus trigger an RIG‐I‐mediated innate immune response. Other than host RBP binding, m6A RNA modification of the circRNA sequence also helps to differentiate host and exogenous circRNAs [58]. In this case, m6A reader protein binds to the m6A modification on the host circRNA to prevent activation of innate immunity.
Host sensing and immunity against nonself circRNA may be useful to protect against viral circRNAs, for example, KSHV circRNAs from virions or even circular virus RNA genomes, such as hepatitis delta virus (HDV) and Lassa fever virus. However, viral circRNAs transcribed inside the infected cells using the host cell machinery may avoid being sensed as foreign circRNAs by using the same mechanism ascribed to host‐derived circRNAs. In fact, m6A modification is prevalent in viruses and a recent study reported that viruses avoided detection by the innate immune system by acquiring m6A in their RNA, which mimics the cellular RNA [59, 60].
Controversially, Wesselhoeft et al. [61] reported that the immunogenicity of exogenously introduced circRNAs was due to impurities in the synthetic circRNA preparations. RNase R‐treated, HPLC‐purified, and phosphatase‐treated unmodified exogenous circRNAs were relatively less immunogenic compared with those having contaminating RNA species. This evasion of cellular RNA sensors was diminished with linearized circRNA. However, in contrast to the findings for application of exogenous mRNAs in similar studies, nucleoside modification including N1‐methylpseudouridine (m1ψ) of exogenous circRNAs was not necessary to prevent the activation of innate immune responses. The encapsulation of exogenous circRNAs into lipid nanoparticles (LNPs) has also been shown to prevent activation of RIG‐I or Toll‐like receptor (TLR) in vitro and in vivo.
Based on the above, further studies will be needed to address the discordant results on the immunogenicity of exogenous circRNAs (Fig. 4). Notable differences between the two studies are the sequence of circRNA, methodology, and cell type tested. Therefore, in agreement with Basavappa and Cherry, potential contaminants during circRNA preparation, the cell‐type studied, transfection, and inoculation route should be taken into consideration for in vitro and in vivo studies as immunogenicity of a circRNA might be context‐ or cell‐type‐dependent [62].
Fig. 4.
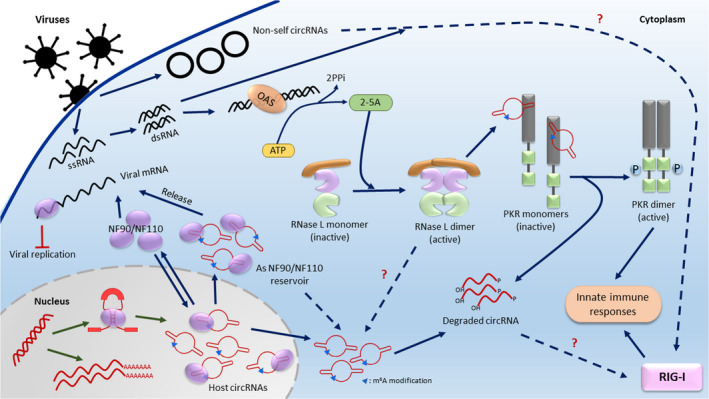
Regulation of host and viral circRNAs by host immune response during viral infection. In noninfected cells, NF90/NF110 localized to the nuclear to promote circRNA biogenesis [63]. Upon viral infection, NF90/NF110 is exported to the cytoplasm to suppress viral replication by targeting viral mRNA. OAS‐activated RNase L would degrade cellular dsRNA‐containing circRNAs bound to PKR that eventually lead to the release and activation of PKR. Both exogenous (not m6A‐modified) and degraded circRNAs are postulated to activate RIG‐I [58, 64]. Activation of PKR and RIG‐1 ultimately leads to innate immune response to eliminate virus in the host. However, it remains inconclusive if exogenous circRNA is immunogenic [57, 58, 61].
Virus infection alters host circRNAs
The host circRNAs play a role in immunosurveillance which was demonstrated by recent studies. Chen Ling‐Ling’s group demonstrated that NF90/NF110, the immune response factors against viral infection, plays a role in facilitating RNA circularization in the nucleus [63]. In the nucleus, binding of NF90/NF110 to the double‐stranded RNAs (dsRNAs) formed by inverted repeat elements in the introns flanking the two splice sites during nascent pre‐mRNA transcription enhances circRNA biogenesis by stabilizing such intronic RNA hairpin structures. Upon viral infection, PKR activation caused NF90/NF110 to be exported to the cytoplasm to suppress viral replication. This reduces the abundance of NF90/NF110 available for pre‐mRNA binding in the nucleus and eventually leads to the reduction in circRNA formation. In addition, some of these mature exonic circRNAs form NF90/NF110‐circRNA protein (circRNP) complexes in the cytoplasm. Interestingly, NF90/NF110 was released from these circRNP complexes to bind viral mRNAs as an immune response for efficient host defense. It is thought that the binding of circRNAs to immune factors in the cytoplasm act as an immediate reservoir for the release of immune factors upon activation but are sequestered away from undesirable immune response. Future work on how other host immune factors identified in the same study, including RIG‐I, contributes to circRNA formation would enable us to discover the function of RNA circularization. It is also possible that viruses utilize the host antiviral response for viral circRNA formation.
In a subsequent study by the same group, Liu et al. reported a significant drastic reduction in global host circRNA levels in cells upon poly(I:C) (a synthetic double‐stranded RNA that is used experimentally to model viral infections in vivo) transfection or encephalomyocarditis virus (EMCV) infection [64]. Intriguingly, they found that a proportion of circRNAs tends to form imperfect short 16–26 bp intra‐RNA duplexes in cells and preferentially interacted with nucleic acid receptors such as PKR. The finding that certain host circRNAs can form a dsRNA structure may explain why NF90/NF110 and RIG‐I are able to bind host circRNAs. Strikingly, interaction of these dsRNA‐containing circRNAs with PKR suppresses the phosphorylation of PKR and activation under normal situations. A similar mechanism could potentially explain impairment of cell‐intrinsic immunity to viruses in the brainstem of DBR1‐mutated patients, which have cellular accumulation of RNA lariat circles [65]. When exogenous dsRNA is introduced into cells through either poly(I:C) transfection or viral infection, OAS1 is activated to generate 2′,5′‐linked oligoadenylates (2‐5A) which further activate RNase L, a cytoplasmic endoribonuclease that target host and viral RNAs for degradation. Interestingly, activated RNase L would target these dsRNA‐containing host circRNAs binding to PKR for degradation. This leads to the release of PKR for activation, to initiate a signal transduction cascade for immune function upon recognizing pathogenic dsRNAs. Studies have reported that RNase L‐cleaved products generated from either self (cellular) or nonself (viral) RNA can activate the RIG‐I–MDA5–IPS‐1 cascade to enhance IFN‐β gene transcription [66]. It is thought that this phenomenon circumvents the need for nonself viral RNAs in perpetuating and amplifying innate immunity. As such, it is possible that the host self circRNAs may also serve as an additional reservoir of self RNA for degradation by RNase L to amplify antiviral innate immunity against viral infections.
While the above studies summarized in Fig. 4 suggest that multiple host circRNAs expressed in low copy numbers may function together as a group during an immune response, other studies have shown that individual host circRNAs play specific pro‐ or antiviral roles during infection of different viruses (see Fig. 3). Upon de novo KSHV infection, Tagawa et al. [40] have shown that many human host circRNAs are differentially expressed, with hsa_circ_0001400 among the most upregulated. Transient and stable expression of hsa_circ_0001400 significantly reduced KSHV LANA‐1 and RTA transcripts without blocking viral entry into cells, suggesting that it acts as an antiviral factor. KSHV LANA‐1 is an important latent protein for persistence and replication of KSHV, whereas RTA protein acts as the master latent‐to‐lytic switch that triggers KSHV to enter into the productive transcriptional program required for viral spread and KS pathogenesis [67]. Therefore, reduction in both LANA‐1 and RTA could prevent persistence and replication of KSHV in the host, thus protecting host cells from KSHV‐related pathogenesis. Other than repressing viral protein, hsa_circ_0001400 may exert its antiviral effect through stimulating some host defense pathways, for example, upregulation of TNF‐α in cells stably expressing hsa_circ_0001400 upon KSHV infection.
In another study by Le Qin and team, it was shown that host circRNAs defend against viral infections by functioning as miRNA sponges against repressors of innate immunity sensors. Upon poly(I:C) treatment in testicular Sertoli and Leydig cells, upregulation of circRNA‐9119 sponged miR‐26a and miR‐136 that interact with the 3′‐UTR of TLR3 and RIG‐I, respectively, leading to the upregulation of inflammatory cytokines such as IFN‐β, IL‐1β, and MCP‐1 [68].
Unexpectedly, host circRNAs can be hijacked by the virus to enhance viral replication and pathogenesis, or to counteract innate immune responses, as revealed by recent studies. For instance, circGATAD2A (ID: hsa_circ_30753) was upregulated upon H1N1 influenza virus (IAV) infection [69]. Overexpression of circGATAD2A greatly increased the viral titer and nucleic proteins (NPs) of H1N1 IAV, whereas knockdown of circGATAD2A showed an opposite effect, suggesting that circGATAD2A expression promotes H1N1 IAV virus replication. The study showed that circGATAD2A negatively regulates VPS34‐dependent autophagy which is important for removal of intracellular viruses, by inhibiting LC3‐II formation and increasing the level of p62. The increase in p62 level suggests that circGATAD2A might also be responsible for suppressing the maturation of autophagosome in parallel. Similarly, Chen et al. showed that host circRNAs could display proviral activities in hepatitis C virus (HCV)‐infected cells [70]. In particular, host circRNA encoded by Pleckstrin and Sec7 domain‐containing 3 (circPSD3) that is upregulated upon HCV infection inhibits cellular nonsense‐mediated decay (NMD) pathway, a general virus restriction pathway. They further showed that this inhibition occurs independent of translation factor eIF4A3, a factor that is essential in the execution of NMD (circPSD3 RNA harbors six predicted eIF4A3 binding sites). Interestingly, circPSD3 also exerts a similar proviral effect in dengue virus but not in chikungunya virus‐infected cells, suggesting distinct effects on different RNA viruses. Other host circRNAs with proviral function are circFNDC3B and circCNOT1, which are upregulated upon infection of lung cells with Middle East respiratory syndrome coronavirus (MERS‐CoV). MERS‐CoV viral load was significantly downregulated in circFNDC3B and circCNOT1‐knowdown cells, presumably due to the decrease in levels of circFNDC3B‐ and circCNOT1‐regulated target genes in mitogen‐activated protein kinase (MAPK) and ubiquitination pathways [71].
Lastly, RNA‐seq data analysis of cells infected with other viruses such as porcine endemic diarrhea virus (PEDV) [72], grass carp reovirus [73], avian leukosis virus [74], simian virus 40 (SV40) [75], transmissible gastroenteritis virus (TGEV) [76], and canine influenza viruses [77] has also revealed host circRNAs changes upon virus infection. The potential function of these host circRNAs has been predicted based on GO enrichment and KEGG pathway analysis, as well as by the concept of ceRNA networks. Although some of the miRNAs predicted to be sponged by host circRNAs might be involved in viral infection and antiviral effect, the role of these circRNAs still remains to be proven.
Viral and host antiviral circRNA as diagnostic and prognostic marker
Continued studies on the implications of host or viral‐encoded circRNAs during host–virus interaction not only enhance our understanding of currently unknown physiological functions of circRNAs and their potential role in disease pathogenesis, but also lead toward potential novel diagnostic and/or prognostic as well as therapeutic strategies against virus‐associated diseases.
The fact that circRNAs were reported to be stably detected in human peripheral whole blood [78], plasma [79], and saliva [80] offers an easily accessible yet noninvasive method for their use in the detection of a variety of diseases. The high stability along with cell‐type and developmental stage specificity of host and viral circRNAs shows immense potential for their development into useful diagnostic and prognostic biomarkers for virus‐related diseases. The development of a reliable yet rapid detection method to measure the level of viral circRNAs in serum, plasma, or blood could help to assess the risk of a condition associated with pathogenic virus infection. Similar methods can also be employed to monitor organ or tissue transplant recipients, for example, the presence of viral circRNAs in the transplanted organ or tissue. Positive identification of viral circRNAs can indicate a heightened risk for contracting virus‐associated diseases.
Various methods including in situ hybridization and divergent primer PCR can be utilized to determine the presence of viral circRNA in a patient’s tissue or fluid samples. For example, viral circRNAs such as EBV circRPMS1_e4_e3a, KSHV circPAN/K7.3, circvIRF4, and HPV circE7 have been successfully detected in patient samples. It would be interesting to compare the specificity and sensitivity of EBV circRPMS1_e4_e3a to the existing clinical diagnostic marker, EBV‐encoded RNA transcripts (EBER), for EBV detection in tumors. A similar approach can be used to compare the potential of HPV circE7 to the existing HPV DNA testing method as a diagnostic marker for cervical cancer screening. In addition, Chamseddin et al. suggest that HPV circE7 could be used as a sensitive ‘passenger’ biomarker to differentiate the tumor state between basaloid (a more benign state) and keratinizing as its level reduced after calcium‐induced differentiation of keratinocytes [50].
Likewise, the level of immune‐related circRNAs is a potential molecular indicator for antiviral immune responses. Recent advances in high‐throughput techniques are beneficial for the identification of such promising circRNA biomarkers.
Viral and host antiviral circRNAs as therapeutic targets
Based on the current understanding of circRNA functions, host and viral circRNAs could serve as a target for precise therapies. Possible concepts for therapeutic modulation include overexpressing protective or antiviral circRNAs or depleting proviral circRNAs. Mounting evidence has shown that miRNAs are aberrantly expressed and contribute to the disease development of different types of virus‐related diseases. Therefore, the use of circRNAs as inhibitors of miRNA activity in disease therapeutics has great potential. Host circRNAs in antiviral immunity identified through study can be made into synthetic circRNA sponges or super sponges which can be introduced into patients to absorb one or multiple specific overexpressed viral disease‐linked miRNAs and/or RBPs as disease suppressors. A proof‐of‐principle study by Oliver Rossbach’s group reported the first artificial circRNA that acts as a microRNA sponge targeting a specific microRNA, miR‐122, that is required for the life cycle of the hepatitis C virus (HCV) [81]. The study demonstrated that the artificial circRNA sponges sequestered miRNA‐122, depriving HCV of miRNA‐122, and had comparable efficiency as Miravirsen. Miravirsen was the first anti‐microRNA drug that managed to decrease HCV titer in patients by functionally sequestering miRNA‐122. Drugs targeting circRNA biogenesis mechanisms could also be designed to alter the generation of disease‐linked viral or host circRNAs that are antiviral or proviral. Alternatively, synthetic purified circRNAs could be used as an immune adjuvant to purposefully boost innate and adaptive immune signaling against viral infections [58, 82]. Future research efforts focused on identifying circRNA targets and improved patient delivery methods are important for the development of safe and effective circRNA‐based therapies for virus‐related diseases.
Perspectives and Future directions
The field of circRNA in host–virus interaction is still in its infancy but is exciting. The finding that viruses also encode circRNAs adds another layer of complexity to the biology of viruses and host–virus interaction. However, many questions and challenges remain. In order to better understand the role of circRNAs (viral and host) in disease pathogenesis, more studies are needed for the discovery of novel viral circRNAs and to identify circRNAs that are differentially expressed (DE) during host–virus interaction. Utilization of newer algorithms such as CIRIquant that includes a normalization step is beneficial to obtain more accurate expression values of circRNA and DE circRNAs in the virus or host cell [83].
Importantly, do all circRNAs and in particular viral circRNAs have biological functions during viral life cycles, host–virus interactions, and contribute to the pathogenesis of disease, or are these splicing by‐products? Till today, most of the circRNAs that have been described are localized to the cytoplasm and are involved in the post‐transcriptional regulation of gene expression, with many reported to act as miRNA sponges. However, caution should be taken, as many circRNAs are reported to be in low copy numbers thus might be insufficient to act effectively as miRNA sponges in a physiological setting.
In contrast, nuclear circRNAs including intronic circRNA [12] and exon–intron circRNA [84] are less studied. Importantly, circRNA with introns retained could share the same BSJ with the exonic circRNA encoded by the same exons. circRPMS1_e4_e3a, circRPMS1_e4_e2, circLMP2_e8_e2 [56], and circvIRF4 [47] with intron retention were subsequently found to be predominantly localized in the nucleus, which is different from their exonic isoforms that localized to the cytoplasm. Whether or not these nuclear viral circRNA isoforms are functional remains to be investigated. New algorithms such as CIRI‐full [85] and CircAST [86] would be useful for circRNA full‐length assembly and isoform quantification. Full‐length circRNA assembly is important in determination of sequence conservation and identification of RBP/miRNA‐binding sites and diseases‐associated SNP variation in circRNAs.
More studies are also needed to understand the biogenesis and regulation of viral circRNA expression, which would hint on its possible biological function. Given that viral circRNAs are packaged into KSHV virions, it would also be interesting to determine whether the same occurs in other herpesvirus virions and in other virus families. Understanding the importance of virion incorporation may help to uncover the biological functions of viral circRNAs.
Finally, the new concept that multiple host circRNAs may act in groups during an antiviral response provides new insight into how low copy number circRNAs can function and is definitely worth pursuing further.
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
All authors wrote the manuscript.
Acknowledgements
We thank Dr. Ea Chee Kwee (University of Texas Southwestern Medical Center) and Dr. Jennifer Ann Harikrishna (Universiti Malaya) for critical review of the manuscript. This work was supported by grants from Fundamental Research Grant Scheme (FRGS/1/2017/SKK08/UM/02/11) and University of Malaya High Impact Research Grant (UM.C/625/1/HIR/MOE/CHAN/02/07).
Contributor Information
Ke‐En Tan, Email: yatyuen.lim@um.edu.my.
Yat‐Yuen Lim, Email: yatyuen.lim@um.edu.my.
References
- 1. Bieniasz PD (2004) Intrinsic immunity: a front‐line defense against viral attack. Nat Immunol 5, 1109–1115. [DOI] [PubMed] [Google Scholar]
- 2. Salzman J, Gawad C, Wang PL, Lacayo N & Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF & Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al, (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. [DOI] [PubMed] [Google Scholar]
- 5. Jeck WR & Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rybak‐Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal‐Fluss R et al, (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58, 870–885. [DOI] [PubMed] [Google Scholar]
- 7. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G et al, (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7, 11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA & Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134. [DOI] [PubMed] [Google Scholar]
- 9. Ashwal‐Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N & Kadener S (2014) circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell 56, 55–66. [DOI] [PubMed] [Google Scholar]
- 10. Liang D & Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28, 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dubin RA, Kazmi MA & Ostrer H (1995) Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167, 245–248. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y, Zhang X‐O, Chen T, Xiang J‐F, Yin Q‐F, Xing Y‐H, Zhu S, Yang L & Chen L‐L (2013) Circular intronic long noncoding RNAs. Mol Cell 51, 792–806. [DOI] [PubMed] [Google Scholar]
- 13. Barrett SP, Wang PL & Salzman J (2015) Circular RNA biogenesis can proceed through an exon‐containing lariat precursor. eLife 4, e07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mayer A, Mosler G, Just W, Pilgrim C & Reisert I (2000) Developmental profile of Sry transcripts in mouse brain. Neurogenetics 3, 25–30. [DOI] [PubMed] [Google Scholar]
- 15. Huang S, Yang B, Chen BJ, Bliim N, Ueberham U, Arendt T & Janitz M (2017) The emerging role of circular RNAs in transcriptome regulation. Genomics 109, 401–407. [DOI] [PubMed] [Google Scholar]
- 16. Huang H, Wei L, Qin T, Yang N, Li Z & Xu Z (2019) Circular RNA ciRS‐7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR‐7/KLF4 and NF‐κB signals. Cancer Biol Ther 20, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Piwecka M, Glažar P, Hernandez‐Miranda LR, Memczak S, Wolf SA, Rybak‐Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P et al, (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526. [DOI] [PubMed] [Google Scholar]
- 18. Kleaveland B, Shi CY, Stefano J & Bartel DP (2018) A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174, 350–362.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J & Zhou Y (2015) Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β‐catenin pathway. Oncotarget 6, 6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang G, Zhu H, Shi Y, Wu W, Cai H & Chen X (2015) cir‐ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β‐catenin pathway. PLoS One 10, e0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hentze MW & Preiss T (2013) Circular RNAs: splicing's enigma variations. EMBO J 32, 923–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, Song W, Li X, Li L, Du Z et al, (2018) A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol 19, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P & Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44, 2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Du WW, Yang W, Chen Y, Wu Z‐K, Foster FS, Yang Z, Li X & Yang BB (2016) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38, 1402–1412. [DOI] [PubMed] [Google Scholar]
- 25. Rossi F, Legnini I, Megiorni F, Colantoni A, Santini T, Morlando M, Di Timoteo G, Dattilo D, Dominici C & Bozzoni I (2019) Circ‐ZNF609 regulates G1‐S progression in rhabdomyosarcoma. Oncogene 38, 3843–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liang W‐C, Wong C‐W, Liang P‐P, Shi M, Cao Y, Rao S‐T, Tsui SK‐W, Waye MM‐Y, Zhang Q, Fu W‐M & et al, (2019) Translation of the circular RNA circβ‐catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol 20, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M et al, (2017) Circ‐ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66, 22–37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li X, Yang L & Chen LL (2018) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71, 428–442. [DOI] [PubMed] [Google Scholar]
- 29. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB & Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20, 675–691. [DOI] [PubMed] [Google Scholar]
- 30. Xiao M‐S, Ai Y & Wilusz JE (2020) Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol 30, 226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilusz JE (2018) A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA 9, e1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanger HL, Klotz G, Riesner D, Gross HJ & Kleinschmidt AK (1976) Viroids are single‐stranded covalently closed circular RNA molecules existing as highly base‐paired rod‐like structures. Proc Natl Acad Sci USA 73, 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW & Vogelstein B (1991) Scrambled exons. Cell 64, 607–613. [DOI] [PubMed] [Google Scholar]
- 34. Zhang Z, Qi S, Tang N, Zhang X, Chen S, Zhu P, Ma L, Cheng J, Xu Y, Lu M et al, (2014) Discovery of replicating circular RNAs by RNA‐seq and computational algorithms. PLoS Pathog 10, e1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steger G & Riesner D (2018) Viroid research and its significance for RNA technology and basic biochemistry. Nucleic Acids Res 46, 10563–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Young LS, Yap LF & Murray PG (2016) Epstein‐Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 16, 789–802. [DOI] [PubMed] [Google Scholar]
- 37. Skalsky RL & Cullen BR (2015) EBV noncoding RNAs. Curr Top Microbiol Immunol 391, 181–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S, Baddoo M, Moss WN, Yu Y, Seddon M et al, (2018) The Epstein Barr virus circRNAome. PLoS Pathog 14, e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, Lee N, Shair KH, Moore PS & Chang Y (2018) Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci USA 115, E8737–E8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tagawa T, Gao S, Koparde VN, Gonzalez M, Spouge JL, Serquiña AP, Lurain K, Ramaswami R, Uldrick TS, Yarchoan R et al, (2018) Discovery of Kaposi's sarcoma herpesvirus‐encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci USA 115, 12805–12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ungerleider N, Jain V, Wang Y, Maness NJ, Blair RV, Alvarez X, Midkiff C, Kolson D, Bai S, Roberts C et al, (2019) Comparative analysis of gammaherpesvirus circular RNA repertoires: conserved and unique viral circular RNAs. J Virol 93, e01952‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Q, Shuai M & Xia Y (2019) Knockdown of EBV‐encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag Res 11, 8023–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang J‐T, Chen J‐n, Gong L‐p, Bi Y‐h, Liang J, Zhou L, He D & Shao C‐k (2019) Identification of virus‐encoded circular RNA. Virology 529, 144–151. [DOI] [PubMed] [Google Scholar]
- 44. Gong L‐P, Chen J‐N, Dong M, Xiao Z‐D, Feng Z‐Y, Pan Y‐H, Zhang Y, Du Y, Zhang J‐Y, Bi Y‐H et al, (2020) Epstein‐Barr virus‐derived circular RNA LMP2A induces stemness in EBV‐associated gastric cancer. EMBO Rep 21, e49689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohtsuka J, Oshima H, Ezawa I, Abe R, Oshima M & Ohki R (2018) Functional loss of p53 cooperates with the in vivo microenvironment to promote malignant progression of gastric cancers. Sci Rep 8, 2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cesarman E, Damania B, Krown SE, Martin J, Bower M & Whitby D (2019) Kaposi sarcoma. Nat Rev Dis Primers 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abere B, Li J, Zhou H, Toptan T, Moore PS & Chang Y (2020) Kaposi’s sarcoma‐associated herpesvirus‐encoded circRNAs are expressed in infected tumor tissues and are incorporated into virions. MBio 11, e03027‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berman TA & Schiller JT (2017) Human papillomavirus in cervical cancer and oropharyngeal cancer: one cause, two diseases. Cancer 123, 2219–2229. [DOI] [PubMed] [Google Scholar]
- 49. Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, Gusho E, Xie Y, Chiang C‐M, Buszczak M et al, (2019) Transforming activity of an oncoprotein‐encoding circular RNA from human papillomavirus. Nat Commun 10, 2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chamseddin BH, Lee EE, Kim J, Zhan X, Yang R, Murphy KM, Lewis C, Hosler GA, Hammer ST, Wang RC et al, (2019) Assessment of circularized E7 RNA, GLUT1, and PD‐L1 in anal squamous cell carcinoma. Oncotarget 10, 5958–5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Torresi J, Tran BM, Christiansen D, Earnest‐Silveira L, Schwab RHM & Vincan E (2019) HBV‐related hepatocarcinogenesis: the role of signalling pathways and innovative ex vivo research models. BMC Cancer 19, 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu M, Liang Z, Pan J, Hu X, Zhang X, Xue R, Cao G & Gong C (2020) HBV pgRNA can generate a circRNA with two junction sites. bioRxiv, p. 2020.05.14.095273. [PREPRINT] [Google Scholar]
- 53. Sekiba K, Otsuka M, Ohno M, Kishikawa T, Yamagami M, Suzuki T, Ishibashi R, Seimiya T, Tanaka E & Koike K (2018) DHX9 regulates production of hepatitis B virus‐derived circular RNA and viral protein levels. Oncotarget 9, 20953–20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aktaş T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R & Akhtar A (2017) DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544, 115–119. [DOI] [PubMed] [Google Scholar]
- 55. Cai Z, Fan Y, Zhang Z, Lu C, Zhu Z, Jiang T, Shan T & Peng Y (2020) VirusCircBase: a database of virus circular RNAs. Brief Bioinform. online ahead of print. [DOI] [PubMed] [Google Scholar]
- 56. Ungerleider NA, Tibbetts SA, Renne R & Flemington EK (2019) Gammaherpesvirus RNAs come full circle. MBio 10, e00071‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A & Chang HY (2017) Sensing self and foreign circular RNAs by intron identity. Mol Cell 67, 228–238.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B et al, (2019) N6‐methyladenosine modification controls circular RNA immunity. Mol Cell 76, 96–109.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, Liang X, Gao TZ, Xu Y, Zhou J et al, (2020) N6‐methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG‐I. Nat Microbiol 5, 584–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu F, Cheng W, Zhao F, Tang M, Diao Y & Xu R (2019) Association of N6‐methyladenosine with viruses and related diseases. Virol J 16, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wesselhoeft RA, Kowalski PS, Parker‐Hale FC, Huang Y, Bisaria N & Anderson DG (2019) RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol Cell 74, 508–520.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Basavappa MG & Cherry S (2019) Going in circles: the black box of circular RNA immunogenicity. Mol Cell 76, 3–5. [DOI] [PubMed] [Google Scholar]
- 63. Li X, Liu C‐X, Xue W, Zhang Y, Jiang S, Yin Q‐F, Wei J, Yao R‐W, Yang L & Chen L‐L (2017) Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell 67, 214–227.e7. [DOI] [PubMed] [Google Scholar]
- 64. Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H et al, (2019) Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880.e21. [DOI] [PubMed] [Google Scholar]
- 65. Zhang SY, Clark NE, Freije CA, Pauwels E, Taggart AJ, Okada S, Mandel H, Garcia P, Ciancanelli MJ, Biran A et al, (2018) Inborn errors of RNA lariat metabolism in humans with brainstem viral infection. Cell 172, 952–965.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Malathi K, Dong B, Gale M Jr & Silverman RH (2007) Small self‐RNA generated by RNase L amplifies antiviral innate immunity. Nature 448, 816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Uppal T, Banerjee S, Sun Z, Verma SC & Robertson ES (2014) KSHV LANA–the master regulator of KSHV latency. Viruses 6, 4961–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Qin L, Lin J & Xie X (2019) CircRNA‐9119 suppresses poly I: C induced inflammation in Leydig and Sertoli cells via TLR3 and RIG‐I signal pathways. Mol Med 25, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu T, Ding Y, Zhang Y, Liu Y, Li Y, Lei J, Zhou J, Song S & Hu B (2019) Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet Microbiol 231, 238–245. [DOI] [PubMed] [Google Scholar]
- 70. Chen TC, Tallo‐Parra M, Cao QM, Kadener S, Böttcher R, Pérez‐Vilaró G, Boonchuen P, Somboonwiwat K, Díez J & Sarnow P (2020) Host‐derived circular RNAs display proviral activities in Hepatitis C virus‐infected cells. PLoS Pathog 16, e1008346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang X, Chu H, Wen L, Shuai H, Yang D, Wang Y, Hou Y, Zhu Z, Yuan S, Yin F et al, (2020) Competing endogenous RNA network profiling reveals novel host dependency factors required for MERS‐CoV propagation. Emerg Microbes Infect 9, 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen J, Wang H, Jin L, Wang L, Huang X, Chen W, Yan M & Liu G (2019) Profile analysis of circRNAs induced by porcine endemic diarrhea virus infection in porcine intestinal epithelial cells. Virology 527, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. He L, Zhang A, Xiong L, Li Y, Huang R, Liao L, Zhu Z & Wang AY (2017) Deep circular RNA sequencing provides insights into the mechanism underlying grass carp reovirus infection. Int J Mol Sci 18, 1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang X, Yan Y, Lei X, Li A, Zhang H, Dai Z, Li X, Chen W, Lin W, Chen F et al, (2017) Circular RNA alterations are involved in resistance to avian leukosis virus subgroup‐J‐induced tumor formation in chickens. Oncotarget 8, 34961–34970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shi J, Hu N, Li J, Zeng Z, Mo L, Sun J, Wu M & Hu Y (2017) Unique expression signatures of circular RNAs in response to DNA tumor virus SV40 infection. Oncotarget 8, 98609–98622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ma X, Zhao X, Zhang Z, Guo J, Guan L, Li J, Mi M, Huang Y & Tong D (2018) Differentially expressed non‐coding RNAs induced by transmissible gastroenteritis virus potentially regulate inflammation and NF‐κB pathway in porcine intestinal epithelial cell line. BMC Genom 19, 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tao P, Ning Z, Hao X, Lin X, Zheng Q & Li S (2019) Comparative analysis of whole‐transcriptome RNA expression in MDCK cells infected with the H3N2 and H5N1 canine influenza viruses. Front Cell Infect Microbiol 9, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Memczak S, Papavasileiou P, Peters O & Rajewsky N (2015) Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 10, e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, Wyss‐Coray T, Blumenfeld YJ, El‐Sayed YY & Quake SR (2014) Noninvasive in vivo monitoring of tissue‐specific global gene expression in humans. Proc Natl Acad Sci USA 111, 7361–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bahn JH, Zhang Q, Li F, Chan T‐M, Lin X, Kim Y, Wong DTW & Xiao X (2015) The landscape of microRNA, Piwi‐interacting RNA, and circular RNA in human saliva. Clin Chem 61, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jost I, Shalamova LA, Gerresheim GK, Niepmann M, Bindereif A & Rossbach O (2018) Functional sequestration of microRNA‐122 from Hepatitis C Virus by circular RNA sponges. RNA Biol 15, 1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Holdt LM, Kohlmaier A & Teupser D (2018) Circular RNAs as therapeutic agents and targets. Front Physiol 9, 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang J, Chen S, Yang J & Zhao F (2020) Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat Commun 11, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L et al, (2015) Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22, 256–264. [DOI] [PubMed] [Google Scholar]
- 85. Zheng Y, Ji P, Chen S, Hou L & Zhao F (2019) Reconstruction of full‐length circular RNAs enables isoform‐level quantification. Genome Med 11, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wu J, Li Y, Wang C, Cui Y, Xu T, Wang C, Wang X, Sha J, Jiang B, Wang K et al, (2019) CircAST: full‐length assembly and quantification of alternatively spliced isoforms in circular RNAs. Genomics Proteomics Bioinformatics 17, 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]


