Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) entry factors, ACE2 and TMPRSS2, are highly expressed in nasal epithelial cells. However, the association between SARS‐CoV‐2 and nasal inflammation in chronic rhinosinusitis with nasal polyps (CRSwNP) has not been investigated. We thus investigated the expression of SARS‐CoV‐2 entry factors in nasal tissues of CRSwNP patients, and their associations with inflammatory endotypes of CRSwNP.
Methods
The expression of ACE2 and TMPRSS2 was assessed in nasal tissues of control subjects and eosinophilic CRSwNP (ECRSwNP) and nonECRSwNP patients. The correlations between ACE2/TMPRSS2 expression and inflammatory indices of CRSwNP endotypes were evaluated. Regulation of ACE2/TMPRSS2 expression by inflammatory cytokines and glucocorticoids was investigated.
Results
ACE2 expression was significantly increased in nasal tissues of nonECRSwNP patients compared to ECRSwNP patients and control subjects, and positively correlated with the expression of IFN‐γ, but negatively correlated with tissue infiltrated eosinophils, and expression of IL5 and IL13. IFN‐γ up‐regulated ACE2 expression while glucocorticoid attenuated this increase in cultured nasal epithelial cells. Genes co‐expressed with ACE2 were enriched in pathways relating to defence response to virus in nasal tissue. TMPRSS2 expression was decreased in nasal tissues of CRSwNP patients compared to control subjects and not correlated with the inflammatory endotypes of CRSwNP. Glucocorticoid treatment decreased ACE2 expression in nasal tissues of nonECRSwNP patients, but not in ECRSwNP patients, whereas TMPRSS2 expression was not affected.
Conclusion
These findings indicate that ACE2 expression, regulated by IFN‐γ, is increased in nasal tissues of nonECRSwNP patients and positively correlates with type 1 inflammation.
Keywords: ACE2, chronic rhinosinusitis with nasal polyps, inflammatory endotype, SARS‐CoV‐2, TMPRSS2
The expression of SARS‐CoV‐2 receptor ACE2 is significantly increased in nasal tissues of nonECRSwNP patients compared to ECRSwNP patients and control subjects. ACE2 expression is increased in nasal tissues of type 1 endotype of CRSwNP and positively correlated with the expression of IFN‐γ. IFN‐γ up‐regulates ACE2 expression while glucocorticoids attenuate this increase in cultured nasal epithelial cells.
Abbreviations: ACE2, angiotensin‐converting enzyme 2; ECRSwNP, eosinophilic chronic rhinosinusitis with nasal polyps; non‐ECRSwNP, noneosinophilic chronic rhinosinusitis with nasal polyps; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), a virus which causes the coronavirus disease‐2019 (COVID‐19), was first reported in December, 2019. 1 SARS‐CoV‐2 has spread worldwide and is presently considered as a pandemic, leading to confirmed infection of more than 6 million individuals and 365 thousand deaths globally, as of May 31, 2020. SARS‐CoV‐2 typically affects the respiratory system, causing pneumonia, fever, cough and shortness of breath. 2 , 3 Individuals with pre‐existing health conditions such as hypertension, chronic lung disease and diabetes, as well as the elderly, are particularly at greater risk for developing severe COVID‐19. 4 , 5 , 6 Interestingly, studies have indicated that the prevalence of asthma and allergic rhinitis in patients with COVID‐19 was markedly lower than that reported in the adult population, suggesting that asthma and respiratory allergy might be not significant risk factors for COVID‐19. 4 , 7 , 8 However, at present, it is important and urgent to identify potential risk factors for COVID‐19.
SARS‐CoV‐2 employs angiotensin‐converting enzyme 2 (ACE2) as the receptor for cell entry and transmembrane protease serine 2 (TMPRSS2) for S protein priming. 9 The expression of ACE2 has generally been found to be low in multiple tissues, whereas TMPRSS2 has been shown to be highly expressed with a broader distribution, suggesting that ACE2, rather than TMPRSS2, may be a limiting factor for viral entry at the initial infection stage. 10 , 11 Recently, Sungnak and colleagues reported that genes associated with SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells. 10 Moreover, Wang and colleagues 12 investigated the presence of SARS‐CoV‐2 in different types of clinical specimens, including bronchoalveolar fluid, sputum, nasal swabs, faeces, blood, and urine; from patients with COVID‐19 infection in China, and indicated that the viral load was much higher in nasal swabs than in all the other types of clinical specimens. Thus, it is likely that the nasal mucosa may be an important site of infection. Indeed, a systematic review of existing scientific evidence for sinonasal pathophysiology in COVID‐19 has concluded that sinonasal tract may be an important site of infection, and sinonasal viral shedding may be an important transmission mechanism. 13 However, the association between nasal inflammation and COVID‐19 has not been investigated to date.
Chronic rhinosinusitis (CRS) is a common inflammatory disease of nasal and sinus mucosa, which affects 5%‐15% of the general population. 14 CRS with nasal polyps (CRSwNP) accounts for approximately 20% of all CRS and has greater severity of clinical disease. 15 Based on the degree of eosinophil infiltration in nasal tissue, CRSwNP can further be classified into eosinophilic CRSwNP (ECRSwNP) and noneosinophilic CRSwNP (nonECRSwNP), which show distinct immune‐inflammatory characteristics. 16 Specifically, ECRSwNP has a predominantly type 2 inflammation characterized by pronounced eosinophilia and high levels of interleukin‐4 (IL‐4), IL‐5 and IL‐13 cytokines, while nonECRSwNP is characterized by type 1 and/or type 3 inflammations. 17 In the present study, we investigated the expression of SARS‐CoV‐2 entry factors, ACE2 and TMPRSS2, in nasal tissues of ECRSwNP and nonECRSwNP. The correlations between the expression of SARS‐CoV‐2 entry factors and inflammatory indices of CRSwNP endotypes were evaluated. Regulation of ACE2/TMPRSS2 expression by inflammatory cytokines and glucocorticoid therapy was further investigated.
2. MATERIALS AND METHODS
2.1. Subjects
Patients with CRSwNP were diagnosed according to the European Position Paper on Rhinosinusitis and Nasal Polyps 2020 guidelines. 14 Patients undergoing septoplasty because of anatomic variations and without other sinonasal diseases were recruited as control subjects. Sixteen ECRSwNP patients, 10 nonECRSwNP patients and 19 healthy control subjects were enrolled in series from the Rhinology Department of Beijing TongRen Hospital. Subjects with immunodeficiency, fungal sinusitis, coagulation disorder, neoplasia, pregnancy and nonsteroidal anti‐inflammatory drugs exacerbated respiratory disease (NERD) were excluded. None of the patients had been treated with corticosteroids, antibiotics or biologics within the 4‐week period before enrolment. On admission to the hospital, nasal polyp tissues were collected from CRSwNP patients and these patients were prescribed a 2‐week therapy of oral glucocorticoid (24 mg of methylprednisolone by mouth every morning). At the end of this treatment, all patients underwent endoscopic surgery, and nasal polyp tissues were collected again during surgery. Nasal tissues of inferior turbinate, middle turbinate and uncinate process from healthy subjects were collected for comparison as controls.
In order to better understand any association between CRS and COVID‐19, a cohort of 92 hospitalized patients with COVID‐19 infection admitted from June 13 to June 25, 2020 in the Otolaryngology Department of Beijing Ditan Hospital, a designated hospital to treat patients infected with SARS‐CoV‐2, were further investigated in this study. SARS‐CoV‐2 infection and diagnosis of COVID‐19 were confirmed according to the Diagnosis and Treatment Guidance of Corona Virus Diseases 2019 (Seventh Edition) document published by the National Health Commission of China (http://www.gov.cn/zhengce/zhengceku/2020‐03/04/ content_5486705.htm). All COVID‐19 patients had mild and non‐pneumonia disease, as defined previously. 18 Underlying CRS in any patient was documented based on history of doctor‐diagnosed CRS in the last three months before SARS‐CoV‐2 infection, and percentage and absolute counts of blood eosinophils were determined in these CRS patients with COVID‐19. The study protocol was approved by the Ethics Committees of Beijing Tongren Hospital and Beijing Ditan Hospital, and all subjects provided written informed consent prior to enrolment in the study.
2.2. Histologic analysis
Nasal polyp tissues collected at surgery were immediately formalin fixed, and then processed further by dehydration and embedding in paraffin wax, according to standardized histology protocols. Paraffin sections were stained with haematoxylin and eosin (H&E) and evaluated for the presence of different inflammatory cell types by optical microscopy at ×400 magnification. The absolute numbers and percentages of infiltrating inflammatory cells, including eosinophils, neutrophils, plasma cells and lymphocytes, were recorded as mean of six non‐overlapping regions in each section by two independent pathologists, who were blinded to the study design and clinical background of the patients. The tissues from the CRSwNP patients were further classified as ECRSwNP or nonECRSwNP, based on whether or not the percentage of infiltrating eosinophils exceeded 10% of the total inflammatory cells. 16 , 19
2.3. Cell cultures
Primary human nasal epithelial cells (HNECs) were derived from nasal polyp tissue samples of CRSwNP patients obtained during surgery, as previously described. 20 Briefly, HNECs were isolated by incubation of the polyp tissue overnight at 4°C in Dulbecco's modified Eagle medium (DMEM) containing 0.1% protease (Sigma‐Aldrich, St Louis, MO). At the end of this incubation, separated HNECs were collected and cultured in Bronchial Epithelial Growth Medium (BEGM; Lonza, Walkersville, MD). HNECs at the 2nd passage were collected and seeded onto 6.5‐mm‐diameter Transwells, with polyester membranes of pore size 0.4 mm (COSTAR; Corning, NY). The cells were grown submerged in culture medium for 4 days until they reached complete confluence. At this stage, the culture medium was remove, and DMEM: BEGM (1:1) containing 50 nmol/L all‐trans retinoic acid was added only basolaterally to allow the cells to differentiate and establish as air‐liquid interface (ALI) cultures. The differentiated HNECs were harvested on day 21 of ALI culture and processed for RNA sequencing as described below.
Cultured primary HNECs were also stimulated with IFN‐γ (10ng/mL), IL‐4 (50ng/mL), IL‐5 (50ng/mL), IL‐13 (50ng/mL) and IL‐17 (50ng/mL), or with a combination of IFN‐γ (10ng/mL) and budesonide (0.4 μg/mL or 0.8 μg/mL) or IFN‐γ (10 ng/mL) and dexamethasone (0.5 μg/mL or 1.0 μg/mL) for 24 hours. At the end of this incubation, the cells were harvested and processed for RNA sequencing or real‐time PCR for ACE2 and TMPRSS2 expression as described below.
2.4. Quantitative real‐time PCR
Total RNA was extracted using Trizol reagent (Thermo Fisher, Waltham, MA) from HNECs according to the manufacturer's instructions, and cDNA was subsequently synthesized using the PrimeScript RT Master Mix (TaKaRa Biotechnology, China). Quantitative real‐time PCR was then performed with TB Green Fast qPCR kit (TaKaRa Biotechnology, China) to evaluate the amount of mRNA expression according to the manufacturer's instructions. The sequences of primers used are shown in Table S3.
2.5. Western blot
Total protein was extracted from nasal tissues of ECRSwNP patients, nonECRSwNP patients and healthy control subjects using RIPA lysate with proteinase inhibitor cocktail, and the concentration in each sample was measured using BCA protein assay kit (Pierce, IL, USA). Following heat denaturation, 40 μg of the total protein was loaded onto 10% sodium dodecyl sulphate‐polyacrylamide gels, and the different proteins separated electrophoretically. The separated protein bands were transferred to a polyvinylidene difluoride membrane and blocked with 5% non‐fat dry milk, before being probed with anti‐ACE2 (1:1000 dilution, R&D Systems, USA), anti‐TMPRSS2 (1:500 dilution, Proteintech, China) and anti‐β‐actin (1:10 000, Thermo Fisher, USA). After washing in TBST buffer, the membrane was then incubated with secondary antibody. Blots were visualized using enhanced chemiluminescence and the intensity of each band was quantified by densitometry using ImageJ software (National Institute of Health, USA). The intensities of ACE2 and TMPRSS2 bands were expressed relative to the density of β‐actin standard.
2.6. RNA sequencing and data analysis
RNA sequencing was performed on both nasal tissue samples collected during surgery and cultured HNECs. Fresh samples were immediately suspended in RNA later solution (Qiagen, Hilden, Germany) and processed for extraction and purification of total RNA. RNA quantity and quality were determined using the NanoDrop 2000 Spectrophotometer (Thermo Fischer Scientific) and 2100 TapeStation Automated Electrophoresis System (Agilent Technologies). Ribosomal RNA was removed and sequencing libraries were prepared using the rRNA‐depleted RNA by NEBNext UltraTM Directional RNA Library Prep Kit (New England Biolabs, USA), following the manufacturer's instructions. RNA sequencing was performed on the Illumina Hiseq platform and 150 bp paired‐end reads were generated by Novogene Bioinformatics Technology Cooperation (Beijing, China).
Adapters and low‐quality tail were trimmed from reads prior to read alignment. Clean sequence reads were aligned to the human genome with Hisat2 (v2.0.5). Cufflinks (v2.2.1) was used to assemble transcripts, estimate the abundance of these transcripts and detect differential expression among samples. The reference genome build GRCh37 was used as the annotation references for mRNAs. Fragments per kilo‐base of exon per million fragments mapped (FPKM) of mRNAs in each sample was calculated based on the length of the fragments and reads count mapped to this fragment. Differential expression analysis was performed using Cuffdiff software (v2.2.1). P‐value < .05 plus fold change > 2 was used as the cut‐off for significantly differentially expressed genes (DEGs).
2.7. Expression analysis of SARS‐CoV‐2 entry factors in nasal tissue
The expression of ACE2 and TMPRSS2 was assessed based on RNA sequencing data from nasal tissues of control subjects (n = 19), ECRSwNP patients (n = 16) and nonECRSwNP patients (n = 10), HNECs of CRSwNP patients (n = 11), and primary human bronchial epithelial cells (HBECs) of control subjects (n = 5), asthma patients (n = 6) and chronic obstructive pulmonary disease (COPD) patients (n = 5). RNA sequencing data of nasal tissues and HNECs were generated as described above. RNA sequencing data of HBECs were generated in previous study. 21 One published RNA sequencing database of nasal tissues from control, ECRSwNP and nonECRSwNP (n = 3, each group) 22 was obtained from the Gene Expression Omnibus (GSE72713) and used for verifying the expression of ACE2 and TMPRSS2. To adjust batch‐specific systematic variations, the expression levels of ACE2 and TMPRSS2 were normalized using reference genes. For example, the expression level of ACE2 in each sample was calculated as: ACE2 (FPKM)/reference gene (FPKM). However, in case of the RNA sequencing data, ACE2 (FPKM) was used to represent the expression level of ACE2 directly from one single database (GSE72713), which was not affected by batch effects. Ribosomal protein lateral stalk subunit P0 (RPLP0) was used as reference gene owing to its stable expression in nasal tissue of CRS patients. 23 Co‐expressed genes of ACE2 in nasal tissue were determined by Pearson correlation analysis, based on with a finding of P < .01 plus Correlation Coefficient > .5 or <−.5; and Gene Ontology (GO) enrichment analysis was performed on the Co‐expressed genes using DAVID (https://david.ncifcrf.gov). 24
2.8. Cell type deconvolution
To determine the fractions of cell types, especially the immunocytes, infiltrating the nasal tissues, RNA sequencing data from nasal tissues of control subjects, ECRSwNP patients and nonECRSwNP patients were subjected to cell type deconvolution analysis using the xCell tool (https://xcell.ucsf.edu/). 25 xCell is a gene signatures‐based method which performs cell type enrichment analysis from gene expression data for 64 immune and stroma cell types. The cell type enrichment score (xCell scores) was calculated based on the gene expression data. The associations between ACE2 expression and xCell scores of each cell type were assessed by Spearman correlation analysis.
2.9. Endotyping of CRSwNP
Based on RNA sequencing data, CRSwNP patients were classified into type 1, type 2 and type 3 endotypes according to the mRNA expression of IFNG, IL5 and IL17A, respectively, in nasal tissues. 26 , 27 Gene expression was normalized to reference gene RPLP0, and the endotypes were defined by using a cut‐off of greater than 90th percentile of the expression in control tissue. 26 The cut‐off values for IFNG/RPLP0, IL5/RPLP0 and IL17A/RPLP0 were 0.00054, 0.00000 and 0.00031, respectively.
2.10. Statistical analysis
Statistical analysis for expression of the genes investigated (ACE2, TMPRSS2, IFNG, IL4, IL5, IL13 and IL17A) and non‐RNA‐sequencing data was performed using GraphPad Prism Version 8.0 (GraphPad Software, La Jolla, Calif). Data are presented as medians and interquartile range (IQR), except for age, which is presented as mean ± SD. Differences between groups were analysed using the Kruskal‐Wallis ANOVA with post hoc Dunn's multiple comparisons test (for multiple group comparisons) and Mann‐Whitney U test (for two group comparisons). For paired data, Wilcoxon matched‐pairs signed rank test was performed. The χ2 or Fisher exact test was used for qualitative data. Associations between variables were evaluated using Spearman and Pearson correlation analysis. Differences were considered significant at P‐value < .05.
3. RESULTS
3.1. Demographic and clinical characteristics of the subjects
Demographic and clinic characteristics of subjects whose samples were used for RNA sequencing are presented in Table S1. Groups were comparable in terms of age, female/male ratio, atopic status and smokers or non‐smokers. Assessment of the nasal polyp tissues from CRSwNP patients for local inflammatory patterns demonstrated that patients with nonECRSwNP had significantly fewer eosinophils (7.08%; P < .01) and more lymphocytes (68.95%; P < .01) than patients with ECRSwNP (41.25% eosinophils and 36.24% lymphocytes).
3.2. Clinical characteristics of CRS patients with COVID‐19
Among the 92 patients with mild and non‐pneumonia COVID‐19 enrolled in the study, 8 patients (8.7%) were found to have underlying CRS condition before SARS‐CoV‐2 infection. The two groups of COVID‐19 patients with and without CRS were comparable in terms of age, sex and disease severity (data not shown). There was also no significant difference in blood eosinophil counts between the two groups (COVID‐19 patients, median: 0.09; IQR: 0.01‐0.21; CRS patients with COVID‐19, median: 0.03; IQR: 0.01‐0.09). The absolute blood eosinophil counts in the 8 CRS patients with COVID‐19 were 0.00, 0.01, 0.01, 0.01, 0.05, 0.08, 0.09 and 0.32 × 109/L, respectively (Table S2). Based on the finding that eosinophilic CRS is associated with elevated blood eosinophils (>0.215 × 109/L), 28 7 of the 8 (87.5%) CRS with COVID‐19 patients were classified as noneosinophilic CRS patients.
3.3. Highest expression of ACE2 in human nasal epithelial cells
Based on RNA sequencing data obtained from cultured nasal and bronchial epithelial cells, the expression of ACE2 was found to be significantly higher in HNECs than HBECs of control subjects and asthma and COPD patients (P < .01, respectively; Figure S1A). However, there was no significant difference in TMPRSS2 expression between HNECs and HBECs from control subjects and asthma and COPD patients (Figure S1B).
3.4. Increased expression of ACE2 in nasal tissues of nonECRSwNP patients
RNA sequencing data of control subjects, ECRSwNP patients and nonECRSwNP patients, were subjected to differential expression analysis. There were 4698 DEGs (including TMPRSS2) between ECRSwNP and controls, 4447 DEGs (including ACE2 and TMPRSS2) between nonECRSwNP and control, and 618 DEGs (including ACE2) between nonECRSwNP and ECRSwNP (Figure 1A). The 618 DEGs between nonECRSwNP and ECRSwNP patients consisted 285 down‐regulated genes and 333 up‐regulated genes including ACE2 (Figure 1B).
Figure 1.
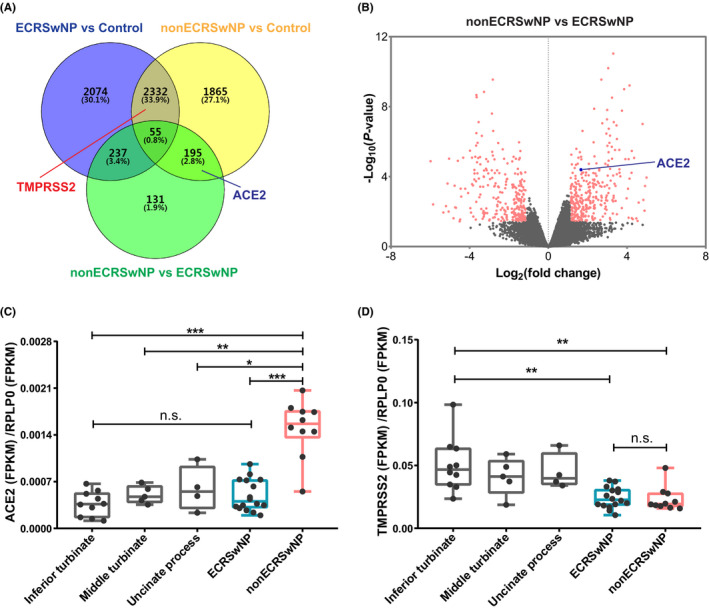
Differentially expressed SARS‐CoV‐2 entry factors in nasal tissues. RNA sequencing was performed on nasal tissues from control subjects (n = 19), ECRSwNP patients (n = 16) and nonECRSwNP patients (n = 10). A, Venn diagrams depicting DEGs of ECRSwNP versus control, nonECRSwNP versus control and nonECRSwNP versus ECRSwNP. ACE2, TMPRSS2 and the numbers of DEGs are marked in the corresponding areas. B, Volcano plots illustrating DEGs of nonECRSwNP versus ECRSwNP. C,D, Expression of ACE2 and TMPRSS2 in nasal tissues from inferior turbinate, middle turbinate and uncinate process of control subjects, and nasal polyp tissues from ECRSwNP patients and nonECRSwNP patients. Gene expression was normalized to RPLP0. * P < .05, ** P < .01, *** P < .001. CRSwNP, chronic rhinosinusitis with nasal polyps; ECRSwNP, eosinophilic CRSwNP; nonECRSwNP, noneosinophilic CRSwNP; ACE2, angiotensin‐converting enzyme 2; TMPRSS2, transmembrane protease serine 2; RPLP0, ribosomal protein lateral stalk subunit P0; DEGs, differentially expressed genes; FPKM, fragments per kilo‐base of exon per million fragments mapped; n.s., no significance
We further analysed the expression of ACE2 and TMPRSS2 in control tissues collected from different sites in the nose (inferior turbinate, middle turbinate and uncinate process) and nasal polyp tissue from CRSwNP patients. CRSwNP was further classified into ECRSwNP and nonECRSwNP based on tissue infiltrating eosinophils. The expression of these genes was normalized to reference gene RPLP0 and ACTB (actin beta). ACE2 expression was significantly up‐regulated in nasal tissues of nonECRSwNP compared to ECRSwNP (P < .001) and the three control groups (P < .001, respectively) (Figures 1C and S2A). In contrast, TMPRSS2 was significantly down‐regulated in both ECRSwNP group (P < .01) and nonECRSwNP group (P < .01) compared to inferior turbinate controls (Figures 1D and S2B). The expression of ACE2 and TMPRSS2 was not significantly different in inferior turbinate, middle turbinate and uncinate process controls.
As peripheral blood eosinophil counts (with a cut‐off value of 0.215 × 109/L in Chinese patients 28 ) can also distinguish ECRSwNP and nonECRSwNP, CRSwNP was further classified into ECRSwNP and nonECRSwNP based on blood eosinophil counts. Similarly, assessment of ACE2 expressed in CRSwNP patients classified according to blood eosinophil count demonstrated that this was significantly higher in nasal tissues of nonECRSwNP than ECRSwNP and control group (Figure S3A). Furthermore, ACE2 expression was significantly positively correlated with blood eosinophil count (r = .510, P < .001; Figure S3B). Further evaluation of the expression of ACE2 in nasal tissues of smoker and non‐smoker CRSwNP patients showed that this was not significantly different between the smokers and non‐smokers (Figure S4).
Cross‐reference with a published RNA sequencing database in the Gene Expression Omnibus database 22 confirmed the up‐regulated expression of ACE2 in nasal tissues of nonECRSwNP compared to ECRSwNP (Figure S5A); however, expression of TMPRSS2 was not found to be significantly different between nasal tissues from control subjects, ECRSwNP patients and nonECRSwNP patients (Figure S5B).
To further investigate the expression of ACE2 and TMPRSS2 at protein level, Western blot assay was performed on nasal tissues of control subjects, ECRSwNP patients and nonECRSwNP patients. Similar to findings by RNA sequencing, the Western blot analysis also showed that ACE2 expression was significantly higher in nonECRSwNP than in ECRSwNP (P < .01) and control (P < .001) groups (Figure 2A and C), while TMPRSS2 expression was significantly lower in ECRSwNP (P < .05) and nonECRSwNP (P < .05) groups than in control group (Figure 2B and D).
Figure 2.
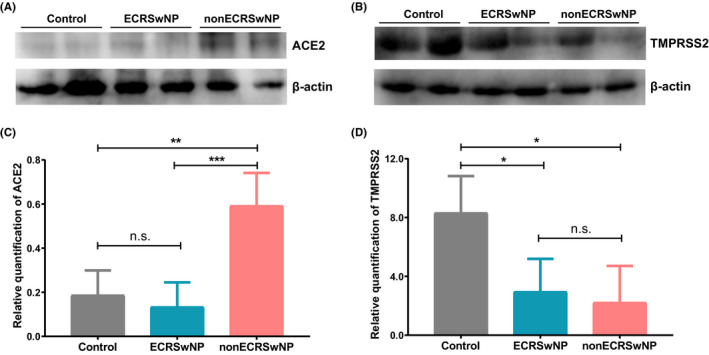
Expression of (A) ACE2 and (B) TMPRSS2 detected by Western blot assay in nasal tissues of control subjects, ECRSwNP patients and nonECRSwNP patients. C,D, The intensity of protein bands was quantified by densitometry and normalized to β‐actin. Data are presented as means ± SDs (n = 4 for each group). *P < .05, **P < .01, ***P < .001, Kruskal‐Wallis ANOVA with post hoc Dunn's multiple comparisons test. CRSwNP, chronic rhinosinusitis with nasal polyps; ECRSwNP, eosinophilic CRSwNP; nonECRSwNP, noneosinophilic CRSwNP; ACE2, angiotensin‐converting enzyme 2; TMPRSS2, transmembrane protease serine 2; n.s., no significance
3.5. ACE2 expression is associated with endotypes of CRSwNP
As ECRSwNP and nonECRSwNP are characterized by different patterns of immune cell infiltration, the association between ACE2 and/or TMPRSS2 expression and tissue inflammatory cells was explored. ACE2 expression was negatively correlated with tissue eosinophils (Spearman r = −.756, P < .001; Pearson r = −0.564, P = .002), and positively correlated with tissue lymphocytes (Spearman r = 0.476, P = .014; Pearson r = 0.610, P < .001) in nasal mucosa of CRSwNP patients (Figure 3A and B). Cell type enrichment analysis, based on xCell scores determined from RNA sequencing data, showed that the infiltrations of immune cells in nasal tissue were different between ECRSwNP and nonECRSwNP patients. In particular, nasal tissue from ECRSwNP patients had significantly more infiltrating eosinophils (P < .01) and dendritic cells (P < .01), than nasal tissue from nonECRSwNP patients (Figure 3C). We further assessed the association between ACE2 expression and xCell scores of each cell type and found that both eosinophils (r = −.711, P < .001) and dendritic cells (r = −.685, P = .003) were negatively correlated with the expression of ACE2 (Figure 3D and E). However, no significant correlation was found between the expression of TMPRSS2 and any of the immune cells.
Figure 3.
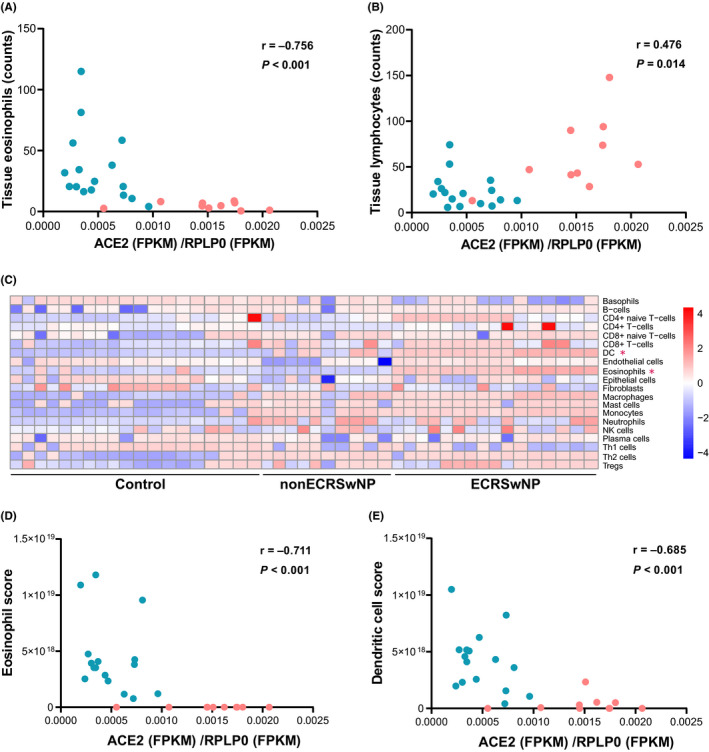
Association between ACE2 expression and differentially infiltrating immune cells in nasal tissue of ECRSwNP and nonECRSwNP patients. A,B, Correlations between ACE2 expression and tissue eosinophils and lymphocytes. ECRSwNP patients are depicted by turquoise dots and nonECRSwNP patients by red dots. C, Cell type deconvolution analysis on gene expression in ECRSwNP and nonECRSwNP patients and control subjects, using xCell. Heatmap depicts the cell type enrichment scores (xCell scores) calculated based on the gene expression data in each sample. *Significantly different between ECRSwNP and nonECRSwNP. D,E, Association between ACE2 expression and xCell scores for eosinophils and dendritic cells, assessed by Spearman correlation analysis. N = 19 for control subjects, n = 16 for ECRSwNP patients and n = 10 for nonECRSwNP patients. CRSwNP, chronic rhinosinusitis with nasal polyps; ECRSwNP, eosinophilic CRSwNP; nonECRSwNP, noneosinophilic CRSwNP; ACE2, angiotensin‐converting enzyme 2; RPLP0, ribosomal protein lateral stalk subunit P0; FPKM, fragments per kilo‐base of exon per million fragments mapped; DC, dendritic cells
Endotypes of CRSwNP are mainly characterized by type 1, type 2 and type 3 inflammatory patterns; with increased levels of (1) IFNG indicating predominantly type 1 inflammation; (2) IL4, IL5, IL13 indicating type 2 inflammation; and (3) IL17 indicating type 3 inflammation. Thus, CRSwNP patients were further classified into type 1, type 2 and type 3 endotypes according to the mRNA expression of IFNG, IL5 and IL17A in nasal tissues, 26 , 27 respectively. Of the 26 CRSwNP patients in this study, 6, 9 and 1 patients were characterized by single type 1, type 2 and type 3 endotype, respectively; 5 patients had a combination of type 1 and type 2 endotype; 1 patient had a combination of type 1 and type 3 endotype; while 4 patients had an untypeable endotype with no increase in the expressions of IFNG, IL5 or IL17A. ACE2 showed significantly increased expression in type 1 endotype of CRSwNP compared to type 2 endotype of CRSwNP (P < .001) and control subjects (P < .001), and comparable expression between type 2 endotype of CRSwNP and control subjects (Figure 4A).
Figure 4.
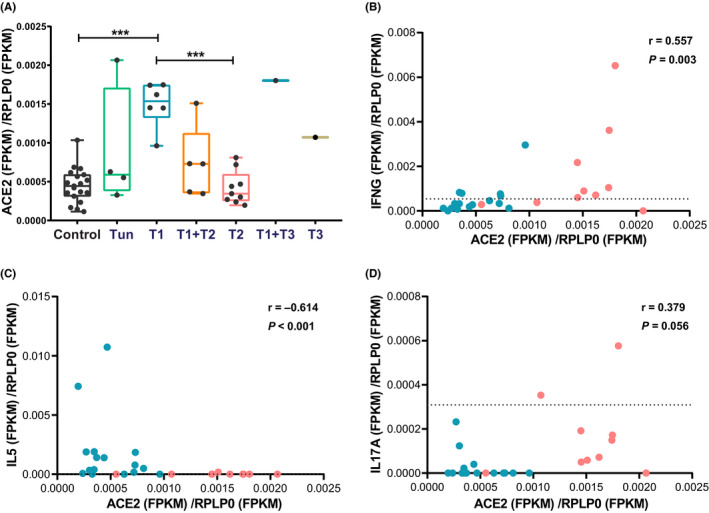
Association between ACE2 expression and different inflammatory endotypes of CRSwNP. A, The ACE2 expression in different inflammatory endotypes of CRSwNP. Endotypes of CRSwNP were defined by the expression of IFNG, IL5 and IL17A, respectively. *** P < .001, Kruskal‐Wallis ANOVA with post hoc Dunn's multiple comparisons test. B‐D, RNA sequencing data obtained for nasal tissues from ECRSwNP (n = 16) and nonECRSwNP patients (n = 16) were subjected to Spearman correlation analysis for associations between ACE2 expression and the expression of IFNG, IL5 and IL17A. ECRSwNP patients are depicted by turquoise dots and nonECRSwNP patients by red dots. The cut‐off values of type 1 (IFNG), type 2 (IL5) and type 3 (IL17A) endotypes are indicated by a dashed line. Correlation coefficient and P‐value are shown in the upper right hand corner of each graph. Gene expression was normalized to RPLP0. T, type; Tun, untypeable; ACE2, angiotensin‐converting enzyme 2; IL, interleukin; IFNG, interferon gamma; RPLP0, ribosomal protein lateral stalk subunit P0; FPKM, fragments per kilo‐base of exon per million fragments mapped
Assessment of the association between expression of ACE2 and/or TMPRSS2 and different inflammatory cytokines indicated that ACE2 expression was positively correlated with the expression of IFNG (Spearman r = .557, P = .003; Pearson r = .514, P = .007); negatively correlated with the expression of IL5 (Spearman r = −.614, P < .001; Pearson r = −.401, P = .043) and IL13 (Spearman r = −0.537, P = .004; Pearson r = −.472, P = .015); and not correlated with IL17A (Figures 4B‐D and S6). TMPRSS2 showed lower expression in type 1, type 2 and their mixed endotypes of CRSwNP, compared to control group (Figure S7). However, TMPRSS2 expression was not correlated with the expression of IFNG, IL5 and IL17A.
3.6. Influence of glucocorticoid treatment on ACE2 expression
Assessment of the effect of glucocorticoid treatment on the expression of ACE2 and TMPRSS2 indicated that the expression of ACE2 was significantly decreased in nasal polyp tissues of nonECRSwNP patients (P < .05), but was not altered in nasal polyp tissues of ECRSwNP patients after 2 weeks’ glucocorticoid treatment (Figure 5A). In contrast, the expression of TMPRSS2 was not significantly altered in nasal tissues of either ECRSwNP patients or nonECRSwNP patients in response to glucocorticoid treatment (Figure S8).
Figure 5.
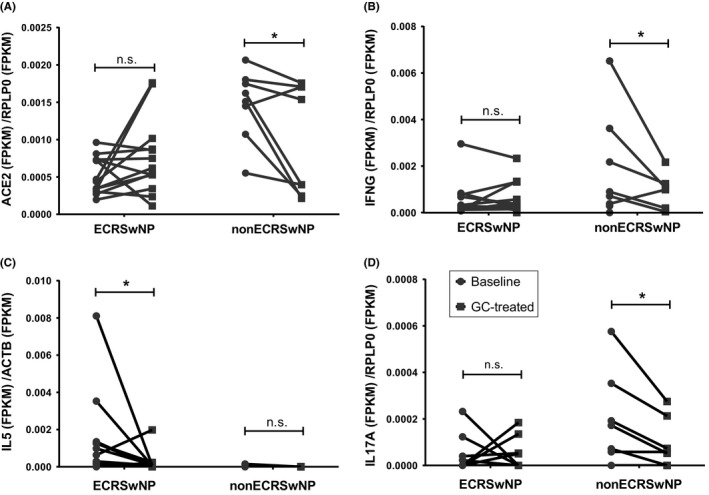
Influence of glucocorticoid treatment on the expression of ACE2 and inflammatory cytokines. CRSwNP patients received a 2‐week course of oral glucocorticoid. Nasal polyp tissues were collected before (Baseline) and after glucocorticoid treatment (GC‐treated). The expression of ACE2 (A), IFNG (B), IL5 (C) and IL17A (D) in the tissue was detected by RNA sequencing. N = 13 for ECRSwNP patients, n = 8 for nonECRSwNP patients. *P < .05. CRSwNP, chronic rhinosinusitis with nasal polyps; ECRSwNP, eosinophilic CRSwNP; nonECRSwNP, noneosinophilic CRSwNP; ACE2, angiotensin‐converting enzyme 2; RPLP0, ribosomal protein lateral stalk subunit P0; IL, interleukin; IFNG, interferon gamma; FPKM, fragments per kilo‐base of exon per million fragments mapped; GC, glucocorticoid; n.s., no significance
The effect of glucocorticoid treatment on type 1, type 2 and type 3 cytokines in nasal tissue was also explored. Similar to ACE2, IFNG and IL17A mRNA expression was significantly decreased by glucocorticoid treatment in nonECRSwNP (P < .05, respectively), but not influenced by glucocorticoid in ECRSwNP (Figure 5B and D). Conversely, IL5 mRNA expression was significantly down‐regulated in ECRSwNP (P < .05), but not influenced in nonECRSwNP (Figure 5C).
3.7. The regulation of ACE2 expression in nasal epithelial cells
We further explored the regulation of ACE2 expression by cytokines and glucocorticoids in cultured primary nasal epithelial cells. ACE2 expression was detected by RNA sequencing or real‐time PCR after incubation for 24 hours with type 1, type 2 and type 3 cytokines, and IFN‐γ ± budesonide or dexamethasone. The expression of ACE2 was significantly up‐regulated by IFN‐γ (10ng/mL) (P < .001), but not altered by any of the other cytokines (IL‐4, IL‐5, IL‐13 and IL‐17) investigated (Figure 6A). In contrast, TMPRSS2 expression was significantly increased by IL‐4 (P < .01) and IL‐13 (P < .001), but not altered by any of the other cytokines investigated (Figure S9). Although budesonide and dexamethasone did not directly influence ACE2 expression, both compounds significantly attenuated IFN‐γ‐induced expression of ACE2 (Figure 6B).
Figure 6.
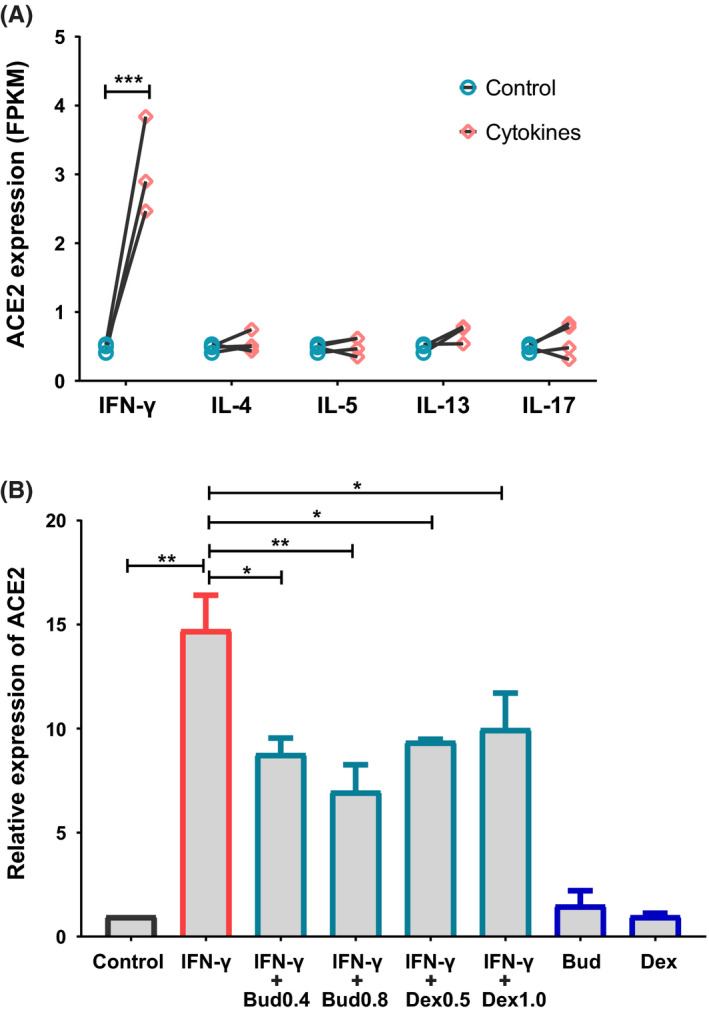
IFN‐γ‐induced ACE2 expression is attenuated by glucocorticoids. A, Cultured primary nasal epithelial cells were incubated with IFN‐γ (10 ng/mL), IL‐4 (50 ng/mL), IL‐5 (50 ng/mL), IL‐13 (50 ng/mL) or IL‐17 (50 ng/mL) for 24 h, and then analysed for expression of ACE2 by RNA sequencing. N = 3 for IFN‐γ and IL‐13 group, n = 4 for IL‐4, IL‐5 and IL‐17 group. B, Cultured primary nasal epithelial cells (n = 3) were incubated with IFN‐γ (10 ng/mL) ± budesonide (0.4 μg/mL, 0.8 μg/mL) or dexamethasone (0.5 μg/mL, 1.0 μg/mL) for 24 h, and then analysed for expression of ACE2 by real‐time PCR. ACE2, angiotensin‐converting enzyme 2; IL, interleukin; FPKM, fragments per kilo‐base of exon per million fragments mapped; Bud, budesonide; Dex, dexamethasone
3.8. Potential function of genes co‐expressed with ACE2 gene
Pearson correlation analysis of RNA sequencing data for nasal tissues from CRSwNP patients and control subjects, to determine the genes co‐expressed with ACE2 gene, showed that 1286 genes were significantly co‐expressed with ACE2. GO pathway enrichment analysis of the ACE2 co‐expressed genes further demonstrated that the ACE2 co‐expressed genes were mostly associated with defence response to virus, negative regulation of viral genome replication, type I interferon signalling pathway, NIK/NF‐kappaB signalling, cilium assembly, intraciliary transport and other biological processes (Figure 7A). In contrast, the significantly enriched pathways by TMPRSS2 co‐expressed genes were mainly associated with cell cycle, RNA biosynthetic process, intracellular transport, protein localization and other similar biological processes, which are likely to be generally irrelevant in eliciting response to virus (Figure S10).
Figure 7.
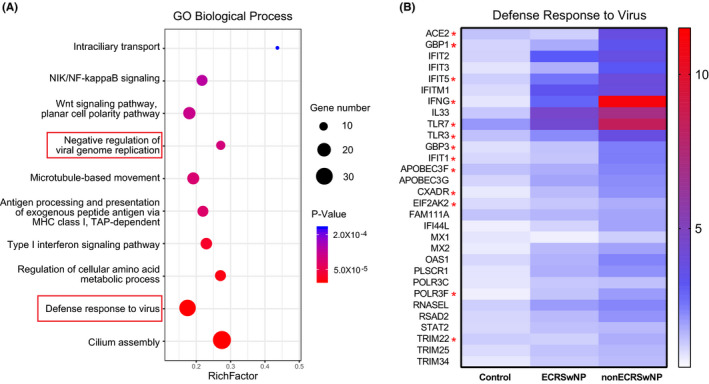
Expression and potential function of genes co‐expressed with ACE2 gene in nasal tissue. Co‐expressed genes of ACE2 were assessed in nasal tissues from control subjects, ECRSwNP patients and nonECRSwNP, and performed GO enrichment analysis. A, Bubble chart depicting top 10 significantly enriched GO biological processes by co‐expressed genes of ACE2. B, Expression pattern of ACE2 co‐expressed genes that involved the GO term ‘defence response to virus’. The colour coding of heat maps represents the level of gene expression normalized to control group, calculated based on FPKM. *Significantly increased expression in nonECRSwNP compared to ECRSwNP. CRSwNP, chronic rhinosinusitis with nasal polyps; ECRSwNP, eosinophilic CRSwNP; nonECRSwNP, noneosinophilic CRSwNP; GO, gene ontology; FPKM, fragments per kilo‐base of exon per million fragments mapped
Figure 7B shows the ACE2 co‐expressed genes, which are involved in the defence response to virus. All of these genes were found to be positively correlated with the expression of ACE2 in nasal tissues and showed increased expression trends in particularly nonECRSwNP patients; with 12 of these including GBP1, GBP3, IFNG, TLR3, TLR7, IFIT1, IFIT5, APOBEC3F, CXADR, EIF2AK2, POLR3F and TRIM22 being significantly increased in nonECRSwNP compared to ECRSwNP.
4. DISCUSSION
COVID‐19 caused by SARS‐CoV‐2 has become a worldwide pandemic and a major public health emergency. The SARS‐CoV‐2 receptor ACE2 plays a crucial role in cellular entry leading to infection. 29 Thus, high expression of ACE2 has been suggested to be a potential risk factor for virus infection and disease severity. 11 , 30 , 31 Our study has demonstrated that expression of ACE2 was significantly increased in nasal tissues of nonECRSwNP patients compared to ECRSwNP patients and control subjects; with no significant difference between ECRSwNP patients and control subjects. The expression of ACE2 was positively correlated with type 1 inflammation indicators and conversely negatively correlated with type 2 inflammation indicators. Furthermore, ACE2 expression was up‐regulated by type 1 cytokine IFN‐γ. To our knowledge, this is the first study to elucidate the associations between ACE2 expression and different inflammatory endotypes of CRSwNP.
High expression of ACE2 has been identified in airway epithelial cells, which might explain the fact that SARS‐CoV‐2 infection is primarily a respiratory illness. 32 Employing RNA sequencing and data analysis, we have demonstrated that ACE2 is highly expressed in epithelial cells of both the lower and upper airways, and that the expression of ACE2 was highest in nasal epithelial cells. This finding is consistent with a recent study which has demonstrated that SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. 10 Considering the primary mode of viral transmission, our findings suggest that the nasal mucosa may be an important most readily available route for SARS‐CoV‐2 infection.
It is important to urgently identify the potential risk factors, which facilitate SARS‐CoV‐2 infection. In particular, as SARS‐CoV‐2 depends on ACE2 for cell entry, 33 various pathological conditions that could increase the expression of ACE2 has been identified as risk factors for COVID‐19. Fang and colleagues 31 have suggested that patients with cardiac diseases, hypertension or diabetes, who are treated with ACE2‐increasing drugs, are at higher risk for severe COVID‐19. Similarly, Leung and colleagues 30 have demonstrated that COPD and active cigarette smoking could up‐regulate ACE2 expression in the lower airways, thus in part explaining the increased risk of severe COVID‐19 in these groups of individuals. Moreover, Higham and colleagues 34 have observed increased ACE2 expression in COPD patients who are overweight compared to those not overweight, which suggests that overweight COPD patients might be at greater risk of developing severe COVID‐19. Our findings demonstrated that patients with nonECRSwNP have increased expression of ACE2 in nasal tissues compared to ECRSwNP patients and control subjects, indicating that this might lead to increased susceptibility to SARS‐CoV‐2 infection in nonECRSwNP patients. Analysis with published gene expression data sets also confirmed the up‐regulated expression of ACE2 in nasal tissues of nonECRSwNP compared to ECRSwNP. 35
Very recently, Wang and colleagues 36 reported that the prevalence of CRS in hospitalized patients with COVID‐19 in cohort from Wuhan, China was 6.1%, which is close to the prevalence in our cohort from Beijing, China (8.7%). In addition, our finding that there was no significant difference in blood eosinophil counts between COVID‐19 patients with and without CRS is in line with Wang and colleagues’ findings, which might suggest a low rate of SARS‐CoV‐2 infection for eosinophilic CRS or a relative high rate of SARS‐CoV‐2 infection for noneosinophilic CRS. The same research group also reported that ACE2 expression level was comparable in the inferior turbinate and middle turbinate from the control subjects, 37 which is consistent with our findings. Additionally, Wang and colleagues reported lower ACE2 expression level in nasal polyp samples of CRSwNP patients than in their inferior turbinate samples. 37 Our study did not inquire the regional differences in ACE2 expression in the sinonasal mucosa of patients with CRSwNP, which could be a limitation. However, our study made a thorough inquiry into the associations between ACE2 expression and different inflammatory endotypes of CRSwNP.
CRSwNP is a heterogeneous disease with several endotypes, which are mainly characterized by type 1, type 2 and type 3 inflammatory patterns. 26 , 38 ECRSwNP has a predominantly type 2 inflammation characterized by pronounced eosinophilia and high levels of IL‐4, IL‐5 and IL‐13, while nonECRSwNP is characterized by type 1 inflammation with high levels of IFN‐γ and/or type 3 inflammation with high levels of IL‐17. 17 In the present study, we did not observe significant changes of ACE2 expression in ECRSwNP patients compared to control subjects. In view of the fact that type 2 inflammation is also the predominant inflammatory endotype of asthma, our findings are in agreement with the previous reports that asthma might be not a significant risk factor for COVID‐19. 4 , 39 Although the most prevalent endotype in CRSwNP is characterized by type 2 inflammation, 40 it is well known that compared to Western CRSwNP patients, East Asian CRSwNP patients have substantially more type 1 and/or type 3 inflammation. 27 , 41 The positive correlation between ACE2 expression and type 1 inflammation suggests that this may possibly influence susceptibility to SARS‐CoV‐2.
Indeed, our study has further demonstrated that ACE2 expression was significantly up‐regulated by IFN‐γ, but not influenced by IL‐4, IL‐5, IL‐13 and IL‐17; lending further support to the notion that type 1 inflammation is likely to positively influence susceptibility to SARS‐CoV‐2. This finding is consistent also with the findings of Ziegler and colleagues, 42 who recently demonstrated that IFN‐α2 and IFN‐γ, but not IL‐4, IL‐13 and IL‐17A, significantly up‐regulated ACE2 expression in human nasal epithelial cell cultures. However, whether type 2 inflammation influences ACE2 expression in nasal epithelial cells remains controversial. In this study, we showed comparable expression of ACE2 between Chinese ECRSwNP patients and control subjects. In addition, Wang and colleagues 43 have shown that both mRNA and protein expression of ACE2 were comparable in nasal tissues between control subjects and patients with allergic rhinitis, a disease driven by type 2 inflammation. Conversely, unpublished data from Krysko and colleagues 44 suggest that expression of ACE2 may be reduced in nasal polyp compared to control tissue, while Kimura and colleagues have reported that IL‐13 reduces ACE2 expression in airway epithelial cells. 45 In this regard, considering race and regional differences, 46 we speculate that the severity of type 2 inflammation might partly contribute to these controversial findings of ACE2 expression in ECRSwNP patients.
NonECRSwNP shows prominent infiltration of inflammatory cells, which are mostly lymphocytes and plasma cells, whereas ECRSwNP shows a large number of eosinophils. 19 , 47 In accordance with these findings, we have also shown that patients with nonECRSwNP had significantly less eosinophilic and more lymphocytic infiltration in nasal tissues than ECRSwNP patients. Moreover, ACE2 expression was positively correlated with the numbers of lymphocytes infiltrating the nasal tissue, and negatively correlated with infiltrating eosinophils in nasal tissue. Baba and colleagues have observed that there are significantly more CD4+ and CD8+T lymphocyte in nasal tissues of nonECRSwNP, 48 which might contribute to the high production of IFN‐γ 49 and subsequently to the increased expression of ACE2.
Intranasal or systemic use of glucocorticoid is the first‐line pharmaceutical treatment for CRSwNP; significantly improving polyp size, nasal symptoms and quality of life. 50 , 51 An important mechanism for glucocorticoid treatment is to block inflammatory mediators and thus regulate the immune response. The present study demonstrated that glucocorticoids had inhibitory effects on both type 1 and type 2 inflammations. However, we did not observe a direct regulation on ACE2 expression by glucocorticoid treatment. The decreased expression of ACE2 in nasal tissues of nonECRSwNP in response to glucocorticoid treatment might be due to the inhibition of type 1 inflammatory mediators such as IFN‐γ. Recently, experts from the European Academy of Allergy and Clinical Immunology (EAACI) suggested that intranasal corticosteroids remain the standard treatment for CRS in patients with SARS‐CoV‐2 infection. 52 Our findings also suggest that use of glucocorticoid therapy is not a risk factor for SARS‐CoV‐2 infection in CRSwNP patients and should be continued as prescribed.
Co‐expressed genes are more likely to be co‐regulated and share similar functions. 53 In view of the multiple biological functions of ACE2; including vasodilatory effects, regulation of renin‐angiotensin system and receptor for coronavirus, 54 the present study further investigated and identified the genes co‐expressed with ACE2 gene in nasal tissues, in order to predict its function. We found that the genes co‐expressed with ACE2 gene in nasal tissue were mostly associated with the biological process of defence response to virus, suggesting that the main biological function of ACE2 in nasal tissues may be associated with enhancing virus infection. We further observed generally increased expression patterns of ACE2 co‐expressed genes related to defence response to virus in nonECRSwNP compared to ECRSwNP, with 12 of these being significantly increased. Interestingly, 5 (GBP1, TLR3, TLR7, IFIT1 and TRIM22) of these 12 genes have been shown to be IFN‐γ stimulated genes, 55 , 56 , 57 further indicating a relationship between type 1 inflammation and response to virus. Our findings complement the findings by Hwang and colleagues, 58 who showed reduced expression of IFN‐β, IFN‐λ1 and IFN‐λ2 in nasal tissues of CRSwNP and down‐regulation of antiviral factors by type 2 cytokines in cultured epithelial cells. These authors further demonstrated that although TLR3 and TLR7 mRNA expression was not significantly different between nasal tissues of controls and CRSwNP patients, there was a trend for increased expression of both TLR3 and TLR7 mRNA expression in nonECRSwNP compared to control and ECRSwNP. Thus, it is possible that the inflammatory pattern in nasal tissue of CRSwNP might be associated with different potential of antiviral response. 56
In addition to using ACE2 as a cell entry receptor, SARS‐CoV‐2 employs human proteases as entry activators. In this regard, TMPRSS2 is a host cell serine protease that primes the spike protein of SARS‐CoV‐2, which is essential for viral spread and pathogenesis in the infected host. 9 , 59 Compared with ACE2, TMPRSS2 has been shown to be highly expressed with a broader distribution in multiple tissues and cell types. 10 In accordance with the findings of Kimura and colleagues, 45 we found that expression of TMPRSS2 was also increased by IL‐4 and IL‐13 stimulation in cultured nasal epithelial cells. However, our study has demonstrated that the expression of TMPRSS2 in nasal tissues of ECRSwNP patients was decreased compared to control subjects, which might be due to the presence of various factors which suppress TMPRSS2 expression; for example tumour necrosis factor (TNF) and hypoxia. 60 Additionally, SARS‐CoV‐2 can use other proteases such as CTSB and CTSL for priming of the SARS‐CoV‐2 and cell infection. 9 , 61 However, as the present study demonstrated that expression of CTSB and CTSL was comparable between control subjects and CRSwNP patients (Figure S11), it is presently not possible to deduce whether differential expression of TMPRSS2 in CRSwNP is a protective or risk factor of SARS‐CoV‐2 infection.
The present study is somewhat limited. Firstly, there is lack of clinical information regarding the patients with COVID‐19 and CRSwNP, which may directly link ACE2 expression to SARS‐CoV‐2 infection. Secondly, the sample size used for RNA sequencing is relatively small. Thirdly, the correlation coefficient between ACE2 expression and inflammatory cytokines is relatively low. One important reason for this low correlation might be the low expression of cytokines under the detection limits by RNA sequencing in many samples.
In summary, the present study has demonstrated that the expression of SARS‐CoV‐2 receptor ACE2 is significantly increased in nasal tissues of nonECRSwNP patients compared to ECRSwNP patients and control subjects. Furthermore, ACE2 expression is positively correlated with the expression of type 1 inflammatory cytokines, particularly IFN‐γ, and negatively correlated with type 2 inflammatory indicators. The increased expression of ACE2 is most likely regulated by IFN‐γ.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
MW, CW and LZ designed the study. MW, XT, GL and YH performed the experiments. G.F collected the clinical data of COVID‐19 patients. MW analysed the data and wrote the manuscript. CA, CW and LZ critically revised the manuscript. All authors reviewed the manuscript.
Supporting information
Supplementary Material
Wang M, Bu X, Fang G, et al. Distinct expression of SARS‐CoV‐2 receptor ACE2 correlates with endotypes of chronic rhinosinusitis with nasal polyps. Allergy.2021;76:789–803. 10.1111/all.14665
Funding informationThis work was supported by grants from the national key R&D programme of China (2018YFC0116800, 2016YFC0905200), the programme for the Changjiang scholars and innovative research team (IRT13082), the national natural science foundation of China (81800882, 81630023, 81870698), Beijing municipal administration of hospitals’ mission plan (SML20150203), Beijing municipal administration of hospitals’ Dengfeng plan (DFL20190202), Beijing municipal administration of hospitals clinical medicine development of special funding support (XMLX201816).
Contributor Information
Chengshuo Wang, Email: wangcs830@126.com.
Luo Zhang, Email: dr.luozhang@139.com.
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from patients with pneumonia in China, 2019. N Eng J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of Coronavirus disease 2019 in China. N Eng J Med. 2020;382(18):1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang W, Cao Q, Qin L, et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID‐19): A multi‐center study in Wenzhou city, Zhejiang, China. J Infect. 2020;80(4):388‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li X, Xu S, Yu M, et al. Risk factors for severity and mortality in adult COVID‐19 inpatients in Wuhan. J Allergy Clin Immunol. 2020;146(1):110‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marhl M, Grubelnik V, Magdic M, Markovic R. Diabetes and metabolic syndrome as risk factors for COVID‐19. Diabetes Metab Syndr. 2020;14(4):671‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang B, Li R, Lu Z, Huang Y. Does comorbidity increase the risk of patients with COVID‐19: evidence from meta‐analysis. Aging 2020;12(7):6049‐6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy 2020;75(7):1730‐1741. [DOI] [PubMed] [Google Scholar]
- 8. Dong X, Cao YY, Lu XX, et al. Eleven faces of coronavirus disease 2019. Allergy 2020;75(7):1699‐1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181(2):271‐280 e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sungnak W, Huang N, Becavin C, et al. SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single‐cell RNA‐seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019‐nCoV infection. Front Med. 2020;14(2):185‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang W, Xu Y, Gao R, et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA 2020;323(18):1843‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gengler I, Wang JC, Speth MA, Sedaghat AR. Sinonasal pathophysiology of SARS‐CoV‐2 and COVID‐19: A systematic review of the current evidence. Laryngoscope Investig Otolaryngo. 2020;5(3):354‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fokkens WJ, Lund VJ, Hopkins C, et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2020. Rhinology. 2020;58(Suppl S29):1‐464. [DOI] [PubMed] [Google Scholar]
- 15. Stevens WW, Peters AT, Hirsch AG, et al. Clinical characteristics of patients with chronic rhinosinusitis with nasal polyps, asthma, and aspirin‐exacerbated respiratory disease. J Allergy Clin Immunol Pract. 2017;5(4):1061‐1070 e1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cao PP, Li HB, Wang BF, et al. Distinct immunopathologic characteristics of various types of chronic rhinosinusitis in adult Chinese. J Allergy Clin Immunol. 2009;124(3):478‐484, 484 e471–472. [DOI] [PubMed] [Google Scholar]
- 17. Zhang N, Van Zele T, Perez‐Novo C, et al. Different types of T‐effector cells orchestrate mucosal inflammation in chronic sinus disease. J Allergy Clin Immunol. 2008;122(5):961‐968. [DOI] [PubMed] [Google Scholar]
- 18. Tian S, Hu N, Lou J, et al. Characteristics of COVID‐19 infection in Beijing. J Infect. 2020;80(4):401‐406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lou H, Zhang N, Bachert C, Zhang L. Highlights of eosinophilic chronic rhinosinusitis with nasal polyps in definition, prognosis, and advancement. Int Forum Allergy Rhinol. 2018;8(11):1218‐1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiao J, Wang M, Duan S, et al. Transforming growth factor‐beta1 decreases epithelial tight junction integrity in chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2018;141(3):1160‐1163 e1169. [DOI] [PubMed] [Google Scholar]
- 21. Wang M, Tan G, Eljaszewicz A, et al. Laundry detergents and detergent residue after rinsing directly disrupt tight junction barrier integrity in human bronchial epithelial cells. J Allergy Clin Immunol. 2019;143(5):1892‐1903. [DOI] [PubMed] [Google Scholar]
- 22. Wang W, Gao Z, Wang H, et al. Transcriptome analysis reveals distinct gene expression profiles in eosinophilic and noneosinophilic chronic rhinosinusitis with nasal polyps. Sci Rep. 2016;6:26604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakayama T, Okada N, Yoshikawa M, et al. Assessment of suitable reference genes for RT‐qPCR studies in chronic rhinosinusitis. Sci Rep. 2018;8(1):1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44‐57. [DOI] [PubMed] [Google Scholar]
- 25. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stevens WW, Peters AT, Tan BK, et al. Associations between inflammatory endotypes and clinical presentations in chronic rhinosinusitis. J Allergy Clin Immunol Pract. 2019;7(8):2812‐2820 e2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang X, Zhang N, Bo M, et al. Diversity of TH cytokine profiles in patients with chronic rhinosinusitis: A multicenter study in Europe, Asia, and Oceania. J Allergy Clin Immunol. 2016;138(5):1344‐1353. [DOI] [PubMed] [Google Scholar]
- 28. Hu Y, Cao PP, Liang GT, Cui YH, Liu Z. Diagnostic significance of blood eosinophil count in eosinophilic chronic rhinosinusitis with nasal polyps in Chinese adults. Laryngoscope. 2012;122(3):498‐503. [DOI] [PubMed] [Google Scholar]
- 29. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leung JM, Yang CX, Tam A, et al. ACE‐2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID‐19. Eur Respir J. 2020;55(5):2000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID‐19 infection? Lancet Respir Med. 2020;8(4):e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang H, Rostami MR, Leopold PL, et al. Expression of the SARS‐CoV‐2 ACE2 receptor in the human airway epithelium. Am J Respir Crit Care Med. 2020;202(2):219‐229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Higham A, Singh D. Increased ACE2 expression in the bronchial epithelium of COPD patients who are overweight. Obesity. 2020;28(9):1586‐1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saheb Sharif‐Askari F, Saheb Sharif‐Askari N, Goel S, et al. Are patients with chronic rhinosinusitis with nasal polyps at a decreased risk of COVID‐19 infection? Int Forum Allergy Rhinol. 2020;10(10):1182‐1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang H, Song J, Pan L, et al. The characterization of chronic rhinosinusitis in hospitalized patients with COVID‐19. J Allergy Clin Immunol Pract. 2020;8(10):3597‐3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang H, Song J, Pan L, et al. Regional differences in ACE2 expression in the sinonasal mucosa of adult Chinese patients with chronic rhinosinusitis. Allergy 2020. 10.1111/all.14623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tomassen P, Vandeplas G, Van Zele T, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016;137(5):1449‐1456 e1444. [DOI] [PubMed] [Google Scholar]
- 39. Svenningsen S, Nair P. Asthma endotypes and an overview of targeted therapy for asthma. Front Med. 2017;4:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Avdeeva K, Fokkens W. Precision medicine in chronic rhinosinusitis with nasal polyps. Curr Allergy Asthma Rep. 2018;18(4):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang ET, Zheng Y, Liu PF, Guo LJ. Eosinophilic chronic rhinosinusitis in East Asians. World J Clin Cases. 2014;2(12):873‐882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS‐CoV‐2 receptor ACE2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020;181(5):1016‐1035 e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang H, Song J, Yao Y, et al. Angiotensin‐converting enzyme II expression and its implication in the association between COVID‐19 and allergic rhinitis. Allergy 2020. 10.1111/all.14569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jian L, Yi W, Zhang N, et al. Perspective: COVID‐19, implications of nasal diseases and consequences for their management. J Allergy Clin Immunol. 2020;146(1):67‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kimura H, Francisco D, Conway M, et al. Type 2 inflammation modulates ACE2 and TMPRSS2 in airway epithelial cells. J Allergy Clin Immunol. 2020;146(1):80‐88.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim DK, Kim DW. Does inflammatory endotype change in patients with chronic rhinosinusitis? Allergy Asthma Immunol Res. 2019;11(2):153‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim JW, Hong SL, Kim YK, Lee CH, Min YG, Rhee CS. Histological and immunological features of non‐eosinophilic nasal polyps. Otolaryngol Head Neck Surg. 2007;137(6):925‐930. [DOI] [PubMed] [Google Scholar]
- 48. Baba S, Kagoya R, Kondo K, Suzukawa M, Ohta K, Yamasoba T. T‐cell phenotypes in chronic rhinosinusitis with nasal polyps in Japanese patients. Allergy Asthma Clin Immunol. 2015;11:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cao PP, Wang ZC, Schleimer RP, Liu Z. Pathophysiologic mechanisms of chronic rhinosinusitis and their roles in emerging disease endotypes. Ann Allergy Asthma immunol. 2019;122(1):33‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hopkins C. Chronic rhinosinusitis with nasal polyps. N Eng J Med. 2019;381(1):55‐63. [DOI] [PubMed] [Google Scholar]
- 51. Wang C, Lou H, Wang X, et al. Effect of budesonide transnasal nebulization in patients with eosinophilic chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2015;135(4):922‐929 e926. [DOI] [PubMed] [Google Scholar]
- 52. Klimek L, Jutel M, Bousquet J, et al. Management of patients with chronic rhinosinusitis during the COVID‐19 pandemic ‐ An EAACI Position Paper. Allergy 2021;76:677‐688. 10.1111/all.14629 [DOI] [PubMed] [Google Scholar]
- 53. Stuart JM, Segal E, Koller D, Kim SK. A gene‐coexpression network for global discovery of conserved genetic modules. Science 2003;302(5643):249‐255. [DOI] [PubMed] [Google Scholar]
- 54. Gargaglioni LH, Marques DA. Let's talk about sex in the context of COVID‐19. J Appl Physiol. 2020;128(6):1533‐1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kajita AI, Morizane S, Takiguchi T, Yamamoto T, Yamada M, Iwatsuki K. Interferon‐gamma enhances TLR3 expression and anti‐viral activity in keratinocytes. J Investig Dermatol. 2015;135(8):2005‐2011. [DOI] [PubMed] [Google Scholar]
- 56. Chen Y, Kumar RK, Thomas PS, Herbert C. Th1/17‐Biased inflammatory environment associated with COPD alters the response of airway epithelial cells to viral and bacterial stimuli. Mediators Inflamm. 2019;2019:7281462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pan W, Zuo X, Feng T, Shi X, Dai J. Guanylate‐binding protein 1 participates in cellular antiviral response to dengue virus. Virol J. 2012;9:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hwang JW, Lee KJ, Choi IH, Han HM, Kim TH, Lee SH. Decreased expression of type I (IFN‐beta) and type III (IFN‐lambda) interferons and interferon‐stimulated genes in patients with chronic rhinosinusitis with and without nasal polyps. J Allergy Clin Immunol. 2019;144(6):1551‐1565 e1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zang R, Gomez Castro MF, McCune BT, et al. TMPRSS2 and TMPRSS4 promote SARS‐CoV‐2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5(47):eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gkogkou E, Barnasas G, Vougas K, Trougakos IP. Expression profiling meta‐analysis of ACE2 and TMPRSS2, the putative anti‐inflammatory receptor and priming protease of SARS‐CoV‐2 in human cells, and identification of putative modulators. Redox Biol. 2020;36:101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019‐nCoV contains a furin‐like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


