Abstract
The new coronavirus (2019‐nCoV) or the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) was officially declared by the World Health Organization (WHO) as a pandemic in March 2020. To date, there are no specific antiviral drugs proven to be effective in treating SARS‐CoV‐2, requiring joint efforts from different research fronts to discover the best route of treatment. The first decisions in drug discovery are based on 2D cell culture using high‐throughput screening. In this context, spheroids and organoids emerge as a reliable alternative. Both are scaffold‐free 3D engineered constructs that recapitulate key cellular and molecular events of tissue physiology. Different studies have already shown their advantages as a model for different infectious diseases, including SARS‐CoV‐2 and for drug screening. The use of these 3D engineered tissues as an in vitro model can fill the gap between 2D cell culture and in vivo preclinical assays (animal models) as they could recapitulate the entire viral life cycle. The main objective of this review is to understand spheroid and organoid biology, highlighting their advantages and disadvantages, and how these scaffold‐free engineered tissues can contribute to a better comprehension of viral infection by SARS‐CoV‐2 and to the development of in vitro high‐throughput models for drug screening.
Keywords: 3D in vitro models, COVID‐19, drug screening, high‐throughput, infection model, organoids, SARS‐CoV‐2, scaffold‐free tissue engineering, spheroids
1. INTRODUCTION
For centuries, infectious diseases have been a major challenge to the existence of humanity. 1 Together, these diseases represent the annual mortality of more than 10 million people worldwide. 2 Even with all the progress in the prevention and control of infectious diseases, pathogens still represent a major threat to human health. 3 Every year, the world is somehow affected by a new disease and new infectious agents being inserted into our environment, representing a continuous challenge, especially those caused by viruses, such as the H7N9 influenza virus in 2013, 4 Zika virus outbreak in 2016, 5 and Chikungunya outbreak in 2019. 6
Currently, we are experiencing a pandemic caused by the SARS‐CoV‐2 virus, a new type of coronavirus, which was reported earlier this year in Wuhan, Hubei Province, China. The first case occurred on December 12, 2019, and since then, the numbers of notifications of contagion and deaths attributed to the disease, called COVID‐19, have been growing and spreading across the world rapidly. 7 Epidemiological investigations have suggested that the outbreak was associated with a seafood market in Wuhan. 8 With the exponential growth of cases, as of February 14, 2020, the WHO declared it a state of a public health emergency, and then, moved on to a pandemic state. COVID‐19 had already spread all over the world, killing more than 700,000 people from different countries. To achieve all these outbreaks, it is mandatory to understand the mechanisms of infection by viral agents, leading to the production of vaccines and antivirals that are increasingly effective and specific to respond to these diseases.
Pharmaceutical industry in vitro methods for drug screening, metabolism, and toxicity are based on bidimensional (2D) cell culture of animal lineages. However, 2D cell cultures and cell lineages from animal sources are not able to mimic the tissue microenvironment found in vivo, mainly due to the limitations of cell‐cell and cell‐extracellular matrix interactions. In this context, three‐dimensional (3D) cell culture models emerge as a relevant support tool, as they can provide reproducible results and adaptation of experiments to high throughput performance. Humanized 3D cell culture may fill the gap between 2D cell cultures and animal models, contributing to an understanding of cell and molecular mechanisms closer to human tissue physiology. Tissue engineering is essentially a 3D cell culture approach, and its recent advances have contributed to the development of complex and highly organized 3D cell culture models mimicking tissue microenvironments at the protein and gene levels.
The majority of tissue engineering approaches are based on the use of scaffolds as a substitute for extracellular matrices. However, scaffold‐free approaches are increasingly being recognized as humanized 3D cell culture models capable of recapitulating cell‐cell and cell‐extracellular matrix interactions mimicking cell differentiation and physiology. The main objective of this review is to understand spheroid and organoid biology, highlighting their advantages and disadvantages, and how these scaffold‐free engineered tissues can contribute to a better comprehension of viral infection by SARS‐CoV‐2 and to in vitro high‐throughput models of drug screening.
2. SARS‐COV‐2
SARS‐CoV‐2 belongs to the Coronaviridae family, order Nidovirales. They have the largest genome of all RNA viruses, usually ranging from 27 to 32 kb. The single‐stranded positive‐sense RNA is packaged inside a helical capsid formed by the nucleocapsid protein (N) surrounded by an envelope. Associated with the viral envelope are at least three structural proteins: the membrane protein (M) and the envelope protein (E) that are involved in the assembly of the virus, while the glycoprotein spike (S) is involved in the entry of the virus into cell hosts. These structural proteins form spikes on the surface of the virus, giving the appearance of having crowns, hence, its name. In addition to mediating the entry of the virus, protein S is determinant in reaching the viral host, tissue tropism and an important inducer of the host immune responses. 9 The protein spike (S) is divided into two domains: S1 and S2. The first is responsible for the interaction with the ACE2 receptor (angiotensin‐converting enzyme 2), initiating the infection of the host cell by SARS‐CoV‐2.
Since coronavirus was discovered as the causative agent of COVID‐19, scientists have been attempting to better understand the genetic makeup of the virus and to discover how to effectively treat the infection. Until now, no anti‐SARS‐CoV‐2 drug or vaccine has been officially approved due to the absence of adequate evidence, and medical experts can only treat the symptoms of the disease. The individual, when infected, must remain isolated to prevent the disease from spreading for 14 days. 8 One strategy that has been applied is convalescent plasma transfusion, which may be beneficial in the treatment of critically ill patients with severe infection. 10 Many drugs, such as ivermectin, 11 hydroxychloroquine, 12 azithromycin, 12 and dexamethasone 13 are being speculated to treat the infection.
Drug screening based on in silico analysis has been used to predict the pathogenic mechanisms and some drug targets of SARS‐CoV‐2. The structure determination by cryo‐electron microscopy (cryo‐EM) and X‐ray crystallography of the viral proteins is essential for this kind of approach, where drug therapies can be useful affecting the interaction with the host cells or blocking virus replication. After in silico analysis, the potential drugs can be tested using 3D cell culture models analyzing the virus replication and drug candidates to inhibit the SARS‐CoV‐2 infection. 14
Another recent approach to combat SARS‐CoV‐2 is the therapeutic use of extracellular vesicles derived from adult stem cells, due to their effect on the modulation of anti‐inflammatory and antiapoptotic pathways. The extracellular vesicles were already tested in acute respiratory distress syndrome and promise a novel treatment for the patients with COVID‐19. 15 Furthermore, the efficacy of extracellular vesicles could be tested using 3D cell culture models.
The long‐term strategy to combat COVID‐19 is the development of a vaccine. Currently, approximately 250 candidate vaccines against SARS‐CoV‐2 are in development worldwide, 16 including mRNA vaccines, replicating or nonreplicating viral vectored vaccines, DNA vaccines, autologous dendritic cell‐based vaccines, and inactive virus vaccines. 17 To date, at least 27 of these vaccine candidates are under evaluation in clinical trials. Two replicating or nonreplicating viral vectored vaccines are being tested in phase 3, namely, ChAdOx1 nCoV‐19 (Oxford University, UK), 18 Ad5‐vectored COVID‐19 (CanSino Biologics, China), 16 and one mRNA vaccine called mRNA‐1273 (Moderna, USA). 19 These vaccines have been described as capable of producing neutralizing antibodies and humoral and cellular responses.
The race for rapid detection of the disease has caused many laboratories and companies work on rapid, sensitive, and low‐cost tests, in addition to vaccines and treatments. The rapid tests can detect antigen or antibodies. Rapid antigen tests detect the presence of viral proteins and return positive results during infection. Antigen tests provide results in less than 30 minutes, do not need to be processed in a laboratory and are inexpensive to produce, but this speed comes at a cost in sensitivity. If a person has a low amount of virus in their body, the test can give a false negative result. 20 The antibody test detects the body's immune system response to the virus but is not effective in the early stages of infection. Some companies that produce this type of test are: Panbio Abbott test with sensitivity of 93.3% and specificity of 99.4%, this test can detect antigens and antibodies in swab and blood samples; One Step COVID‐19 test from Celer with sensitivity of 86.43% and specificity of 99.57%, this test uses only a blood sample and gives the result in 15 minutes, for example.
However, to combat COVID‐19 in a specific way, a greater understanding of the molecular mechanisms of infection is necessary, emphasizing the importance of the 3D cell culture models developed by the tissue engineering field. Based on these studies, more effective treatment and diagnoses can be produced.
3. UNDERSTANDING HUMAN VIRUS INFECTION FROM IN VITRO MODELS
To understand human virus infections, different methodologies are used to study the entry of viruses into animal and human cells. For example, scanning electron microscopy was recently used by Caldas and collaborators 21 to identify for the first time the exact moment when SARS‐CoV‐2 infects a cell and was used in the past to study the morphological structure of SARS‐CoV. 22 Other microscopy analyses, such as confocal and epifluorescence analyses, can also be used to track the cytoskeletal movements of viruses during cell infection. This fluorescence technique was already used for HIV, dengue, and hepatitis C virus. 23
In some cases, viral nucleic acids or proteins are abundant and can be easily detected by in situ hybridization or immunohistochemistry. Currently, the most common analysis performed to identify SARS‐CoV‐2 infection and diagnosis 24 is polymerase chain reaction (PCR), 25 which amplifies nucleic acids to detect the virus at a molecular level. In addition, high‐throughput proteomic analysis was also performed to identify the set of proteins modulated in Chikungunya infection. Among the 1047 proteins expressed in infective cells, 209 proteins were related to transcription, translation, apoptosis and stress response of the Chikungunya virus. 26 Functional analysis performed by supernatant harvesting as ELISA, multiplex and proteomics might be useful to quantify inflammatory mediators directly related to human virus infections. This approach is particularly interesting for infections caused by SARS‐CoV‐2, as it has already been shown that the virus is capable of establishing an inflammatory condition known as a “cytokine storm”. 27
Proteome analysis has been applied to better understand the changes in the total protein repertoire suffered by the host cell during viral infection. Using two‐dimensional difference in gel electrophoresis (2‐D DIGE), eight significant changes in host proteins have already been identified (MDCK and A549 cells) in H1N1 infection. 28 A proteomic approach was also used to detect virion‐associated viral and cellular proteins during HCV infection, and because of that, new host factors, including a nuclear pore complex (NPC) protein that participates in HCV infection, were characterized. 29
The measurement of the release of infectious particles per cell is important because they are related to applications ranging from the manufacture of vaccines to serum neutralization tests for clinical effectiveness. The most commonly accepted methods include endpoint dilution (TCID50) and plate assays. 30 , 31 However, depending on the type of virus, the result can take more than two weeks, in addition to requiring intensive work of handling cell culture. The interpretation of the results of these tests is subjective and highly variable, requiring many replicates to obtain a reliable statistical result.
In conclusion, different analyses to identify human virus infections in animal and human cells are available, and many of them are helpful for clinical diagnosis. Furthermore, the use of high‐throughput methods is considered promising for drug screening tests in in vitro models of virus infection.
4. SCAFFOLD‐FREE 3D ENGINEERED TISSUES AS INFECTION MODELS
2D cell culture is commonly used in research laboratories and pharmaceutical industries. It is considered cost‐effective and easy, and most of the analysis protocols are already well established and validated. The overwhelming majority of our knowledge of how viruses cause infection is based upon studies with 2D culture using animal cell lines. However, 2D cell cultures do not resemble the tissue microenvironment found in vivo (Table 1). Engineered scaffold‐free approaches as 3D cell cultures, mostly represented by spheroids and organoids, have gained attention in the last few years due the capacity to closely mimic cell and tissue physiology. In 3D cell cultures, cells are architecturally organized in a compact structure, where a dynamic equilibrium is reached over time in culture of gene expression and protein production, mimicking the performance of tissues in vivo. Besides, the extracellular matrix found in spheroids are produced by their own cells only, so there is no interference of a hydrogel or scaffold or the plastic of culture flasks. 35 , 40
TABLE 1.
Main differences between 2D and 3D cell culture models
| 2D cell culture | 3D cell culture | References | |
|---|---|---|---|
| Cell‐to‐cell contact | + | +++ | 32 |
| Extracellular matrix production | + | +++ | 33 |
| Higher cell density in vitro | ++ | +++ | 34 |
| Production of pro angiogenic factors | + | ++ | 35 |
| Capacity of large‐scale tissue production | + | ++ | 36 |
| Use for high‐throughput systems and drug screening tests | ++ | +++ | 37 |
| Mimicry in vivo tissue microenvironments | + | +++ | 38 |
| Mimicry of embryogenesis and organogenesis processes | + | +++ | 39 |
Since the 2000s, the number of scientific studies using spheroids and organoids to understand the host cell's response to viral infections has been increasing (Figure 1). 23 , 41 , 42 The use of these scaffold‐free 3D models can provide information about host‐pathogen interactions, which are necessary for the formulation of more effective antivirals and vaccines. Recently, Takayama reviewed cell culture models in the literature that can faithfully reproduce the viral life cycle and pathology of SARS‐Cov‐2. The work presents relevant published studies with cell lines, organoids and animal models that were performed this year to better understand COVID‐19 infection. 1 , 43
FIGURE 1.
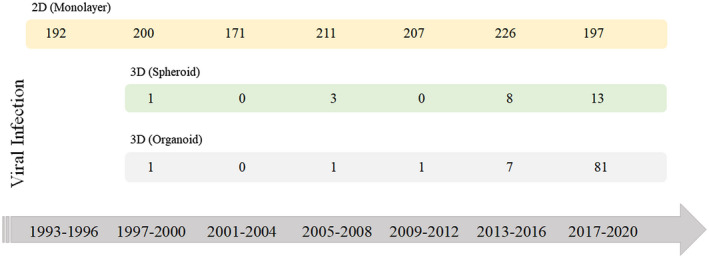
Timeline of numbers of articles published related to the use of 2D and 3D cell culture techniques as viral infection models. The search was performed on the PubMed database, by entering the combination of the following words: “virus infection” or “viral infection,” “spheroid,” and “virus infection” or “viral infection” and “organoid.” Review articles were not included. The search was conducted on November 6, 2020 [Color figure can be viewed at wileyonlinelibrary.com]
4.1. 3D scaffold‐free engineered spheroids
Spheroids can be formed from differentiated and undifferentiated cells, including adult and multipotent stem cells. 35 Cells are architecturally organized in a compact structure, where a dynamic equilibrium of gene expression and protein production is reached over culture, mimicking the physiology of tissues in vivo. 44 More importantly, the extracellular matrix found in spheroids is produced by their own cells once there is no interference of a hydrogel or scaffold. Cells inside spheroids are responsive to external stimuli, including differentiation events at the molecular level. 45 , 46
In the past few years, many studies have been performed using spheroids for tissue engineering approaches. To engineer these 3D models, spheroids were produced from different cell sources, such as human bone marrow mesenchymal stromal/stem cells (MSCs) and human adipose‐derived stem cells (ASCs), to engineer bone 46 , 47 , 48 , 49 , 50 and cartilage 44 , 50 , 51 microtissues. In addition, human chondrocytes were also successfully used to engineer cartilage in vitro. 52 , 53 Spheroids were also produced from induced pluripotent stem cells (iPSCs), mostly to engineer the liver, 54 neural 55 and cardiovascular tissues. 56 However, compared with the number of studies performed with MSCs or ASCs, studies with iPSCs to produce spheroids remain limited. Human umbilical vein endothelial cells (HUVECs) were also already used to produce spheroids for endothelial tissue engineering approaches, especially in coculture with MSCs. 57 , 58 In addition, a high number of studies with cancer lineages were also published to produce reproducible tumor spheroid models for drug screening tests in vitro. 59 , 60
Spheroids can be produced by different 3D cell culture techniques. 35 , 40 Different studies have already been published showing high‐throughput production of spheroids from different cell types by using the hanging‐drop technique. 61 , 62 , 63 , 64 , 65 Other techniques already describe high‐throughput spheroid production consisting of using nonadhesive agarose microwell systems, 66 cell culture plates with a higher number of wells, as described by Grinner et al, 67 which used a 1536‐well plate format to engineer tumor spheroids for drug screening tests. Other studies were already performed using well plates for high‐throughput applications, as performed by Liao and collaborators 68 ; however, these authors used a 96‐well plate to cast agarose microwells. Alginate hydrogels produced from casted PDMS molds were also used to produce a large number of MSC spheroids with homogeneous size and shape. 69 Another technique consists of using magnetic nanoparticles to assemble spheroids, as performed by Kim and colleagues. 70 The authors used a magnetic pin‐array system to concentrate magnetic nanoparticle‐incorporated cells and form spheroids. The main advantage is the possibility of assembling 96 spheroids at once with homogeneous size and shape.
4.2. 3D scaffold‐free engineered organoids
Prior to 2005, the term organoid was referred to as small tissue fragments taken from organs. Most studies were performed with epithelial tissues, separated from stroma by mechanical and/or enzymatic digestion and cultured in different types of 3D gels to produce organ‐like structures in vitro. 71 Currently, an organoid is defined as “a 3D structure grown from stem cells and consisting of organ‐specific cell types that self‐organizes through cell sorting and spatially restricted lineage commitment”. 72 Organoids must be formed from embryonic stem cells (ESCs), iPSCs, and adult stem cells that spatially organize into a restricted lineage functional structure. 72 , 73 , 74 Organoids must develop functions of that specific organ to be considered organoids, and for that reason, they are commonly used to recapitulate morphogenesis in vitro.
During the past few years, different organoid models have been successfully developed in vitro, such as the brain, 75 , 76 intestine, 77 , 78 , 79 lungs, 80 pancreas, 81 stomach, 82 and liver. 83 Recent successful models of the intestine 84 , 85 derived from adult stem cells and brain derived from iPSC 86 organoids showed that the constructs were able to mimic morphological and functional properties of both native tissues. To allow organoid formation in vitro, several growth factors are needed to control the self‐renewal, self‐organization and differentiation of stem cells. 74 The use of Matrigel, a gelatinous protein mixture secreted by mouse sarcoma cells, is required, providing key biomechanical cues to organoid formation. 74 , 87 However, their inherent lot‐to‐lot variability and tumor‐derived nature impair the use of organoids as humanized platforms for drug screening. 88
4.3. Spheroids and organoids for viral infection
Spheroids and organoids have already been explored as a model of viral cycle infection by human viruses (Figure 1). 23 , 41 , 42 A scalable model of spheroids derived from human hepatocytes to study hepatitis C virus (HCV) infection and replication was developed by Ananthanarayanan and collaborators. 41 The main results showed that spheroids presented liver‐specific functions and a higher level of HCV infection compared with 2D cell culture. Hepatocyte spheroids infected with HCV were produced embedded in Matrigel and showed functional liver‐like structures in addition to releasing infectious virions from HCV. 89 The life cycle of blood‐borne HCV was also investigated on a 3D nonadherent gel showing high levels of the enzyme thromboxane A2 synthase (TXAS). This enzyme is required for the production of infectious HCV. The result was corroborated in a preclinical assay. Using a TXAS inhibitor (prostaglandin I2 receptor agonist), in an animal model, the levels of HCV infection were reduced. 90
To explore the viral tropism of SARS‐CoV‐2, Yang and collaborators have used a human pluripotent stem cells (hPSCs) platform to generate multiple different cells and organoids. To determine the permissiveness of hPSC‐derived cells, different MOI of SARS‐CoV‐2 was used for infection and the results showed the liver organoids, cardiomyocytes, pancreatic alpha and beta cells and dopaminergic neurons are permissive to SARS‐CoV‐2 infections. 91 Lamers and collaborators 92 infected gut enterocyte organoids with SARS‐CoV and SARS‐CoV‐2, revealing upregulation of typical genes related to SARS‐CoV‐2 infection. Another recent study published by Monteil and collaborators 93 investigated the impact of human recombinant soluble angiotensin‐converting enzyme 2 (hrsACE2) on SARS‐CoV‐2 growth. In summary, the authors used blood vessels and kidney organoids as a 3D model of SARS‐CoV‐2 infection, and hrsACE2 had a positive effect in blocking the infection in both organoid models. However, although hrsACE2 could block the early stages of SARS‐Cov‐2 infection, new studies with lung organoids must be performed to better evaluate its effects in vitro. Recently, Susuki and colleagues 94 cultivated normal human bronchial epithelial cells in Matrigel drops as a 3D scaffold‐free model for SARS‐CoV‐2 infection. The 3D culture system was responsive to infection, showing fibrose areas. The authors named the strategy lung organoids; however, the culture was not derived from stem cells.
All over the world, there is a concern with diseases transmitted by viruses transmitted by arthropods, such as the Dengue (DENV), Chikungunya (CHIKV) and Zika (ZIKV) viruses, but there is still no treatment for this type of infection. Garcez and collaborators 95 investigated the effects of Zika virus (ZIKV) infection in human neural stem cells growing as neurospheres and brain organoids. The results showed that the virus was able to infect the cells and, in the organoids, reduced the rate of growth after 11 days in culture. The use of a 3D model was able to show more reliable evidence of ZIKV behavior in vivo, contributing to a better comprehension of microcephaly cases in newborn children during ZIKV epidemics in Brazil.
5. SCAFFOLD‐FREE 3D ENGINEERED TISSUE PLATFORM FOR DRUG SCREENING
High‐throughput systems have the potential to contribute to reducing the number of preclinical studies because they can test a larger set of compounds faster. An efficient scaffold‐free 3D engineered tissue platform for drug screening must show scalability and have well‐established end‐points. More importantly, the platform must be amenable to the high‐throughput system needed for the fast screening of large numbers of drugs. The data generated by high‐throughput systems need to be interpreted by adequately designed software, making bioinformatics an essential tool. An important technological bottleneck in the adaptation of these systems is the size and shape of the 3D scaffold‐free tissues, because the readout is commonly based on fluorescence emission.
5.1. High‐throughput systems for spheroids and organoids
Currently, the protocols used for spheroid production are more amenable to high‐throughput drug screening compared with organoids (Figure 2). Ivanov and Grabowska 96 presented a device that allowed the formation and arrangement of 66 spheroids in a unique platform for high‐throughput biomarker analysis of spheroids. Recently, Li and collaborators 97 developed a micro‐scaffold array showing advantages related to the reduction of cell number, assay time, culture media, and drug consumption that can be applied for different cell types, tissue engineering studies, and new drug discoveries.
FIGURE 2.
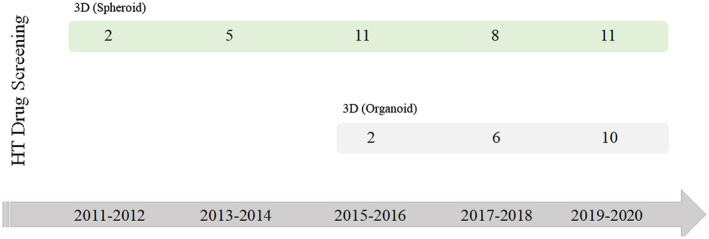
Timeline of numbers of articles published related to the established protocols of spheroids and organoids for high‐throughput drug screening. The search was performed on the PubMed database, by entering the combination of the following words: “drug screening,” “high‐throughput” and “organoid,” and “drug screening,” “high‐ throughput” and “spheroid.” Review articles were not included. The search was conducted on November 6, 2020. HT: High‐throughput [Color figure can be viewed at wileyonlinelibrary.com]
Cytotoxicity test results showed that the higher drug resistance of the 3D spheroids from cancer cells was independent of cell density. In this line, the spheroid model for high‐throughput hepatotoxic studies discussed that drug resistance can be explained by the higher amount of extracellular matrix production, proving its functionality and sensitivity to drug response. 98 Hepatocyte spheroids are sensitive to inducers and inhibitors of liver metabolizing enzymes and are maintained for up to 20 days of culture. 99 An innovative method suitable for high‐throughput drug screening was developed by Knowlton and collaborators. 100 The method relies on the bioprinting of encapsulated hepatic spheroids into a microfluidic device, showing the maintenance of spheroid morphology after bioprinting.
Interestingly, metabolism can be measured in hepatocyte spheroids using a microsensor platform integrated into a 96‐well plate. 101 The main results showed alterations in lactate production under exposure to different drug concentrations. The perspective is that the platform can be used for complex organ‐on‐chip studies. Another method was developed by Abe‐Fukasawa and collaborators 102 and consists of the use of a low‐molecular‐weight agar, which allows cells to grow as dispersed spheroids suitable for drug assessment. The main advantage of the method is the possibility to create numbers of spheroid clones and image them with quality through the period of culture. Lim and Park 103 developed a microfluidic device to produce spheroids of cancer lineage and tested the responsiveness of the spheroids to anticancer drugs. The spheroids were formed even with a few cells, showing remarkable changes in their morphology in the presence of these drugs.
Recent studies have begun to test drugs in 3D culture models infected with SARS‐CoV‐2 (Figure 3). Katsura and collaborators reported an alveolosphere culture system for the propagation and differentiation of human alveolar type 2 cells derived from primary lung tissue. The pneumocytes expressed the SARS‐CoV‐2 receptor ACE2 and the infection with the virus was confirmed by transcriptome and histological analysis. The authors showed that the treatment with low doses of interferons (IFNs) promoted a reduction in viral replication. 104 Han and collaborators produced human pluripotent stem cells derived colonic organoids as a model of SARS‐CoV‐2 infection. The authors performed a high throughput screen of FDA‐approved drugs and identified entry inhibitors of SARS‐CoV‐2 in the colonic organoids. 105
FIGURE 3.
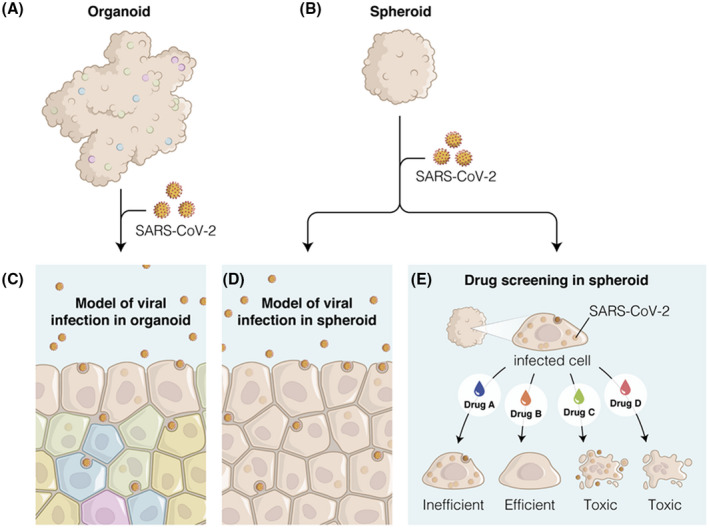
Spheroid and organoid can be used as a model of SARS‐Cov‐2 infection (A, B) to understand the mechanism of virus entry into cells (C, D). (E) An efficient drug candidate should eliminate the virus while maintaining cell viability inside spheroids. Toxic drug candidates show no antiviral effect or even toxic effects in both cells and virus [Color figure can be viewed at wileyonlinelibrary.com]
The number of studies dedicated to high‐throughput systems using organoids has increased in the last decade; however, the study of these systems are still low compared with spheroids (Figure 2). The majority of studies use organoids derived from tumor biopsies as a model for cancer, 106 , 107 , 108 , 109 and recent studies are investigating the development of organ‐on‐chip devices with organoids for drug screening. 110 , 111 The tumor organoids usually have a diameter of 200 micrometers, making them appropriate for high‐throughput systems. The other types of organoids, including the brain and intestine, can reach up to 1 millimeter in diameter, in addition to having various shapes, because they can recapitulate tissue morphogenesis, resulting in tissue microenvironments similar to those in vivo.
Driehuis and collaborators 106 used a biobank of 30 pancreatic tumor organoids from patients. Phan and collaborators 109 developed a high‐throughput approach to establish and perform drug screening of tumor organoids. A total of 240 tumor inhibitors were used, and organoids were sensitive to changes in size and morphology. Recently, a method to produce liver organoids by using Matrigel suspension instead of drops was developed by Garnier et al. 112 The use of Matrigel suspension allows automation and high‐throughput applications, in addition to showing better stability in the expression of typical liver genes, a usual challenge to be addressed in the development of liver models. This study may represent an important step toward the change of organoid protocols for high‐throughput systems. Qian and colleagues also developed a scalable brain organoid platform to model ZIKV exposure. 113
The complexity of organoids also results in a lack of reproducibility and different shapes and sizes, and the same batch impairing image assays are usually used in high‐throughput systems for drug screening. 114 As an in vitro viral infection model, the organoid becomes appealing for understanding the impact of infection on cell differentiation and in subpopulations of a given tissue. For example, SARS‐CoV‐2 seems to infect a transient population of bronchial secretory cells. 115
6. CONCLUSION AND PERSPECTIVES
The pandemic caused by SARS‐CoV‐2 and other human viruses, such as Zika, Dengue and Chikungunya, present worldwide could be better understood through the development of reproducible 3D tissue models. The use of human cells in complex 3D scaffold‐free culture systems that reproduce tissue microenvironments and cellular interactions is the future for a better understanding of the mechanisms of viral infection, the discovery of therapeutic targets and high‐throughput platforms for drug screening. In this context, high‐throughput platforms have already been developed using spheroids and some types of organoids, especially tumor organoids. Organoids represent an interesting humanized 3D model for a deeper understanding of human virus infection due to their complexity of cell subpopulations. The use of CRISPR, transduction and other technologies of gene editing can expand the application of these 3D scaffold‐free models for drug screening and discovery. In the near future high‐throughput platforms using spheroids or organoids should add microphysiological systems (organ‐on‐a‐chip) to reach a more accurate cell physiological response, under dynamic conditions and real‐time monitoring.
CONFLICT OF INTEREST
LS B is the unpaid scientific advisor in GCell 3D cell culture. DF R is a fellowship of GCell 3D cell culture through the call 06/2019 ‐ FAPERJ. All other authors have no conflicts of interest.
ACKNOWLEDGMENTS
We would like to thank the Coordination for the Improvement of Higher Education Personnel (CAPES), No. 88881.362199/2019‐01; the Carlos Chagas Filho Foundation for Research Support of the State of Rio de Janeiro (FAPERJ), No. E26/202.682/2018, No. E26/260.172/2019; National Council for Scientific and Technological Development (CNPq), No.307460/2019‐3.
Kronemberger GS, Carneiro FA, Rezende DF, Baptista LS. Spheroids and organoids as humanized 3D scaffold‐free engineered tissues for SARS‐CoV‐2 viral infection and drug screening. Artif Organs. 2021;45:547–557. 10.1111/aor.13880
REFERENCES
- 1. Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re‐emerging infectious diseases. Nature. 2004;430:242–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization . Mortality and global health estimates 2016. Available from: https://www.who.int/data/gho/data/themes/mortality‐and‐global‐health‐estimates/GHO/mortality‐and‐global‐health‐estimates [Google Scholar]
- 3. Heesterbeek H, Anderson RM, Andreasen V, Bansal S, De Angelis D, Dye C, et al. Modeling infectious disease dynamics in the complex landscape of global health. Science. 2015;347:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lam TT, Wang J, Shen Y, Zhou B, Duan L, Cheung CL, et al. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature. 2013;502:241–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lessler J, Chaisson LH, Kucirka LM, Bi Q, Grantz K, Salje H, et al. Assessing the global threat from Zika virus. Science. 2016;353:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. World Health Organization . Outbreak update – Chikungunya in Sudan, 21 December 2019. Available from: http://www.emro.who.int/pandemic‐epidemic‐diseases/chikungunya/outbreak‐update‐chikungunya‐in‐sudan‐21‐december‐2019.html [Google Scholar]
- 7. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2019;579:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. World Health Organization . WHO Novel Coronavirus (2019‐nCoV) situation reports 2020. Available from: https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019/situation‐reports/ [Google Scholar]
- 9. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;2:281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, et al. Treatment of 5 critically Ill patients with COVID‐19 with convalescent plasma. JAMA. 2020;27:1582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA‐approved drug ivermectin inhibits the replication of SARS‐CoV‐2 in vitro. Antiviral Res. 2020;178:104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fantini J, Chahinian H, Yahi N. Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS‐CoV‐2: What molecular dynamics studies of virus‐host interactions reveal. Int J Antimicrob Agents. 2020;56:106020. [DOI] [PubMed] [Google Scholar]
- 13. Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582:469. [DOI] [PubMed] [Google Scholar]
- 14. Raimondi MT, Donnaloja F, Barzaghini B, Bocconi A, Conci C, Parodi V, et al. Bioengineering tools to speed up the discovery and preclinical testing of vaccines for SARS‐CoV‐2 and therapeutic agents for COVID‐19. Theranostics. 2020;10:7034–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Öztürk S, Elçin AE, Koca A, Elçin YM. Therapeutic applications of stem cells and extracellular vesicles in emergency care: futuristic perspectives. Stem Cell Rev Rep. 2020;1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu FC, Guan XH, Li YH, et al. Immunogenicity and safety of a recombinant adenovirus type‐5‐vectored COVID‐19 vaccine in healthy adults aged 18 years or older: a randomised, double‐blind, placebo‐controlled, phase 2 trial. Lancet. 2020;396:479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amanat F, Krammer F. SARS‐CoV‐2 vaccines: status report. Immunity. 2020;52:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij‐Rammerstorfer S, et al. Safety and immunogenicity of the ChAdOx1 nCoV‐19 vaccine against SARS‐CoV‐2: a preliminary report of a phase 1/2, single‐blind, randomised controlled trial. Lancet. 2020;396:467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA vaccine against SARS‐CoV‐2 ‐ preliminary report. N Engl J Med. 2020;383:1920–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gluglielmi G. Fast coronavirus tests: what they can and can’t do. Nature. 2020;585:496–8. [DOI] [PubMed] [Google Scholar]
- 21. Caldas LA, Carneiro FA, Higa LM, Monteiro FL, Silva GP, Costa LJ, et al. Ultrastructural analysis of SARS‐CoV‐2 interactions with the host cell via high resolution scanning electron microscopy. bioRxiv 2020;10:144642. Available from: https://www.biorxiv.org/content/10.1101/2020.06.10.144642v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goldsmith CS, Tatti KM, Ksiazek TG. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10:320–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang I‐H, Burckhardt C, Yakimovich A, Greber U. Imaging, tracking and computational analyses of virus entry and egress with the cytoskeleton. Viruses. 2018;4:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J Med Virol. 2020;92:1518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nuovo GJ. The utility of in situ‐based methodologies including in situ polymerase chain reaction for the diagnosis and study of viral infections. Human Pathol. 2007;8:1123–36. [DOI] [PubMed] [Google Scholar]
- 26. Abraham R, Mudaliar P, Jaleel A, Srikanth J, Sreekumar E. High throughput proteomic analysis and a comparative review identify the nuclear chaperone, Nucleophosmin among the common set of proteins modulated in Chikungunya virus infection. J Proteomics. 2015;120:126–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coperchini F, Chiovatoa L, Crocea L, Magri F, Rotondi M. The cytokine storm in COVID‐19: An overview of the involvement of the chemokine/chemokine‐receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vester D, Rapp E, Gade D, Genzel Y, Reichl U. Quantitative analysis of cellular proteome alterations in human influenza A virus‐infected mammalian cell lines. Proteomics. 2009;12:3316–27. [DOI] [PubMed] [Google Scholar]
- 29. Lussignol M, Kopp M, Molloy K, Vizcay‐Barrena G, Fleck RA, Dorner M, et al. Proteomics of HCV virions reveals an essential role for the nucleoporin Nup98 in virus morphogenesis. Proc Natl Acad Sci U S A. 2016;113:2484–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baer A, Kehn‐Hall K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. J Vis Exp. 2014;93:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hygiene. 1938;27:493–7. [Google Scholar]
- 32. Sánchez‐Romero N, Schophuizen CM, Giménez I, Masereeuw R. In vitro systems to study nephropharmacology: 2D versus 3D models. Eur J Pharmacol. 2016;790:36–45. [DOI] [PubMed] [Google Scholar]
- 33. Kim SJ, Kim EM, Yamamoto M, Park H, Shin H. Engineering multi‐cellular spheroids for tissue engineering and regenerative medicine. Adv Healthc Mater. 2020;9:e2000608. [DOI] [PubMed] [Google Scholar]
- 34. Ovsianikov A, Khademhosseini A, Mironov V. The synergy of scaffold‐based and scaffold‐free tissue engineering strategies. Trends Biotechnol. 2018;36:348–57. [DOI] [PubMed] [Google Scholar]
- 35. Baptista LS, Kronemberger GS, Côrtes I, Charelli LE, Matsui RAM, Palhares TN, et al. Adult stem cells spheroids to optimize cell colonization in scaffolds for cartilage and bone tissue engineering. Int J Mol Sci. 2018;19:1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laschke MW, Menger MD. Life is 3D: boosting spheroid function for tissue engineering. Trends Biotechnol. 2017;35:133–44. [DOI] [PubMed] [Google Scholar]
- 37. Fitzgerald KA, Malhotra M, Curtin CM, O'Brien FJ, O'Driscoll CM. Life in 3D is never flat: 3D models to optimise drug delivery. J Control Release. 2015;215:39–54. [DOI] [PubMed] [Google Scholar]
- 38. Baker BM, Chen CS. Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125(Pt 13):3015–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grapin‐Botton A. Three‐dimensional pancreas organogenesis models. Diabetes Obes Metab. 2016;18(Suppl 1):33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Achilli TM, Meyer J, Morgan JR. Advances in the formation, use and understanding of multi‐cellular spheroids. Expert Opin Biol Ther. 2012;12:1347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ananthanarayanan A, Nugraha B, Triyatni M, Hart S, Sankuratri S, Yu H. Scalable spheroid model of human hepatocytes for hepatitis C infection and replication. Mol Pharm. 2014;7:2106–14. [DOI] [PubMed] [Google Scholar]
- 42. Israelsson S, Jonsson N, Gullberg M, Lindberg AM. Cytolytic replication of echoviruses in colon cancer cell lines. Virol J. 2011;8:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takayama K. In Vitro and Animal Models for SARS‐CoV‐2 research. Trends Pharmacol Sci. 2020;41:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wrzesinski K, Rogowska‐Wrzesinska A, Kanlaya R, Borkowski K, Schwämmle V, Dai J, et al. The cultural divide: exponential growth in classical 2D and metabolic equilibrium in 3D environments. PLoS ONE. 2014;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Côrtes I, Matsui RAM, Azevedo MS, Beatrici A, Souza KLA, Launay G, et al. A scaffold‐ and serum‐free method to mimic human stable cartilage validated by secretome. Tissue Eng Part A. 2019. 10.1089/ten.TEA.2018.0311 [DOI] [PubMed] [Google Scholar]
- 46. Kronemberger GS, Dalmônico GML, Rossi AL, Leite PEC, Saraiva AM, Beatrici A. Scaffold‐ and serum‐free hypertrophic cartilage tissue engineering as an alternative approach for bone repair. Artif Organs. 2020;44:E288–99. [DOI] [PubMed] [Google Scholar]
- 47. Suenaga H, Furukawa K, Suzuki Y, Takato T, Ushida T. Bone regeneration in calvarial defects in a rat model by implantation of human bone marrow‐derived mesenchymal stromal cell spheroids. J Mat Sci Mater Med. 2015;26:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murphy KC, Fang S, Leach J. Human mesenchymal stem cell spheroids in fibrin hydrogels exhibit improved cell survival and potential for bone healing. Cell Tissue Res. 2014;357:91–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wessely A, Waltera A, Reichert TE, Stöckl S, Grässel S, Bauer RJ. Induction of ALP and MMP9 activity facilitates invasive behavior in heterogeneous human BMSC and HNSCC 3D spheroids. FASEB J. 2019;33:11884–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sankar S, Sharma CS, Rath SN. Enhanced osteodifferentiation of MSC spheroids on patterned electrospun fiber mats ‐ an advanced 3D double strategy for bone tissue regeneration. Mater Sci Eng C Mater Biol Appl. 2019;94:703–12. [DOI] [PubMed] [Google Scholar]
- 51. Yoon HH, Bhang SH, Shin JY, Shin J, Kim BS. Enhanced cartilage formation via three‐dimensional cell engineering of human adipose‐derived stem cells. Tissue Eng Part A. 2012;18:1949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stuart MP, Matsui RAM, Santos MFS, Côrtes I, Azevedo MS, Silva KR. Successful low‐cost scaffold‐free cartilage tissue engineering using human cartilage progenitor cell spheroids formed by micromolded nonadhesive hydrogel. Stem Cells Int. 2017;2017:7053465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jeon JH, Yun BG, Lim MJ, Kim SJ, Lim MH, Lim JY, et al. Rapid cartilage regeneration of spheroids composed of human nasal septum‐derived chondrocyte in rat osteochondral defect model. Tissue Eng Regen Med. 2020;17:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goulart E, de Caires‐Junior LC, Telles‐Silva KA, Araujo BHS, Rocco SA, Sforca M, et al. 3D bioprinting of liver spheroids derived from human induced pluripotent stem cells sustain liver function and viability in vitro. Biofabrication. 2019;12:015010. [DOI] [PubMed] [Google Scholar]
- 55. Song L, Tsai AC, Yuan X, Bejoy J, Sart S, Ma T, et al. Neural differentiation of spheroids derived from human induced pluripotent stem cells‐mesenchymal stem cells coculture. Tissue Eng Part A. 2018;24:915–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yan Y, Bejoy J, Xia J, Griffin K, Guan J, Li Y. Cell population balance of cardiovascular spheroids derived from human induced pluripotent stem cells. Sci Rep. 2019;9:1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Heo D, Hospodiuk M, Ozbolat I. Synergistic interplay between human MSCs and HUVECs in 3D spheroids laden in collagen/fibrin hydrogels for bone tissue engineering. Acta Biomaterialia. 2019;95:348–56. [DOI] [PubMed] [Google Scholar]
- 58. Inglis S, Kanczler JM, Oreffo ROC. 3D human bone marrow stromal and endothelial cell spheres promote bone healing in an osteogenic niche. FASEB J. 2019;33:3279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen B, Wu Y, Ao Z, Cai H, Nunez A, Liu Y, et al. High‐throughput acoustofluidic fabrication of tumor spheroids. Lab Chip. 2019;19:1755–63. [DOI] [PubMed] [Google Scholar]
- 60. Ganpule A, Gui Z, Almuteri MA, D'Souza GGM. In vitro testing of anticancer nanotherapeutics using tumor spheroids. Methods Mol Biol. 2019;2000:387–93. [DOI] [PubMed] [Google Scholar]
- 61. Hurrell T, Ellero AA, Masso ZF, Cromarty AD. Characterization and reproducibility of HepG2 hanging drop spheroids toxicology in vitro. Toxicol in Vitro. 2018;50:86–94. [DOI] [PubMed] [Google Scholar]
- 62. Kim H, Cho CH, Park J‐K. High‐throughput culture and embedment of spheroid array using droplet contact‐based spheroid transfer. Biomicrofluidics. 2018;4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Langer K, Joensson HN. Rapid production and recovery of cell spheroids by automated droplet microfluidics. SLAS Technol. 2020;2:111–22. [DOI] [PubMed] [Google Scholar]
- 64. Neto AI, Correia CR, Oliveira MB, Rial‐Hermida MI, Alvarez‐Lorenzo C, Reis RL, et al. A novel hanging spherical drop system for the generation of cellular spheroids and high throughput combinatorial drug screening. Biomater Sci. 2015;4:581–5. [DOI] [PubMed] [Google Scholar]
- 65. Widder M, Lemke K, Kekeç B, Förster T, Grodrian A, Gastrock G. A modified 384‐well‐device for versatile use in 3D cancer cell (co‐)cultivation and screening for investigations of tumor biology in vitro. Eng Life Sci. 2018;2:132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. De Moor L, Merovci I, Baetens S, Verstraeten J, Kowalska P, Krysko DV. High‐throughput fabrication of vascularized spheroids for bioprinting. Biofabrication. 2018;10:035009. [DOI] [PubMed] [Google Scholar]
- 67. Griner LM, Gampa K, Do T, Nguyen H, Farley D, Hogan CJ, et al. Generation of high‐throughput three‐dimensional tumor spheroids for drug screening. J Vis Exp. 2018;(139):57476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liao W, Wang J, Xu J, You F, Pan M, Xu X, et al. High‐throughput three‐dimensional spheroid tumor model using a novel stamp‐like tool. J Tissue Eng. 2019;10:2041731419889184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vorwald CE, Ho SS, Whitehead J, Leach JK. High‐throughput formation of mesenchymal stem cell spheroids and entrapment in alginate hydrogels. Methods Mol Biol. 2018;1758:139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim JA, Choi JH, Kim M, Rhee WJ, Son B, Jung HK, et al. High‐throughput generation of spheroids using magnetic nanoparticles for three‐dimensional cell culture. Biomaterials 2013;34:8555–63. [DOI] [PubMed] [Google Scholar]
- 71. Simian M, Bissell MJ. Organoids: a historical perspective of thinking in three dimensions. J Cell Biol. 2017;216:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Clevers H. Modeling development and disease with organoids. Cell. 2016;7:1586–97. [DOI] [PubMed] [Google Scholar]
- 73. Lou Y‐R, Leung AW. Next generation organoids for biomedical research and applications. Biotechnol Adv. 2018;1:132–49. [DOI] [PubMed] [Google Scholar]
- 74. Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering stem cell organoids. Cell Stem Cell. 2016;18:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Benito‐Kwiecinski S, Lancaster MA. Brain Organoids: Human neurodevelopment in a dish. Cold Spring Harb Perspect Biol. 2019;5:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol. 2018;5:432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kasendra M, Tovaglieri A, Sontheimer‐Phelps A, Jalili‐Firoozinezhad S, Bein A, Chalkiadaki A, et al. Development of a primary human small intestine‐on‐a‐Chip using biopsy‐derived organoids. Sci Rep. 2018;8:2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sachs N, Tsukamoto Y, Kujala P, Peters PJ, Clevers H. Intestinal epithelial organoids fuse to form self‐organizing tubes in floating collagen gels. Development. 2017;6:1107–12. [DOI] [PubMed] [Google Scholar]
- 79. Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long‐term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–72. [DOI] [PubMed] [Google Scholar]
- 80. Miller AJ, Dye BR, Ferrer‐Torres D, Hill DR, Overeem AW, Shea LD, et al. Generation of lung organoids from human pluripotent stem cells in vitro. Nat Protoc. 2019;14:518–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hohwieler M, Illing A, Hermann PC, Mayer T, Stockmann M, Perkhofer L, et al. Human pluripotent stem cell‐derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 2017;3:473–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bartfeld S, Clevers H. Organoids as model for infectious diseases: culture of human and murine stomach organoids and microinjection of helicobacter pylori. J Vis Exp. 2015;105:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vyas D, Baptista PM, Brovold M, Moran E, Gaston B, Booth C, et al. Self‐assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology. 2018;2:750–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fujii M, Matano M, Toshimitsu K, Takano A, Mikami Y, Nishikori S, et al. Human intestinal organoids maintain self‐renewal capacity and cellular diversity in niche‐inspired culture condition. Cell Stem Cell. 2018;23:787–93.e6. [DOI] [PubMed] [Google Scholar]
- 86. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, et al. Feeder‐free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–4. [DOI] [PubMed] [Google Scholar]
- 88. Cruz‐Acuña R, García AJ. Engineered materials to model human intestinal development and cancer using organoids. Exp Cell Res. 2019;377:109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Molina‐Jimenez F, Benedicto I, Thi VLD, Gondar V, Lavillette D, Marin JJ, et al. Matrigel‐embedded 3D culture of Huh‐7 cells as a hepatocyte‐like polarized system to study hepatitis C virus cycle. Virology. 2012;1:31–9. [DOI] [PubMed] [Google Scholar]
- 90. Abe Y, Aly HH, Hiraga N, Imamura M, Wakita T, Shimotohno K, et al. Thromboxane A2 Synthase inhibitors prevent production of infectious hepatitis C virus in mice with humanized livers. Gastroenterology. 2013;3:658–67. [DOI] [PubMed] [Google Scholar]
- 91. Yang L, Han Y, Nilsson‐Payant BE, Gupta V, Wang P, Duan X, et al. A human pluripotent stem cell‐based platform to study SARS‐CoV‐2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–36.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS‐CoV‐2 productively infects human gut enterocytes. Science. 2020;369:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell. 2020;181:905–13.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Suzuki T, Itoh Y, Sakai Y, Saito A, Okuzaki D, Motooka D, et al. Generation of human bronchial organoids for SARS‐CoV‐2 research. bioRxiv 2020;5(25):115600. Available from: https://www.biorxiv.org/content/10.1101/2020.05.25.115600v2 [Google Scholar]
- 95. Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–8. [DOI] [PubMed] [Google Scholar]
- 96. Ivanov DP, Grabowska AM. Spheroid arrays for high‐throughput single‐cell analysis of spatial patterns and biomarker expression in 3D. Sci Rep. 2017;1:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Li X, Zhang X, Zhao S, Wang J, Liu G, Du Y. Micro‐scaffold array chip for upgrading cell‐based high‐throughput drug testing to 3D using benchtop equipment. Lab Chip. 2014;3:471–81. [DOI] [PubMed] [Google Scholar]
- 98. Ramaiahgari SC, den Braver MW, Herpers B, Terpstra V, et al. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver‐like properties for repeated dose high‐throughput toxicity studies. Arch Toxicol. 2014;5:1083–95. [DOI] [PubMed] [Google Scholar]
- 99. Wei J, Lu J, Liu Y, Yan S, Li X. Spheroid culture of primary hepatocytes with short fibers as a predictable in vitro model for drug screening. J Mater Chem B. 2016;44:7155–67. [DOI] [PubMed] [Google Scholar]
- 100. Knowlton S, Tasoglu S. A bioprinted liver‐on‐a‐chip for drug screening applications. Trends Biotechnol. 2016;9:681–82. [DOI] [PubMed] [Google Scholar]
- 101. Weltin A, Hammer S, Noor F, Kaminski Y, Kieninger J, Urban GA. Accessing 3D microtissue metabolism: Lactate and oxygen monitoring in hepatocyte spheroids. Biosens Bioelectron. 2017;87:941–8. [DOI] [PubMed] [Google Scholar]
- 102. Abe‐Fukasawa N, Otsuka K, Aihara A, Itasaki N, Nishino T. Novel 3D liquid cell culture method for anchorage‐independent cell growth, cell imaging and automated drug screening. Sci Rep. 2018;8:3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lim W, Park S. A microfluidic spheroid culture device with a concentration gradient generator for high‐throughput screening of drug efficacy. Molecules. 2018;12:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Katsura H, Sontake V, Tata A, Kobayashi Y, Edwards CE, Heaton BE, et al. Human lung stem cell‐based alveolospheres provide insights into SARS‐CoV‐2‐mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27:890–904.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Han Y, Duan X, Yang L, Nilsson‐Payant BE, Wang P, Duan F, et al. Identification of SARS‐CoV‐2 Inhibitors using Lung and Colonic Organoids. Nature. 2020. 10.1038/s41586-020-2901-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Driehuis E, van Hoeck A, Moore K, Kolders S, Francies HE, Gulersonmez MC, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A. 2019;116:26580–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Francies HE, Barthorpe A, McLaren‐Douglas A, Barendt WJ, Garnett MJ. Drug sensitivity assays of human cancer organoid cultures. Methods Mol Biol. 2016;1576:339–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li YF, Gao Y, Liang BW, Cao XQ, Sun ZJ, Yu JH, et al. Patient‐derived organoids of non‐small cells lung cancer and their application for drug screening. Neoplasma. 2020;67:430–7. [DOI] [PubMed] [Google Scholar]
- 109. Phan N, Hong JJ, Tofig B, Mapua M, Elashoff D, Moatamed NA, et al. A simple high‐throughput approach identifies actionable drug sensitivities in patient‐derived tumor organoids. Commun Biol. 2019;2:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jung DJ, Shin TH, Kim M, Sung CO, Jang SJ, Jeong GS. A One‐Stop microfluidic‐based lung cancer organoid culture platform for testing drug sensitivity. Lab Chip. 2019;17:2854–65. [DOI] [PubMed] [Google Scholar]
- 111. Mittal R, Woo FW, Castro CS, Cohen MA, Karanxha J, Mittal J, et al. Organ‐on‐chip models: implications in drug discovery and clinical applications. J Cell Physiol. 2019;234:8352–80. [DOI] [PubMed] [Google Scholar]
- 112. Garnier D, Li R, Delbos F, Fourrier A, Collet C, Guguen‐Guillouzo C, et al. Expansion of human primary hepatocytes in vitro through their amplification as liver progenitors in a 3D organoid system. Sci Rep. 2018;8:8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain‐region‐specific organoids using mini‐bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Liu C, Oikonomopoulos A, Sayed N, Wu JC. Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development. 2018;145:dev156166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, et al. SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]


