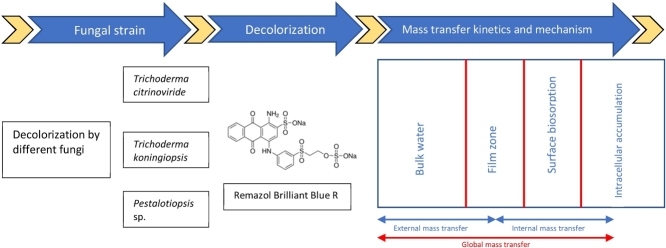
Graphical abstract
Keywords: Decolorization performance, External mass transfer, Internal mass transfer, Rainforest fungus, Synthetic dye
Highlights
-
•
Decolorization of RBBR dye mediated by different fungi was investigated.
-
•
Experimental data were analyzed using the modified mass transfer factor models.
-
•
The resistance of mass transfer for decolorization of RBBR dye was determined.
-
•
Analysis of RBBR decolorization performance gives new insight on the role of fungus.
Abstract
The release of synthetic dye into the environment causing abnormal growth of phytoplankton may lead to a decline in the photosynthetic performance of aquatic ecosystem. Scientific knowledge of Remazol Brilliant Blue R (RBBR) decolorization is essential for designing the engineered bioremediation systems of employing fungal mycelium. The biodegradation of RBBR dye mediated by an appropriate fungus was analyzed using the modified mass transfer factor models to get better understanding on the decolorization kinetics and mechanisms of external and internal mass transfer. The results showed that the limited capacities of the kinetic and isotherm models are still not able to comprehensively explain many important phenomena of RBBR decolorization mediated by the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains. The rate-limiting step of RBBR decolorization depends on the EMT resistance and the vegetative growth rates of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains can be described by second-order polynomial equation. The analysis of decolorization performance may provide a new insight on the role of fungus in the degradation of RBBR dye.
1. Introduction
Synthetic dyes are widely used in the various industries such as the textile, cosmetics, pharmaceutical, plastics, leather, papermaking, rubber and food industries. Several synthetic dyes used in the textile industries such as heterocyclic, anthraquinone, azo, phthalocyanine and triphenylmethane are usually designed to increase the quality of textile to be resistant to light, biological activity, ozone and other environmental conditions. More than 105 synthetic dyes have been produced with approximately 7 × 105 metric tons of annual production [1]. It is estimated the loss of around 10–15 % colorants during the dyeing process leading to a variety of environmental problems [2]. A high amount of dye released into the aquatic environment may cause abnormal growth of phytoplankton and leads to a decline in the ecosystem photosynthetic performance due to the lack of light penetration limits the algal reproduction [3]. The degradation of synthetic dyes is important to avoid ruinous effects on the environment since the disposal of textile wastewater can cause the depletion of dissolved oxygen and leads to ecological hypoxia of causing fish kills [4]. Preventing environmental pollution and conserving water are important to assure a continuing availability of water to maintain sustainable economic development.
The Remazol Brilliant Blue R (RBBR) that contains anthracene derivatives as a persistent and recalcitrant organic compound is commonly used as starting material in the production of polymeric dyes [5]. The analysis of several mechanisms involved in the various methods of RBBR dye removal has been performed to explain the conveniences and disadvantages of the physical, chemical and biological processes offering an insight into the removal of color from wastewater [6]. The decolorization of RBBR dye by the various methods of physical, chemical and combined physicochemical treatments have many disadvantages and limitations [7,8]. Photoelectrocatalysis combined with a single-step H2O2 dosage on TiO2 meshes has an effective configuration optimizing with a low energy cost for the decolorization and biodegradability of RBBR dye from a pharmaceutical wastewater [9]. Biological based methods for the RBBR degradation could be more environmentally friendly than chemical and physical methods due to the use of biological reductant may result in the formation of nonhazardous bioproduct and lowers the energy requirements [10,11].
Many different types of microorganism including bacteria, fungi and algae are capable of decolorizing synthetic dyes [12] but the biodegradation process by microorganism would be more challenging for the decolorization of dye molecules contained five-membered rings or more [13]. The biosorption isotherm of anthraquinone dye by different fungi has been investigated by selecting the optimal conditions relying on the initial biomass, pH and temperature [14]. Combining active and passive transports of molecules during the biosorption process could be favorable for a rapid biological degradation of the colorant compounds in the natural environment. Active transport is defined as the movement of molecules from an area of lower to higher concentration against a concentration gradient and requires enzymes and an input of energy [15]. Passive transport is defined as the movement of molecules from an area of higher to lower concentration in a liquid medium and is controlled by driving force [16]. Even though the biosorption kinetics and isotherm studies of synthetic dyes by the various fungal strains have been investigated from different environments [17,18], the decolorization mechanisms and mass transfer kinetics of dye molecules transported from liquid medium to accumulate into the rigid layers of fungal cell walls are not fully understood. The objectives of this study are: (1) to screen and select the best fungal strains from specific incubation condition and then to identify and characterize the selected fungal strains for use in the rest of the experiments, (2) to study the effects of carbon source, nitrogen source, agitation and pH on the performance of RBBR biodegradation for finding the optimal conditions of incubation process and (3) to verify the decolorization kinetics and isotherms by using several models and then to evaluate the decolorization kinetics and mass transfer mechanisms of RBBR dye by using the modified mass transfer factor (MMTF) models.
2. Materials and methods
2.1. Chemicals and culture media
This study used the RBBR dye originally coming from Sigma-Aldrich Produktions GmbH (Steinheim am Albuch, Germany). The malt extract agar (MEA) and silica gel were obtained from Merck (Darmstadt, Germany). The potato dextrose agar (PDA) was obtained from Lotte Chemical Titan Holding Sdn Bhd (Kuala Lumpur, Malaysia). The reagents of acetone, ammonium chloride (nitrogen source), ammonium nitrate (nitrogen source), chloramphenicol, dichloromethane, ethanol, ethyl acetate of chemical grade, hydrochloric acid and sodium sulfate anhydrous were obtained from QReC (New Zealand). The carbon sources of d-(-)-Fructose, d-(+)-Galactose and d-(+)-Glucose monohydrate extra pure were obtained from Systerm (Selangor, Malaysia), Acros Organics - Fisher Scientific Belgium and QReC (New Zealand), respectively. The yeast extract (nitrogen source) and Tween 80 of synthesis grade were obtained from Scharlab (Barcelona, Spain).
2.2. Sampling and screening of fungi
This study selected 20 species among the various species of fungi collected from decaying pieces of wood and soil in a tropical rainforest around the campus of Universiti Teknologi Malaysia. Each fungus was put in the labeled plastic bag and then kept at 0 °C in a freezer of labo-720-chromat (Philipp Kirsch GmbH, Germay) and then analyzed under the Vilber Lourmat UV lamp (Illkirch, France) in a dark room to select the representative samples of fungal strains for use in the next steps of the experiments. Each species of fungus was isolated and cultured on the PDA (39 g L−1) medium in a glass petri dish containing of 50-mg/L RBBR dye. The glassware and PDA medium were put into an autoclave of EF18967, 50X-120 V Model (Daigger Scientific, New Jersey, USA) at 121 °C and 101 kPa for 45 min to eliminate any bacteria and then cooled at room temperature. Then 20 mL of PDA as culture medium was transferred into a petri dish and then added 1 mg of RBBR dye. Small vegetative part (0.2 g) of luminescent fungus was cut and then cultured on the PDA medium at room temperature. For the screening purpose, all unidentified species of fungi were cultured on the PDA growth media and incubated at 27 °C for 7 d and then measured the average mycelial diameter of each fungus to find the most efficient fungi for use in the decolorization of RBBR dye. Phylogenetic tree analysis of fungal identification was performed to depict the lines of evolutionary descent of different fungal species from a common ancestor. The fungus was named by referring to the GenBank database of the National Center for Biotechnology Information (NCBI).
2.3. Biodegradation experiments
A liquid medium containing of 20-g L−1 malt extract agar, 20-g L−1 glucose, 5-g L−1 yeast extract, 300-mg L−1 chloramphenicol and 1-L distilled water was prepared in a beaker of 1500 mL and then sterilized in an autoclave at 121 °C and 101 kPa for 45 min. Then approximately 1 cm (3 plugs) of the best growth performance of fungi on the PDA medium was punched out using a cork borer and transferred into 100-mL Erlenmeyer flask containing of 20-mL liquid medium and then incubated using the BBD 6220 CO2 incubator (Thermo Fisher Scientific, Massachusetts, United States) at 27 °C to allow new mycelium to grow for 3 d. Then 0.5 mL of RBBR dye was slowly added to the bottom of Erlenmeyer flask and incubated again at 27 °C to record the degradation of RBBR dye at 7th and 14th d of incubation. After the incubation period of 14 d, the liquid culture was centrifuged using the LC-8 Series Laboratory Centrifuge (Benchmark Scientific, New Jersey, United States) at 2000 rpm for 15 min to produce the supernatant. The UV–vis absorbance of supernatant collected was measured using the UV–vis Spectrophotometer (Manual NONOCOLOR® UV/VIS, Macherey-Nagel GmbH & Co., Germany) at λ (max) of 595 nm to analyze the occurrence of RBBR dye decolorization mediated by different fungal strains in the liquid medium containing of glucose as carbon source.
For the purposes of studying the RBBR decolorization performance of fungi, first, 20 g L−1 of either glucose, galactose or fructose as carbon source was added together with 5 g L−1 of yeast extract and 300 mg L−1 of chloramphenicol into 20 mL of liquid medium in 100-mL Erlenmeyer flask and then incubated at 27 °C for 7 d. Second, 20 g L−1 of either ammonium chloride, ammonium nitrate or yeast extract as nitrogen source was added together with 5 g L−1 of yeast extract and 300 mg L−1 of chloramphenicol into 20 mL of liquid medium in 100-mL Erlenmeyer flask and then incubated at 27 °C for 7 d. Third, 20 g L-1 of glucose, 5 g L-1 of yeast extract and 300 mg L−1 of chloramphenicol and 50 mg L−1 of RBBR dye were added into 20 mL of liquid medium in 100-mL Erlenmeyer flask of being wrapped with aluminium foil and then shaken on a rotary shaker at 120 rpm for 28 d of incubation. Fourth, 20 g L−1 of glucose, 5 g L−1 of yeast extract and 300 mg L-1 of chloramphenicol and 50 mg L−1 of RBBR dye were added into 20 mL of liquid medium in 100-mL Erlenmeyer flask and used hydrochloric acid for the adjustment of pH at 1, 3 or 5 and then incubated at 27 °C for 7 d. All the liquid media of first, second, third and fourth experiments were then centrifuged at 2000 rpm for 15 min to produce supernatant. All the treatments of biodegradation were performed in triplicate to analyze the effects of carbon source in the first, nitrogen source in the second, operating condition in the third, and pH in the fourth experiments on the efficiency of RBBR dye decolorization.
2.4. Analytical methods
The concentration of RBBR dye in the supernatant was analyzed using the Gas chromatography (GC) (Agilent 5975E GC/FID, California, USA) with a DB-1 GC column (30 m x0.25 mm ID 0.25 μm film thickness) with the helium carrier gas of 40 mL min−1. The operation of GC was started with a temperature gradient of 100 °C for 2 min. Then the temperature was increased at a rate of 15 °C min−1 until 160 °C and then increased again at a rate of 25 °C min−1 until to 290 °C. The temperatures of injector, column and detector were kept at 260 °C.
The efficiency of RBBR decolorization was determined by computing the percentage of degradation using the formula:
| (1) |
where E is the efficiency of RBBR decolorization by fungus (%), C0 is the initial concentration of RBBR dye in liquid medium (mg L−1), Ct is the concentration of RBBR dye in liquid medium at t time (mg L−1).
2.5. Characterization of fungi
The characterization of morphology was carried out by transferring a small vegetative part of the best growth performance of fungi under the optimum experimental conditions and then observed using the Monocular microscope (Kyowa Biolux-12 Series, model 41–400, Sagamihara, Japan) after 7 d of incubation. The functional groups in supernatant containing of RBBR and degraded RBBR dye were analyzed using the Fourier transform infrared spectroscopy (FTIR) (Perkin Elmer Spectrum One IR, Massachusetts, USA). The supernatant FTIR spectrum was recorded in the spectral region of 400 - 4000 cm−1 for understanding covalent bonding information. After centrifugation at 2000 rpm for 15 min, the liquid medium containing of RBBR or degraded RBBR dye was filtered to separate the precipitates from the supernatant solution. The precipitates were scanned using the Field emission scanning electron microscopy (FESEM) (Supra 40, Carl Zeiss AG, Oberkochen, Germany) for analyzing the surface characteristics of fungi before and after used to degrade the RBBR dye.
2.6. Numerical simulation
2.6.1. Kinetic models
The quantity of RBBR dye biosorbed by the different fungal strains can be calculated using the equation of:
| (2) |
where qt is the quantity of RBBR dye biosorbed by fungus at t time (mg g−1), C0 is the initial concentration of RBBR dye in liquid medium (mg L−1), Ct is the concentration of RBBR dye at t time (mg L−1), V is the volume of liquid medium (L) and W is the mass of fungus (g).
The incubation experiment was carried out by adding three fungal mycelium plugs into 20 mL of liquid medium contained of 20-g L−1 glucose, 5-g L−1 yeast extract, 300-mg L−1 chloramphenicol and 50-mg L−1 RBBR dye to collect the experimental data for the kinetic evaluation of RBBR decolorization. The concentration of RBBR dye in liquid medium was monitored within an incubation period of 30 d with 3-d time intervals. The adsorption kinetic models of pseudo-first order, pseudo-second order and intraparticle diffusion (IPD) [19] were used to describe the decolorization of RBBR dye mediated by different species of fungi and expressed by the following equations:
| (3) |
where qe is the quantity of RBBR dye biosorbed by fungus at equilibrium (mg g−1), k1 is the pseudo-first order rate constant (d−1)
| (4) |
where k2 is the pseudo-second order rate constant (d−1)
| (5) |
where kd is the measure of diffusion coefficient (mg g−1 min-1(1/2)) and cd is the intraparticle diffusion constant (mg g−1).
2.6.2. Isotherm models
The isothermal decolorization experiments were carried out by adding three fungal mycelium plugs into 20 mL of liquid medium consisting of 20-g L−1 glucose, 5-g L−1 yeast extract, and 300-mg L−1 chloramphenicol and by varying the concentration of RBBR dye in the range of 10–60 mg L−1. The isotherm models [19] of Langmuir, Freundlich and Temkin were used to describe the decolorization of RBBR dye mediated by different species of fungi. The Langmuir isotherm model can written as:
| (6) |
where qe is the quantity of RBBR dye biosorbed by fungus at equilibrium (mg g−1), qm is the maximum quantity of RBBR dye biosorbed by fungus (mg g−1), KL is the Langmuir constant (dimensionless).
The Freundlich isotherm model can written as:
| (7) |
where KF is the Langmuir constant (mg g−1) and is the correction factor (L g−1).
The Temkin isotherm model can written as:
| (8) |
with where B is the constant related to heat of biosorption (J mol−1), AT is the Temkin isotherm equilibrium binding constant (L g−1), R is the universal gas constant (J mol−1 K−1), T is the temperature (K), bT is the Temkin isotherm constant (dimensionless).
2.6.3. Mass transfer kinetic models
Reflections on the history of the mass transfer factor (MTF) models were firstly developed by Fulazzaky [16,20] to describe the adsorption of single solute from aqueous solution onto granular activated carbon (GAC) processed in a hydrodynamic column. Using the MTF models permits us to describe the mass transfer kinetics of Cd(II) ions adsorption by titania polyvinylalcohol-alginate beads from aqueous solution in a batch reactor [21]. The MTF models were then modified to extend their applicability for describing the adsorption of multi-solute from surface water onto GAC applied to a hydrodynamic column [22]. The use of the MMTF models could useful to describe the biosorption of oil and grease from agro-industrial processing effluent applied for growing biomass in a packed-bed column reactor [23] and the biosorption of carbonaceous, nitrogenous and phosphorous matters from palm oil mill effluent applied to the development of aerobic granules in a sequencing batch reactor [[24], [25], [26]]. The purpose of this study was to scrutinize the usability of the MMTF models for use in the investigation of RBBR decolorization by different fungal strains performed in liquid medium, aiming at wide applicability of the models. The cumulative amount of RBBR dye biosorbed by fungal strain can be calculated using the formula [26] of:
| (9) |
where q is the cumulative amount of RBBR dye biosorbed by fungal strain (mg g−1), V is the volume of liquid medium (L) and m is the mass of fungus (g).
This study assumed that the decolorization of RBBR dye by a fungal strain successively passes through three different processes of extracellular deposition, cell surface sorption and intracellular accumulation. The first process of extracellular deposition relates to an external mass transfer (EMT) and both the second and third processes of cell surface sorption and intracellular accumulation relate to an internal mass transfer (IMT). The use of the MMTF models may be expanded to investigate the decolorization mechanisms and mass transfer kinetics of RBBR dye mediated by fungus. The MMTF models have been formulated [22] as follows:
| (10) |
where [kLa]g is the global mass transfer factor of RBBR decolorization mediated by fungus (d−1), β is the absorbate-adsorbent affinity parameter (g d mg−1) and t is the time of incubation (d).
A deduction of Eq. (10) yields the linear equation [22] of:
| (11) |
with
| (12) |
where B is the potential mass transfer index related to driving force and intracellular accumulation of mass transfer during the decolorization of RBBR dye mediated by fungus (mg g−1).
The correlation of EMT factor to global mass transfer (GMT) factor can be written by the mathematical equation [22] of:
| (13) |
where [kLa]f is the external mass transfer factor of RBBR decolorization mediated by fungus (d−1).
Firstly, the variation of [kLa]g following the C0/Cs ratio can be determined using Eq. (12) since the values of β and B have been verified from the curve of plotting ln(q) versus ln(t) as shown in Eq. (11). Then the variation of [kLa]f to follow q value can be determined using Eq. (13) since the variation of [kLa]g has been verified accordingly.
The IMT factor is the difference between GMT and EMT and can be expressed in the form [22] of:
| (14) |
where [kLa]d is the internal mass transfer factor of RBBR decolorization mediated by fungus (d−1).
Since the [kLa]d value can be determined as the [kLa]g value minus the [kLa]f value according to Eq. (14), the variations of [kLa]g, [kLa]f and [kLa]d can finally be traced over time of the incubation. The investigation of decolorization kinetics and mass transfer mechanisms were carried out using the experimental data obtained from the degradation of RBBR dye mediated by different fungi under optimal conditions during the incubation period of 21 d with 3-d time intervals.
3. Results and discussion
3.1. Screening and identification
3.1.1. Fungal screening results
The screening of fungal strains was carried out for 20 species of fungi labeled with the codes of W01, W02, W03, W04, W05, W06, W07, W08, W09, W10, W11, W12, W13, W14, W15, W16, W17, W18, W19 and W20. Even though the degradation of RBBR dye cannot be induced by the action of light [27], all the liquid cultures contained of RBBR dye were kept in a very dark room to avoid any light assist in the decolorization process of RBBR dye. The results showed that three fungal strains of W04, W14 and W15 provide a quantification of the best growth performance identified by faster growth rates of 4, 5 and 5 d, respectively, compared to other fungal strains needed for fully decolorized RBBR dye in the PDA media at least 10 d of incubation. The occurrence of liquid culture decolorization was monitored through visual observation. Verifying the occurrence of RBBR decolorization in the liquid medium using the UV–vis Spectrophotometer shows the different peaks of absorbance at λ (max) of 595 nm for the supernatants of W04, W14 and W15.
3.1.2. Phylogenetic tree identification
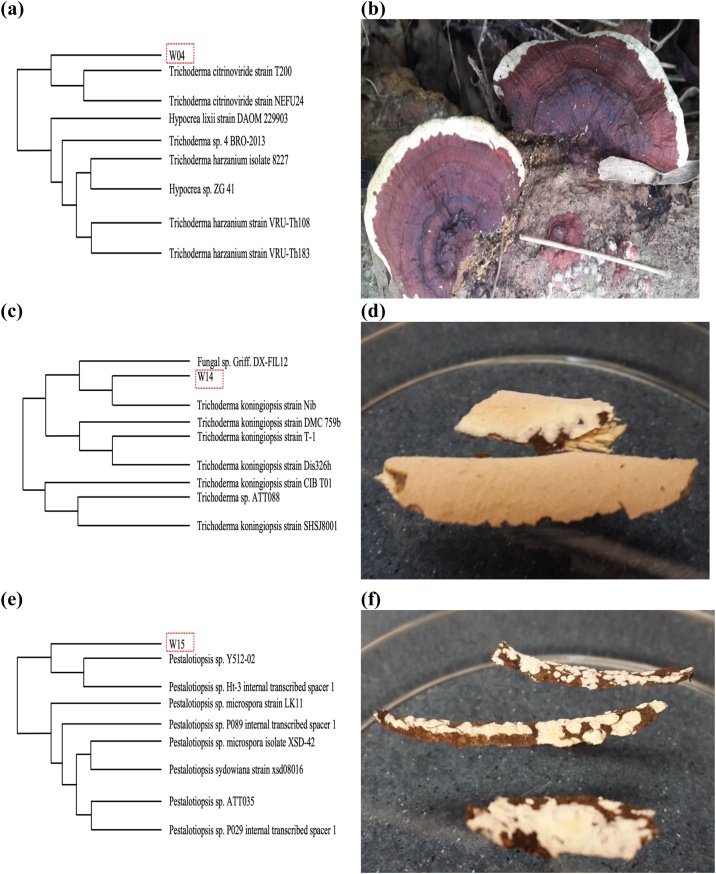
The identification and phylogenetic analysis of W04, W14 and W15 was conducted at the First BASE Laboratories Sdn Bhd in the Selangor state of Malaysia using the MEGA 4.0 software with the neighbor-joining method. The gel electrophoresis was carried out to separate the macromolecules based on series of bands, which contain only particular size fragments of DNA. An extrinsic band could be related to the interaction of macromolecule with chromophore in every fungal strain due to the structural transition of macromolecule may occur during the incubation process [28]. The fungal strains of W04, W14 and W15 were verified having 622 bp (base pairs of DNA), 606 bp and 552 bp, respectively and then referred according to the GenBank database of the NCBI belonging to the fungal species of Trichoderma citrinoviride (T. citrinoviride), Trichoderma koningiopsis (T. koningiopsis) and Pestalotiopsis sp., respectively, as shown in Fig. 1. Numerous fungal strains from the Trichoderma and Pestalotiopsis species could be characterized by a high potential of lignocellulose degradation [29] and identified by an incubation period in the region of 4–5 d. A fungal genus name of Trichoderma is seemly similar between the fungal species of T. citrinoviride and T. koningiopsis because of the similar physical properties of mycelium that are confined essentially to the genus of Trichoderma [30].
Fig. 1.
Phylogenetic tree identification with (a) the identification of fungal strain coded with W04, (b) the identification of fungal strain coded with W14, (c) the identification of fungal strain coded with W15, (d) the picture of fungal strain coded with W04, (e) the picture of fungal strain coded with W14 and (f) the picture of fungal strain coded with W15.
3.1.3. Morphology of the fungi
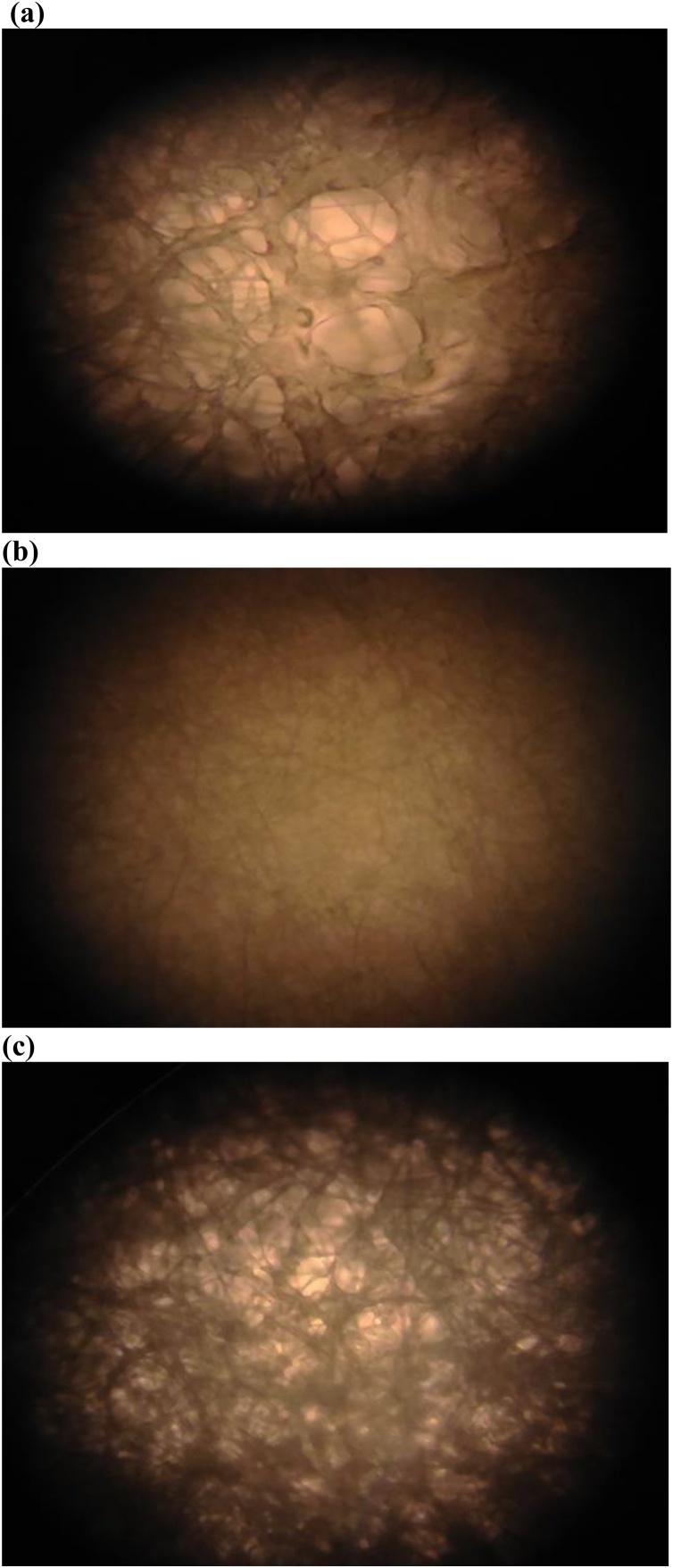
The morphology of the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains was observed using the Monocular microscope when fungal mycelium that represents the essence of each fungal lifestyle has fully occupied petri dish after 7 d of incubation. The results (Fig. 2) show that the fungal strain of T. citrinoviride characterized by a surface morphology of thick layer may be related to growth rate of fully decolorized RBBR dye within 4 d of incubation (see Fig. 2a). Fungal strains of T. koningiopsis and Pestalotiopsis sp. being characterized by their surface morphologies of thin layer may be related to growth rates of fully decolorized RBBR dye within 5 d of incubation (see Figs. 2b, c). Microscopic examination confirms the strain of T. citrinoviride characterized by the morphological feature of rough surface (see Fig. 2a). The strain of T. koningiopsis has an amorphous buffer layer of thin film surface morphology (see Fig. 2b). The distribution of rough surface morphology combined with an amorphous thin film for the Pestalotiopsis sp. strain has the different modes of morphological feature (see Fig. 2c). The decolorization capacity of T. citrinoviride strain with its rough surface and porous structure could be more effective than that of T. koningiopsis or Pestalotiopsis sp. strain. Decolorization of textile dyes by the isolated bacterial strains can be processed by the biological degradation [31]. The pore channels of the T. citrinoviride, T. koningiopsis or Pestalotiopsis sp. strain can typically absorb the RBBR dye through capillary action affecting the efficiency of RBBR decolorization.
Fig. 2.
Morphological character of (a) the T. citrinoviride strain, (b) the T. koningiopsis strain and (c) the Pestalotiopsis sp. strain.
3.2. Study on the RBBR decolorization performance of fungi
3.2.1. Effect of carbon source
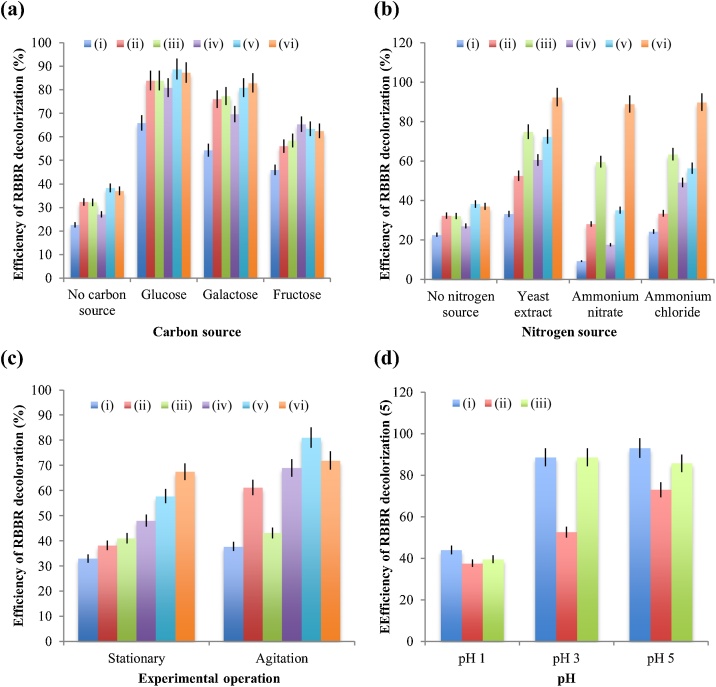
The carbon sources of glucose, galactose and fructose were used to enhance the decolorization of RBBR dye. The results (Fig. 3a) show that the use of glucose as carbon source can increase the decolorization efficiencies of RBBR dye by 43.3 % (from 22.6 to 65.9%), 51.6 % (from 32.3 to 83.9%) and 51.6 % (from 32.2 to 83.8%) after 7 d of incubation and by 53.8 % (from 27.1 to 80.9%), 50.6 % (from 38.2 to 88.8%) and 50.2 % (from 37.0 to 87.2%) after 14 d of incubation since the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp., respectively, mediates the degradation of RBBR dye. In spite of the glucose converted by the isolated bacterial strains to organic acids may inhibit the decolorization of textile dyes [31], the glucose spike can accelerate the decolorization process of RBBR dye by different fungi. An increase in the incubation period of 7 d for the decolorization process of RBBR dye mediated by the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. may increase the performances of RBBR degradation by 15.0 % (from 65.9 to 80.9%), 4.9 % (from 83.9 to 88.8%) and 3.4 % (from 83.8 to 87.2%), respectively. It seems that prolonging the incubation time from 7 to 14 d may slightly increase the sensitivity of RBBR decolorization. This result is consistent with a previous study that the activity of laccase from Ganoderma lucidum in PD medium for decolorizing the RBBR dye slightly increases after 6 d of the incubation [32].
Fig. 3.
Effects of (a) carbon source, (b) nitrogen source, (c) operating condition and (d) pH on the decolorization efficiency of RBBR dye by different fungal strains. Noted that the efficiency of RBBR dye decolorized by the strains of (i) T. citrinoviride, (ii) T. koningiopsis and (iii) Pestalotiopsis sp. analyzed after 7 d of incubation and by the strains of (iv) T. citrinoviride, (v) T. koningiopsis and (vi) Pestalotiopsis sp. analyzed after 14 d of incubation were based on the triplicate data for each experiment.
The decolorization efficiencies increased by 31.7 % (from 22.6 to 54.3%), 43.7 % (from 32.3 to 76.0%) and 45.1 % (from 32.2 to 77.3%) after 7 d of incubation and by 42.6 % (from 27.1 to 69.7%), 42.6 % (from 38.2 to 80.8%) and 45.9 % (from 37.0 to 82.9%) after 14 d of incubation can be verified from the degradation of RBBR dye mediated by the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp., respectively, with added galactose. The efficiencies of RBBR decolorization with added fructose increased by 23.3 % (from 22.6 to 45.9%), 23.7 % (from 32.3 to 56.0%) and 26.2 % (from 32.2 to 58.4%) after 7 d of incubation and by 38.2 % (from 27.1 to 65.3%), 25.2 % (from 38.2 to 63.4%) and 25.5 % (from 37.0 to 62.5%) after 14 d of incubation can be obtained for the degradation of RBBR dye mediated by the fungal strain of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp., respectively. This revealed that glucose is the best carbon source for the degradation of RBBR dye by different fungi due to it provides higher decolorization efficiency than other sugars of galactose and fructose.
3.2.2. Effect of nitrogen source
The nitrogen sources of yeast extract, ammonium nitrate and ammonium chloride were used to boost the decolorization of RBBR dye. The results by using yeast extract as nitrogen source (Fig. 3b) show that the decolorization efficiencies of RBBR dye can increase by 10.5 % (from 22.6 to 33.1%), 20.2 % (from 32.3 to 52.5%) and 42.6 % (from 32.2 to 74.8%) after 7 d of incubation and by 33.4 % (from 27.1 to 60.5%), 34.1 % (from 38.2 to 72.3%) and 55.4 % (from 37.0 to 92.4%) after 14 d of incubation due to the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp., respectively, mediates the decolorization of RBBR dye. Fig. 3b shows that the yeast extract is the best nitrogen source for degradation of RBBR dye by different fungi since it provides higher decolorization efficiency than other nitrogen sources of ammonium nitrate and ammonium chloride. In a previous study has reported that the highest laccase activity of Trametes ljubarskyi can be obtained by using urea as the nitrogen source during the degradation process of RBBR dye [33]. The RBBR decolorization efficiencies of 92.4, 88.9 and 89.8 % after 14 d of incubation can be achieved by using the nitrogen sources of yeast extract, ammonium nitrate and ammonium chloride, respectively, since the fungal strain of Pestalotiopsis sp. is used to mediate the biodegradation of RBBR dye. The fungal strain of Pestalotiopsis sp. is the most efficient fungus in term of the RBBR dye decolorization affected by the addition of different nitrogen sources (see Fig. 3b).
3.2.3. Effect of agitation
The effects of stationary and agitation on the decolorization of RBBR dye were investigated to get better understanding of different conditions of the incubation. The results (Fig. 3c) show that the RBBR decolorization efficiencies of 32.9, 38.1 and 41.0 % after 7 d of incubation and those of 48.0, 57.7 and 67.4 % after 14 d of incubation can be obtained by using the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp., respectively, to degrade the RBBR dye under stationary conditions. The decolorization efficiencies of RBBR dye by agitating the liquid medium may reach 37.7, 61.1 and 43.1 % after 7 d of incubation and 68.9, 81.0 and 71.9 % after 14 d of incubation since to the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp., respectively, were used to mediate the biodegradation of RBBR dye. Effect of agitation on the fungal growth rates of the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains may increase the decolorization efficiencies of RBBR dye by 4.8 % (from 32.9 to 37.7%), 23.0 % (from 38.1 to 61.1%) and 2.0 % (from 41.1 to 43.1%), respectively, after 7 d of incubation and by 20.9 % (from 48.0 to 68.9%), 23.3 % (from 57.7 to 81.0%) and 4.5 % (from 67.4 to 71.9%), respectively, after 14 d of incubation. The agitation of liquid medium during the incubation periods of 7 and 14 d increases approximately 23 % of the efficiency for the decolorization of RBBR dye mediated by the T. koningiopsis strain, which is the best fungal strain likely adapted after 7 d of incubation with or without agitation. This result reinforces the point that the agitation condition of stirred-tank bioreactor could be preferable for the production of mycelial biomass and exopolysaccharide in Paecilomyces sinclairii [34].
3.2.4. Effect of pH
The results (Fig. 3d) show that the decolorization efficiencies of 43.9, 88.5 and 93.0 % can be achieved with aid of the T. citrinoviride strain to degrade the RBBR dye since the pH of the liquid cultures was adjusted at 1, 3 and 5, respectively. The degradation of RBBR dye mediated by the T. koningiopsis strain can achieve at 37.6, 57.5 and 72.9 % of the efficiency when the liquid medium was conditioned at the pHs of 1, 3 and 5, respectively. The decolorization efficiencies of 39.5, 88.5 and 85.7 % can be achieved for the degradation of RBBR dye mediated by the strain of Pestalotiopsis sp. conditioned the pH of liquid medium at 1, 3 and 5, respectively. Even though the decolorization efficiency for degrading the RBBR dye with aid of T. citrinoviride strain at pH 5 is slightly higher than that at pH 3, the decolorization efficiency of RBBR dye mediated by the Pestalotiopsis sp. strain at pH 3 is slightly higher than that at pH 5. This suggests that the most favorable pH for decolorizing the RBBR dye could be at the pH range of 3−5. This result supports the previous observation that the highest decolorization efficiency of RBBR dye mediated by the white rot fungus KRUS-G strain is when the culture medium was conditioned at pH 4 [35].
3.3. Characteristics of the RBBR dye
3.3.1. FTIR analysis
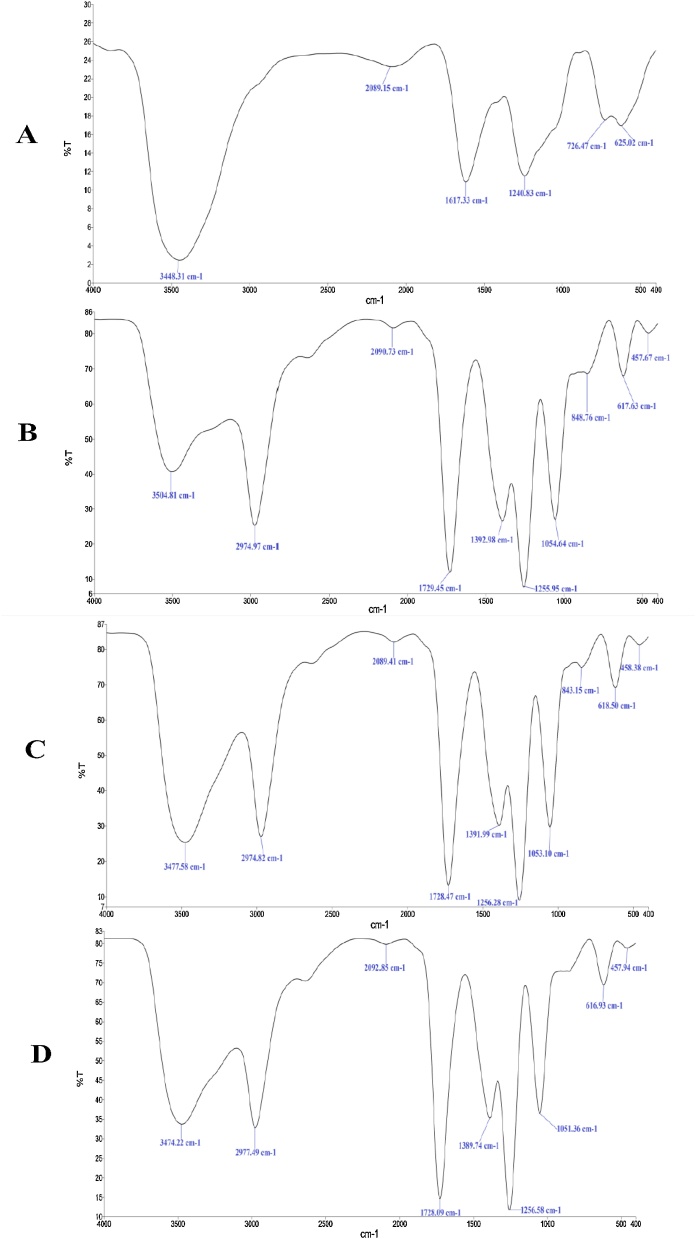
The FTIR analysis (Fig. 4) of investigating the functional groups of RBBR dye was performed for (A) the RBBR dye decolorized without fungus as control, (B) the RBBR dye decolorized with aid of the T. citrinoviride strain, (C) the RBBR dye decolorized with aid of the T. koningiopsis strain and (D) the RBBR dye decolorized with aid of the Pestalotiopsis sp. strain. Fig. 4A shows the FTIR spectrum of broad overlapping absorption bands indicated N—H and O—H stretching vibrations at 3448.31 cm−1 [36,37] and signals attributed to N—H vibrations appeared at 2089.15 cm−1 [36]. The appearance of sharp and medium peak at 1617.33 cm−1 may be attributed to the formation of C C, C O and C—N functional groups [38,39]. The 1240.83, 726.47 and 625.02 cm−1 bands are due to stretching vibrations of C—O, S O and C—CO—C group characteristics of the RBBR dye [39,40].
Fig. 4.
FTIR analysis of functional groups for the RBBR dye decolorized (A) without fungus, (B) with aid of the T. citrinoviride strain, (C) with aid of the T. koningiopsis strain and (D) with aid of the Pestalotiopsis sp. strain.
The appearance of the new bands in Figs. 4B, C and D can be attributed to a change in the characteristics of RBBR dye because of the decolorization of RBBR dye mediated by the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains may result in the fragmentation of RBBR dye to produce the RBBR derivative molecules. The absorption bands at the wavenumbers around of 3504.81, 2974.97 and 2090.73 cm−1 as shown in Fig. 4B can be attributed to stretching vibrations of N—H group characteristic of the decolorized RBBR dye [39,41]. The absorption bands appearing at around 1729.45 and 1392.98 cm−1 can indicate the presence of C O and SO2 functional groups [38,39]. The position of absorption bands at around of 1255.95 and 1054.64 cm−1 can be assigned to stretching vibrations of C—O and the P—O—C asymmetric stretching vibration, respectively [39]. The absorption bands at the wavenumbers of around 848.76, 617.63 and 457.67 cm−1 could be probably due to an asymmetric P—O—S stretching vibration, C—CO—C bond formation and Si-O-Si bond formation, respectively, in the decolorized RBBR dye [39,42]. The absorption bands at 3477.58, 2974.82, 2089.41, 1728.47, 1391.99, 1256.28, 1053.10, 843.15, 618.50 and 458.38 cm−1 appeared in Fig. 4C are almost similar to those appeared in Fig. 4B and therefore can be suggested to the presence of the similar functional groups of decolorized RBBR dye. The absorption bands at 3474.22, 2977.49, 2092.85, 1728.09, 1389.74, 1256.58, 1051.36, 616.93 and 457.94 cm−1 appeared in Fig. 4D are slightly different from those appeared in Figs. B and C due to the absorption band of P—O—S stretching vibration does not appear. The results of FTIR analysis showed that the characteristics of decolorized RBBR dye mediated by the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. are different from the genuine RBBR dye.
3.3.2. FESEM analysis
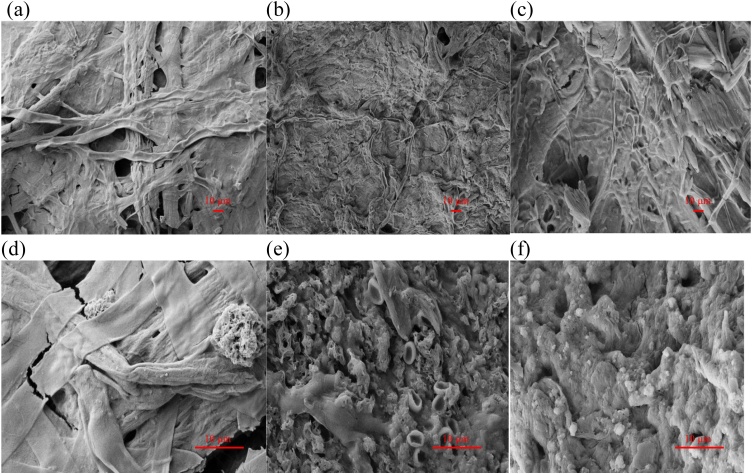
The FESEM analysis of the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains was carried out to get better understanding on the material degradation and mode of the RBBR decolorization. Before using these fungal strains for decolorizing the RBBR dye, the FESEM image of T. citrinoviride strain under magnification of 1.00 kX (Fig. 5a) shows a matt non-adhesive patterned surface of the fungal mycelium. The FESEM image at 1.00-kX magnification (Fig. 5b) of T. koningiopsis strain before used to degrade the RBBR dye shows smooth thin layer surface without any hydrophilic RBBR encapsulation. The FESEM image of Pestalotiopsis sp. strain under 1.00 kX of magnification (Fig. 5c) shows smooth thin layer surface made up of fungal mycelium without attracted hydrophilic RBBR dye. After the decolorization reaction of RBBR dye, the FESEM image of Fig. 5d with a magnification of 5.00 kX shows the hollow spheres of RBBR dye attached on the surface of T. citrinoviride strain. Decolorization of RBBR dye in the liquid medium can lead to an increment in the laccase activity of T. citrinoviride strain [14]. The FESEM image of Fig. 5e with a magnification of 5.00 kX looks like the blood pellets of RBBR dye and shows the rough and thick surfaces of T. koningiopsis strain after used to degrade the RBBR dye. The biosorption process of RBBR decolorization mediated by the T. koningiopsis strain can enhance the transformation of RBBR by the immobilized laccase [43]. The FESEM image at 5.00-kX magnification (Fig. 5f) of Pestalotiopsis sp. strain looks like the long thick pieces of RBBR dye bonded small balls and appears the aggregate facets showing rough surfaces of Pestalotiopsis sp. strain and the asperities after the incubation period of 7 d. The decolorization of RBBR dye mediated by the Pestalotiopsis sp. strain produces a color change from dark blue before to light color of liquid culture medium after 7 d of incubation. The decolorization ability of ligninolytic enzymes produced by Marasmius cladophyllus can be proposed for the bioremediation of RBBR dye contained in the textile industry wastewater [44]. The results (Figs. 5d, e, f) show the agglomeration and aggregation of the RBBR dye on the surface of every fungus after an incubation period of 7 d. Fungal cell walls are responsible for the binding of RBBR dye to form the thick layers strongly attached at certain parts of hollow spheres [32,45].
Fig. 5.
FESEM images of the surface structures for studying the surface patterns of the T. citrinoviride (see Fig. 5a), T. koningiopsis (see Fig. 5b) and Pestalotiopsis sp. (see Fig. 5c) strains with a magnification of 1.00 kX and the surface patterns of the T. citrinoviride (see Fig. 5d), T. koningiopsis (see Fig. 5e) and Pestalotiopsis sp. (see Fig. 5f) strains with a magnification of 5.00 kX.
3.4. Kinetics and isotherm studies
3.4.1. Decolorization kinetics
The results (Table 1) show that the kinetics of RBBR decolorization mediated by the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains follow pseudo-second order kinetic reaction since to the R2 values of 0.90471, 0.97263 and 0.93419, respectively, are all near unity. This means that the rate-limiting step of RBBR decolorization involves a chemisorption, where the removal of RBBR dye from liquid medium is due to the physicochemical interaction between fungal mycelium of solid phase and the RBBR molecules in liquid phase [46].
Table 1.
Value of the parameters in the kinetic models.
| Decolorization kinetic models | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Fungal strain | Pseudo-first order | Pseudo-second order | Intraparticle diffusion | ||||||
| qe (mg/g) | k1 (min−1) | R2 | qe (mg/g) | k2 (min−1) | R2 | Cd (mg/g) | kd (min−1) | R2 | |
| T. citrinoviride | 2.1345 | 0.1914 | 0.70824 | 0.679 | 0.1622 | 0.90471 | 0.3262 | 0.8835 | 0.80612 |
| T. koningiopsis | 1.8223 | 0.1805 | 0.64336 | 0.951 | 0.1389 | 0.97263 | 0.0629 | 0.7799 | 0.94219 |
| Pestalotiopsis sp. | 1.3581 | 0.1077 | 0.94130 | 1.1415 | 0.1386 | 0.93419 | 0.2204 | 0.7809 | 0.92153 |
The kinetics of RBBR decolorization mediated by the T. koningiopsis and Pestalotiopsis sp. strains can be described by the IPD model due to the R2 values of 0.94219 and 0.92153, respectively, are also near unity. Because of the initial biosorption factor of IPD model can be classified into four zones: weakly initial biosorption, intermediately initial biosorption, strongly initial biosorption and approaching completely initial biosorption [47], the primary IPD zones of RBBR decolorization mediated by the T. koningiopsis and Pestalotiopsis sp. strains likely associated with the mycelium surface of an amorphous thin film are both intermediately initial biosorption and strongly initial biosorption. The kinetics of RBBR decolorization mediated by the Pestalotiopsis sp. strain may also follow the pseudo-first order kinetic model because of the R2 value of 0.94130 is near unity. The chemical combination of RBBR dye and fungal mycelium of Pestalotiopsis sp. strain in the reaction is made to behave like a first order reaction [48] and occurs by a great excess of available fungal mycelium compared to the supply of RBBR dye from liquid medium. The influence of temperature on the decolorization kinetics of RBBR dye has been investigated using the bismuth oxychloride nanosphere material and (K0.5Na0.5)NbO3 crystal as pyrocatalysis in the treatment of wastewater [49,50]. The limited capacity of these kinetic models cannot explain many important phenomena of RBBR decolorization mediated by the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains. Therefore, the aim of this study is to extend the application of the MMTF models for investigating the decolorization mechanisms and mass transfer kinetics of RBBR dye associated with the notable roles played by different fungi.
3.4.2. Decolorization isotherm
The isotherm models of Langmuir, Freundlich and Temkin were used to describe the decolorization of RBBR dye mediated by the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains. The results (Table 2) show that the decolorization of RBBR dye mediated by the T. koningiopsis strain is the only best fitted in Langmuir isotherm since the R2 value of 0.92246 is near unity. A thin layer morphological trait (see Fig. 2b) of the T. koningiopsis strain is assumed to be an ideal solid surface composed of distinct sites capable of binding the RBBR molecules to form a continuous monolayer surrounding a smooth homogeneous solid surface [51,52].
Table 2.
Value of the parameters in the isotherm models.
| Adsorption isotherm models | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Fungal strain | Langmuir | Freundlich | Temkin | ||||||
| qe (mg g−1) | k1 (min−1) | R2 | qe (mg g−1) | k1 (min−1) | R2 | qe (mg g−1) | k1 (min−1) | R2 | |
| T. citrinoviride | 405.98 | 18.226 | 0.78438 | 5.099 | 0.4854 | 0.78042 | 0.0135 | 0.0135 | 0.72060 |
| T. koningiopsis | 44.268 | 5.8793 | 0.92246 | 2.4348 | 0.1316 | 0.49795 | 0.0720 | 0.0191 | 0.51076 |
| Pestalotiopsis sp. | 400.11 | 12.685 | 0.42827 | 5.1338 | 0.5570 | 0.70728 | 0.0302 | 0.0211 | 0.56825 |
The decolorization of RBBR dye mediated by the T. citrinoviride strain may follow the isotherm models of Langmuir, Freundlich and Temkin since the R2 value of 0.78438, 0.78042 and 0.72060, respectively, shows a moderately strong positive linear and is close from each other. This describes the relation of the available RBBR dye on the surface of T. citrinoviride mycelium to the concentration of RBBR dye in the liquid medium attributed to a change in the equilibrium constant of the binding process to the heterogeneity (see Fig. 2a) of the fungal mycelium surface [53]. This result cannot be clearly supported for the design of RBBR decolorization and the interpretation of biological recognition phenomena [54]. The decolorization of RBBR dye mediated by the Pestalotiopsis sp. strain may follow the Freundlich isotherm since the R2 value of 0.70728 fitted in Freundlich isotherm is the highest R2 value of fitting the isotherm models. The decolorization of RBBR dye mediated by the Pestalotiopsis sp. strain depends on the rough surface of sites occupation and heterogeneous pore distribution.
3.5. Decolorization mechanisms and mass transfer kinetics
3.5.1. Linear curve fitting
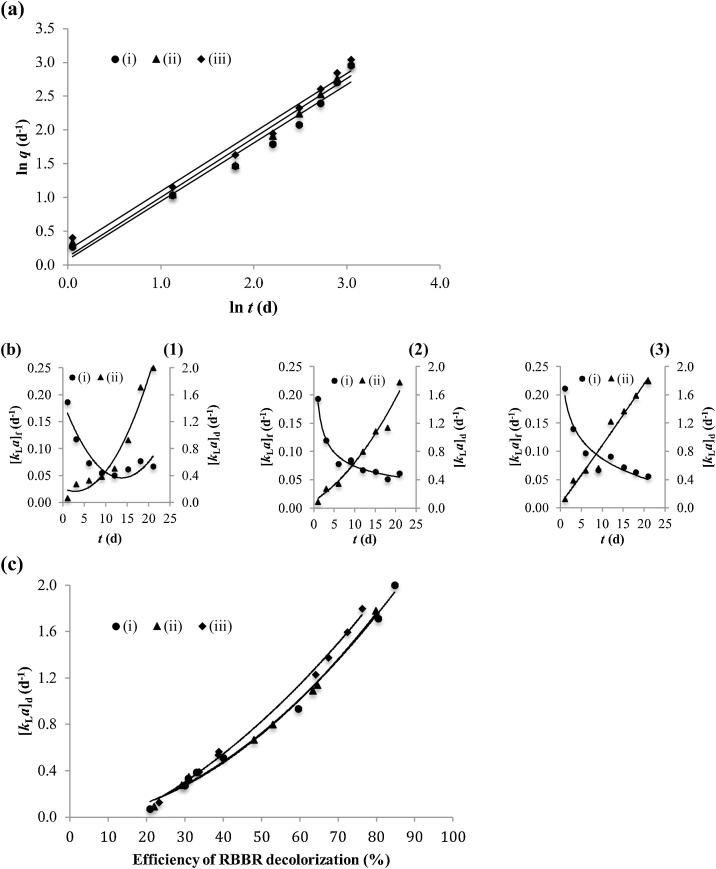
The novelty and originality of this study compared to the previous studies [16,[20], [21], [22], [23], [24], [25], [26]] is the use of the MMTF models to scrutinize the kinetics and decolorization mechanisms of the RBBR dye mediated by different fungi. This may provide better understanding on the mass transfer of dye molecules by employing fungal mycelium for use in designing the further engineered bioremediation systems. By plotting ln(q) versus ln(t) (see Fig. 6a) yields the linear curves with 1/β as slope and B as the y-intercept. Correlation for the parameters β and B in Eq. (11) providing a very good fit to the experimental data is due to the values of R2 are all near unity (R2 > 0.96675; see Table 3). The mass transfer kinetics of RBBR decolorization mediated by different fungi can be determined on the basis of the [kLa]g, [kLa]f and [kLa]d values. The mechanisms of RBBR decolorization mediated by fungal strain depend on the behaviors of dye molecules passing through three successive points of extracellular deposition located at film surface outside of the mycelium surface, cell surface sorption located at interfacial liquid-fungal mycelium and intracellular accumulation located within fungal mycelium. It is suggested that every fungal strain may have the typical mass transfer kinetics of RBBR decolorization and affects the efficiency of RBBR biodegradation. The β values of 1.1589, 1.1375 and 1.1456 g d mg−1 and the B values of 0.0805, 0.1231 and 0.2202 mg g−1 were verified from the linear curve of ln(q) versus ln(t) for the decolorization of RBBR dye mediated by the for the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp., respectively (see Table 3). An increase in the B value of transporting dye molecules from liquid medium to diffuse through the plasma membrane to aggregate into the cells could be due to an increase in the number of cells with a rough surface pattern of the fungal mycelium from the T. citrinoviride to T. koningiopsis and to Pestalotiopsis sp. strain (see Figs. 5d, e, f). This may have related to an increased driving force [22] and intracellular accumulation of removing the RBBR molecules from liquid medium during the incubation period of 21 d.
Fig. 6.
Analysis of the mass transfer kinetics and mechanisms of RBBR decolorization mediated by different fungi with (a) the linear curve fitting, (b) the curves of plotting [kLa]f or [kLa]d versus t where (1) the decolorization of RBBR mediated by the T. citrinoviride mycelium, (2) the decolorization of RBBR mediated by the T. koningiopsis mycelium and (3) the decolorization of RBBR mediated by the Pestalotiopsis sp. mycelium and (c) the the curves of plotting [kLa]d versus E.
Table 3.
Values of β and B obtained from slope and interception, respectively, of plotting ln(q) versus ln(t) for the experiments of RBBR decolorization.
| Fungal strain | β (g d mg−1) | B (mg g−1) | R2 |
|---|---|---|---|
| T. citrinoviride | 1.1589 | 0.0805 | 0.96677 |
| T. koningiopsis | 1.1375 | 0.1231 | 0.97375 |
| Pestalotiopsis sp. | 1.1456 | 0.2202 | 0.97760 |
The decolorization mechanisms of RBBR dye mediated by fungus may associate with active chemical groups of mycelium surface and intracellular accumulation due to the behaviors of physical sorption, ion exchange and chemical bond are dependent on the availability of different surface functional groups [55,56]. Fungal mycelium composed of different natural polymers such as cellulose, chitin and proteins has the possibility of tailoring a unique structure and composition of the cell walls [57]. This paves the way for environmental security and efficiency in the various scale applications of fungus by properly choosing the toxic organic substrates. The decolorization behaviors of RBBR dye mediated by the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains can differ from each other due to the various mechanisms of molecular transformation occur naturally in the different species of fungi during the incubation period of 21 d. The mechanisms and mass transfer kinetics of the RBBR decolorization can be predicted from the values of [kLa]g, [kLa]f and [kLa]d to provide a comprehensive insight into the overall external and internal mechanisms of mass transfer involved molecular diffusion and convection [58].
3.5.2. Mass transfer kinetics and decolorization mechanisms
The variation trends of [kLa]g and [kLa]d values increased with increasing of the incubation period are almost similar with each other. A decrease in the variation of [kLa]f value is counterbalanced by an increased variation of both the [kLa]g and [kLa]d values over the course of 21 d of incubation. Studying the decolorization kinetics of RBBR dye can be limited by the analysis of external and internal mass transfer. Fig. 6b shows the conformational variation of [kLa]f or [kLa]d pursuant to the time of incubation. The most appropriate curve types of [kLa]f versus t traced over 21 d of incubation are the polynomial, power and logarithmic equations for the decolorization of RBBR dye mediated by the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp., respectively, while the curve types of [kLa]d versus t are all most appropriate for the polynomial equations. The [kLa]f values of 0.1865, 0.1934 and 0.2118 d−1 at the 1st d of incubation are all higher than the [kLa]d values of 0.0705, 0.0917 and 0.1276 d−1, respectively. This reveals that the decolorization kinetics of RBBR dye mediated by three different fungal strains are mainly controlled by intracellular accumulation at the 1st d of incubation and then mainly controlled by film mass transfer along the period of 20 d from the 2nd to 21st day of incubation. The resistance of mass transfer could be prominently dependent on EMT since the [kLa]f values are all lower than the [kLa]d values except at the beginning of incubation. The rate-limiting step of RBBR decolorization depends on the EMT resistance due to the IMT resistance and kinetic limitations for the migration of RBBR molecules from bulk solution to intracellular accumulation are negligible. It has been reported that the generation of clean hydrogen energy converted by vibration energy through the Bi2WO6 layered-perovskite may enhance the process of RBBR dye decolorization [59]. Detail decolorization mechanisms of RBBR dye mediated by fungus affecting the EMT and IMT rates can be scrutinized by plotting the most appropriate equations of [kLa]f or [kLa]d versus t, as shown in Table 4.
Table 4.
Curve of [kLa]f or [kLa]d versus t for describing the RBBR decolorization by different fungi.
| Fungal strain | Type of the equation |
R2 | ||
|---|---|---|---|---|
| [kLa]f versus t | R2 | [kLa]d versus t | ||
| T. citrinoviride | [kLa]f = 0.0008 t2 - 0.0207 t + 0.1875 | 0.87837 | [kLa]d = 0.0058 t2 - 0.033 t + 0.2124 | 0.96722 |
| T. koningiopsis | [kLa]f = 0.1899 t0.409 | 0.95188 | [kLa]d = 0.0018 t2 - 0.0361 t + 0.1005 | 0.96729 |
| Pestalotiopsis sp. | [kLa]f = -0.05ln(t) + 0.2012 | 0.93098 | [kLa]d = 0.0002 t2 - 0.0814 t + 0.0525 | 0.96329 |
The primary concern of this study was to develop scientific knowledge from using the MMTF models for describing the decolorization behaviors of RBBR dye mediated by different fungi. This attempts to extend the applicability of the models and to gain a new insight on the mass transfer process and RBBR dye degradation. An investigation of the response mechanisms by fungal strain in the RBBR decolorization is important to get an in depth understanding of the mass transfer dynamic behaviors of reduced color by fungal mycelium. A polynomial model (Fig. 6b1 line-i) of external mass transfer mechanisms mediated by the T. citrinoviride strain could be influenced by the variables of driving force and intracellular accumulation. The curve of Fig. 6b1 line-i shows the [kLa]f value decreased during an incubation period of 15 d and then increased from the 15th to 21st d of incubation. This could be due to the rapid intracellular accumulation caused vacant sites on the surface of T. citrinoviride mycelium cannot be immediately occupied by the RBBR molecules and leads to an increase in the driving force for mass transfer. A power model (Fig. 6b2 line-i) of transporting the RBBR dye from bulk liquid to the extracellular deposition points mediated by the T. koningiopsis strain may be mainly affected by driving force for molecular diffusion rapidly transported at the beginning of incubation. A logarithmic model (Fig. 6b3 line-i) of mass transfer is mainly dependent on the logarithmically decreased driving force of transporting the RBBR dye from the bulk liquid to the extracellular enzymes of Pestalotiopsis sp. mycelium. In a previous study reported that the vibration-catalysis mediated by ZnO might be a potential method to decolorize the RBBR dye during the treatment of wastewater decolorization [60].
The polynomial models (Fig. 6b1 line-ii, b2 line-ii, b3 line-ii) of internal mass transfer diffusion mechanisms mediated by the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. could be due to the movement of RBBR dye is dependent on the variables of driving force and intracellular accumulation. It seems that an increase in the intracellular accumulation is counterbalanced by a decrease in the film mass transfer. This indicates that the shortcoming of RBBR dye as a tracer for mycelium growth cannot support fungus for very long. The vegetative growth rates of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains are all faster than the reduced rates of RBBR dye moved from bulk liquid to extracellular deposition points. The efficiency of RBBR decolorization affected by different fungi can be evaluated by tracing the appropriate models. Empirical modeling and data analysis for the prediction of RBBR decolorization performance mediated by the T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. strains are based on the [kLa]d values plotted versus the RBBR degradation efficiency calculated using Eq. (1).
3.5.3. Prediction of decolorization efficiency
A plot (Fig. 6c) of [kLa]d versus E displays the curves fitting with a second-order polynomial trend line while the efficiency of RBBR decolorization increases with increasing of the IMT factor. The polynomial regression models of best fit (R2 > 0.9944; see Table 5) represent strong relationship between [kLa]d and E for use in the prediction of RBBR decolorization performance. The results (Fig. 6c) show that the efficiency of RBBR decolorization mediated by the T. citrinoviride strain is almost similar to that mediated by the T. koningiopsis strain and this may be related to the same fungal genus of Trichoderma (see Fig. 6c lines-i,-ii). The morphological identification of Trichoderma genus shows great homoplasy in the conidial structures [61]. The internal mass transfer rate of RBBR decolorization mediated by the Pestalotiopsis sp. strain (see Fig. 6c line-iii) requires faster for achieving the same percentage of degradation efficiency as achieved by the Trichoderma genus (see Fig. 6c lines-i,-ii). The selection of the relevant fungal species during the incubation process can do the role to optimize the RBBR decolorization and consequently leads to a polynomial growth of fungal mycelium for increasing the efficiency of RBBR dye degradation during the experimental period of 21 d. It requires the [kLa]d values of 1.9997, 1.7787 and 1.7980 d−1 to reach at approximately 84.78, 79.80 and 76.33 % efficiency of RBBR decolorization mediated by the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp., respectively. The application of photo-/piezo-bi-catalysis of ZnO by harvesting the light and vibration energy of the natural environment may enhance catalytic efficiency of RBBR dye decomposition [62]. The high efficiency of RBBR decolorization can be designed from the selection of appropriate fungal species [63], which can further aid in the engineered bioremediation systems of employing fungal mycelium. This study reveals that the performance of RBBR decolorization depends on the metabolic potential of fungi to degrade the RBBR dye molecules.
Table 5.
Modeling of [kLa]d value to predict the efficiency of RBBR decolorization.
| Fungal strain | Polynomial equation of [kLa]d versus E | R2 |
|---|---|---|
| T. citrinoviride | [kLa]d = 0.0002E2 + 0.0038E - 0.0461 | 0.99442 |
| T. koningiopsis | [kLa]d = 0.0002E2 + 0.0023E - 0.0231 | 0.99547 |
| Pestalotiopsis sp. | [kLa]d = 0.0002E2 + 0.0108E - 0.1931 | 0.99580 |
4. Conclusions
The decolorization of RBBR dye mediated by the fungal strains of T. citrinoviride, T. koningiopsis and Pestalotiopsis sp. was performed to get better understanding into the kinetics and mechanisms of mass transfer. The experimental data of 21-d incubation time were analyzed using the MMTF models to determine the GMT, EMT and IMT factors. The variation trends of [kLa]g and [kLa]d values are almost similar to most likely due to an increased variation trend of both [kLa]g and [kLa]d value is regularly counterbalanced by a decreased variation trend of [kLa]f value over the course of 21-d incubation. The decolorization of RBBR dye mediated by different fungal strains in liquid medium is mainly controlled due to the EMT resistance. The results findings show that the efficiency of RBBR decolorization analyzed based on the variation of [kLa]d value could be useful for researchers in predicting the performance of sythetic dye decolorized by incubation on the liquid media. The performance of RBBR decolorization polynomially increased depends on the growth rate of fungal mycelium and this may contribute to designing the further engineered bioremediation systems of employing fungal mycelium. This study suggested that the use of the MMTF models could be useful to describe the decolorization kinetics of synthetic dye mediated by different fungal strains in the future researches.
Author statement
AS and MAF conceived and designed the study including conceptualization, data curation, project administration, resources and software. AS and MAF performed the formal analysis, investigation, methodology, validation and visualization. MAF supervised the research project. AS and MAF acquired the funding acquisition. AS wrote the original draft of the manuscript. MAF reviewed and edited the final draft of the manuscript. All authors critically reviewed and revised the manuscript, and they have read and approved the final version.
Declaration of Competing Interest
The authors reported no declarations of interest.
Acknowledgements
The authors acknowledge financial support from the Malaysian Ministry of Education under the Exploratory Research Grant Scheme for Vote No. 4L102 and the Ton Duc Thang University for Contract No. 551/2019/TĐT-HĐLV-NCV.
References
- 1.Jadhav S.B., Chougule A.S., Shah D.P., Pereira C.S., Jadhav J.P. Application of response surface methodology for the optimization of textile effluent biodecolorization and its toxicity perspectives using plant toxicity, plasmid nicking assays. Clean Technol. Environ. Policy. 2015;17:709–720. [Google Scholar]
- 2.Lellis B., Fávaro-Polonio C.Z., Pamphile J.A., Polonio J.C. Effects of textile dyes on health and the environment and bioremediation potential of living organisms. Biotechnol. Res. Innov. 2019;3:275–290. [Google Scholar]
- 3.Morgan-Kiss R.M., Priscu J.C., Pocock T., Gudynaite-Savitch L., Huner N.P.A. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol. Mol. Biol. Rev. 2006;70:222–252. doi: 10.1128/MMBR.70.1.222-252.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lade H., Kadam A., Paul D., Govindwar S. Biodegradation and detoxification of textile azo dyes by bacterial consortium under sequential microaerophilic/ aerobic processes. EXCLI J. 2015;14:158–174. doi: 10.17179/excli2014-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichlerová I., Homolka L., Benada O., Kofroňová O., Hubálek T., Nerud F. Decolorization of Orange G and Remazol Brilliant Blue R by the white rot fungus Dichomitus squalens: toxicological evaluation and morphological study. Chemosphere. 2007;69:795–802. doi: 10.1016/j.chemosphere.2007.04.083. [DOI] [PubMed] [Google Scholar]
- 6.Collivignarelli M.C., Abbà A., Miino M.C., Damiani S. Treatments for color removal from wastewater: state of the art. J. Environ. Manage. 2019;236:727–745. doi: 10.1016/j.jenvman.2018.11.094. [DOI] [PubMed] [Google Scholar]
- 7.Hayat H., Mahmood Q., Pervez A., Bhatti Z.A., Baig S.A. Comparative decolorization of dyes in textile wastewater using biological and chemical treatment. Separ. Purif. Technol. 2015;154:149–153. [Google Scholar]
- 8.Routoula E., Patwardhan S.V. Degradation of anthraquinone dyes from effluents: a review focusing on enzymatic dye degradation with industrial potential. Environ. Sci. Technol. 2020;54:647–664. doi: 10.1021/acs.est.9b03737. [DOI] [PubMed] [Google Scholar]
- 9.Collivignarelli M.C., Abbà A., Miino M.C., Arab H., Bestetti M., Franz S. Decolorization and biodegradability of a real pharmaceutical wastewater treated by H2O2-assisted photoelectrocatalysis on TiO2 meshes. J. Hazard. Mater. 2020;387 doi: 10.1016/j.jhazmat.2019.121668. [DOI] [PubMed] [Google Scholar]
- 10.Mallet J. Group selection and the development of the biological species concept. Philos. Trans. R. Soc. Lond B: Biol. Sci. 2010;365:1853–1863. doi: 10.1098/rstb.2010.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paz A., Carballo J., Pérez M.J., Domínguez J.M. Biological treatment of model dyes and textile wastewaters. Chemosphere. 2017;181:168–177. doi: 10.1016/j.chemosphere.2017.04.046. [DOI] [PubMed] [Google Scholar]
- 12.Jamee R., Siddique R. Biodegradation of synthetic dyes of textile effluent by microorganisms: an environmentally and economically sustainable approach. Eur. J. Microbiol. Immunol. 2019;9:114–118. doi: 10.1556/1886.2019.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H.-h., Wang Y.-t., Wang Y., Wang H.-x., Sun K.-k., Lu Z.-m. Bacterial degradation of anthraquinone dyes. J. Zhejiang Univ. Sci. B. 2019;20:528–540. doi: 10.1631/jzus.B1900165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rybczyńska-Tkaczyk K., Korniłłowicz-Kowalska T. Biosorption optimization and equilibrium isotherm of industrial dye compounds in novel strains of microscopic fungi. Int. J. Environ. Sci. Technol. (Tehran) 2016;13:2837–2846. [Google Scholar]
- 15.Klingenberg M. Transport catalysis. Biochim. Biophys. Acta - Bioenergetics. 2006;1757:1229–1236. doi: 10.1016/j.bbabio.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Fulazzaky M.A. Determining the resistance of mass transfer for adsorption of the surfactants onto granular activated carbons from hydrodynamic column. Chem. Eng. J. 2011;166:832–840. [Google Scholar]
- 17.Aksu Z., Tezer S. Equilibrium and kinetic modelling of biosorption of Remazol Black B by Rhizopus arrhizus in a batch system: effect of temperature. Process Biochem. 2000;36:431–439. [Google Scholar]
- 18.Dotto G., Lima E., Pinto L. Biosorption of food dyes onto Spirulina platensis nanoparticles: equilibrium isotherm and thermodynamic analysis. Bioresour. Technol. 2012;103:123–130. doi: 10.1016/j.biortech.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 19.Syafiuddin A., Salmiati S., Jonbi J., Fulazzaky M.A. Application of the kinetic and isotherm models for better understanding of the behaviors of silver nanoparticles adsorption onto different adsorbents. J. Environ. Manage. 2018;218:59–70. doi: 10.1016/j.jenvman.2018.03.066. [DOI] [PubMed] [Google Scholar]
- 20.Fulazzaky M.A. Analysis of global and sequential mass transfers for the adsorption of atrazine and simazine onto granular activated carbons from hydrodynamic column. Anal. Methods. 2012;4:2396–2403. [Google Scholar]
- 21.Fulazzaky M.A., Majidnia Z., Idris A. Mass transfer kinetics of Cd (II) ions adsorption by titania polyvinylalcohol-alginate beads from aqueous solution. Chem. Eng. J. 2017;308:700–709. [Google Scholar]
- 22.Fulazzaky M.A., Khamidun M.H., Omar R. Understanding of mass transfer resistance for the adsorption of solute onto porous material from the modified mass transfer factor models. Chem. Eng. J. 2013;228:1023–1029. [Google Scholar]
- 23.Fulazzaky M.A., Abdullah S., Salim M.R. Fundamentals of mass transfer and kinetics for biosorption of oil and grease from agro-food industrial effluent by Serratia marcescens SA30. RSC Adv. 2015;5:104666–104673. [Google Scholar]
- 24.Fulazzaky M.A., Nuid M., Aris A., Muda K. Kinetics and mass transfer studies on the biosorption of organic matter from palm oil mill effluent by aerobic granules in a sequencing batch reactor. Process Saf. Environ. Prot. 2017;107:259–268. doi: 10.1080/09593330.2017.1351494. [DOI] [PubMed] [Google Scholar]
- 25.Fulazzaky M.A., Nuid M., Aris A., Muda K. Mass transfer kinetics of biosorption of nitrogenous matter from palm oil mill effluent by aerobic granules in sequencing batch reactor. Environ. Technol. 2018;39:2151–2161. doi: 10.1080/09593330.2017.1351494. [DOI] [PubMed] [Google Scholar]
- 26.Fulazzaky M.A., Nuid M., Aris A., Fulazzaky M., Sumeru K., Muda K. Mass transfer kinetics of phosphorus biosorption by aerobic granules. J. Water Process Eng. 2019;31 [Google Scholar]
- 27.O’mahony T., Guibal E., Tobin J. Reactive dye biosorption by Rhizopus arrhizus biomas. Enzyme Microbial Technol. 2002;31:456–463. [Google Scholar]
- 28.Chiu M.H., Prenner E.J. Differential scanning calorimetry: an invaluable tool for a detailed thermodynamic characterization of macromolecules and their interactions. J. Pharm. Bioallied Sci. 2011;3:39–59. doi: 10.4103/0975-7406.76463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marecik R., Błaszczyk L., Biegańska-Marecik R., Piotrowska-Cyplik A. Screening and identification of Trichoderma strains isolated from natural habitats with potential to cellulose and xylan degrading enzymes production. Pol. J. Microbiol. 2018;67:181–190. doi: 10.21307/pjm-2018-021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyde K.D., Xu J., Rapior S., et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019;97:1–136. [Google Scholar]
- 31.Chen K.-C., Wu J.-Y., Liou D.-J., Hwang S.-C.J. Decolorization of the textile dyes by newly isolated bacterial strains. J. Biotechnol. 2003;101:57–68. doi: 10.1016/s0168-1656(02)00303-6. [DOI] [PubMed] [Google Scholar]
- 32.Qin P., Wu Y., Adil B., Wang J., Gu Y., Yu X., Zhao K., Zhang X., Ma M., Chen Q., Chen X., Zhang Z., Xiang Q. Optimization of laccase from Ganoderma lucidum decolorizing Remazol Brilliant Blue R and Glac1 as main laccase-contributing gene. Molecules. 2019;24:3914. doi: 10.3390/molecules24213914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goh S.M., Chan M.Y., Ong L.G.A. Degradation potential of basidiomycetes Trametes ljubarskyi on Reactive Violet 5 (RV 5) using urea as optimum nitrogen source. Biotechnol. Biotechnol. Equip. 2017;31:743–748. [Google Scholar]
- 34.Kim S.W., Hwang H.J., Xu C.P., Choi J.W., Yun J.W. Effect of aeration and agitation on the production of mycelial biomass and exopolysaccharides in an enthomopathogenic fungus Paecilomyces Sinclairii. Lett. Appl. Microbiol. 2003;36:321–326. doi: 10.1046/j.1472-765x.2003.01318.x. [DOI] [PubMed] [Google Scholar]
- 35.Sumandono T., Saragih H., Migirin T., Watanabe R..Amirta. Decolorization of Remazol Brilliant Blue R by new isolated white rot Fungus collected from tropical rain forest in East Kalimantan and its ligninolytic enzymes activity. Proc. Environ. Sci. 2015;28:45–51. [Google Scholar]
- 36.Sahoo S., Chakraborti C.K., Behera P.K., Mishra S.C. FTIR and raman spectroscopic investigations of a norfloxacin/carbopol934 polymeric suspension. J. Young Pharm. 2012;4:138–145. doi: 10.4103/0975-1483.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z.P., Rong M.Z., Zhang M.Q., Yuana C. Alkoxyamine with reduced homolysis temperature and its application in repeated autonomous self-healing of stiff polymers. Polym. Chem. 2013;4:4648–4654. [Google Scholar]
- 38.Balan V., Mihai C.-T., Cojocaru F.-D., Uritu C.-M., Dodi G., Botezat D., Gardikiotis I. Vibrational spectroscopy fingerprinting in medicine: from molecular to clinical practice. Materials. 2019;12:2884. doi: 10.3390/ma12182884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coates J. Encyclopedia Analytical Chemistry: Applications, Theory and Instrumentation. John Wiley & Sons; New Jersey: 2006. Interpretation of infrared spectra, a practical approach; pp. 10815–10837. [Google Scholar]
- 40.Moosavinejad S.M., Madhoushi M., Vakili M., Rasouli D. Evaluation of degradation in chemical compounds of wood in historical buildings using FT-IR and FT-Raman vibrational spectroscopy. Maderas Cienc. Y Tecnol. 2019;21:381–392. [Google Scholar]
- 41.Maegawa Y., Mizoshita N., Taniab T., Inagaki S. Transparent and visible-light harvestingacridone-bridged mesostructured organosilica film. J. Mater. Chem. 2010;20:4399–4403. [Google Scholar]
- 42.Huang X., Chen Z. Preparation and characterization of CoFe2O4/SiO2 nanocomposites. Chin. Sci. Bull. 2006;51:2529–2534. [Google Scholar]
- 43.Osma J.F., Toca-Herrera J.L., Rodríguez-Couto S. Transformation pathway of remazol brilliant blue r by immobilised laccase. Bioresour. Technol. 2010;101:8509–8514. doi: 10.1016/j.biortech.2010.06.074. [DOI] [PubMed] [Google Scholar]
- 44.Sing N.N., Husaini A., Zulkharnain A., Roslan H.A. Decolourisation capabilities of ligninolytic enzymes produced by Marasmius cladophyllus UMAS MS8 on Remazol Brilliant Blue R and other azo dyes. Biomed Res. Int. 2017;2017 doi: 10.1155/2017/1325754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houtman C.J., Kitin P., Houtman J.C.D., Hammel K.E., Hunt C.G. Acridine orange indicates early oxidation of wood cell walls by fungi. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngulube T., Gumbo J.R., Masindi V., Maity A. Calcined magnesite as an adsorbent for cationic and anionic dyes: characterization, adsorption parameters, isotherms and kinetics study. Heliyon. 2018;4 doi: 10.1016/j.heliyon.2018.e00838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu F.-C., Tseng R.-L., Juang R.-S. Initial behavior of intraparticle diffusion model used in the description of adsorption kinetics. Chem. Eng. J. 2009;153:1–8. [Google Scholar]
- 48.Jawad A.H., Mubarak N.S.A., Ishak M.A.M., Ismail K., Nawawi W.I. Kinetics of photocatalytic decolourization of cationic dye using porous TiO2 film. J. Taibah Univ. Sci. 2016;10:352–362. [Google Scholar]
- 49.Ma J., Jia Y., Chen L., Zheng Y., Wu Z., Luo W., Jiang M., Cui X., Li Y. Dye wastewater treatment driven by cyclically heating/cooling the poled (K0.5Na0.5)NbO3 pyroelectric crystal catalyst. J. Clean. Prod. 2020;276 [Google Scholar]
- 50.Wu Z., Luo W., Zhang H., Jia Y. Strong pyro-catalysis of shape-controllable bismuth oxychloride nanomaterial for wastewater remediation. Appl. Surf. Sci. 2020;513 [Google Scholar]
- 51.Wang S., Zhu Y. Facile method to prepare smooth and homogeneous polymer brush surfaces of varied brush thickness and grafting density. Langmuir. 2009;25:13448–13455. doi: 10.1021/la901785t. [DOI] [PubMed] [Google Scholar]
- 52.Zhang B., Wang J., Liu Z., Zhanr X. Beyond Cassie equation: local structure of heterogeneous surfaces determines the contact angles of microdroplets. Sci. Rep. 2015;4:5822. doi: 10.1038/srep05822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vijaykumar A., Bolhuis P.G., Rein ten Wolde P. The intrinsic rate constants in diffusion-influenced reactions. Faraday Discuss. 2016;195:421–441. doi: 10.1039/c6fd00104a. [DOI] [PubMed] [Google Scholar]
- 54.Eichlerová I., Homolka L., Benada O., Kofroňová O., Hubálek T., Nerud F. Decolorization of Orange G and Remazol Brilliant Blue R by the white rot fungus Dichomitus squalens: toxicological evaluation and morphological study. Chemosphere. 2007;69:795–802. doi: 10.1016/j.chemosphere.2007.04.083. [DOI] [PubMed] [Google Scholar]
- 55.Dhankhar R., Hooda A. Fungal biosorption - an alternative to meet the challenges of heavy metal pollution in aqueous solutions. Environ. Technol. 2011;32:467–491. doi: 10.1080/09593330.2011.572922. [DOI] [PubMed] [Google Scholar]
- 56.Fulazzaky M.A. Study of the dispersion and specific interactions affected by chemical functions of the granular activated carbons. Environ. Nanotechnol. Monit. Manage. 2019;12 [Google Scholar]
- 57.Haneef M., Ceseracciu L., Canale C., Bayer I.S., Heredia-Guerrero J.A., Athanassiou A. Advanced materials from fungal mycelium: fabrication and tuning of physical properties. Sci. Rep. 2017;7:41292. doi: 10.1038/srep41292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joshi S.Y., Harold M.P., Balakotaiah V. Overall mass transfer coefficients and controlling regimes in catalytic monoliths. Chem. Eng. Sci. 2010;65:1729–1747. [Google Scholar]
- 59.Xu X., Xiao L., Wu Z., Jia Y., Ye X., Wang F., Yuan B., Yu Y., Huang H., Zou G. Harvesting vibration energy to piezo-catalytically generate hydrogen through Bi2WO6 layered-perovskite. Nano Energy. 2020;78 [Google Scholar]
- 60.Xu X., Jia Y., Xiao L., Wu Z. Strong vibration-catalysis of ZnO nanorods for dye wastewater decolorization via piezo-electro-chemical coupling. Chemosphere. 2018;193:1143–1148. doi: 10.1016/j.chemosphere.2017.11.116. [DOI] [PubMed] [Google Scholar]
- 61.Sandoval-Denis M., Sutton D.A., Cano-Lira J.F., Gené J., Fothergill A.W., Wiederhold N.P., Guarro J. Phylogeny of the clinically relevant species of the emerging fungus Trichoderma and their antifungal susceptibilities. J. Clin. Microbiol. 2014;52:2112–2125. doi: 10.1128/JCM.00429-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma J., Ren J., Jia Y., Wu Z., Chen L., Haugen N.O., Huang H., Liu Y. High efficiency bi-harvesting light/vibration energy using piezoelectric zinc oxide nanorods for dye decomposition. Nano Energy. 2019;62:376–383. [Google Scholar]
- 63.Deshmukh R., Khardenavis A.A., Purohit H.J. Diverse metabolic capacities of fungi for bioremediation. Indian J. Microbiol. 2016;56:247–264. doi: 10.1007/s12088-016-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]