Abstract
Antagonism of ROS signaling can inhibit cell apoptosis and autophagy, thus favoring the maintenance and expansion of hematopoietic stem cells. Alpha lipoic acid (ALA), a small antioxidant molecule, affects cell apoptosis by lowering the ROS level. In this study, we show that ALA promoted production of human pluripotent stem cells (hPSCs) derived hemogenic endothelial cells and hematopoietic stem/progenitor cells in vitro. Transcriptome analysis of hPSCs derived hemogenic endothelial cells showed that ALA promoted endothelial‐to‐hematopoietic transition by up‐regulating RUNX1, GFI1, GFI1B, MEIS2, and HIF1A and down‐regulating SOX17, TGFB1, TGFB2, TGFB3, TGFBR1, and TGFBR2. ALA also up‐regulated sensor genes of ROS signals, including HIF1A, FOXO1, FOXO3, ATM, PETEN, SIRT1, and SIRT3, during the process of hPSCs derived hemogenic endothelial cells generation. However, in more mature hPSC‐derived hematopoietic stem/progenitor cells, ALA reduced ROS levels and inhibited apoptosis. In particular, ALA enhanced development of hPSCs derived hematopoietic stem/progenitor cells by up‐regulating HIF1A in response to a hypoxic environment. Furthermore, addition of ALA in ex vivo culture greatly improved the maintenance of functional cord blood HSCs by in vivo transplantation assay. Our findings support the conjecture that ALA plays an important role in efficient regeneration of hematopoietic stem/progenitor cells from hPSCs and maintenance of functional HSCs, providing insight into understanding of regeneration of early hematopoiesis for engineering clinically useful hPSCs derived hematopoietic stem/progenitor cells transplantation. Thus, ALA can be used in the study of hPSCs derived HSCs.
Keywords: alpha lipoic acid, endothelial‐to‐hematopoietic transition (EHT), hematopoiesis, hematopoietic stem/progenitor cells, human pluripotent stem cells (hPSCs), ROS
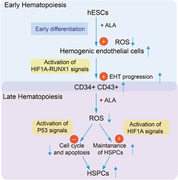
Graphical Abstract
Examines the effect of alpha lipoic acid on hematopoiesis in human pluripotent stem cells and hematopoietic stem/progenitor cells.
Abbreviations
- AGM‐S3
aorta‐gonad‐mesonephros (AGM)‐S3 cells
- ALA
alpha lipoic acid
- hESCs
human embryonic stem cells
- hPSCs
human pluripotent stem cells
- HSCs
hematopoietic stem cells
- ROS
reactive oxygen species
1. INTRODUCTION
Besides intrinsic transcriptional regulation, the birth of hematopoietic stem cells (HSCs) is regulated by multiple exogenous factors, such as hypoxic and extracellular components. Hypoxia and a low ROS level are required for HSCs self‐renewal and maintenance. 1 , 2 , 3 , 4 Optimization of the levels of environmental factors such as oxygen, cytokines, and fatty acids enables maintenance of HSCs quiescence ex vivo. 5 Small molecules that can maintain low ROS levels 6 and inhibit cell apoptosis or autophagy 7 are also useful for maintenance or expansion of HSCs. Elevated ROS production, generated by genetic defects, aging, and ionizing radiation, impairs the function of hematopoietic stem/progenitor cells. 8 Antioxidants are used widely to modulate ROS levels and to rescue loss of function caused by high ROS levels. For example, alpha lipoic acid (ALA) 9 and N‐acetyl‐cysteine (NAC) 10 are used to rescue or preserve the function of hematopoietic stem/progenitor cells.
ROS balance is important for the differentiation of stem cells. 11 Small molecules that can regulate ROS levels play an important role in differentiation of hematopoietic stem/progenitor cells from human pluripotent stem cells (hPSCs), especially in the serum‐ and stroma cell‐free hematopoietic differentiation system. 12 Modulation of ROS level enhances development of CD34+ cells derived from hPSCs, 13 indicating that high ROS levels might cause apoptosis of hemogenic endothelial cells. In addition, ROS accumulation and cell apoptosis inhibit development of primitive hematopoietic stem/progenitor cells undergoing further differentiation from hPSCs. 14 Modulation of ROS levels during differentiation of hPSCs might promote production of hemogenic endothelial cells and consequentially promote development of hematopoietic stem/progenitor cells from hPSCs.
While regeneration of functional hematopoietic cells derived from hPSCs such as NK cells, 15 T lymphocytes, 16 erythrocytes, 17 neutrophils, 18 , 19 and Mϕs 20 has been reported widely, recent research has focused on the molecular regulation and applications of hematopoietic cell differentiation from hPSCs. Two main methods are used for hematopoietic differentiation of human pluripotent stem cells: coculture with stroma cells and embryoid body formation. We have used cocultures of hPSCs with AGM‐S3 stroma cells to explore the hematopoiesis of hPSCs. 17 , 21 , 22 Previous reports show that ALA affects intracellular ROS levels and apoptosis. 23 , 24 Upon irradiation, ALA protects HSCs in bone marrow and improves survival of animals 25 ; in addition, it can partially rescue the loss of HSCs caused by high ROS levels in GRK6− /− mice. 9 ALA has very strong antioxidant properties, and it can protect venous endothelial cells from apoptosis and autophagy caused by hypoxia and ROS. 23 Therefore, we hypothesized that addition of ALA to our coculture system might affect hematopoiesis of hPSCs as well as survival of hematopoietic stem/progenitor cells, thereby allowing us to explore further the subtle regulation of hPSCs during early hematopoietic differentiation.
Our results revealed that ALA plays an important role in the hematopoiesis of hPSCs. Production of hemogenic endothelial cells and hematopoietic stem/progenitor cells in ALA‐treated cultures was higher than that in normal cultures. Low ROS levels were observed in both hemogenic endothelial cells and hematopoietic stem/progenitor cells. Transcriptome analysis of hemogenic endothelial cells revealed that ALA affected pathways influenced by hematopoiesis signals and myeloid development signals, ROS signals, and hypoxia. In addition, our data show that ALA inhibits apoptosis of hematopoietic stem/progenitor cells without changing their differentiation potential. We propose a new function for ALA in human early hematopoiesis and suggest that ALA has an anti‐apoptosis effect on HSCs in vitro.
2. MATERIALS AND METHODS
2.1. Coculture of human embryonic stem cells with AGM‐S3 or OP9 stroma cells
H1 human embryonic stem cells (hESCs) were provided by Professor Tao Cheng and maintained on Matrigel‐coated plates in mTeSR medium (STEMCELL Technologies). The cells were dissociated into clumps using ReLeSR (STEMCELL Technologies). The procedure for inducing hESCs into hematopoietic stem/progenitor cells was as described previously. 21 Briefly, hESCs were sliced into pieces containing about 500 cells, and then seeded onto irradiated AGM‐S3 (13.0 Gy) or OP9 (18.0 Gy) stroma cells.
2.2. CFU assay and liquid differentiation culture
The colony‐forming potential of hematopoietic stem/progenitor cells was assessed by culture on methylcellulose (H4320, STEMCELL Technologies) supplemented with 1% penicillin/streptomycin (Invitrogen) and cytokines, as described previously. 17 BFU‐E, CFU‐Mix, CFU‐GM, CFU‐G, and CFU‐M colonies were assessed on day 14. The lineage differentiation potential of hematopoietic stem/progenitor cells was assayed in IMDM basic medium containing 10% FBS and hematopoietic cytokines (100 ng/ml SCF, 50 ng/ml IL‐6, 20 ng/ml FLT3L, 20 ng/ml IL‐3, 10 ng/ml GM‐CSF, 10 ng/ml G‐CSF, 10 ng/ml TPO, and 4 U/ml EPO). Anti‐human CD45‐APC/Cy7 (BioLegend, Catalog No.: 368516), anti‐human CD71‐FITC (BD Biosciences, Catalog No.: 555536), anti‐human GPA(235a)‐APC (BD Biosciences, Catalog No.: 551336), anti‐human CD66B‐PE (BioLegend, Catalog No.: 305105), and anti‐human CD14‐PE/Cy7 (BD Biosciences, Catalog No.: 557742) were used to detect erythrocytes, granulocytes, and monocytes on day 10 of lineage induction.
2.3. Flow cytometry and cell sorting
To sort and analyze hemogenic endothelial cells, cocultured cells on day 8 were dissociated for 5 min at 37°C with 0.1% trypsin‐EDTA solution and filtered through a 70 μm nylon mesh to obtain a single‐cell suspension. Single cells were incubated with anti‐human Fc block (BD Biosciences, Catalog No.: 564219) for 15 min and then incubated for 40 min with an Ab mixture: anti‐human CD34‐APC (BD Biosciences, Catalog No.: 555824), anti‐human KDR‐PE/CY7 (BioLegend, Catalog No.: 359912), anti‐human CD43‐PE (BD Biosciences, Catalog No.: 560199), and anti‐human CD144‐FITC (BD Biosciences, Catalog No.: 560411). Analysis or sorting of hematopoietic stem/progenitor cells was performed on day 14 of coculture. Cocultured cells on day 14 were dissociated for 5 min at 37°C with 0.25% trypsin‐EDTA solution and filtered through a 70‐μm nylon mesh to obtain a single‐cell suspension. Single cells were incubated for 15 min with anti‐human Fc block (BD Biosciences, Catalog No.: 564219), followed by incubation for 40 min with an Ab mixture: anti‐human CD34‐APC (BD Biosciences, Catalog No.: 555824), anti‐human CD43‐PE (BD Biosciences, Catalog No.: 560199), and anti‐human CD45‐APC/CY7 (BioLegend, Catalog No.: 368516). Finally, all cells were resuspended in staining medium containing 7‐AAD. Flow cytometry was performed on a FACSCanto II system (BD Biosciences) and data were analyzed using FlowJo software (v.10.0.8.). Hematopoietic stem/progenitor cells and hemogenic endothelial cells were sorted using a FACSJazz Cell Sorter (BD Biosciences).
2.4. Apoptosis assays
Apoptotic cells were stained with annexin V–APC and 7‐AAD according to the manufacturer's instructions (BioLegend, Catalog No.: 640930). Cell apoptosis was detected by using a FACSCanto II system (BD Biosciences).
2.5. ROS analysis
Total cells were stained with a surface marker first, and then incubated with 10 μm DCFH‐DA (Nanjing Jiancheng Bioengineering Institute, Catalog No.: E004) at 37°C for 40 min. All cells were detected using a FACSCanto II system (BD Biosciences).
2.6. Cell cycle analysis
Cell cycle analysis was performed using the APC‐BrdU Flow Kit (BD, Catalog No.: 552598). Coculture cells or cord blood total nucleated cells were treated with BrdU for 6 h before analysis. All cells were first stained for surface Ags, and then processed using an APC‐BrdU Flow Kit (BD) according to the manufacturer's instructions. All cells were detected using a FACSCanto II system (BD Biosciences).
2.7. Intracellular analysis
Intracellular staining was used to measure the protein expression level of HIF1A and P53 in CD34+CD43+CD45low/+ hematopoietic stem/progenitor cells of cocultures at day14. All cells were first stained for surface Ags, fixation and permeabilization were done using Fixation/Permeabilization Solution Kit (BD, Catalog No.: 554714) according to the manufacturer's instructions. Finally, fixed/permeabilized cells were stained with the flow Abs of HIF1A (Biolegend, Catalog No.: 359705) and P53 (Biolegend, Catalog No.: 645803) for 40 min. All cells were detected using a FACSCanto II system (BD Biosciences).
2.8. RNA‐Seq and data analysis
One thousand CD34+ CD43− KDR+ CD144+ hemogenic endothelium cells were sorted into a 1.5 ml Eppendorf tube containing 200 µl DPBS‐BSA buffer (0.5% BSA) using a FACSJazz Cell Sorter (BD Biosciences). The dsDNA of sorted 1000‐cell aliquots was generated and amplified as described previously. 26 , 27 Then, aliquots of the amplified cDNA were subjected to qPCR analysis of housekeeping genes (B2M, ACTB, and GAPDH). Samples that passed quality control were used for subsequent sequencing library preparation. dsDNA (1 ng) was used to generate the library using Vazyme TruePrepTM DNA library Prep Kit V2 from Illumina (TD‐503). All libraries were sequenced using the Illumina NovaSeq 6000 system (Novogene Co., Ltd.). The clean fastq raw data files were uploaded to the Gene Expression Omnibus public database (Series GSE144307). All fastq sequences were aligned to the hg38 human genome using Hisat2 software. Normalization of gene expression and differential expression genes (DEGs) analysis were performed using DESeq2. Ego analysis of differential expression genes was performed using clusterProfiler R packages. 28 Heatmaps were plotted using gplots (heatmap.2). Gene set enrichment analysis (GSEA) was performed as described. 29
2.9. HSCs transplantation
Mice were housed in the SPF‐grade animal facility of the Center for Stem Cell Research and Application, Institute of Blood Transfusion. All animal experiments were approved by the Institutional Ethics Review Committee of Institute of Blood Transfusion (IERC‐IBT). Adult B‐NDG mice was purchased from Beijing Biocytogen Co., Ltd. Fresh or cultured human cord blood (CB) nucleated cells were transplanted through retro‐orbital venous sinus into irradiated (1.0 Gy) 8–10 weeks old B‐NDG mice. Transplantation dose of total fresh cord blood nucleated cells contained 1 × 104 CD34+ cells, and the culture cells derived from the same dose of fresh cord blood nucleated cells were cultured for 7 days (Ctrl or ALA treated cultures, respectively). The transplanted mice were maintained on Gentamicin sulfate containing water for 2 weeks. Peripheral blood was obtained from the retro‐orbital venous sinus for flow cytometric analysis 3 months post transplantation. Cord blood nucleated cells were cultured in StemSpan™ SFEM II (Stemcell Technologies, Catalog No.: 09655) containing 50 ng/ml SCF, 10 ng/ml IL‐11, 25 ng/ml Flt3, and 5 ng/ml TPO.
2.10. Quantitative RT‐PCR analysis
Expression of apoptosis‐ and ROS‐related genes was analyzed by quantitative RT‐PCR (qRT‐PCR). Template dsDNA from 1000 CD34+ CD43+ CD45+ hematopoietic stem/progenitor cells was synthesized as described above. Primers specific for HIF1A, FOXO3, P53, P21, P16, ATG5, ATG7, CASP6, CASP7, BCL2, and B2M were selected from the primer bank (https://pga.mgh.harvard.edu/primerbank/) (Table 1.). The reactions were performed in a Thermal cycler (Bio‐Rad). Gene expression levels in each sample were calculated using minimal cycle threshold (Ct) values normalized to expression of the housekeeping gene B2M. All reactions were performed in triplicate.
TABLE 1.
Primers used for RT‐PCR
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| TP53 | CAGCACATGACGGAGGTTGT | TCATCCAAATACTCCACACGC |
| CDKN1A(P21) | TGTCCGTCAGAACCCATGC | AAAGTCGAAGTTCCATCGCTC |
| CDKN2A(P16) | GATCCAGGTGGGTAGAAGGTC | CCCCTGCAAACTTCGTCCT |
| BCL2 | GGTGGGGTCATGTGTGTGG | CGGTTCAGGTACTCAGTCATCC |
| CASP6 | ATGGCGAAGGCAATCACATTT | GTGCTGGTTTCCCCGACAT |
| CASP7 | AGTGACAGGTATGGGCGTTC | CGGCATTTGTATGGTCCTCTT |
| FOXO3 | CGGACAAACGGCTCACTCT | GGACCCGCATGAATCGACTAT |
| ATG5 | AGAAGCTGTTTCGTCCTGTGG | AGGTGTTTCCAACATTGGCTC |
| ATG7 | ATGATCCCTGTAACTTAGCCCA | CACGGAAGCAAACAACTTCAAC |
| HIF1A | CACCACAGGACAGTACAGGAT | CGTGCTGAATAATACCACTCACA |
| B2M | GAGGCTATCCAGCGTACTCCA | CGGCAGGCATACTCATCTTTT |
2.11. Statistical analysis
Data in the bar graphs are presented as the mean ± sd. Statistical analysis was performed using an unpaired Student's t‐test (for comparison of 2 groups). P‐values < 0.05 were considered statistically significant (* P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001).
2.12. Data availability
Data supporting the findings of this study are available from the corresponding author upon request. All RNA‐Seq data can be found in the GEO database under accession code GSE144307.
3. RESULTS
3.1. ALA promotes production of hemogenic endothelium cells and hematopoietic stem/progenitor cells derived from hESCs
The antioxidant ALA protects HSCs and improves survival of animals under irradiation stress when added to chow. 25 , 30 However, these reports only show indirect effects of ALA on HSCs; it is not known whether ALA affects HSCs development directly. To discover the effect of ALA on hematopoiesis, we added ALA to the in vitro hESCs/AGM‐S3 coculture hematopoietic induction system to examine whether ALA affected development and survival of hematopoietic stem/progenitor cells (Fig. 1A). We used differentiation medium without (Ctrl) or with different concentrations (50, 100, 200, 400, and 800 µg/ml) of ALA (ALA group) to induce hematopoietic differentiation. To analyze the effect of ALA on hemogenic endothelium cells, we used flow cytometry to determine the percentages of CD34+ CD43− KDR+ CD144+ hemogenic endothelium cells and absolute cell numbers. Flow cytometry analysis revealed that different concentrations of ALA had different effects on hemogenic endothelium cells differentiation. Low concentrations (<200 µg/ml) had a positive effect on hemogenic endothelium cells production, while high concentrations (≥400 µg/ml) seriously inhibited hemogenic endothelium cells production. At 100 µg/ml, ALA greatly increased the percentage and absolute cell number of hemogenic endothelium cells on day 8 (Fig. 1B and C). To measure the effect of ALA on development of hematopoietic stem/progenitor cells, we used differentiation medium containing 100 µg/ml ALA or normal differentiation medium (Ctrl) to induce hematopoietic differentiation, and hematopoietic stem/progenitor cells were detected on day 14. Flow cytometry analysis indicated that ALA improved the production of CD34+ CD43+ CD45+ hematopoietic stem/progenitor cells significantly (P < 0.0001; Fig. 1D and E).
FIGURE 1.
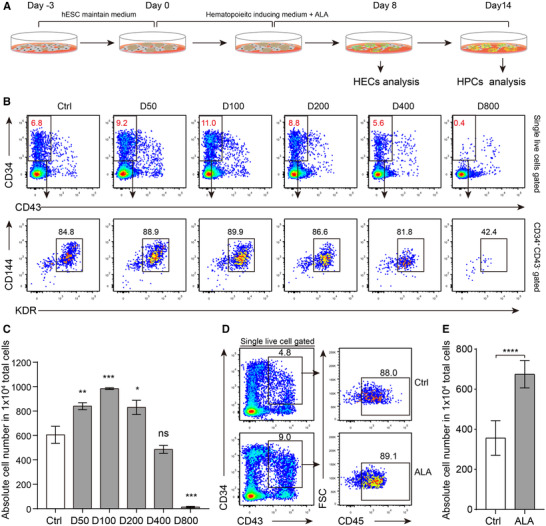
ALA promotes production of hemogenic endothelium cells and hematopoietic stem/progenitor cells. (A) Strategy used to analyze the impact of ALA on hematopoietic differentiation. (B) Flow cytometry analysis of CD34+ CD43− KDR+ CD144+ hemogenic endothelium cells on day 8 after addition of different doses of ALA to the H1/AGM‐S3 hematopoietic differentiation coculture wells. D50, D100, D200, D40, and D800 indicate final ALA concentrations of 50, 100, 200, 400, and 800 µg/ml, respectively. (C) Absolute number of CD34+ CD43− KDR+ CD144+ hemogenic endothelium cells in 1 × 104 total live cells were calculated. (D) Flow cytometry analysis of CD34+ CD43+ CD45+ hematopoietic stem/progenitor cells on day 14 with and without (Ctrl) ALA treatment; 100 µg/ml ALA was added to the H1/AGM‐S3 hematopoietic differentiation coculture wells. (E) Absolute number of CD34+ CD43+ CD45+ hematopoietic stem/progenitor cells in 1 × 104 total live cells were calculated. Data in the bar graphs in (C) and (E) are presented as the mean ± sd. An unpaired Student's t‐test (two‐tailed) was performed. N = 3 replicates; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001; ns, not significant
To demonstrate that the positive effect of ALA on hematopoietic stem/progenitor cells production was not specific to one particular cell type, we measured the effect of ALA in the hESCs/OP9 coculture hematopoietic induction system. The results showed that ALA also significantly promoted production of hemogenic endothelium cells and hematopoietic stem/progenitor cells at concentrations of 50 µg/ml (P < 0.0001) and 100 µg/ml (P < 0.001) (Supplementary Fig. S1A and B). The number of hematopoietic stem/progenitor cells derived from the hESCs/OP9 system on day 12 was also greatly increased (P < 0.001) after addition of 100 µg/ml ALA (Supplementary Fig. S1C and D). In addition, we found that day 2 was the optimal time to add ALA to hematopoietic‐inducing medium to improve production of CD34+ CD43+ hematopoietic stem/progenitor cells (P < 0.0001) (Supplementary Fig. S1E and F). Together, these findings indicate that ALA promotes production of hematopoietic stem/progenitor cells from more than 1 cell type.
3.2. ALA preserves the differentiation potential of hematopoietic stem/progenitor cells
Considering that ALA improved production of hematopoietic stem/progenitor cells, we next investigated whether ALA affected the subpopulations or functions of CD34+ CD43+ hematopoietic stem/progenitor cells. Erythroid or myeloid progenitor cells in bone marrow or cord blood are defined by their expression of the surface markers Lin (CD3, CD4, CD8, CD33, CD19, GAP), CD45RA, CD34, CD38, CD123, and CD71. 31 Given that in the coculture hematopoiesis‐induction system, CD34+ CD43+ hematopoietic stem/progenitor cells on day 14 were almost all CD45low/+ (Fig. 1D), we analyzed the subpopulations of hematopoietic stem/progenitor cells within the CD34+ CD43+ CD45+ population on day 14 by flow cytometry. The results showed that ALA‐treated cocultures had more CD38− CD90+ HSC phenotype‐like cells 22 within the CD34+ CD43+ CD45+ populations (Fig. 2A and C). Common myeloid progenitors (CMPs) are defined as CD34+ CD43+ CD38+ CD123low CD71, and megakaryocyte/erythrocyte progenitors (MEPs) are defined as CD34+ CD43+ CD38+ CD71+ CD123−. Flow cytometry analysis of myeloid progenitors and erythroid progenitors indicated that ALA‐treated cocultures contained lower percentages of CD34+ CD43+ CD38+ CD71− CD123low CMP‐like cells, and lower percentages of CD34+ CD43+ CD38+ CD71+ CD123− MEP‐like cells (Fig. 2B). ALA treatment resulted in significantly lower numbers of CMPs (P < 0.05) and MEPs (P < 0.01) (Fig. 2D). Cell cycle analysis of CD34+CD43+CD45+/low indicated that ALA inhibited the cell cycle progress of CD34+CD43+CD45+/low hematopoietic stem/progenitor cells with significant higher G0/G1 phase population (77.95 ± 0.58% vs. 67.68 ± 2.36%, P < 0.001) and lower percentage of S phase population (14.80 ± 0.54% vs. 7.60 ± 0.19%, P < 0.0001) (Fig. 2E). To further explore the effects of ALA on lineage differentiation of hematopoietic stem/progenitor cells, we performed a colony assay and lineage differentiation in liquid suspension cultures with 8 cytokines (SCF, FLT3L, IL‐6, IL‐3, TPO, EPO, GM‐CSF, and G‐CSF), but without ALA. One thousand CD34+ CD45+ hematopoietic stem/progenitor cells were sorted for each CFU assay per replicate. CFU assay results indicated that ALA‐treated and ALA‐untreated cocultures produced equal numbers of CFU colonies (Fig. 2F and G). Furthermore, 1000 CD34+ CD45+ hematopoietic stem/progenitor cells were seeded into one 96‐well plate containing lineage differentiation culture medium. Lineages cells of CD66B+ CD14− granulocytes, CD71−/+ GPA+ erythrocytes, and CD66B+ CD14+ monocytes within CD45+ populations were all detected in the liquid differentiation system on day 10. The results indicated that ALA did not affect the percentage of granulocytes significantly, but slightly changed the percentages of erythrocytes (P < 0.05) and monocytes (P < 0.05) (Fig. 2H and I). These results suggest that ALA does not change the lineage differentiation potential at the stem/progenitor level.
FIGURE 2.
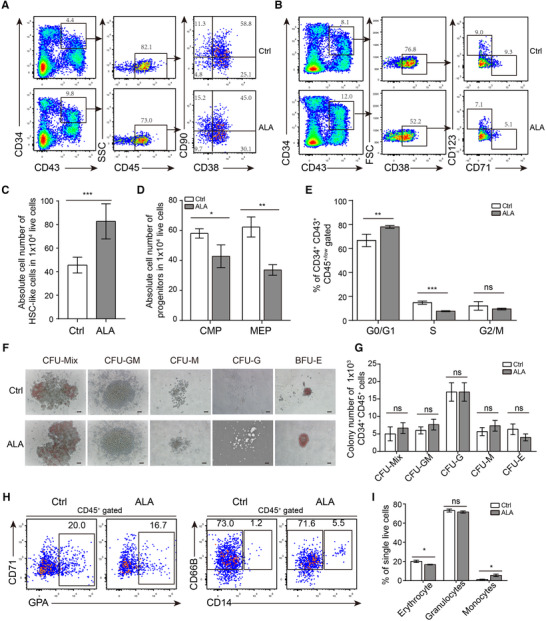
ALA preserves the function of hematopoietic stem/progenitor cells. (A and B) Flow cytometry analysis of CD34+ CD43+ CD45+ CD38− CD90+ hematopoietic stem cell‐like (HSC‐like) cells (A), CD34+ CD43+ CD38+ CD123low CD71− common myeloid progenitors (CMP) like cells, and CD34+ CD43+ CD38+ CD123− CD71+ megakaryocyte–erythroid progenitor (MEP) like cells (B) on day 14 of coculture. (C and D) Absolute numbers of HSC‐like cells, CMPs and MEPs progenitors in 104 live cells were calculated based their respective percentages, as measured by flow cytometry analysis. (E) Flow cytometry analysis was performed to analyze the cell cycle of coculture derived CD34+CD43+CD45+/low hematopoietic stem/progenitor cells at Day14. (F and G) CFU assay of CD34+ CD43+ hematopoietic stem/progenitor cells. (F) Representative images of CFU colonies were presented. (G) The colonies number of CFU‐Mix/GEMM, CFU‐GM, CFU‐G, CFU‐M, and CFU‐E (BFU‐E and CFU‐E) were counted. One thousand CD34+ CD43+ hematopoietic stem/progenitor cells on Day 14 were sorted into each replicate. Absolute colony numbers were counted on day 14. (F), (H) Flow cytometry analysis of CD45+ CD71+/− GPA+ erythrocytes, CD45+ CD66B+ CD14− granulocytes, and CD45+ CD14+ monocytes derived from hematopoietic stem/progenitor cells. (I) Percentages of CD45+ CD71+/‐ GPA+ erythrocytes, CD45+ CD66B+ CD14− granulocytes, and CD45+ CD14+ monocytes were calculated. The data in the bar graphs in (C, D, E, G, and I) are presented as the mean ± sd. An unpaired Student's t‐test (2‐tailed) was performed. N = 3 replicates; * P < 0.05, and ** P < 0.01. ns, not significant
3.3. ALA up‐regulates hematopoiesis‐related transcription factors
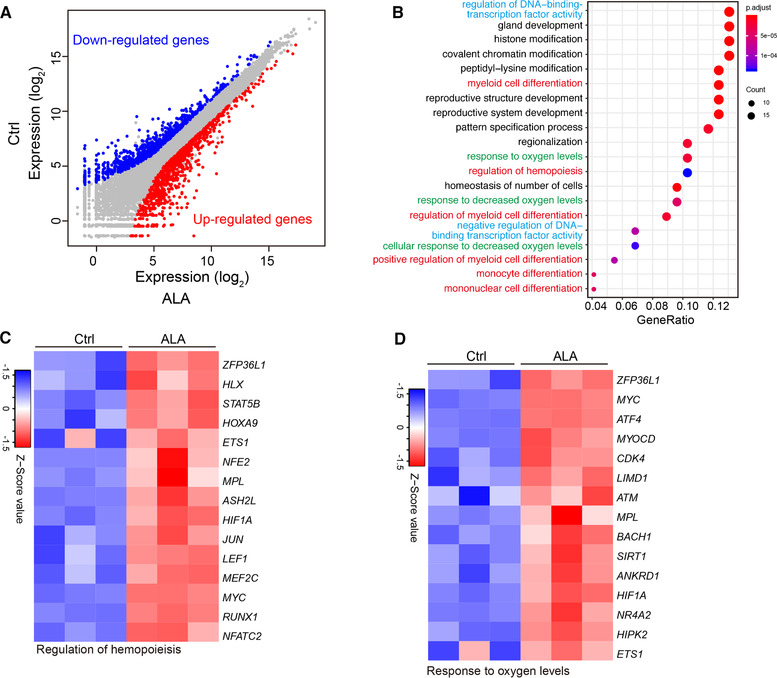
To explore the molecular mechanism responsible for the increase in hematopoietic stem/progenitor cells production by ALA, we sorted hemogenic endothelium cells for RNA‐seq analysis. Given that 1000 cells is enough to cover all transcripts uniformly, 32 we sorted 1000 hemogenic endothelium cells on day 8 to generate RNA‐Seq libraries as described previously 26 , 27 and sequenced the libraries on the NovaSeq platform. Differential expression analysis indicated that 1960 genes were up‐regulated significantly, and 1720 genes were down‐regulated significantly, in the ALA‐treated group compared with the Ctrl group. Differential expression genes were defined as genes with a 1.5‐fold change in expression with an adjusted P‐value < 0.05 (DESeq2 R package) (Fig. 3A). Geno ontology (GO) enrichment analysis 28 (clusterProfiler R package) of total differential expression genes showed that differential expression genes were more likely to be related to catabolic processes, cell adhesion, and biosynthetic processes (Supplementary Fig. S2A). GO enrichment analysis of up‐regulated genes showed that ALA up‐regulated NF‐KB signaling and endothelium and defense response‐related pathways (Supplementary Fig. S2B). We used a lower threshold (a difference in expression of >1.2‐fold; adjusted P‐value < 0.01 [DESeq2 R package]) to analyze changes in transcription factor expression. We identified 145 up‐regulated (Supplementary File S1) and 113 downregulated (Supplementary File S2) differentially expressed transcription factors. GO enrichment analysis of the up‐regulated transcription factors showed that ALA up‐regulated hematopoiesis (marked in red), myeloid development (marked in red), and DNA‐binding transcription factors (marked in blue), as well as oxygen response transcription factors (marked in green) (Fig. 3B). Among the up‐regulated transcription factors, ALA up‐regulated the hematopoiesis‐related genes RUNX1, MYC, NFE2, MPL, JUN, LEF1, MEF2C, HOXA9, STAT5, and ETS1 (Fig. 3C), which were also related to myeloid development (Supplementary Fig. S2C). ALA also up‐regulated oxygen response transcription factors HIF1A, ETS1, NR4A2, MPL, ATM, MYOCD. BACH1, HIPK2, ANKRD1, ZFP36L1, and SIRT1 (Fig. 3D). In addition, GO enrichment analysis of ALA down‐regulated transcription factors showed that ALA mainly down‐regulated transcription factors related to neuron development and development of other non‐hematopoietic tissues (Supplementary Fig. S2D).
FIGURE 3.
ALA up‐regulates hematopoiesis and oxygen level responses related transcription factors. (A) Expression of various genes in hemogenic endothelium cells in normal cocultures against the expression of those in hemogenic endothelium cells in ALA‐treated cocultures on day 8 (horizon axis) (n = 3 replicates), presented as normalized expression values (mean) and plotted as FPKM values; colors (key) indicate genes significantly upregulated (red) or downregulated (blue) in hemogenic endothelium cells by ALA (a difference in expression of <2‐fold; adjusted P‐value < 0.05 (DESeq2 R package)). (B) Gene ontology (GO)–enrichment analysis of the 145 differentially up‐regulated transcription factors using the clusterProfiler R package: each symbol represents a GO term (noted in the plot); color indicates the adjusted P‐value (P‐adj (significance of the GO term); bottom key), and symbol size is proportional to the number of genes (top key). (C and D) Expression of genes encoding regulators of hematopoiesis‐ (C) and oxygen level response‐ (D) related transcription factors identified from the results of GO enrichments analysis. Columns represent the indicated replicates of each population. (E–G) GSEA of hematopoiesis stem cells (E) and targets of NUP98‐HOXA9 fusion genes. (F) Pathways from the dataset of c2.all.v5.2.symbols downloaded from the GSEA website. Heatmaps represent the expression of leading‐edge genes found in the hematopoietic stem cell gene cluster (G) and targets of the NUP98‐HOXA9 fusion gene (H). Columns represent the indicated replicates of each population (C, D, G, and H)
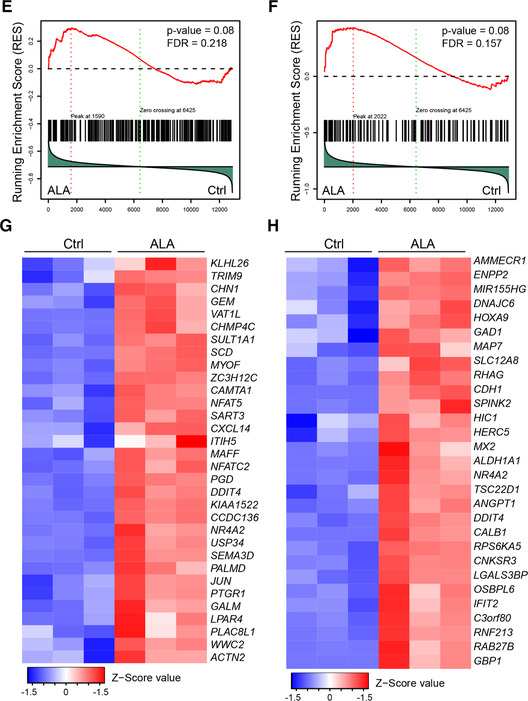
Furthermore, GSEA analysis of the whole transcriptome showed that ALA up‐regulated genes related to HSC self‐renewal and proliferation‐related pathways, including genes found in the HSC gene cluster 33 (Fig. 3E), and targets of NUP98‐HOXA9 fusion 34 (Fig. 3F). Heatmaps of the leading‐edge genes of the HSC cluster genes showed that ALA up‐regulated HSC maintenance‐related gene NR4A2 35 and lymphocyte development‐related genes NFAT5 36 and NFATC2 (NFAT1) 37 (Fig. 3G). While the NUP98‐HOXA9 pathway is more related to myeloid leukemia development, heatmaps of leading‐edge genes of NUPU98‐HOXA9 target genes showed that ALA did not up‐regulate the leukemia development‐related genes MESI1, PBX3, 38 and MLL (KMT2A), 39 but upregulated HOXA9, which plays an important role in the hematopoietic commitment of hESCs 40 and in HSC self‐renewal 41 (Fig. 3H). These results indicated that ALA might promote hematopoiesis.
3.4. ALA promotes the commitment of hematopoietic cells
ALA upregulates hematopoiesis‐ and oxygen response‐related genes and inhibits apoptosis of human umbilical vein endothelial cells induced by high glucose. 42 First, we analyzed the effect of ALA on apoptosis of hemogenic endothelium cells. The results indicated that ALA did not improve the percentage of viable hemogenic endothelium cells (Supplementary Fig. S3A and B). To determine how ALA improves production of hematopoietic stem/progenitor cells, we checked the expression levels of genes that positively regulate endothelial‐to‐hematopoietic transition (EHT) CBFB, RUNX1, 43 , 44 GFI1, GFI1B, 45 GATA2, 22 HIF1A, HIF2A, 46 MEIS2, TAL1, 47 and HOXB4 48 , 49 and those that negatively regulate the transition SOX7, 50 SOX17, NOTCH1, 51 TGFBs (TGFB1, TGFB2, TGFB3), and TGFBRs (TGFBR1, TGFBR2, and TGFBR3)] 52 in ALA‐treated and normal cultures derived from hemogenic endothelium cells. The results indicated that the positive regulating endothelial‐to‐hematopoietic transition genes RUNX1, GFI1, GFI1B, HIFIA, MEIS2, and CBFB were up‐regulated (Fig. 4A). The endothelial‐to‐hematopoietic transition negative regulating genes SOX17, NOTCH1, TGFB1, TGFB2, TGFB3, TGFBR1, and TGFBR2 were down‐regulated (Fig. 4B). These results indicate that ALA might promote the endothelial‐to‐hematopoietic transition by changing the expression of endothelial‐to‐hematopoietic transition related genes. To answer this, we sorted CD34+ CD43− KDR+ hemogenic endothelium cells from cocultures on day 8 (Supplementary Fig. 3C), seeded them onto AGM‐S3 stroma cells, and cultured the cells for 4 days in hematopoietic‐inducing medium with or without ALA. Flow cytometry results showed that ALA promoted transformation of CD34+ CD43− KDR+ hemogenic endothelium cells into CD34+ CD43+ (Supplementary Fig. S3D and E) or CD34+ CD45+ hematopoietic cells compared to Ctrl (82.48 ± 0.87% vs. 78.60 ± 1.47%, P < 0.05) (Fig. 4C and D). Together, these results indicate that ALA promotes endothelial‐to‐hematopoietic transition by up‐regulating endothelial‐to‐hematopoietic transition positive regulating genes and down‐regulating endothelial‐to‐hematopoietic transition negative regulating genes.
FIGURE 4.
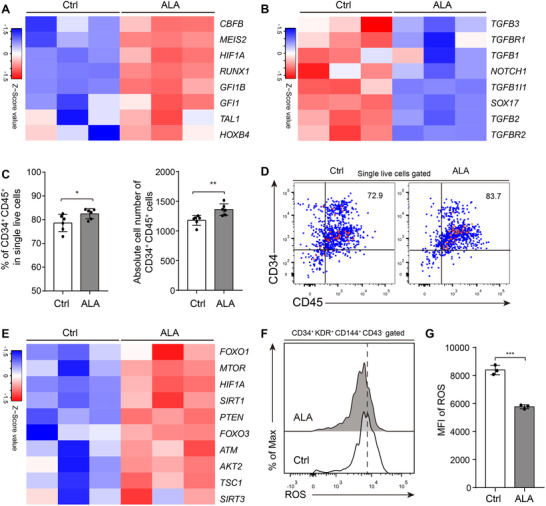
ALA promotes endothelial‐to‐hematopoietic transition progress. Heatmaps show the expression of endothelial‐to‐hematopoietic transition positive regulated genes (A) and endothelial‐to‐hematopoietic transition negative regulated genes (B). (C) CD34+ CD45+ hematopoietic stem/progenitor cells percentages or absolute cell numbers derived from 2500 hemogenic endothelium cells were calculated. Sorted hemogenic endothelium cells were seeded onto AGM‐S3 stroma cells and cultured for 4 days. (D) Representative flow cytometry results of CD34+ CD45+ hematopoietic stem/progenitor cells derived from CD34+ KDR+ CD43− hemogenic endothelium cells. Numbers in quadrants indicate percentages. (E) Heatmaps represent the expression of ROS sensor‐responsive genes. (F) Flow cytometry analysis of ROS levels in CD34+ CD43− KDR+ CD144+ hemogenic endothelium cells from normal or ALA‐treated cocultures on Day 8. (G) Mean fluorescence intensity (MFI) was calculated. Each symbol (C and G) represents an individual replicate; small horizontal lines indicate the mean (±sd). Columns represent the indicated replicates of each population (A, B, and E).The data in the bar graphs in (C), and (G) are presented as the mean ± sd. An unpaired Student's t‐test (2‐tailed) was performed. N = 3 replicates; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001; ns, not significant
In addition, ALA activated the oxidative stress response pathway (Supplementary Fig. S3F), upregulated crucial regulators of ROS and redox sensor molecules that associated with HSC normal function, 11 namely HIF1A, FOXO1, FOXO3, MTOR, ATM, PTEN, TSC1, SIRT1, and AKT2 (Fig. 4E). Flow cytometry analysis of ROS levels showed that the hemogenic endothelium cells cultured with ALA had a significantly lower ROS level (P < 0.001) (Fig. 4F and G). These results indicate that a lower ROS level may increase differentiation of hemogenic endothelium cells into hematopoietic stem/progenitor cells, along with early hematopoiesis signals.
3.5. ALA maintains low ROS levels and inhibits apoptosis of hematopoietic stem/progenitor cells
Because ALA decreased ROS levels in hemogenic endothelium cells, we used DCFH‐DA to measure ROS levels in hematopoietic stem/progenitor cells. Flow cytometry results showed that ALA significantly decreased ROS levels in hematopoietic stem/progenitor cells (P < 0.0001) (Fig. 5A and B). To determine whether ALA affects hematopoietic stem/progenitor cells apoptosis, we performed flow cytometry to analyze apoptosis of CD34+ CD43+ CD45+ hematopoietic stem/progenitor cells on day 14. The results showed that ALA significantly reduced the percentage of early apoptotic cells (4.10 ± 1.21% vs. 19.94 ± 1.92%, P < 0.001) and significantly increased the percentage of live cells (91.43 ± 1.34% vs. 69.30 ± 0.84%, P < 0.001) (Fig. 5C and D). In addition, we used RT‐PCR to check the expression levels of genes related to ROS and apoptosis. First, we performed RT‐PCR to detect ROS‐related genes HIF1A and FOXO3, which were up‐regulated in hemogenic endothelium cells (Fig. 4). The results showed that HIF1A (P < 0.0001) and FOXO3 (P < 0.0001) were significantly up‐regulated in hematopoietic stem/progenitor cells (Fig. 5E). As ROS downstream effect genes, P53, P16, and P21 trigger cell differentiation and senescence in HSCs. 2 RT‐PCR of genes related to apoptosis and autophagy showed that P53 expression increased significantly in ALA‐treated hematopoietic stem/progenitor cells (P < 0.0001), and that P21 (P < 0.01) expression decreased as significantly (Fig. 5E). Cell autophagy‐related genes ATG5 (P < 0.0001) and ATG7 (P < 0.0001) were down‐regulated significantly in ALA‐treated HSCs (Fig. 5F). In addition, the cell apoptosis‐related genes CASP6 (P < 0.0001) and BCL2 (P < 0.0001) were also down‐regulated significantly (Fig. 5E).
FIGURE 5.
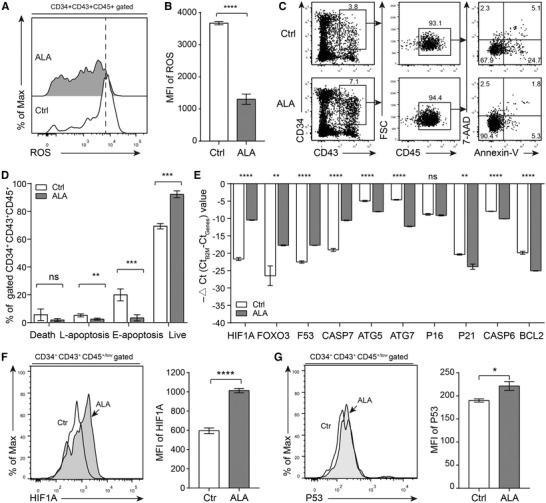
ALA maintains a low ROS level and inhibits apoptosis of hematopoietic stem/progenitor cells. (A) Representative flow cytometry results of ROS levels in hematopoietic stem/progenitor cells from normal or ALA‐treated cocultures on day 14. (B) Mean fluorescence intensity (MFI) of ROS (DCFH‐DA) was calculated. (C) Representative flow cytometry results of apoptosis of hematopoietic stem/progenitor cells from normal and ALA‐treated cocultures on day 14. (D) The percentages of live cells, early apoptosis (E‐apoptosis), late apoptosis (L‐apoptosis), and cell death were calculated. (E) RT‐PCR indicated the mRNA encoding relative expression levels of stem cell maintenance or cell apoptosis related genes in hematopoietic stem/progenitor cells, the data were presented as –△Ct (Ct B2M – Ct Genes) value. (F and G) Intracellular staining assay of was performed to analysis the protein expression level of HIF1A (F) and P53 (G) in CD34+CD43+CD45+/low cells at day14 of coculture. Data in the bar graphs in (B) and (D–F) are presented as the mean ± sd. An unpaired Student's t‐test (2‐tailed) was performed. N = 3 replicates; * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001; ns, not significant
HIF1A and P53 play an important role in the quiescence of hematopoietic stem/progenitor cells 11 , 53 and also play an important role in ROS induced cell apoptosis. 2 To further confirm the pathways that ALA affected hematopoietic stem/progenitor cells apoptosis, we used intracellular staining to assay the protein expression level of HIF1A and P53. The results showed that ALA significantly up‐regulated the protein expression of both HIF1A (Fig. 5F) and P53 (Fig. 5G). All these results suggested that ALA inhibited hematopoietic stem/progenitor cells apoptosis by HIF1A‐ROS‐P53 signals. In short, the decreasing of ROS up‐regulated HIF1A and P53 to inhibit cell cycle and cell apoptosis.
3.6. ALA maintains HSC function partially in ex vivo culture
To further analysis the effect of ALA exerted on hematopoietic stem/progenitor cells function in vivo, we tried to transplant CD34+CD43+CD45+/low hematopoietic stem/progenitor cells into the NSG mice. The results indicated that there were scarcely human blood cells were detected in the peripheral blood of NSG mice (data do not show). Given that co‐culture derived hematopoietic stem/progenitor cells were difficult to be transplanted successfully in the NSG mice. Thus, we analyzed the cell cycle and ROS level of cord blood derived CD34+CD38− hematopoietic stem/progenitor cells. The results indicated that ALA significantly inhibited the cell cycle progress (Supplementary Fig. S4A and B) and reduced ROS level (Supplementary Fig. S4C and D) of CD34+CD38− hematopoietic stem/progenitor cells at Day2 of in vitro culture. ALA inhibited the cell cycle and ROS level both in cord blood derived hematopoietic stem/progenitor cells and hESCs derived hematopoietic stem/progenitor cells, indicating that cord blood might be used to assay the ability of hematopoietic stem/progenitor cells to rebuild hematopoiesis in vivo.
Sequentially, we used CFU assay and in vivo transplantation system to assay the effect of ALA exerting on HSCs (Fig. 6A). We isolated the nucleated cells from cord blood and then cultured for 7 days using SFEMII culture medium with ALA treatment or not. CFU assay was performed to reveal the differentiation potential of Ctrl or ALA‐treated cultures. The results showed that ALA‐treated cultures produced significantly more CFU‐mix (P < 0.01), CFU‐E (P < 0.05), and CFU‐M (P < 0.05) colonies (Fig. 6B). At the same time, the components of hematopoietic stem or progenitor cells were analyzed by flow cytometry. The results indicated that higher proportion or absolute cell numbers of CD34+ CD38− CD90+ CD45RA− long‐term HSC (LT‐HSCs), CD34+ CD38− CD90− CD45RA− short‐term HSC (ST‐HSC), CD34+ CD38− CD90− CD45RA+ committed progenitors (C‐progenitors), and CD34+ CD38+ progenitors were observed in ALA‐treated cultures (P < 0.0001) (Fig. 6C and D). Sequentially, we transplanted Ctrl or ALA‐treated cultures derived from 1 × 104 CD34+ cells into irradiated B‐NDG mice. We also transplanted fresh cord blood nucleated cells that containing 1 × 104 CD34+ cells as positive control. We conducted flow cytometry to analysis the chimerism of human blood cells in the recipient mice peripheral blood at 3 months post transplantation. The results indicated that ALA‐treated cultures contributed significantly higher percentages of chimerism in mice peripheral blood compared to Ctrl cultures (1.96 ± 0.91% vs. 0.13 ± 0.12%, P < 0.05) (Fig. 6E and F). It is worth noting that more than 1% of human blood cells were detected in the recipient mice (3/4) of ALA‐treated cultures, while none of the recipient mice contribute more that 1% of human derived blood cells in the group of Ctrl cultures (Fig. 6E and F). Although there is no significant difference in transplantation efficiency between ALA‐cultured cells and fresh cord blood cells (P = 0.11) (Fig. 6F). However, based on the effects that ALA affects the cell proliferation and ROS level of hematopoietic stem and progenitor cells (Supplementary Fig. S4). All these results suggested that ALA can maintains HSCs function partially through inhibiting cell cycle and decreasing ROS level in ex vivo culture.
FIGURE 6.
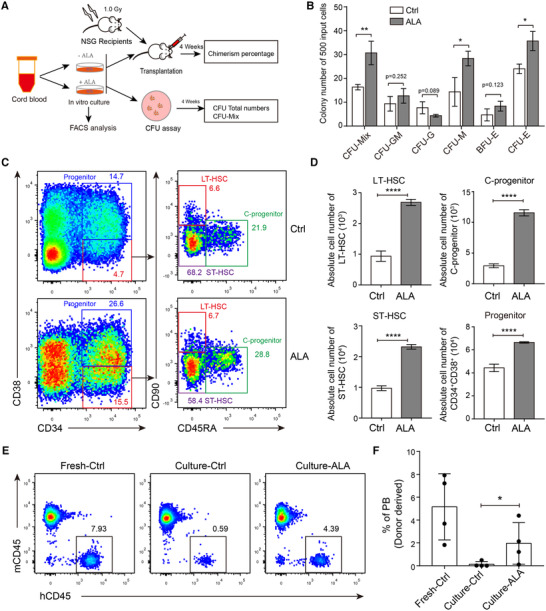
ALA maintained functional HSC in ex vivo culture. (A) Strategy of analysis the effect of ALA exerted on HSC. (B) CFU assay of Ctrl or ALA‐treated cultures with 7 days ex vivo culture. Total nucleated cells of cord blood were used as input cells for ex vivo culture. Five hundred total cells derived from Ctrl or ALA‐treated cultures were used for CFU assay, and absolute colony numbers were counted on day 14. (C) Representative flow cytometry plots of CD34+ CD38− CD90+ CD45RA− LT‐HSCs, CD34+ CD38− CD90− CD45RA− ST‐HSC, CD34+ CD38− CD90− CD45RA+ committed progenitors (C‐progenitors), and CD34+ CD38+ progenitors at day 7 for Ctrl or ALA‐treated cultures. (D) Absolute cell numbers of LT‐HSCs, ST‐HSC, C‐progenitors and CD34+CD38+ progenitors were calculated derived from 1 million total cord blood nucleated cells. (E) Representative flow cytometry plots of human blood cells chimerism at 3 months post‐transplantation. Fresh cord blood nucleated cells, Ctrl cultures, and ALA‐treated cultures were transplanted into 1.0 Gy irradiated recipients. (F) Statistic analysis the percentages of donor derived cells at 3 months post‐transplantation. Each point indicates an individual recipient. The data in the bar graphs in (B, D, and F) are presented as the mean ± sd. An unpaired Student's t‐test (2‐tailed) was performed. N = 3 replicates; * P < 0.05, ** P < 0.01, **** P < 0.0001
4. DISCUSSION
In this study, we used an in vitro coculture system to provide a new insight into the effects of ALA on hPSCs hematopoiesis. We also used cord blood derived hematopoietic stem/progenitor cells to investigate the ex vivo maintenance effect of ALA by transplantation experiments. First, ALA increased production of hPSCs derived early hemogenic endothelium cells and late hematopoietic stem/progenitor cells both in hESCs/AGM‐S3 and hESCs/OP9 coculture systems, indicating that ALA affects hematopoietic development and differentiation irrespective of cell type. Second, ALA helped to maintain cord blood derived hematopoietic stem/progenitor cells in culture.
At early coculture stages, a high proportion of hemogenic endothelium cells with a low level of ROS was observed in ALA‐treated cocultures, suggesting that ALA may promote development of hemogenic endothelium cells by modulating the ROS level. Our findings are consistent with those of a previous report showing that modulating ROS signaling increases the efficacy of CD34+ cell production from hPSCs, 13 although this study did not confirm hematopoietic activity in CD34+ fractions. Transcriptome analysis of hemogenic endothelium cells indicated that ALA up‐regulates genes promoting hematopoietic commitment, such as RUNX1, GFI1, GFI1B, MEIS2, and HOXA9, 43 , 45 , 47 and down‐regulates signals in TGFβ pathways that negatively regulate endothelial‐to‐hematopoietic transition. 52 Interestingly, addition of ALA up‐regulated the ROS sensor genes 11 HIF1A, FOXO1, FOXO3, ATM, PTEN, SIRT1, and AKT2 during early hematopoiesis. Among ALA up‐regulated transcription factors, HIF1A, RUNX1, GFI1, GFI1B, and HOXA9 are crucial factors involved in modulating hemogenic endothelial cells development. As reported previously, HIF1A acts as an upstream regulator of RUNX1 and NOTCH signaling, 46 and GFI1 and GFI1B act as downstream targets of RUNX1. 45 Our findings showed that ALA might promote HEC development by affecting HIF1A‐RUNX1 signaling.
In the late stage cocultures, the number of hPSCs derived CD34+ CD43+ CD45+ hematopoietic stem/progenitor cells increased, and these hematopoietic stem/progenitor cells contained a high percentage of CD34+ CD43+ CD45+ CD90+ CD38− HSC‐like cells 22 and a low percentage of differentiated progenitor cells (CMPs and EMPs) (Fig. 2). A comparison of hematopoietic activity using the semisolid colony assay showed that there was no significant difference in cell production between ALA‐treated and nontreated hematopoietic stem/progenitor cells. Our results indicate that addition of ALA to cocultures efficiently promotes hematopoiesis along with an early wave of endothelial‐to‐hematopoietic transition, while inhibiting further differentiation of hematopoietic stem/progenitor cells by inhibiting cell cycle. In addition, lower levels of ROS and apoptosis were detected in ALA‐treated hematopoietic stem/progenitor cells, which confirms reports that ALA protects HSCs in bone marrow from total body irradiation stress 25 and rescues the dysfunction of HSCs caused by ROS. 9 Furthermore, we found that ALA significantly up‐regulated expression of ROS‐regulating genes HIF1A, FOXO3, and P53, and the genes related to apoptosis or autophagy, including ATG5, ATG7, P21, and BCL2. ALA also significantly up‐regulated the protein expression of both HIF1A (Fig. 5F) and P53 (Fig. 5G). All these results suggested that ALA inhibited hematopoietic stem/progenitor cells apoptosis by HIF1A‐ROS‐P53 signals. In short, ALA decreased of ROS to up‐regulated HIF1A and up‐regulated P53 to inhibit cell cycle and cell apoptosis. 2
Throughout the current study, we showed the role of ALA of affecting hematopoietic development or function by regulating ROS. The effect of ALA exerted on hematopoietic differentiation of human embryonic stem cells in three stages. In the first stage, ALA promoted endothelial‐to‐hematopoietic transition and development of hemogenic endothelium cells from undifferentiated hPSCs by modulating ROS levels, as reported previously for CD34+ cell differentiation from hPSCs. 13 In the second stage, ALA enhanced the commitment of hematopoietic stem/progenitor cells from hemogenic endothelium cells by modulating ROS levels via coordination multiple signaling factors such as HIF1A and RUNX1. This observation is reminiscent of a previous report showing that modulation of ROS signaling affects development of primitive hematopoietic progenitors from hPSCs. 14 In the third stage, ALA inhibited the rapid differentiation of hPSCs derived hematopoietic stem/progenitor cells in the coculture system. In addition, ALA regulated both ROS and apoptosis‐related pathways to inhibit the apoptosis of hPSCs derived mature hematopoietic stem/progenitor cells at comparatively late stages. Thus, ALA serves as a potent promoter of hematopoiesis at the early endothelial‐to‐hematopoietic transition stage, ALA regulated both ROS and apoptosis related pathways to inhibit apoptosis of hPSCs derived mature hematopoietic stem/progenitor cells at late stages. Particularly, ALA treatment allows observation of the mechanism by which ROS signals control early events during developmental hematopoiesis from hPSCs, which can never be mimicked using adult‐type HSCs.
ALA also inhibited cell cycle and ROS of cord blood derived hematopoietic stem/progenitor cells that observed in embryonic stem cells derived hematopoietic stem/progenitor cells. Thus, ALA treatment might have the potential use in promoting in vitro expansion and maintenance of adult HSCs for clinical transplantation. Subsequently, both ex vivo and in vivo experiments were performed to assay the effect of ALA exerted on human cord blood HSCs. These results confirmed that ALA efficiently maintained functional HSCs in ex vivo culture. In summary, ALA could be added to hematopoietic differentiation systems to study development of HSCs from hPSCs.
AUTHORSHIP
Y.D. and J.B. performed the core experiments and data analysis. Y.M.Z., Y.Z., X.L., X.P., Y.C., J.F., F.X., Q.Z., M.L., G.B., B.M., B.C., and Y.G.Z. performed some of the experiments. Y.D. and F.M. designed the project, discussed the data, and wrote the manuscript. F.M. approved the manuscript. Y.D. and J.B. contributed equally to this study.
DISCLOSURE
The authors declare no conflict of interest.
Supporting information
Supporting Information.
Supporting Information.
Supporting Information.
ACKNOWLEDGMENTS
This research was supported by the China Postdoctoral Fund Program (Grand 2018M641265) and the CAMS Initiatives for Innovative Medicine (2016‐I2M‐1‐018, 2019‐I2M‐1‐006, 2017‐I2M‐2005).
Dong Y, Bai J, Zhang Y, et al. Alpha lipoic acid promotes development of hematopoietic progenitors derived from human embryonic stem cells by antagonizing ROS signals. J Leukoc Biol. 2020;108:1711–1725. 10.1002/JLB.1A0520-179R
Contributor Information
Yonggang Zhang, Email: yonggangzhang@ibt.pumc.edu.cn.
Feng Ma, Email: mafeng@ibt.pumc.edu.cn.
REFERENCES
- 1. Wielockx B, Grinenko T, Mirtschink P, et al. Hypoxia pathway proteins in normal and malignant hematopoiesis. Cells. 2019;8:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shao L, Li H, Pazhanisamy SK, et al. Reactive oxygen species and hematopoietic stem cell senescence. Int J Hematol. 2011;94:24‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rehn M, Olsson A, Reckzeh K, et al. Hypoxic induction of vascular endothelial growth factor regulates murine hematopoietic stem cell function in the low‐oxygenic niche. Blood. 2011;118(6):1534‐1543. [DOI] [PubMed] [Google Scholar]
- 4. Tan DQ, Suda T. Reactive oxygen species and mitochondrial homeostasis as regulators of stem cell fate and function. Antioxid Redox Signal. 2018;29:149‐168. [DOI] [PubMed] [Google Scholar]
- 5. Kobayashi H, Morikawa T, Okinaga A, et al. Environmental optimization enables maintenance of quiescent hematopoietic stem cells ex vivo. Cell Rep. 2019;28:145‐158 e9. [DOI] [PubMed] [Google Scholar]
- 6. Bai T, Li J, Sinclair A, et al. Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel. Nat Med. 2019;25:1566‐1575. [DOI] [PubMed] [Google Scholar]
- 7. Xie SZ, Garcia‐Prat L, Voisin V, et al. Sphingolipid modulation activates proteostasis programs to govern human hematopoietic stem cell self‐renewal. Cell Stem Cell. 2019;25:639‐653.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu L, Zhang Y, Miao W, et al. Reactive oxygen species and Nrf2: functional and transcriptional regulators of hematopoiesis. Oxid Med Cell Longev. 2019;2019:5153268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le Q, Yao W, Chen Y, et al. GRK6 regulates ROS response and maintains hematopoietic stem cell self‐renewal. Cell Death Dis. 2016;7:e2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu L, Cheng H, Gao Y, et al. Antioxidant N‐acetyl‐L‐cysteine increases engraftment of human hematopoietic stem cells in immune‐deficient mice. Blood. 2014;124:e45‐e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bigarella CL, Liang R, Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206‐4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Olivier EN, Marenah L, McCahill A, et al. High‐efficiency serum‐free feeder‐free erythroid differentiation of human pluripotent stem cells using small molecules. Stem Cells Transl Med. 2016;5:1394‐1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song SH, Kim K, Park JJ, et al. Reactive oxygen species regulate the quiescence of CD34‐positive cells derived from human embryonic stem cells. Cardiovasc Res. 2014;103:147‐155. [DOI] [PubMed] [Google Scholar]
- 14. Nakata S, Matsumura I, Tanaka H, et al. NF‐kappaB family proteins participate in multiple steps of hematopoiesis through elimination of reactive oxygen species. J Biol Chem. 2004;279:55578‐55586. [DOI] [PubMed] [Google Scholar]
- 15. Li Y, Hermanson DL, Moriarity BS, et al. Human iPSC‐derived natural killer cells engineered with chimeric antigen receptors enhance anti‐tumor activity. Cell Stem Cell. 2018;23:181‐192 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Montel‐Hagen A, Seet CS, Li S, et al. Organoid‐induced differentiation of conventional T cells from human pluripotent stem cells. Cell Stem Cell. 2019;24:376‐389 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma F, Ebihara Y, Umeda K, et al. Generation of functional erythrocytes from human embryonic stem cell‐derived definitive hematopoiesis. Proc Natl Acad Sci USA. 2008;105:13087‐13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trump LR, Nayak RC, Singh AK, et al. Neutrophils derived from genetically modified human induced pluripotent stem cells circulate and phagocytose bacteria in vivo. Stem Cells Transl Med. 2019;8:557‐567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yokoyama Y, Suzuki T, Sakata‐Yanagimoto M, et al. Derivation of functional mature neutrophils from human embryonic stem cells. Blood. 2009;113:6584‐6592. [DOI] [PubMed] [Google Scholar]
- 20. Zhang H, Reilly MP. Human induced pluripotent stem cell‐derived macrophages for unraveling human macrophage biology. Arterioscler Thromb Vasc Biol. 2017;37:2000‐2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mao B, Huang S, Lu X, et al. Early development of definitive erythroblasts from human pluripotent stem cells defined by expression of glycophorin A/CD235a, CD34, and CD36. Stem Cell Reports. 2016;7:869‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Y, Zhang Y, Chen B, et al. Overexpression of GATA2 enhances development and maintenance of human embryonic stem cell‐derived hematopoietic stem cell‐like progenitors. Stem Cell Reports. 2019;13:31‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Deng H, Liu L, et al. alpha‐Lipoic acid protects against hypoxia/reoxygenation‐induced injury in human umbilical vein endothelial cells through suppression of apoptosis and autophagy. Mol Med Rep. 2015;12:180‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simbula G, Columbano A, Ledda‐Columbano GM, et al. Increased ROS generation and p53 activation in alpha‐lipoic acid‐induced apoptosis of hepatoma cells. Apoptosis. 2007;12:113‐123. [DOI] [PubMed] [Google Scholar]
- 25. Wambi C, Sanzari J, Wan XS, et al. Dietary antioxidants protect hematopoietic cells and improve animal survival after total‐body irradiation. Radiat Res. 2008;169:384‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang M, Dong Y, Hu F, et al. Transcription factor Hoxb5 reprograms B cells into functional T lymphocytes. Nat Immunol. 2018;19:279‐290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dong Y, Xia C, Weng Q, et al. Synergy of NUP98‐HOXA10 fusion gene and NrasG12D mutation preserves the stemness of hematopoietic stem cells on culture condition. Cells. 2019;8:951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramakrishnan N, Wolfe WW, Catravas GN. Radioprotection of hematopoietic tissues in mice by lipoic acid. Radiat Res. 1992;130:360‐365. [PubMed] [Google Scholar]
- 31. Mori Y, Chen JY, Pluvinage JV, et al. Prospective isolation of human erythroid lineage‐committed progenitors. Proc Natl Acad Sci USA. 2015;112:9638‐9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pan X, Durrett RE, Zhu H, et al. Two methods for full‐length RNA sequencing for low quantities of cells and single cells. Proc Natl Acad Sci USA. 2013;110:594‐599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ivanova NB, Dimos JT, Schaniel C, et al. A stem cell molecular signature. Science. 2002;298:601‐604. [DOI] [PubMed] [Google Scholar]
- 34. Takeda A, Goolsby C, Yaseen NR. NUP98‐HOXA9 induces long‐term proliferation and blocks differentiation of primary human CD34+ hematopoietic cells. Cancer Res. 2006;66:6628‐6637. [DOI] [PubMed] [Google Scholar]
- 35. Hou Y, Li W, Sheng Y, et al. The transcription factor Foxm1 is essential for the quiescence and maintenance of hematopoietic stem cells. Nat Immunol. 2015;16:810‐818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berga‐Bolanos R, Alberdi M, Buxade M, et al. NFAT5 induction by the pre‐T‐cell receptor serves as a selective survival signal in T‐lymphocyte development. Proc Natl Acad Sci USA. 2013;110:16091‐16096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kiani A, Habermann I, Haase M, et al. Expression and regulation of NFAT (nuclear factors of activated T cells) in human CD34+ cells: down‐regulation upon myeloid differentiation. J Leukoc Biol. 2004;76:1057‐1065. [DOI] [PubMed] [Google Scholar]
- 38. Garcia‐Cuellar MP, Steger J, Fuller E, et al. Pbx3 and Meis1 cooperate through multiple mechanisms to support Hox‐induced murine leukemia. Haematologica 2015;100:905‐913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shima Y, Yumoto M, Katsumoto T, et al. MLL is essential for NUP98‐HOXA9‐induced leukemia. Leukemia. 2017;31:2200‐2210. [DOI] [PubMed] [Google Scholar]
- 40. Ramos‐Mejia V, Navarro‐Montero O, Ayllon V, et al. HOXA9 promotes hematopoietic commitment of human embryonic stem cells. Blood. 2014;124:3065‐3075. [DOI] [PubMed] [Google Scholar]
- 41. Lawrence HJ, Christensen J, Fong S, et al. Loss of expression of the Hoxa‐9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988‐3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meng X, Li ZM, Zhou YJ, et al. Effect of the antioxidant alpha‐lipoic acid on apoptosis in human umbilical vein endothelial cells induced by high glucose. Clin Exp Med. 2008;8:43‐49. [DOI] [PubMed] [Google Scholar]
- 43. Yzaguirre AD, Howell ED, Li Y, et al. Runx1 is sufficient for blood cell formation from non‐hemogenic endothelial cells in vivo only during early embryogenesis. Development. 2018;145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lie ALM, Marinopoulou E, Lilly AJ, et al. Regulation of RUNX1 dosage is crucial for efficient blood formation from hemogenic endothelium. Development. 2018;145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lancrin C, Mazan M, Stefanska M, et al. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 2012;120:314‐322. [DOI] [PubMed] [Google Scholar]
- 46. Gerri C, Marass M, Rossi A, et al. Hif‐1alpha and Hif‐2alpha regulate hemogenic endothelium and hematopoietic stem cell formation in zebrafish. Blood. 2018;131:963‐973. [DOI] [PubMed] [Google Scholar]
- 47. Wang M, Wang H, Wen Y, et al. MEIS2 regulates endothelial to hematopoietic transition of human embryonic stem cells by targeting TAL1. Stem Cell Res Ther. 2018;9:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teichweyde N, Horn PA, Klump H. HOXB4 increases runx1 expression to promote the de novo formation of multipotent hematopoietic cells. Transfus Med Hemother. 2017;44:128‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Teichweyde N, Kasperidus L, Carotta S, et al. HOXB4 promotes hemogenic endothelium formation without perturbing endothelial cell development. Stem Cell Reports. 2018;10:875‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lilly AJ, Costa G, Largeot A, et al. Interplay between SOX7 and RUNX1 regulates hemogenic endothelial fate in the yolk sac. Development. 2016;143:4341‐4351. [DOI] [PubMed] [Google Scholar]
- 51. Lizama CO, Hawkins JS, Schmitt CE, et al. Repression of arterial genes in hemogenic endothelium is sufficient for haematopoietic fate acquisition. Nat Commun. 2015;6:7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vargel O, Zhang Y, Kosim K, et al. Activation of the TGFbeta pathway impairs endothelial to haematopoietic transition. Sci Rep. 2016;6:21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Takubo K, Goda N, Yamada W, et al. Regulation of the HIF‐1α level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391‐402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.
Supporting Information.
Supporting Information.
Data Availability Statement
Data supporting the findings of this study are available from the corresponding author upon request. All RNA‐Seq data can be found in the GEO database under accession code GSE144307.


