Abstract
The central vein sign (CVS) is an efficient imaging biomarker for multiple sclerosis (MS) diagnosis, but its application in clinical routine is limited by inter-rater variability and the expenditure of time associated with manual assessment. We describe a deep-learning-based prototype for automated assessment of the CVS in white matter MS lesions using data from three different imaging centers.
We retrospectively analyzed data from 3T magnetic resonance images acquired on four scanners from two different vendors, including adults with MS (n=42), MS mimics (n=33, encompassing 12 distinct neurological diseases mimicking MS), and uncertain diagnosis (n=5). Brain white matter lesions were manually segmented on FLAIR* images. Perivenular assessment was performed according to consensus guidelines and used as ground truth, yielding 539 CVS-positive (CVS+) and 448 CVS-negative (CVS-) lesions. A 3D convolutional neural network (“CVSnet”) was designed and trained on 47 datasets, keeping 33 for testing. FLAIR* lesion patches of CVS+/CVS- lesions were used for training and validation (n=375/298) and for testing (n=164/150). Performance was evaluated lesion-wise and subject-wise and compared with a state-of-the-art vesselness filtering approach through McNemar’s test.
The proposed CVSnet approached human performance, with lesion-wise median balanced accuracy of 81%, and subject-wise balanced accuracy of 89% on the validation set, and 91% on the test set. The process of CVS assessment, in previously manually segmented lesions, was approximately 600-fold faster using the proposed CVSnet as compared to human visual assessment (test set: four seconds versus forty minutes). On the validation and test sets, the lesion-wise performance outperformed the vesselness filter method (P<0.001).
The proposed deep learning prototype shows promising performance in differentiating MS from its mimics. Our approach was evaluated using data from different hospitals, enabling larger multicenter trials to evaluate the benefit of introducing the CVS marker into MS diagnostic criteria.
Keywords: Multiple Sclerosis, MS mimics, Central vein sign, Deep learning
Graphical Abstract
The presence of a vein at the center of brain white matter (WM) lesions, the central vein sign (CVS), is a novel imaging biomarker able to differentiate multiple sclerosis (MS) from other diseases mimicking MS.
We describe and validate a new deep-learning-based prototype for automated assessment of the CVS in WM lesions using data from three different imaging centers.
The primary advantage of the proposed method (“CVSnet”) when compared to the manual assessment is the high speed and accuracy in differentiating MS from its mimics.
1. Introduction
Current magnetic resonance imaging (MRI) diagnostic criteria for multiple sclerosis (MS) are based on the dissemination in time and space of characteristic imaging features of the disease. Although highly useful, the specificity of these criteria is limited, and the risk of misdiagnoses based on MRI is still substantial.1,2 Consequently, identification of more specific imaging biomarkers for MS is a high-priority area of research.3
The presence of a vein at the center of white matter (WM) lesions, the “central vein sign” (CVS), has been shown to be an efficient imaging biomarker for MS, able to distinguish MS from other conditions exhibiting similar WM abnormalities on MRI.4 In MS, the immunological reaction leading to the formation of inflammatory WM lesions occurs in the perivascular cuff around small parenchymal veins.5–7 Indeed, pathology studies as early as the 19th century described the presence of a vein at the center of inflammatory lesions as a hallmark of the MS plaque.8 Today, the vein at the center of WM lesions can be imaged in vivo using optimized susceptibility-based MRI sequences,9,10 and several studies have shown that the CVS can accurately distinguish MS from other disorders showing similar T2 hyperintensities on MRI.11–15 Results from a recent multicenter study suggest that the diagnostic performance of this CVS biomarker at clinical field strengths is highly dependent on the type of susceptibility-based MRI sequence used.16 In this context, recent data from our group using an optimized three-dimensional (3D) T2*-weighted segmented echo-planar imaging (T2*-EPI) sequence at clinical field strengths,9 have shown that the CVS can efficiently discriminate between MS and inflammatory vasculopathies of the CNS and that this imaging biomarker can accurately predict an MS diagnosis in patients harboring atypical features for the disease.11,17
However, despite the availability of optimized 3D MRI sequences for CVS detection, there remain substantial technical limitations to applying this imaging biomarker in daily clinical practice. These limitations are primarily related to the manual, operator-dependent assessment required to evaluate the CVS in individual scans. Although previously proposed simplified algorithms considerably reduce the time burden associated with CVS evaluation,18,19 manual assessment generally suffers from inter-rater and intra-rater variability, is time-consuming, and limits the generalization of results across different centers and healthcare systems. In order to promote the translation of the CVS imaging biomarker into clinical routine, a reliable and reproducible method to automatically detect the CVS is needed.
Recently, Dworkin et al. proposed an automated method to detect the CVS in WM lesions, based on a vesselness filter combined with a centrality measure.20 The study showed good subject-wise accuracy, but was limited to single-center data collection.11,12 In the present work, we show results on a broad panel of twelve different MS-mimicking diseases, collected on four different MRI scanners. CVSnet, our approach to automatically assess the CVS, is based on a 3D convolutional neural network architecture.21,22 Because it uses deep learning, it is an end-to-end learning approach that does not require handcrafted discriminative features or filters. Instead, it learns the most appropriate representations of the data by minimizing the CVS positive/negative classification error and has the ability to generalize the data. The training of the network performed using data from four different scanners (two different vendors) and three different imaging centers ensures robustness of the algorithm with respect to contrast and hardware-related differences. To evaluate the potential of our approach excluding the influence of segmentation errors, we use manually segmented lesions for validation.
2. Patients and methods
2.1. Patients
This is a retrospective analysis of images from 80 patients recruited between September 2016 and November 2018 at the University Hospital of Lausanne, Switzerland (n=30), at the Erasmus University Hospital, Brussels, Belgium (n=36), and at the National Institutes of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, Maryland, USA (n=14). In all centers, studies were designed to assess the diagnostic utility of the CVS for differential diagnosis of MS. Included patients had either definite MS according to the 2017 McDonald criteria3 (n=42) or a variety of diseases giving white matter abnormalities similar to those observed in MS (hereafter termed “MS mimics”; n=33). Five patients had uncertain diagnosis at the time of this study (“uncertain diagnosis”). Included patients with MS-mimic diseases encompassed systemic lupus erythematosus (SLE; n=7), Sjögren syndrome (Sjögren; n=4), primary angiitis of the central nervous system (PACNS; n=2), antiphospholipid syndrome (APS; n=1), inflammatory amyloid angiopathy (iAA; n=1), Behçet disease (n=2), sarcoidosis (n=1), neuromyelitis optica spectrum disorder (NMOSD; n=6), Susac syndrome (n=2), HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP; n=1), migraine (n=5), and cerebral small vessel disease (n=1).
2.2. MRI acquisition, postprocessing, and manual CVS assessment
All patients underwent a single brain MRI acquisition on a 3T Magnetom Skyra or Prisma scanner (Siemens Healthcare, Erlangen, Germany) in Lausanne, a 3T Ingenia scanner (Philips, Best, The Netherlands) in Brussels, or a 3T Skyra scanner (Siemens Healthcare, Erlangen, Germany) in Bethesda. In all centers, 3D T2*-EPI and 3D T2-weighted fluid-attenuated inversion recovery (FLAIR) images were acquired during and after intravenous injection of a single dose (0.1mmol/kg) of gadolinium-based contrast material, as previously described.11 FLAIR images were obtained with 1-mm isotropic resolution in all centers, whereas 3D T2*-EPI images were obtained with 0.55-mm isotropic resolution in Brussels and 0.65-mm isotropic resolution in Lausanne and Bethesda (Table 1). DICOM images were processed and analyzed using Medical Image Processing, Analysis, and Visualization (MIPAV; NIH; http://mipav.cit.nih.gov). After bias-field correction and co-registration, FLAIR* images were generated by voxel-wise multiplication of the FLAIR and the T2*-EPI images, as previously described.10 For each patient, WM lesions were manually segmented on FLAIR* images following the North American Imaging in Multiple Sclerosis (NAIMS) Cooperative consensus on lesion inclusion/exclusion criteria for central vein analysis.4,11 Two investigators (P.M., M.A.) independently assessed the percentage of CVS+ lesions for inter-rater reliability, blinded to clinical diagnosis. Disagreements were adjudicated by consensus between the two investigators (see examples in Figure 1). Manual segmentation and classification of CVS+ and CVS- lesions were used as ground truth. Patients were dichotomized into MS vs. MS mimics based on three previously proposed criteria: i) the “50% rule,”11 i.e. a 50% CVS+ lesion cut-off to distinguish MS from MS mimics, ii) the “6-lesion rule”18 and iii) the “3-lesion rule,”19 whereby 10 lesions or 3 lesions are randomly selected and MS is diagnosed if at least 6 or 2 lesions are CVS+.
Table 1.
MRI sequence parameters used in the Erasmus University Hospital, Brussels (Philips scanner), in the Lausanne University Hospital, Lausanne (Siemens scanner) and in the National Institutes of Health, Bethesda, USA (Siemens scanner), imaging centers.
| Sequence | 3D T2*-EPI | 3D T2-FLAIR | ||
|---|---|---|---|---|
| Field strength | 3T | 3T | 3T | 3T |
| Manufacturer | Siemens | Philips | Siemens | Philips |
| Site | Lausanne/Bethesda | Brussels | Lausanne/Bethesda | Brussels |
| Receive channels | 64/32 | 16 | 64/32 | 16 |
| Imaging plane | Sagittal | Sagittal | Sagittal | Sagittal |
| Imaging resolution (mm) | 0.65 | 0.55 | 1 | 1 |
| # slices | 288/256 | 336 | 176 | 180 |
| Repetition time (TR, ms) | 64 | 53 | 5000/4800 | 4800 |
| Echo time (TE, ms) | 35 | 29 | 391/353 | 373 |
| Inversion time (TI, ms) | - | - | 1800 | 1600 |
| Flip angle (deg) | 10 | 10 | Variable/120 | 90 |
| Averages | 1 | 1 | 1 | 1 |
| Acquisition time (min:sec) | 6:20/5:44 | 4:40 | 4:47/6:53 | 6:00 |
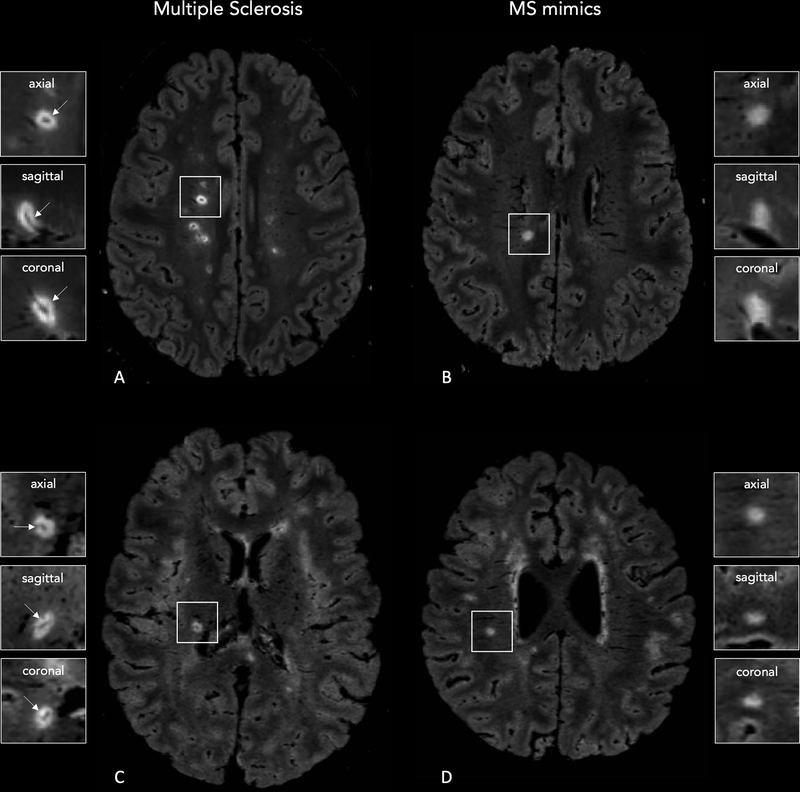
Figure 1 -. 3D FLAIR* images obtained on Siemens (Lausanne University Hospital; A,B) and Philips (Erasmus University Hospital; C,D) MRI scanners showing central vein sign positive (CVS+) and negative (CVS-) lesions.
A central vein running through the lesion (arrows) is visible in the majority of white matter lesions in a 28-year-old man (A) and 27-year-old woman (C) with relapsing-remitting MS. Images from 25-year-old (B) and 46-year-old (D) women with Sjögren syndrome show how the central vein sign is not typical of white matter lesions in MS-mimicking diseases.
In addition, fulfillment of MS MRI criteria for dissemination in space (DIS)3 was assessed for each patient. Average CVS assessment time for one lesion was estimated and used to compute the total evaluation time for each rule used. All the experiments were run on a workstation equipped with an nvidia GTX 1080TI graphics processing unit (GPU) and two intel Xeon Gold 6126 central processing units with 192 GB RAM.
2.3. Automated CVS assessment with a vesselness filter
The “centrality-corrected” vesselness filter method proposed by Dworkin et al.20 was re-implemented and used as baseline for comparison. It combines a classical Frangi multi-scale vesselness filter23 with a map of the distance to the lesion boundary, thereby down-weighting filter responses away from the center. Our implementation differs principally from Dworking et al. in using a standard Euclidean distance transform (Matlab function bwdist) in-house implementation of the multi-scale vesselness filter (parameters: α=0.5, β=0.5, c=0.02), and compilation to optimize execution speed.
A threshold is then applied to the output of the filter. The resulting mask is used as a biomarker for vessel presence and relies on the sum of the probabilities of voxels within the lesion, estimated by the vesselness filter approach. In order to maximize the performance of the vesselness filter, the optimal threshold was computed for each validation fold using the operating point from ROC analysis. True positive and false positive rates in the ROC analysis were estimated according to the ground truth.
2.4. Automated CVS assessment with CVSnet
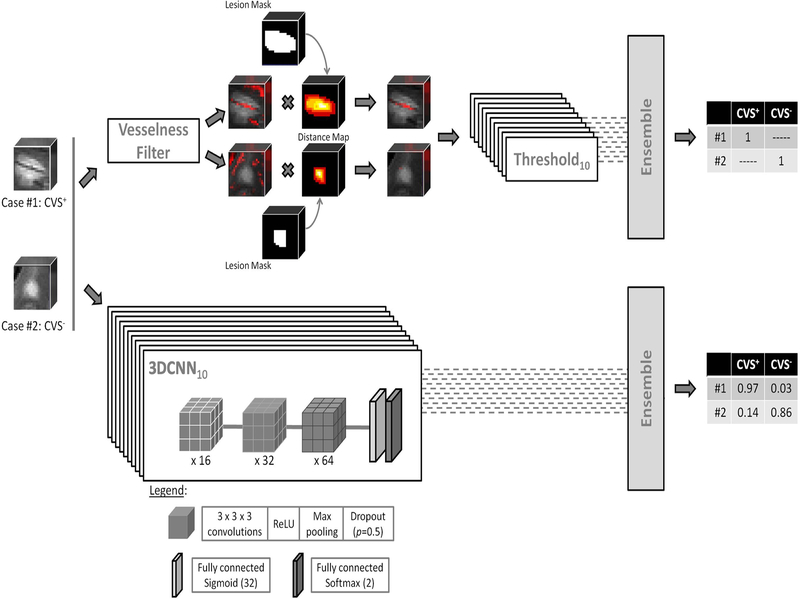
We designed CVSnet as a three layer convolutional neural network, each with a 3D convolution, followed by a rectified linear unit (ReLU) and dropout (p=0.5). This architecture is broadly inspired by the VGGnet, a classical baseline model for computer vision tasks.24 Our network takes patches of size 21×21×21 voxels as input where the center of mass of each lesion was considered the center of the respective patch. This was chosen empirically so that samples of lesion tissue would fit within the patch, and to limit feature map sizes (especially in the first layers) to preserve GPU memory and lower the number of parameters. The convolution kernel sizes were (3×3×3×16), (3×3×3×32), (3×3×3×64). Our choice of small kernel sizes and limited network depth was motivated mainly by the limited amount of data available, as well as the relatively small size of features of interest (vessels). This was followed by a fully connected layer of size 32, then a fully connected layer of size 2 with sigmoid activation (see Figure 2). The architecture comprised 71,810 trainable parameters.25
Figure 2 -. Methods used for central vein sign (CVS) assessment in white matter lesions.
Top stream: a multiscale vesselness enhancement filter combined with a distance map to estimate centrality-corrected vesselness. Bottom stream: CVSnet, the proposed 3D convolutional neural network, consists of a simple, repetitive, and relatively shallow architecture. In both methods, we keep one trained version per cross-validation fold in the cross-validation set. On the test set, we use these ten different versions of the same algorithm to yield ten different predictions for each incoming test patch and average the output (“ensembling”) to yield a single, more stable, prediction.
A ten-fold cross-validation was used to train networks, each using 90% of the lesion patches as training set and 10% as a validation set. Data augmentation based on three 90-degree rotations about one axis was applied to the training and validation sets. After Gaussian initialization, we used categorical cross-entropy loss, training with minibatch stochastic gradient descent (Adam optimizer) for 200 epochs with a minibatch size of 20, at a fixed learning rate of 0.001 checking for approximate class balance for each resampling. In each fold, we saved and used as final model the one providing lowest validation loss in that fold. The weights were initialized with Xavier Gaussian initialization. In each fold, training took an average of 800 (range 600–900) seconds. The classification results for the pure testing set were obtained using an ensemble method by feeding each test patch to all 10 trained networks (1 per cross-validation fold) and averaging their probability outputs (see Figure 2). Even though dropout is used in each of the ensemble constituent networks, fold-to-fold performance is quite variable (see Figure 3), and ensembling allows to lower prediction variance for the test set.
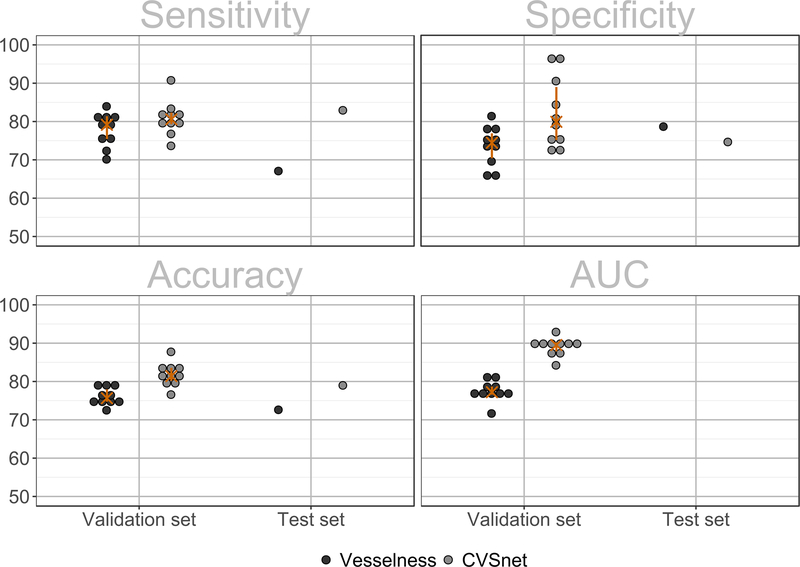
Figure 3 -. Lesion-wise classification using the vesselness filter and the proposed CVSnet.
Lesion-wise classification sensitivity, specificity, accuracy and area-under-the-receiver-operating-characteristic-curve (AUC) for the validation and pure testing sets, using the vesselness filter (dark grey dots) and the new CVSnet (light gray dots). To indicate variability due to data sampling, in the validation set, each dot shows the result of one cross-validation fold, the orange crosses represent median, and the orange bars the range between the 25th and 75th percentiles.
From the multicenter cohort, lesions from 47 (24 MS, 20 MS mimic, 3 uncertain diagnosis) subjects were used to train the network, and lesions from 33 (18 MS, 13 MS mimic, and 2 uncertain diagnosis), selected randomly, were used as a pure-testing set. FLAIR* patches of CVS+ and CVS- lesions were used for training and validation. In total, 673 (375 CVS+, 298 CVS-) and 314 (164 CVS+, 150 CVS-) patches were obtained for the training/validation and pure-testing sets, respectively. In the training/validation set, 338 (158 CVS+, 180 CVS-) patches came from Brussels and 335 (217 CVS+,118 CVS-) came from Lausanne. In the pure-testing set 72 (48 CVS+, 24 CVS-) patches came from Brussels, 103 (40 CVS+,63 CVS-) from Lausanne and 139 (76 CVS+, 63 CVS-) from Bethesda. CVSnet was implemented in Python 3.6.5 (http://www.python.org), with Keras 2.1.5 (http://keras.io) and TensorFlow 1.6.0.26
2.5. Evaluation of the CVS assessment and patient differential diagnosis
The performance was evaluated at two levels:
Lesion-wise: sensitivity, specificity, and balanced accuracy (the average of class-wise accuracies) were computed with respect to the assessment of CVS+ and CVS- lesions on the validation and pure-testing sets. Receiver operating characteristic (ROC) curve analysis was performed, and area under the curve (AUC) values for each of the ten cross-validation folds was computed.
Patient-wise: Intraclass correlation coefficient (ICC(3,1))27 between the percentage of CVS+ lesions estimated automatically and manually were estimated for both methods; sensitivity, specificity, accuracy, and balanced accuracy were computed regarding the differentiation between MS and MS mimics based on the “50% rule,”11 using the classification results of the validation and pure testing sets. The “uncertain diagnosis” cases were excluded from the patient-wise analysis. The total CVS assessment time in the pure testing set was estimated for the vesselness filter, the CVSnet, and the manual CVS assessment rules.
2.6. Statistical analyses
Demographic and MRI differences were tested with Kruskal-Wallis test or Mann-Whitney U test, as appropriate. Difference in perivenular frequency between MS and MS mimics patients was tested using Mann-Whitney U test. Lesion-wise and patient-wise classification results from the proposed CVSnet and the vesselness method were compared using McNemar’s test, and performance was compared to the no information rate (NIR) corresponding to a zero-rule naive classifier that always vote for the majority class, also using McNemar’s test. Statistical analyses were performed using R 3.5.1 (https://www.R-project.org/).
3. Results
3.1. Patient demographics and manual CVS assessment
Clinical and demographic characteristics of MS, MS mimics, and uncertain-diagnosis cases are reported in Table 2. We found no age or gender differences between the groups (P=0.6). The majority of MS mimics scans fulfilled the 2017 MRI diagnostic criteria for MS dissemination in space (20/33 patients, 61%). The median number of brain WM lesions did not differ between MS mimics and MS patients (median=9, range=1–32 vs. median=9, range=1–46, respectively; Mann-Whitney U test=567, P=0.2), whereas median lesion volume was smaller for MS mimics compared to MS patients (median=34mm3, range=14–299 vs. median=52mm3, range=29–217, respectively; Mann-Whitney U test=343, P=0.0001). Expert manual CVS assessment following the NAIMS guidelines for lesion inclusion/exclusion criteria4 showed that the frequency of perivenular lesions was always higher in MS (median=80%, range=52–100%) compared to MS mimics (median=7%, range=0–40%; Mann-Whitney U test = 0, P<0.0001, see Figure 1). In line with previous results using the same optimized high resolution T2*-EPI sequence adopted in this study,11,9 the “50% rule” achieved 100% accuracy in the differentiation between MS and MS mimics, whereas the simplified algorithm “6-lesion rule”18 and the “3-lesion rule”19 showed sensitivity of 95% and 88%, specificity of 100% and 93%, and accuracy of 97% and 90%, respectively. The inter-rater agreement for the percentage of perivenular lesions was “almost perfect” with a Cohen’s k of 0.9 and agreement of 94%. On average, manual CVS assessment of one lesion took 15 seconds, resulting in a total evaluation time on the entire pure-testing set of 40 minutes (“50% rule”), 28 minutes (“6-lesion rule”), and 12 minutes (“3-lesion rule”).
Table 2.
Demographic, clinical, and radiological characteristics of the patient cohort.
| MS | MS mimics | Uncertain | |
|---|---|---|---|
|
Demographics | |||
| # patients | 42 | 33 | 5 |
| Brussels | 15 | 18 | 3 |
| Lausanne | 18 | 10 | 2 |
| Bethesda | 9 | 5 | - |
| Median age (IQR) | 44 (24) | 45 (17) | 48 (8) |
| Sex (female/male) | 33/9 | 23/10 | 4/5 |
|
Disease subtype | |||
| RRMS 34 | SADs 15 | - | |
| SPMS 6 | migraine 5 | - | |
| PPMS 2 | others 13 | - | |
|
DIS MRI criteria | |||
| # (%) patients fulfilling criteria | 42/42 (100%) | 20/33 (61%) | 5/5 (100%) |
|
Manual CVS assessment | |||
| CVS+ lesions, median (range) | 80% (52–100%) | 7% (0–40%) | 62% (20–100%) |
Abbreviations: IQR: interquartile range; RRMS: relapsing-remitting MS; SPMS: secondary-progressive MS; PPMS: primary-progressive MS; SADs: systemic autoimmune disorders (systemic lupus erythematosus, antiphospholipid antibody syndrome, primary angiitis of the central nervous system, Sjögren syndrome, Behçet disease, sarcoidosis); others: neuromyelitis optica spectrum disorder, Susac syndrome, inflammatory amyloid angiopathy, HTLV1 associated myelopathy/tropical spastic paraparesis, small vessel disease); Uncertain: patient having uncertain diagnosis at the time of the study; DIS: dissemination in space.
3.2. Automated CVS assessment
As shown in Figure 3, the CVSnet showed higher performance than the vesselness filter for CVS assessment in both validation (McNemar’s x2=28.96, P=7.40E-08) and pure testing (McNemar’s x2=11.93, P=5.51E-04) sets. In the validation set, median values for the CVSnet were: sensitivity, 81% (range: 74–91%); specificity, 80% (range: 72–97%); balanced accuracy, 81% (range: 77–88%); and AUC, 90% (range: 84–93%). For the vesselness filter: sensitivity, 79% (range: 70–84%); specificity, 74% (range: 66–81%); balanced accuracy, 76% (range: 73–80%); AUC, 77% (range: 72–82%). Similar performance was achieved in the pure testing set, where the CVSnet showed 83%, 75%, and 79%, and the vesselness filter 67%, 79%, and 73% in sensitivity, specificity and accuracy, respectively (see Figure 3). The total CVS assessment time in the pure testing set was 80 seconds for the vesselness filter and 4 seconds for CVSnet on a central processing unit, i.e. not employing accelerated GPU-based implementations. Thus, evaluating the CVS for all lesions was about 600-fold faster using CVSnet as compared to manual assessment (in the test set it was four seconds instead of forty minutes).
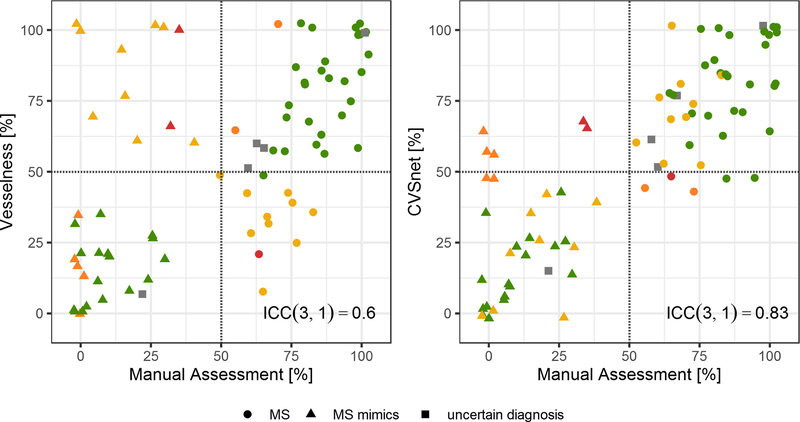
Figure 4 shows a scatter plot of the percentage of CVS+ lesions per patient estimated by both methods against the manual assessment. ICC(3,1)=0.83 is higher for the CVSnet in comparison to the vesselness filter ICC(3,1)=0.60, showing better consistency with manual assessment.
Figure 4 – Patient-wise manual and automated assessment using vesselness filter and CVSnet.
Percentage of CVS+ lesions for each patient, comparing the vesselness filter and CVSnet approaches with respect to manual assessment (ground truth). Different types of diagnoses: MS (circle), MS mimics (triangle), and uncertain diagnosis (square). Colors represent the different scenarios for correct and incorrect disease prediction from both algorithms based on the 50% perivenular rule: green – both algorithms predicted correctly; yellow – only CVSnet predicted correctly; orange – only Vesselness filter predicted correctly; red – both algorithms predicted incorrectly; grey – uncertain diagnosis. The red dashed line indicates the 50% rule. While the percentages diverge somewhat from those estimated manually, the CVSnet approach tends to yield estimates leading to a diagnosis consistent with that derived from manual assessment with an ICC(3,1)=0.83.
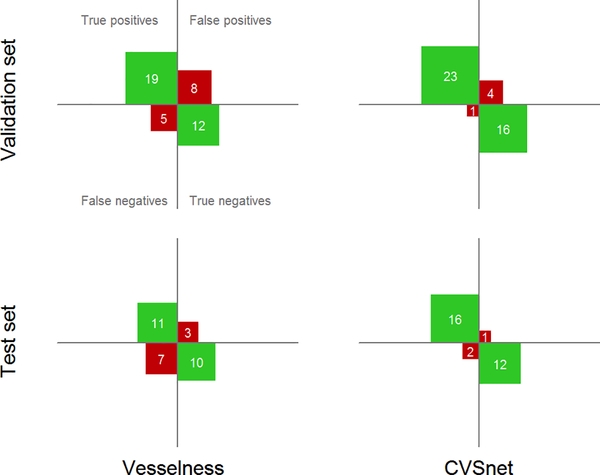
Results of the patient-wise analysis regarding the differentiation between MS and MS mimics in terms of true/false positives and true/false negatives are summarized by the confusion matrices in Figure 5.
Figure 5 -. Patient-wise classification confusion matrices. Differentiation between MS and MS mimics using confusion matrices for the vesselness filter and the proposed CVSnet, on the validation and test sets.
Within each confusion matrix, the top left indicates the number of true positives (actual MS patients predicted as MS), the bottom right true negatives, the top right false positives (MS mimics predicted as MS), and the bottom right false negatives. Note that patients with uncertain diagnoses are not shown on this figure.
Using the 50%-rule, the CVSnet achieved sensitivity of 96%/89%, specificity of 80%/92%, accuracy of 89%/90%, and balanced accuracy of 88%/91% for the validation/testing sets. The vesselness filter-based approach reached 79%/61% sensitivity, 60%/77% specificity, accuracy of 70%/68%, and 70%/69% balanced accuracy for the validation/testing sets. The CVSnet performed better than the vesselness filter, but the head-to-head difference was not statistically significant (Validation: McNemar’s x2 = 3.50, P>0.05, McNemar’s Test x2 = 3.27, P>0.05). However, note that the CVSnet accuracy was statistically different from the NIR (Validation: McNemar’s x2 = 11.53, P=6.85E-04, Test: McNemar’s x2 = 5.79, P=0.02) while the vesselness filter was not (Validation: McNemar’s x2 = 2.12, P=0.15, Test: McNemar’s x2 = 0.24, P=0.63).
4. Discussion
We propose a deep-learning-based automated detection of the CVS differentiating MS from its mimics with high diagnostic sensitivity, specificity, and accuracy. CVSnet performed nearly as well as the previously obtained proportion of CVS+/CVS- lesions based on manual rating, often achieving perfect discrimination between MS and its mimics,11,12,17 and is much faster when compared with manual rating. Importantly, this method showed high performance across different scanner types and hospitals, strengthening its potential for clinical applicability.
Our findings are particularly relevant considering that recent studies using manual CVS classification at clinical field strengths have shown potential for application of the CVS imaging biomarker in clinical practice.11,13,15–18,28 Importantly, the most recent international panel for MS diagnosis called for further research on this topic before considering inclusion of the CVS in future MS diagnostic criteria.3 A substantial limitation to investigating the role of the CVS in large multicenter trials, and in subsequent clinical practice, is the lack of an automated reproducible and non-operator-dependent technique that can determine the frequency of CVS+ lesions in individual scans.
The proposed CVSnet uses deep learning and data augmentation strategies and shows the feasibility to achieve accurate results even in the case of a relatively small dataset for this particular classification problem compared to classical computer vision problems, where samples sizes are in the tens of thousands. While machine learning has already been proposed to predict various aspects of MS diagnosis and prognosis,29,30 to our knowledge, this is the first attempt to use machine learning to automatically assess the CVS as diagnostic biomarker.
Compared to our implementation of a recently proposed approach for automated CVS assessment based on vesselness filter and centrality,20 the CVSnet showed significantly higher lesion-wise performance and was 20 times faster; in addition, considering the slight class imbalance, CVSnet performed significantly above the NIR, while our vesselness filter-based methodology did not.
For several reasons, our results are not directly comparable to those reported in Dworkin et al.20 First, sample size is larger in our study. Second, our data comes from four different MRI machines while Dworkin et al. used data from only one MRI scanner. Third, exclusion criteria are different: Dworkin et al. excluded periventricular lesions, whereas in our study we excluded only lesions not fulfilling NAIMS criteria, that is, periventricular lesions were only excluded in our study if featuring multiple veins or confluent. Lastly, implementation differences discussed in the methods section also likely contribute to some differences. This study presents some limitations. Lesion segmentation was done manually, and thus the overall process (automated lesion segmentation and CVS classification) cannot yet be considered fully automated. Of note, the available automatic lesion segmentation algorithms are often limited by false-positive and false-negative “lesions,”31 and it is possible that the difference in CVS assessment performance obtained in our study with respect to previous work20 rests upon the manually applied lesion inclusion/exclusion criteria in the current study. In addition to including larger numbers of scans from additional centers, future work should implement a fully automated method able to automatically apply lesion inclusion/exclusion criteria for CVS assessment. Moreover, future work could investigate other lesion features (such as shape, ovoid vs. spherical29) that, together with the CVS, may increase the probability that a given WM lesion results from MS-related inflammatory demyelination.
In conclusion, the CVSnet presented in this study showed high speed and accurate performance in differentiating MS from other pathological conditions with similar abnormalities on MRI. Our study represents a step forward toward the translation of this promising imaging biomarker into daily clinical practice.
Acknowledgments
Sources of support:
Dr. Pietro Maggi was supported for this research by the ECTRIMS Clinical Training Fellowship Program and by the University of Lausanne “relève académique” grant.
Dr. Mário João Fartaria is full-time employee of Siemens Healthcare AG
Dr. João Jorge is supported from the “Centre d’Imagerie BioMédicale (CIBM) of the University of Lausanne and the Leenaards and Jeantet Foundations.”
Francesco La Rosa is supported by the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie project TRABIT (agreement No 765148).
Dr. Martina Absinta, Dr. Pascal Sati and Dr. Daniel S. Reich are supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, USA.
Dr. Martina Absinta is supported by the National Multiple Sclerosis Society (NMSS) (FG 2093-A-1) and the Conrad N. Hilton Foundation (grant#17313), unrelated to the present study.
Dr. Jonas Richiardi receives part of his salary from Siemens Healthcare AG and owns shares in Siemens Healthcare AG
Dr. Tobias Kober is full-time employee of Siemens Healthcare AG and owns shares in Siemens Healthcare AG
Abbreviations:
- MS
multiple sclerosis
- CVS
central vein sign
- WM
white matter
- MRI
magnetic resonance imaging
- 3D
three-dimensional
- T2*-EPI
T2*-weighted segmented echo-planar imaging
- FLAIR
3D T2-weighted fluid-attenuated inversion recovery
- ReLU
rectified linear unit
- ROC
Receiver operating characteristic
- AUC
area under the curve
- GPU
graphics processing unit
- NIR
no information rate
Footnotes
Potential Conflicts of Interest: nothing to report
References:
- 1.Filippi M, Preziosa P, Banwell BL, et al. Assessment of lesions on magnetic resonance imaging in multiple sclerosis: practical guidelines. Brain. 2019;142(7):1858–1875; 10.1093/brain/awz144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solomon AJ, Corboy JR. The tension between early diagnosis and misdiagnosis of multiple sclerosis. Nat Rev Neurol. 2017;13(9):567–572; 10.1038/nrneurol.2017.106. [DOI] [PubMed] [Google Scholar]
- 3.Thompson AJ, Banwell BL, Barkhof F, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17(2):162–173; 10.1016/s1474-4422(17)30470-2. [DOI] [PubMed] [Google Scholar]
- 4.Sati P, Oh J, Constable RT, et al. The central vein sign and its clinical evaluation for the diagnosis of multiple sclerosis: a consensus statement from the North American Imaging in Multiple Sclerosis Cooperative. Nat Rev Neurol. 2016;12(12):714–722; 10.1038/nrneurol.2016.166. [DOI] [PubMed] [Google Scholar]
- 5.Adams CW. The onset and progression of the lesion in multiple sclerosis. J Neurol Sci. 1975;25(2):165–182. [DOI] [PubMed] [Google Scholar]
- 6.Barnett MH, Prineas JW. Relapsing and remitting multiple sclerosis: pathology of the newly forming lesion. Ann Neurol. 2004;55(4):458–468; 10.1002/ana.20016. [DOI] [PubMed] [Google Scholar]
- 7.Absinta M, Sati P, Reich DS. Advanced MRI and staging of multiple sclerosis lesions. Nat Rev Neurol. 2016;12(6):358–368; 10.1038/nrneurol.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams CW, Poston Rn Fau - Buk SJ, Buk SJ. Pathology, histochemistry and immunocytochemistry of lesions in acute multiple sclerosis. J Neurol Sci. 1989;92(0022–510):291–306. [DOI] [PubMed] [Google Scholar]
- 9.Sati P, Thomasson D, Li N, et al. Rapid, high-resolution, whole-brain, susceptibility-based MRI of multiple sclerosis. Mult Scler. 2014; 10.1177/1352458514525868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sati P, George IC, Shea CD, Gaitan MI, Reich DS. FLAIR*: a combined MR contrast technique for visualizing white matter lesions and parenchymal veins. Radiology. 2012;265(3):926–932; 10.1148/radiol.12120208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maggi P, Absinta M, Grammatico M, et al. Central vein sign differentiates Multiple Sclerosis from central nervous system inflammatory vasculopathies. Ann Neurol. 2018;83(2):283–294; 10.1002/ana.25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tallantyre EC, Dixon JE, Donaldson I, et al. Ultra-high-field imaging distinguishes MS lesions from asymptomatic white matter lesions. Neurology. 2011;76(6):534–539; 10.1212/WNL.0b013e31820b7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solomon AJ, Schindler MK, Howard DB, et al. “Central vessel sign” on 3T FLAIR* MRI for the differentiation of multiple sclerosis from migraine. 2016;3(2):82–87; 10.1002/acn3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuerfel J, Sinnecker T, Ringelstein EB, et al. Lesion morphology at 7 Tesla MRI differentiates Susac syndrome from multiple sclerosis. Mult Scler. 2012;18(11):1592–1599; 10.1177/1352458512441270. [DOI] [PubMed] [Google Scholar]
- 15.Cortese R, Magnollay L, Tur C, et al. Value of the central vein sign at 3T to differentiate MS from seropositive NMOSD. Neurology. 2018;90(14):e1183–e1190; 10.1212/wnl.0000000000005256. [DOI] [PubMed] [Google Scholar]
- 16.Sinnecker T, Clarke MA, Meier D, et al. Evaluation of the Central Vein Sign as a Diagnostic Imaging Biomarker in Multiple Sclerosis. JAMA Neurol. 2019; 10.1001/jamaneurol.2019.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maggi P, Absinta M, Sati P, et al. The “central vein sign” in patients with diagnostic “red flags” for multiple sclerosis: A prospective multicenter 3T study. Mult Scler. 2019; 10.1177/1352458519876031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mistry N, Abdel-Fahim R, Samaraweera A, et al. Imaging central veins in brain lesions with 3-T T2*-weighted magnetic resonance imaging differentiates multiple sclerosis from microangiopathic brain lesions. Mult Scler. 2016;22(10):1289–1296; 10.1177/1352458515616700. [DOI] [PubMed] [Google Scholar]
- 19.Solomon AJ, Watts R, Ontaneda D, Absinta M, Sati P, Reich DS. Diagnostic performance of central vein sign for multiple sclerosis with a simplified three-lesion algorithm. Mult Scler. 2018;24(6):750–757; 10.1177/1352458517726383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dworkin JD, Sati P, Solomon A, et al. Automated Integration of Multimodal MRI for the Probabilistic Detection of the Central Vein Sign in White Matter Lesions. AJNR Am J Neuroradiol. 2018;39(10):1806–1813; 10.3174/ajnr.A5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erickson BJ, Korfiatis P, Kline TL, Akkus Z, Philbrick K, Weston AD. Deep Learning in Radiology: Does One Size Fit All? J Am Coll Radiol. 2018;15(3 Pt B):521–526; 10.1016/j.jacr.2017.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeCun Y, Kavukcuoglu K, Farabet C. Convolutional networks and applications in vision. Paper presented at: Proceedings of 2010 IEEE International Symposium on Circuits and Systems; 30 May-2 June 2010, 2010. [Google Scholar]
- 23.Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale vessel enhancement filtering. Paper presented at: Medical Image Computing and Computer-Assisted Intervention — MICCAI’98; 1998//, 1998; Berlin, Heidelberg. [Google Scholar]
- 24.Simonyan KZ, Andrew. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv. 2014arXiv1409.1556S. [Google Scholar]
- 25.Mário João Fartaria JR, João Jorge, Pietro Maggi, Pascal Sati, Daniel S. Reich, Reto Meuli, Cristina Granziera, Meritxell Bach Cuadra, Tobias Kober. An ensemble of 3D convolutional neural networks for central vein detection in white matter lesions. Peer-reviewed abstract, 1st Conference on Medical Imaging with Deep Learning (MIDL 2018), Amsterdam, The Netherlandshttps://openreview.net/forum?id=HJu7eTojG. [Google Scholar]
- 26.Abadi M, Agarwal A, Barham P, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv e-prints. 2016:arXiv:1603.04467. [Google Scholar]
- 27.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86(2):420–428; 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 28.Samaraweera AP, Clarke MA, Whitehead A, et al. The Central Vein Sign in Multiple Sclerosis Lesions Is Present Irrespective of the T2* Sequence at 3 T. J Neuroimaging. 2017;27(1):114–121; 10.1111/jon.12367. [DOI] [PubMed] [Google Scholar]
- 29.Haike Zhang EA, Viola Pongratz, Mark Mühlau, Claus Zimmer, Benedikt Wiestler, Paul Eichinger. Predicting conversion from clinically isolated syndrome to multiple sclerosis–An imaging-based machine learning approach. NeuroImage: Clinical. 2018;S2213–1582(18):30341–30343; 10.1016/j.nicl.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marzullo A, Kocevar G, Stamile C, et al. Classification of Multiple Sclerosis Clinical Profiles via Graph Convolutional Neural Networks. Front Neurosci. 2019;13:594; 10.3389/fnins.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fartaria MJ, Bonnier G, Roche A, et al. Automated detection of white matter and cortical lesions in early stages of multiple sclerosis. J Magn Reson Imaging. 2016;43(6):1445–1454; 10.1002/jmri.25095. [DOI] [PubMed] [Google Scholar]