Abstract
Background
The diagnosis of advanced lung cancer is made with minimally invasive procedures. This often results in the availability of cytological material only for subtype determination and companion diagnostic testing, with the latter being technically and clinically validated on histological material only. Thus, the primary objective of the MO29978 clinical study was to assess programmed death ligand 1 (PD‐L1) protein expression on cytology samples as surrogates for histology samples in patients with lung cancer.
Methods
Formalin‐fixed, paraffin‐embedded histological samples and cytological cell blocks from 190 patients were analyzed with immunohistochemical assays using the rabbit monoclonal anti–PD‐L1 antibody clones SP142 and SP263. PD‐L1 expression was quantified on both tumor cells (TC) and tumor‐infiltrating immune cells (IC). Overall concordance, sensitivity, specificity, and accuracy, with a 1% cutoff used for both assays, were assessed for PD‐L1 expression on TC and IC.
Results
In non–small cell lung cancer histology and cytology samples measured with the PD‐L1 (SP142) antibody (n = 173), the intraclass correlation coefficients were 0.40 and 0.06 on TC and IC, respectively. With SP142 and SP263, accuracies of 74.1% for TC and 51.9% for IC and accuracies of 75.2% for TC and 61.2% for IC, respectively, were reported.
Conclusions
Overall, this study has demonstrated that PD‐L1 analysis on TC is feasible in cytological material, but quantification is challenging. Tumor tissue should be preferred over cell block cytology for PD‐L1 immunohistochemical analysis unless laboratories have validated their cytology preanalytical approaches and demonstrated the comparability of histology and cytology for TC PD‐L1 results.
Keywords: cytology, histology, immunohistochemistry, lung cancer, programmed death ligand 1 (PD‐L1), SP142, SP263
Short abstract
Because patients with advanced‐stage, unresectable lung cancer frequently have only cytology samples available for diagnosis, this noninterventional study has assessed whether this sample type can be used as a surrogate for histological material when programmed death ligand 1 (PD‐L1) expression is being measured by immunohistochemistry (IHC) with the antibody clones SP142 and SP263. Results demonstrate that PD‐L1 IHC is feasible on formalin‐fixed, paraffin‐embedded cytological material; however, poor absolute concordance is observed when cytology and histology are compared for both tumor and immune cells, and this challenges the prevailing view of PD‐L1 IHC as a method suitable for evaluating PD‐L1 levels in this sample type.
Introduction
In the majority of patients, lung cancer is diagnosed at advanced, unresectable stages. 1 A diagnosis made through minimally invasive techniques results in the majority of patients' having only small tissue biopsies or cytology samples for subsequent histological and molecular subclassification. 2
The standardization of the preparation of formalin‐fixed, paraffin‐embedded (FFPE) histological material from small biopsies allows for diagnostic assessments that are required for subsequent treatment decisions. However, wide variations in the types, preparatory methods, and diagnostic yields of cytology samples exist. The sample types depend on the accessibility of malignant lesions and include fine‐needle aspiration (FNA) conducted with different needle sizes and imaging guidance, bronchial washing, bronchial brushing, bronchoalveolar lavage, sputum, and pleural effusion. 3 Samples can be prepared as conventional smears, liquid‐based cytology, or cell blocks embedded in paraffin. The variability inherent to cytological material has resulted in its underuse for predictive immunohistochemistry (IHC). This variability includes the range of sample types and preparations, the difficulty of procuring cytology‐specific controls, the use of alcohol‐based fixatives that may interfere with results, and the uncertainty of test validation. 4 In addition, sample preservation also varies with the different types of formalin‐ or alcohol‐based fixatives and storage (eg, air‐dried or stained smears). 5
The diagnosis of non–small cell lung cancer (NSCLC) not only requires the determination of the histopathological subtype (nonsquamous vs squamous) through the use of immunological markers (thyroid transcription factor 1 [TTF1] and p40) but also involves companion diagnostic assessments (eg, epidermal growth factor receptor [EGFR], ALK receptor tyrosine kinase [ALK], ROS proto‐oncogene 1, receptor tyrosine kinase [ROS1], and programmed death ligand 1 [PD‐L1]) that help to inform treatment decisions. 6 , 7 , 8 , 9 , 10 Although companion diagnostic tests have been approved for use with tumor tissue and even plasma, no test has been developed and approved for cytological material. Patients with only cytological material have always been excluded from pivotal clinical studies in NSCLC. 11 To date, small cell lung cancer (SCLC) does not require additional testing because of the absence of approved targeted therapies.
Since the approval of anti–PD‐L1/programmed death 1 (PD‐1) inhibitors (pembrolizumab, atezolizumab, nivolumab, and durvalumab) in NSCLC, PD‐L1 has become an important predictive biomarker for guiding treatment decisions with anti–PD‐L1/PD‐1 therapies. Several PD‐L1 immunohistochemistry (IHC) assays, including the Ventana PD‐L1 SP142 and SP263 assays (Ventana Medical Systems, Inc) and the 22C3 pharmDx and 28‐8 pharmDx assays (Dako/Agilent), have been developed as companion diagnostic tests with histological material. 12
To date, several studies have explored the feasibility of measuring PD‐L1 in cytological material. There is a high degree of agreement of tumor cell (TC) PD‐L1 expression between histology and cell block cytology using PD‐L1 22C3, E1L3N, and SP263 antibody clones. 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 Based on results from a feasibility study on cell lines that explored the various fixatives and preparation types, 23 this prospective, multinational study was aimed at determining which of the many sample types could be used to determine PD‐L1 expression levels with the anti–PD‐L1 antibody clones SP142 and SP263 in patients with lung cancer.
Materials and Methods
Study Design and Objectives
The primary objective of this noninterventional, multinational study (MO29978; NCT03092739) was to assess PD‐L1 expression on TC and tumor‐infiltrating immune cells (IC) in cytological and histological samples prepared from patients with lung cancer with IHC using the SP142 antibody. For a robust evaluation of this objective, 2 readers were employed. Secondary objectives were 1) to explore the concordance of PD‐L1–expressing TC and IC in histological samples in comparison with TC in cytological samples, 2) to explore the feasibility of different PD‐L1 scoring algorithms for cytological specimens, 3) to explore the concordance and robustness of results related to PD‐L1 expression by a second independent reading, and 4) to explore the feasibility of alternative diagnostic technologies. Protocol approval was obtained from the IRB/IEC for each site before participation in the study. The study design is provided in Figure 1.
FIGURE 1.
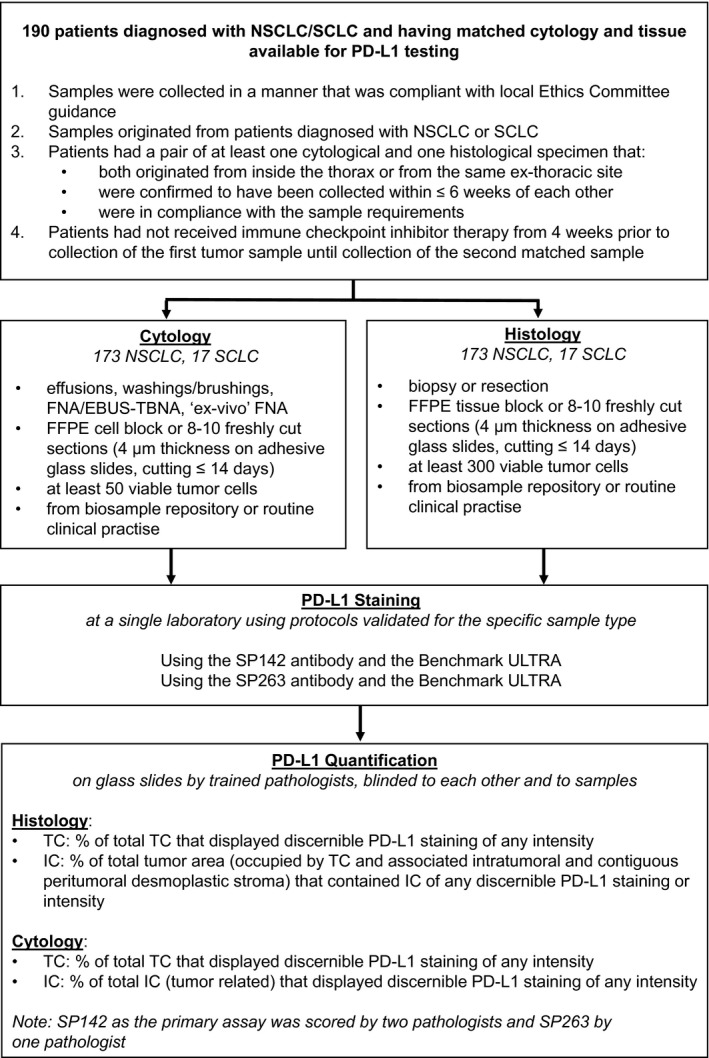
Flowchart of the design of a noninterventional, multinational study to assess PD‐L1 expression on cytological and histological NSCLC and SCLC specimens. EBUS‐TBNA indicates endobronchial ultrasound–guided transbronchial needle aspiration; FFPE, formalin‐fixed, paraffin‐embedded; FNA, fine‐needle aspiration; IC, tumor‐infiltrating immune cells; NSCLC, non–small cell lung cancer; PD‐L1, programmed death ligand 1; SCLC, small cell lung cancer; TC, tumor cells.
Patient Samples
Samples were collected in a manner that was compliant with local ethics committee guidance and originated from patients diagnosed with NSCLC or SCLC. Patients had to have a pair of at least 1 cytological specimen and 1 histological specimen; both specimens must have originated from inside the thorax or the same extrathoracic site, been collected within a window of no more than 6 weeks, and been in compliance with the sample requirements for histological and cytological specimens as defined in this study (Fig. 1). Patients also must have not received immune checkpoint inhibitor therapy from 4 weeks before the collection of the first tumor sample until after the collection of the second sample. All samples were collected from material retained during routine clinical practice or from biosample repositories housed at participating sites.
Sample Processing and Staining Procedures
Cytological samples could have originated from pleural effusions, bronchial washing/brushing, FNA, or ex vivo FNA (see Supporting Table 1 for details of the ex vivo FNA biopsy procedure); they were prepared as FFPE cell blocks with 10% neutral buffered formalin used as either a fixative or a postfixative and were embedded in paraffin according to the local cell block protocol. Exposure to formalin and embedding in paraffin were mandated on the basis of prior research. 23 FFPE histological samples were obtained by biopsy or surgical resection. Blinding of both cohorts was performed so that histology and cytology samples could not be matched by the pathologists. Samples were sent for hematoxylin‐eosin staining and PD‐L1 analysis (Fig. 1). For details of the PD‐L1 staining methodology, please see the supporting information.
Quantification of PD‐L1 Staining
The following parameters were used for histological samples: PD‐L1 expression on TC was indicated by the percentage of total TC that displayed discernible membranous PD‐L1 staining of any intensity (minimum of 300 viable TC), and expression on IC was determined by the percentage of the total tumor area (occupied by TC and associated intratumoral and contiguous peritumoral desmoplastic stroma) that contained IC with discernible PD‐L1 staining of any intensity. For cytological samples, PD‐L1 expression on TC was indicated by the percentage of total TC that displayed discernible membranous PD‐L1 staining of any intensity (minimum of 50 viable TC), and expression on IC was determined by the percentage of total IC that displayed discernible PD‐L1 staining of any intensity. PD‐L1 expression as determined with the anti–PD‐L1 antibody clone SP142 was quantified by 2 independent pathologists. PD‐L1 expression from anti–PD‐L1 antibody clone SP263 expression was quantified by a single pathologist.
Statistical Analysis
This study was designed primarily to assess the concordance of PD‐L1 SP142 staining samples for NSCLC, with SCLC samples analyzed separately in an exploratory manner. PD‐L1 staining was also examined with IHC using the SP263 antibody. Results for TC and IC data are described separately for each PD‐L1 assay. An analysis of variance model was used to calculate intraclass correlation coefficients (ICCs) to summarize the intermethod variability (ie, the variability of PD‐L1 quantification when performed on cytology vs histology) and the interreader variability for NSCLC samples examined with the SP142 antibody. This model included patient and method/reader as random effects. Sensitivity, specificity, and accuracy were determined with PD‐L1 expression measured on histology samples as the reference and with a cutoff of ≥1% for TC or IC for both PD‐L1 assays.
Results
Samples (histology and cytology) from 190 patients with lung cancer (173 patients with NSCLC and 17 patients with SCLC) across 8 sites in 6 countries were analyzed for PD‐L1 expression on TC and IC in this study (Figs. 1 and 2). Baseline characteristics and sample details are provided in Table 1. SCLC results are provided in Supporting Table 2. Overall, most samples for both histology and cytology were lung adenocarcinomas prepared as FFPE blocks and had an ischemia time of 1 to 8 hours, and they were mostly fixed for 6 to 48 hours in formalin. Most samples collected by washing or brushing were squamous cell carcinomas.
FIGURE 2.
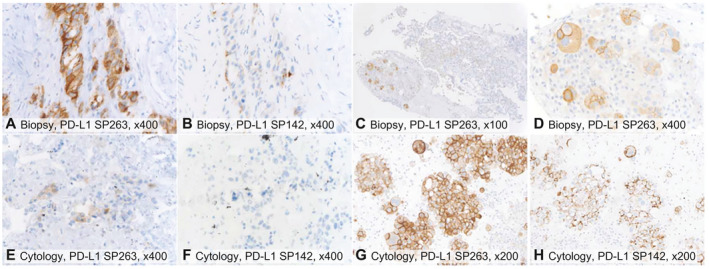
Examples of PD‐L1 immunohistochemistry staining on (A‐D) biopsies and (E‐H) cytological cell block specimens using (A,C‐E,G) the SP263 antibody and (B,F,H) the SP142 antibody, with brown membranous staining indicating PD‐L1 positivity. (A,B,E,F) Case 1: SP263 stained a higher proportion of tumor cells than SP142 and was lower in the fine‐needle aspiration cytology than the biopsy. (F) There was no staining by SP142 in cytology. (C,D,G,H) PD‐L1 staining was heterogeneous in the biopsy ([C,D] SP263) but homogeneous in the cytology of the pleural effusion ([G] SP263 and [H] SP142). The magnifications were (A,B,D‐F) ×400, (G,H) ×200, and (C) ×100. PD‐L1 indicates programmed death ligand 1.
TABLE 1.
Baseline Characteristics of Histological and Cytological Samples From Patients With Lung Cancer
| Histology, No. (%) | Cytology, No. (%) | ||||||
|---|---|---|---|---|---|---|---|
| NSCLC | SCLC | Total | NSCLC | SCLC | Total | ||
| Total | 173 (100.0) | 17 (100.0) | 190 (100.0) | Total | 173 (100.0) | 17 (100.0) | 190 (100.0) |
| Location of lesion | Location of lesion | ||||||
| Thorax | 146 (84.4) | 9 (52.9) | 155 (81.6) | Thorax | 134 (77.5) | 6 (35.3) | 140 (73.7) |
| Bronchus | 27 (15.6) | 8 (47.1) | 35 (18.4) | Lymph node | 8 (4.6) | 4 (23.5) | 12 (6.3) |
| Pleura | 3 (1.7) | — | 3 (1.6) | ||||
| Other | 28 (16.2) | 7 (41.2) | 35 (18.4) | ||||
| Lung cancer subtype | Lung cancer subtype | ||||||
| Small cell carcinoma | — | 17 (100.0) | 17 (8.9) | Small cell carcinoma | — | 17 (100.0) | 17 (8.9) |
| Adenocarcinoma | 98 (56.6) | — | 98 (51.6) | NSCC favor adenocarcinoma | 92 (53.2) | — | 92 (48.4) |
| Squamous cell carcinoma | 58 (33.5) | — | 58 (30.5) | NSCC favor squamous cell carcinoma | 43 (24.9) | — | 43 (22.6) |
| Adenosquamous carcinoma | 3 (1.7) | — | 3 (1.6) | NSCC NOS | 20 (11.6) | — | 20 (10.5) |
| Large cell carcinoma | 4 (2.3) | — | 4 (2.1) | Squamous cell carcinoma | 15 (8.7) | — | 15 (7.9) |
| Other | 10 (5.8) | — | 10 (5.3) | Other | 3 (1.7) | — | 3 (1.6) |
| Method of sample collection | Method of sample collection | ||||||
| Resection | 84 (48.6) | 1 (5.9) | 85 (44.7) | FNA | 50 (28.9) | 7 (41.2) | 57 (30.0) |
| Biopsy | 89 (51.4) | 16 (94.1) | 105 (55.3) | Pleural effusion | 21 (12.1) | 1 (5.9) | 22 (11.6) |
| Washing/brushing | 33 (19.1) | 2 (11.8) | 35 (18.4) | ||||
| EBUS‐TBNA | 7 (4.0) | 3 (17.6) | 10 (5.3) | ||||
| Ex vivo FNA | 62 (35.8) | — | 62 (32.6) | ||||
| Ischemia time | Ischemia time | ||||||
| 1‐8 h | 108 (62.4) | 6 (35.3) | 114 (60.0) | 1‐8 h | 97 (56.1) | 6 (35.3) | 103 (54.2) |
| Unknown | 65 (37.6) | 11 (64.7) | 76 (40.0) | Unknown/missing | 76 (43.9) | 11 (64.7) | 87 (45.8) |
| Fixation time | Fixation time | ||||||
| <6 h | 28 (16.2) | 1 (5.9) | 29 (15.3) | <6 h | 28 (16.2) | 1 (5.9) | 29 (15.3) |
| 6‐48 h | 80 (46.2) | 5 (29.4) | 85 (44.7) | 6‐48 h | 79 (45.7) | 5 (29.4) | 84 (44.2) |
| Unknown | 65 (37.6) | 11 (64.7) | 76 (40.0) | Unknown | 66 (38.2) | 11 (64.7) | 77 (40.5) |
| Fixative | Fixative | ||||||
| 10% neutral buffered formalin | 173 (100.0) | 17 (100.0) | 190 (100.0) | 10% neutral buffered formalin | 113 (65.3) | 6 (35.3) | 119 (62.6) |
| CytoLyt | 58 (33.5) | 11 (64.7) | 69 (36.3) | ||||
| Cytofix | 1 (0.6) | — | 1 (0.5) | ||||
| PreservCyt | 1 (0.6) | — | 1 (0.5) | ||||
| Postfixative | |||||||
| 10% neutral buffered formalin | 158 (91.3) | 16 (94.1) | 174 (91.6) | ||||
| Other | — | 1 (5.9) | 1 (0.5) | ||||
| Missing | 15 (8.7) | — | 15 (7.9) | ||||
| Type of sample sent for analysis | Type of sample sent for analysis | ||||||
| Block | 69 (39.9) | 5 (29.4) | 74 (38.9) | Block | 71 (41.0) | 5 (29.4) | 76 (40.0) |
| Slide | 104 (60.1) | 12 (70.6) | 116 (61.1) | Slide | 102 (59.0) | 12 (70.6) | 114 (60.0) |
Abbreviations: EBUS‐TBNA, endobronchial ultrasound–guided transbronchial needle aspiration; FNA, fine‐needle aspiration; NOS, not otherwise specified; NSCC, non–squamous cell cancer; NSCLC, non–small cell lung cancer; SCLC, small cell lung cancer.
IHC Expression Using the SP142 Antibody
For the NSCLC samples, SP142 IHC TC and IC scoring was successful for 170 histology specimens and 165 cytology specimens. Among these specimens, 62 histology samples (36.5%) and 53 cytology samples (32.1%) were found to express PD‐L1 on TC. PD‐L1 expression on IC was observed for 96 of the 170 histology samples (56.5%) and for 22 of the 165 cytology samples (13.3%; Table 2). Among the 165 evaluable samples, 22 cytological specimens had <100 TC (pleural effusion, 1; FNA, 7; ex vivo FNA, 10; and brushings/washings, 4).
TABLE 2.
PD‐L1 Expression Levels on TC/IC in the NSCLC Cohort
| SP142 Antibody, No. (%) | SP263 Antibody, No. (%) | ||||||
|---|---|---|---|---|---|---|---|
| First Evaluation | Second Evaluation | First Evaluation | |||||
| Cytology (n = 165) | Histology (n = 170) | Cytology (n = 160) | Histology (n = 168) | Cytology (n = 157) | Histology (n = 168) | ||
| TC, % | PD‐L1– (<1%) | 112 (67.9) | 108 (63.5) | 136 (84.5) | 129 (76.3) | 98 (62.4) | 89 (52.7) |
| PD‐L1+ (≥1%) | 53 (32.1) | 62 (36.5) | 25 (15.5) | 40 (23.7) | 59 (37.6) | 80 (47.3) | |
| 1 to <5 | 32 (19.4) | 26 (15.3) | 10 (6.2) | 16 (9.5) | 23 (14.6) | 28 (16.6) | |
| 5 to <50 | 17 (10.3) | 26 (15.3) | 11 (6.8) | 17 (10.1) | 23 (14.6) | 30 (17.8) | |
| ≥50 | 4 (2.4) | 10 (5.9) | 4 (2.5) | 7 (4.1) | 13 (8.3) | 22 (13.0) | |
| IC, % | PD‐L1– (<1%) | 143 (86.7) | 74 (43.5) | 117 (73.1) | 66 (39.3) | 78 (49.7) | 43 (25.6) |
| PD‐L1+ (≥1%) | 22 (13.3) | 96 (56.5) | 43 (26.9) | 102 (60.7) | 79 (50.3) | 125 (74.4) | |
| 1 to <5 | 21 (12.7) | 72 (42.4) | 32 (20.0) | 58 (34.5) | 59 (37.6) | 68 (40.5) | |
| 5 to <10 | 1 (0.6) | 17 (10.0) | 8 (5.0) | 32 (19.0) | 15 (9.6) | 41 (24.4) | |
| ≥10 | 0 (0) | 7 (4.1) | 3 (1.9) | 12 (7.1) | 5 (3.2) | 16 (9.5) | |
Abbreviation: IC, immune cell; PD‐L1, programmed death ligand 1; TC, tumor cell.
PD‐L1 expression on TC according to cytological sample type is described in Figure 3A for NSCLC with the SP142 antibody. For histological samples, the percentage of tumors with PD‐L1 expression (≥1%) on TC was highest in patients who had an FNA cytological sample and lowest for patients with a pleural effusion sample (Fig. 3A).
FIGURE 3.
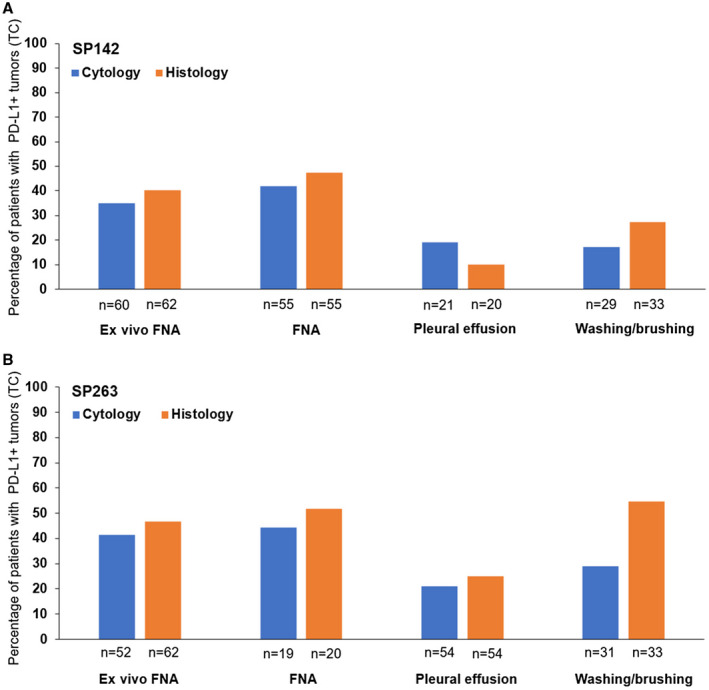
Histograms representing the percentages of patients with TC PD‐L1 expression in histology and cytology samples according to the cytological sample type with (A) the SP142 antibody and (B) the SP263 antibody. FNA also included transbronchial needle aspiration. There were differences in the numbers of evaluable histology and cytology samples between the SP142 and SP263 results. Positive PD‐L1 expression (PD‐L1+) was defined as ≥1% of TC. FNA indicates fine‐needle aspiration; PD‐L1, programmed death ligand 1; TC, tumor cell.
The agreement of histology with cytology was evaluated on a continuous scale. With the SP142 antibody, the ICCs for histology versus cytology in NSCLC were 0.40 (95% CI, 0.26‐0.52) for TC and 0.06 (95% CI, –0.06 to 0.20) for IC (Fig. 4A). ICCs for histology versus cytology of the second independent reading were 0.23 and 0.17, respectively. Agreement was also evaluated by applying a 1% cutoff to both PD‐L1 TC and IC scoring and thereby using PD‐L1 as a binary variable to account for the challenges of performing an absolute quantification on cytological material. For this assay, specificity was high for both TC (84.2%) and IC (94.3%); however, sensitivity was low, especially for IC (19.6%; Table 3). These results were confirmed by the second pathologist's reading. Interreader agreement data are presented in Supporting Figure 1. In addition, post hoc analyses to assess the agreement between artificial ex vivo FNA samples and routinely performed cytological sample types (Supporting Table 3) and between cytology samples with <100 TC and cytology samples with ≥100 TC (Supporting Table 4) were completed.
FIGURE 4.
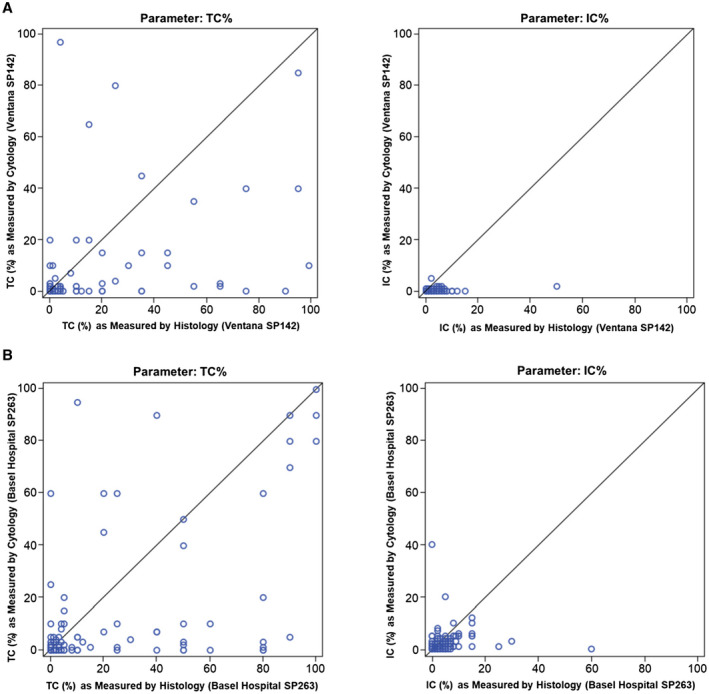
Scatter plots showing (A) the agreement of histology and cytology methods in TC and IC with the PD‐L1 SP142 antibody and (B) the agreement of histology and cytology methods in TC and IC with the PD‐L1 SP263 antibody. IC indicates immune cell; PD‐L1, programmed death ligand 1; TC, tumor cell.
TABLE 3.
Performance of PD‐L1 Scoring Using Cytology Samples and Applying a 1% Cutoff (With Histology as the Reference)
| SP142 Antibody: First Reader | SP142 Antibody: Second Reader | SP263 Antibody | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PD‐L1 TC Status | PD‐L1 TC Status | PD‐L1 TC Status | |||||||||
| Cytology | Histology, No. | Cytology | Histology, No. | Cytology | Histology, No. | ||||||
| <1% | ≥1% | Total | <1% | ≥1% | Total | <1% | ≥1% | Total | |||
| <1% | 85 | 26 | 111 | <1% | 112 | 20 | 132 | <1% | 68 | 27 | 95 |
| ≥1% | 16 | 35 | 51 | ≥1% | 6 | 19 | 25 | ≥1% | 11 | 47 | 58 |
| Total | 101 | 61 | 162 | Total | 118 | 39 | 157 | Total | 79 | 74 | 153 |
| Performance | % (95% CI) | Performance | % (95% CI) | Performance | % (95% CI) | ||||||
| Sensitivity | 57.4 (44.1‐70.0) | Sensitivity | 48.7 (32.4‐65.2) | Sensitivity | 63.5 (51.5‐74.4) | ||||||
| Specificity | 84.2 (75.6‐90.7) | Specificity | 94.9 (89.3‐98.1) | Specificity | 86.1 (76.5‐92.8) | ||||||
| Accuracy | 74.1 (66.6‐80.6) | Accuracy | 83.4 (76.7‐88.9) | Accuracy | 75.2 (67.5‐81.8) | ||||||
| PD‐L1 IC Status | PD‐L1 IC Status | PD‐L1 IC Status | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytology | Histology, No. | Cytology | Histology, No. | Cytology | Histology, No. | ||||||
| <1% | ≥1% | Total | <1% | ≥1% | Total | <1% | ≥1% | Total | |||
| <1% | 66 | 74 | 140 | <1% | 55 | 58 | 113 | <1% | 27 | 47 | 74 |
| ≥1% | 4 | 18 | 22 | ≥1% | 6 | 36 | 42 | ≥1% | 12 | 66 | 78 |
| Total | 70 | 92 | 162 | Total | 61 | 94 | 155 | Total | 39 | 113 | 152 |
| Performance | % (95% CI) | Performance | % (95% CI) | Performance | % (95% CI) | ||||||
| Sensitivity | 19.6 (12.0‐29.2) | Sensitivity | 38.3 (28.5‐48.9) | Sensitivity | 58.4 (48.8‐67.6) | ||||||
| Specificity | 94.3 (86.0‐98.4) | Specificity | 90.2 (79.8‐96.3) | Specificity | 69.2 (52.4‐83.0) | ||||||
| Accuracy | 51.9 (43.9‐59.8) | Accuracy | 58.7 (50.5‐66.6) | Accuracy | 61.2 (53.0‐69.0) | ||||||
Abbreviations: IC, immune cell; PD‐L1, programmed death ligand 1; TC, tumor cell.
IHC Expression Using the SP263 Antibody
PD‐L1 (SP263) TC scoring was successful for 169 histology samples and 157 cytology samples, and IC scoring was successful for 168 histology samples and 157 cytology samples. PD‐L1 staining on TC was present for 80 of 169 histology samples (47.3%) and for 59 of 157 cytology samples (37.6%). PD‐L1 IC staining was observed for 125 of 168 histology samples (74.4%) and for 79 of 157 cytology samples (50.3%). TC staining using the SP263 antibody resulted in a higher percentage of samples with PD‐L1 expression in comparison with SP142 (Table 2).
PD‐L1 expression on TC according to cytological sample type is described in Figure 3B for NSCLC with the SP263 antibody. For histological samples, the percentage of tumors with PD‐L1 expression (≥1%) on TC was lowest when the cytology sample type was a pleural effusion (Fig. 3B).
The agreement of histology with cytology on a continuous scale was also evaluated with the SP263 antibody. Scatterplots of histology and cytology on TC and IC are shown in Figure 4B. The ICCs for cytology versus histology were 0.56 (95% CI, 0.43‐0.65) for TC and 0.10 (95% CI, −0.04 to 0.25) for IC. When a comparison of histology and cytology was performed with a 1% cutoff applied to both PD‐L1 TC and IC scoring, the specificity was 86.1% for TC and 69.2% for IC. Sensitivity was comparable for TC (63.5%) and IC (58.4%), and accuracy was 75.2% for TC and 61.2% for IC with a 1% cutoff (Table 3).
Discussion
This study is the largest fully blinded, prospective analysis to comprehensively assess PD‐L1 expression by using 2 different PD‐L1 antibodies (SP142 and SP263) in lung histology and cell block cytology samples. For the SP142 antibody, this study has demonstrated an interreader agreement for TC scoring consistent with results from the Blueprint2 project in NSCLC. 13 IC scoring had considerably lower interreader agreement, which also confirms the finding from Blueprint2 that IC cannot be reliably quantified in NSCLC histology or cytology material.
When we compared NSCLC histology and cytology, agreement as per the ICC for TC or IC scoring could not be demonstrated with PD‐L1 expression as a continuous variable. For a secondary analysis, a 1% cutoff was applied, and both assays had high specificity but lacked sensitivity. Thus, it may be possible to have false‐negative results in cytological samples because of the low abundancy of IC or TC. In addition, despite being used in routine clinical practice, these cytological samples can be challenging to obtain. A requirement of ≥50 TC was chosen for cytological specimens because routinely obtained materials may contain <50 TC. A subgroup analysis of cytological specimens with ≥100 TC demonstrated that accuracy and specificity were not dissimilar from those for specimens with ≥50 cells. However, sensitivity was lower in samples with 50 to 99 TC, and this suggests that a higher number of TC may be required to achieve better sensitivity in cytological specimens.
In addition, sensitivity was lower for the SP142 antibody than the SP263 antibody, and this confirmed the results from Blueprint1. 20 In addition, some differences might be due to variances between the sample populations (Table 2).
IC scoring of cytological samples was challenging because differentiation of tumor‐associated IC is not possible in samples that lack tissue context (eg, FFPE cell blocks from pleural effusions). Furthermore, samples from endobronchial ultrasound may be contaminated with non–tumor‐associated lymphocytes. Taken together, these data support the common practice to not report IC scores from cytological material. 4 , 16
Overall, TC scoring was considered feasible. With a 1% cutoff, the reported overall agreement for both the SP142 and SP263 antibodies was lower than but comparable to the overall agreement, which has been shown to range from 82% to 97%. 4 , 18 , 20 , 24 These differences may be due to variations in the ways in which samples were obtained in different studies. Prior studies were mostly from a single site, and this allowed for more preanalytical standardization, local validation, and optimization of the IHC protocols for conditions at a single laboratory.
This study also included different cytological sample types, such as washings or brushings, effusions, small aspirates, and artificially generated ex vivo FNA.
Currently, it is unclear to what degree these types of cytological samples are suitable for optimal quantification of PD‐L1 expression. Furthermore, cytological material included in this study was not completely representative of clinical practice because it contained a sizable fraction of artificial ex vivo FNA preparations taken from fresh resection specimens. Such specimens may be less homogeneous and less cellular than endobronchial ultrasound–FNA specimens in clinical practice. Although there is heterogeneity in tumor specimens in clinical practice and in this study, a subgroup analysis evaluating the sensitivity, specificity, and agreement of the NSCLC specimens with artificially generated ex vivo FNA samples from resection specimens did not result in outcomes that differed from the results of samples obtained in routine clinical practice.
This study had several other limitations. Because non–formalin‐fixed samples require a different staining procedure, this study was restricted to formalin‐exposed samples. Nevertheless, there is emerging evidence suggesting that PD‐L1 testing may be applicable to ethanol‐fixed, non–cell block cytology specimens. 24 , 25 Another contributor to variation could be that the histological material used in this study included both biopsy and resection specimens. Furthermore, no meaningful comparisons could be made between histology and cytology for the SCLC cohort because of the small number of samples collected and the overall low PD‐L1 expression. Another limitation caused by the small sample size was the inability to address other expression cutoffs (eg, 25% and 50%) that might be clinically more relevant than the 1% cutoff. 25
Overall, this study has demonstrated that qualitative PD‐L1 analysis is feasible on almost all available cytological sample types when they are processed into cell blocks; however, absolute quantification remains a challenge because of the limited number of TC and IC, the absence of tissue context, and the risk of false‐negative results. Rigorous local validation, quality control, and assurance of tumor content are critical at each laboratory to minimize false‐negative results. Histological tumor specimens should always be preferred over cytology for PD‐L1 assays unless laboratories have validated cytological processes for comparisons. With some limitations and risk of false‐negative results, this study has demonstrated that PD‐L1 expression on TC can be assessed qualitatively (applying a 1% cutoff); however, IC are unable to be robustly scored in cytology and thus should not be reported.
Funding Support
F. Hoffmann‐La Roche Ltd/Genentech, Inc, funded the study; was involved in the study design, data collection, data analysis, data interpretation, and writing of the report; and gave approval to submit the report for publication. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication. Support for third‐party writing assistance for this article by Preshita Gadkari, PhD (Health Interactions), was provided by Genentech, Inc.
Conflict of Interest Disclosures
Lukas Bubendorf received a grant from Roche during the conduct of this study; a grant and personal fees from Bristol Myers Squibb; and personal fees from Boehringer Ingelheim, AstraZeneca, Pfizer, Bayer, AbbVie, and Eli Lilly. Esther Conde received personal fees from Roche and Pfizer and nonfinancial support from Roche, Pfizer, and MSD. Federico Cappuzzo received personal fees from Roche, AstraZeneca, Bristol Myers Squibb, MSD, Pfizer, Takeda, Eli Lilly, and Bayer. Hans‐Ulrich Schildhaus received a grant from Roche during the conduct of this study and personal fees from Roche, Bristol Myers Squibb, and MSD. Isabel Esteban‐Rodriguez received product and technical financing and nonfinancial support from Roche during the conduct of this study and personal fees from Roche, MSD, AstraZeneca, and Pfizer. Janine Feng, Jenny Devenport, Christine Boyiddle, Stefanie Morris, and Kerstin Trunzer are employed by Roche; Feng, Devenport, and Trunzer have received Roche stock. Keith M. Kerr received personal fees from Roche and Roche Diagnostics. The other authors made no disclosures.
Author Contributions
Lukas Bubendorf: Conceptualization, funding acquisition, methodology, investigation, project administration, resources, supervision, writing–original draft, and writing–review and editing. Esther Conde: Resources and writing–review and editing. Federico Cappuzzo: Conceptualization, writing–original draft, and writing–review and editing. Renata Langfort: Investigation, resources, and writing–review and editing. Hans‐Ulrich Schildhaus: Investigation, resources, and writing–review and editing. Jiří Votruba: Investigation, validation, and writing–review and editing. Ángel Concha‐López: Resources and writing–review and editing. Isabel Esteban‐Rodriguez: Investigation, resources, and writing–review and editing. Janine Feng: Investigation and writing–review and editing. Jenny Devenport: Data curation, formal analysis, project administration, visualization, and writing–review and editing. Christine Boyiddle: Methodology and writing–review and editing. Stefanie Morris: Writing–review and editing. Kerstin Trunzer: Conceptualization, investigation, methodology, supervision, visualization, writing–original draft, and writing–review and editing. Keith M. Kerr: Conceptualization, methodology, supervision, writing–original draft, and writing–review and editing.
Supporting information
Supplementary Material
Bubendorf L, Conde E, Cappuzzo F, Langfort R, Schildhaus H-U, Votruba J, Concha-López Á, Esteban-Rodriguez I, Feng J, Devenport J, Boyiddle C, Morris S, Trunzer K, Kerr KM. A noninterventional, multinational study to assess PD-L1 expression in cytological and histological lung cancer specimens. Cancer Cytopathol. 2020:128:928‐938. 10.1002/cncy.22324
We acknowledge K. Schulze, F. Lopez‐Rios, and A. M. Rodríguez for their ideas and advice during the study.
Data Availability
Qualified researchers may request access to individual patient–level data through the clinical study data request platform (https://vivli.org/). Further details on Roche's criteria for eligible studies are available at https://vivli.org/members/ourmembers/. For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, go to https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm.
References
- 1. Walters S, Maringe C, Coleman M, et al. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: a population‐based study, 2004‐2007. Thorax. 2013;68:551‐564. [DOI] [PubMed] [Google Scholar]
- 2. Travis W, Brambilla E, Noguchi M, et al. Diagnosis of lung cancer in small biopsies and cytology: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Arch Pathol Lab Med. 2013;137:668‐684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Santos G, Saieg M. Preanalytic specimen triage: smears, cell blocks, cytospin preparations, transport media, and cytobanking. Cancer Cytopathol. 2017;125:455‐464. [DOI] [PubMed] [Google Scholar]
- 4. Jain D, Nambirajan A, Borczuk A, et al. Immunocytochemistry for predictive biomarker testing in lung cancer cytology. Cancer Cytopathol. 2019;127:325‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischer A, Schwartz M, Moriarty A, et al. Immunohistochemistry practices of cytopathology laboratories. Arch Pathol Lab Med. 2014;138:1167‐1172. [DOI] [PubMed] [Google Scholar]
- 6. National Comprehensive Cancer Network . NCCN Clinical Practice Guidelines in Oncology: Non–Small Cell Lung Cancer. V4.2019. National Comprehensive Cancer Network; 2019. [Google Scholar]
- 7. Planchard D, Popat S, Kerr K, et al. Metastatic non–small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow‐up. Ann Oncol. 2018;29(suppl 4):iv192‐iv237. [DOI] [PubMed] [Google Scholar]
- 8. Yatabe Y, Kerr KM, Utomo A, et al. EGFR mutation testing practices within the Asia Pacific region: results of a multicenter diagnostic survey. J Thorac Oncol. 2015;10:438‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malapelle U, Sirera R, Jantus‐Lewintre E, et al. Profile of the Roche Cobas® EGFR mutation test v2 for non–small cell lung cancer. Expert Rev Mol Diagn. 2017;17:209‐215. [DOI] [PubMed] [Google Scholar]
- 10. Sholl L. Molecular diagnostics of lung cancer in the clinic. Transl Lung Cancer Res. 2017;6:560‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gosney JR, Boothman AM, Ratcliffe M, Kerr KM. Cytology for PD‐L1 testing: a systematic review. Lung Cancer. 2020;141:101‐106. [DOI] [PubMed] [Google Scholar]
- 12. Udall M, Rizzo M, Kenny J, et al. PD‐L1 diagnostic tests: a systematic literature review of scoring algorithms and test‐validation metrics. Diagn Pathol. 2018;13:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsao MS, Kerr KM, Kockx M, et al. PD‐L1 immunohistochemistry comparability study in real‐life clinical samples: results of Blueprint phase 2 project. J Thorac Oncol. 2018;13:1302‐1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heymann JJ, Bulman WA, Swinarski D, et al. PD‐L1 expression in non–small cell lung carcinoma: comparison among cytology, small biopsy, and surgical resection specimens. Cancer Cytopathol. 2017;125:896‐907. [DOI] [PubMed] [Google Scholar]
- 15. Ilie M, Juco J, Huang L, Hofman V, Khambata‐Ford S, Hofman P. Use of the 22C3 anti–programmed death‐ligand 1 antibody to determine programmed death‐ligand 1 expression in cytology samples obtained from non–small cell lung cancer patients. Cancer Cytopathol. 2018;126:264‐274. [DOI] [PubMed] [Google Scholar]
- 16. Munari E, Zamboni G, Sighele G, et al. Expression of programmed cell death ligand 1 in non–small cell lung cancer: comparison between cytologic smears, core biopsies, and whole sections using the SP263 assay. Cancer Cytopathol. 2019;127:52‐61. [DOI] [PubMed] [Google Scholar]
- 17. Skov B, Skov T. Paired comparison of PD‐L1 expression on cytologic and histologic specimens from malignancies in the lung assessed with PD‐L1 IHC 28‐8pharmDx and PD‐L1 IHC 22C3pharmDx. Appl Immunohistochem Mol Morphol. 2017;25:453‐459. [DOI] [PubMed] [Google Scholar]
- 18. Torous V, Rangachari D, Gallant B, Shea M, Costa D, VanderLaan P. PD‐L1 testing using the clone 22C3 pharmDx kit for selection of patients with non–small cell lung cancer to receive immune checkpoint inhibitor therapy: are cytology cell blocks a viable option? J Am Soc Cytopathol. 2018;7:133‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang G, Ionescu DN, Lee CH, et al. PD‐L1 testing on the EBUS‐FNA cytology specimens of non–small cell lung cancer. Lung Cancer. 2019;136:1‐5. [DOI] [PubMed] [Google Scholar]
- 20. Hirsch FR, McElhinny A, Stanforth D, et al. PD‐L1 immunohistochemistry assays for lung cancer: results from phase 1 of the Blueprint PD‐L1 IHC assay comparison project. J Thorac Oncol. 2017;12:208‐222. [DOI] [PubMed] [Google Scholar]
- 21. Russell‐Goldman E, Kravets S, Dahlberg S, Sholl L, Vivero M. Cytologic‐histologic correlation of programmed death‐ligand 1 immunohistochemistry in lung carcinomas. Cancer Cytopathol. 2018;126:253‐263. [DOI] [PubMed] [Google Scholar]
- 22. Lozano MD, Abengozar‐Muela M, Echeveste JI, et al. Programmed death–ligand 1 expression on direct Pap‐stained cytology smears from non–small cell lung cancer: comparison with cell blocks and surgical resection specimens. Cancer Cytopathol. 2019;127:470‐480. [DOI] [PubMed] [Google Scholar]
- 23. Boyiddle C, Ruboyianes M, Del Valle E, et al. Development of IHC staining protocols for assessment of PD‐L1 expression in cytological samples [abstract 655]. Cancer Res. 2017;77(13 suppl):655. [Google Scholar]
- 24. Noll B, Wang WL, Gong Y, et al. Programmed death ligand 1 testing in non–small cell lung carcinoma cytology cell block and aspirate smear preparations. Cancer Cytopathol. 2018;126:342‐352. [DOI] [PubMed] [Google Scholar]
- 25. Shen X, Zhao B. Efficacy of PD‐1 or PD‐L1 inhibitors and PD‐L1 expression status in cancer: meta‐analysis. BMJ. 2018;362:k3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Qualified researchers may request access to individual patient–level data through the clinical study data request platform (https://vivli.org/). Further details on Roche's criteria for eligible studies are available at https://vivli.org/members/ourmembers/. For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, go to https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm.


