Abstract
Background
The Verax PGD rapid test for bacteria in platelets (PLTs) has been updated to simplify workflow and improve specificity and sensitivity by employing a novel sequential format. The performance of this updated version, called PGDprime, was evaluated to determine its suitability for use as an FDA‐cleared “safety measure” to supplant the current PGD test.
Study design and methods
Three consecutive cGMP‐manufactured lots of PGDprime were evaluated for specificity (at three separate sites), sensitivity, reproducibility, interfering substances, assay robustness, and detection in analytical growth and ultralow‐inoculum growth studies. PGDprime's performance was compared to that of PGD.
Results
Specificity studies yielded no false‐positive results among 3802 individual indate PLTs of seven different types (observed specificity, 100%). PGDprime detected all 10 PGD claim bacteria at the same limit of detection or better. Wild‐type Gram‐negative bacteria growing in PLTs were detected at earlier elapsed times than PGD by 12 to 30 hours. In growth studies, PGDprime detected bacteria growing in PLTs within the same 12‐hour interval as PGD or 12 to 48 hours earlier. Assay reproducibility was not affected by operator, day of test, or manufacturing lot. PGDprime tolerated a wide variation in volume transfers, timing, temperature, and relative humidity and was not affected by 15 of 16 potential interferents found in samples at extremely high or low levels.
Conclusion
The PGD test has been successfully updated to PGDprime with an innovative sequential assay format to deliver a robust simplified workflow and improved specificity and sensitivity.
Keywords: bacteria, platelet transfusion, transfusion‐transmitted disease
Abbreviations
- IR
initially reactive
- LoD
limit of detection
- LR
leukoreduced
- LRAP(s)
leukoreduced apheresis platelet(s)
- nLR
nonleukoreduced
- PSP
pre‐storage pools of LR random‐donor platelets
- WBDPs
whole blood–derived platelets
- WBDPp
whole blood‐derived platelet pool
1. INTRODUCTION
Bacterial contamination of platelet (PLT) components remains the primary infectious risk of blood component transfusion. The current practice of performing only a primary aerobic culture detects only 11% to 47% of contaminated components. The FDA issued a final guidance in September 2019 to provide pathways to further mitigate this risk. 1 The guidance provides options to extend expiration dating of leukoreduced apheresis PLTs (LRAPs) in 100% plasma to 7 days by testing for bacterial contamination with a device cleared as a “safety measure.” One such safety measure is the Verax Biomedical PLT PGD test. After a decade in service, the test has been redesigned and updated. The updated test is referred to as PGDprime. This new version of the test remains a rapid, qualitative multiplexed immunoassay for the detection of aerobic and anaerobic Gram‐positive and Gram‐negative bacteria in
LRAP suspended in plasma, LRAP suspended in PAS‐C and plasma, and prestorage pools of up to six leukoreduced (LRs) whole blood–derived PLTs (WBDPs) suspended in plasma, within 24 hours before PLT transfusion as a safety measure after testing with a growth‐based quality control test cleared by the FDA for PLT components;
Poststorage pools (pooled within 4 hours of transfusion) of up to 6 units of LR and nonleukoreduced (nLR) WBDPs suspended in plasma; and
Single units of LR and nLR WBDP suspended in plasma and tested within 4 hours before PLT transfusion as individual PLT units or as components of a poststorage pool.
PGDprime was designed to simplify workflow, improve specificity and sensitivity, and shorten the time to first detection of many wild‐type strains.
1.1. Design
1.1.1. Design factors for improvement of sensitivity and breadth of detection
To achieve simplification of workflow and an expanded breadth of bacterial detection, an innovative sequential lateral flow design was optimized. Conventional lateral‐flow rapid assays such as PGD use detector antibodies directly attached to signal particles such as colloidal gold. The sensitivity of such assays are directly dependent upon the number of signal particles that can be loaded onto a test strip. The test strip has a finite capacity for relatively large signal particles and when the targets of detection reach dozens in number for a pan genera method, the limit of sensitivity for each target is quickly reached.
The PGDprime test uses a sequential format (Figure 1) wherein detector antibodies are not attached to signal particles. 2 These detector antibodies, which are much smaller than the gold signal particles used, are labeled with biotin, a small molecule that attaches to streptavidin, its binding partner. In addition, these detector antibodies can be dissolved in aqueous solutions at far greater molarity than the capacity of the strip to contain them in a dry state. In PGDprime, streptavidin is attached to gold signal particles, which are separately contained and dried on a separate pad on the test strip and are released only after the detector and capture antibodies have formed their sandwich complexes with bacteria and bacterial fragments.
FIGURE 1.
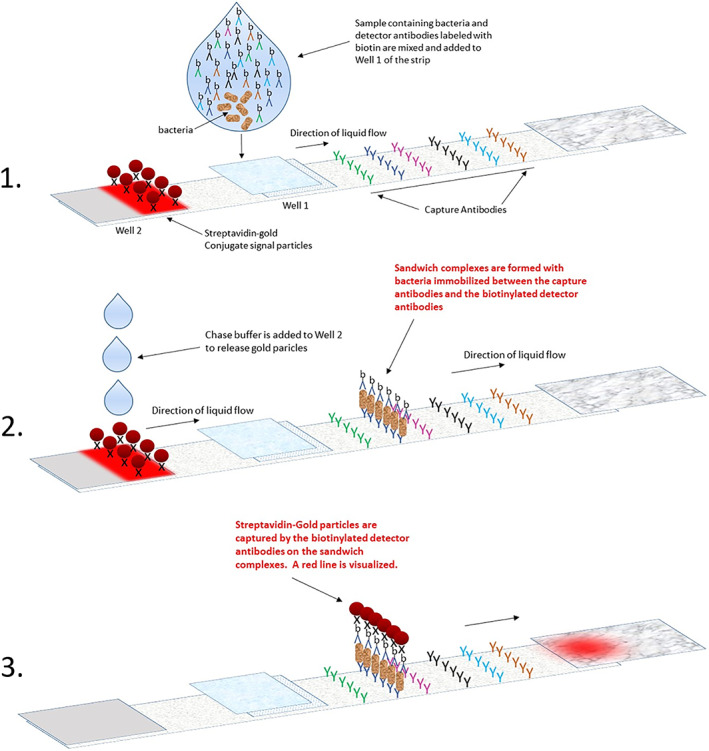
Mechanism of the sequential assay format employed in PGDprime
Detector antibodies can therefore be loaded onto the test strip or contained in a liquid reagent in log fold more quantities than the test strip's capacity for gold particles. This increases the sensitivity and the breadth of detection of the assay, enabling more bacterial targets to be detected. After the sandwich complexes with bacteria are formed, the gold signal particles are released to label the complexes and enable visualization of the test result. The quantity of gold particles required is not high since the signal particles that need to be captured by the formed sandwich complexes are a very small fraction of the total number of signal particles available. Sensitivity is no longer dependent on the number of signal particles. Sensitivity of the assay is driven by the vastly greater quantities of dissolved detector antibodies.
Figure 2 shows the form factor and layout of the updated PGDprime. PGDprime uses a smaller sample size than PGD (150 μL vs. 500 μL) and requires no centrifugation, no pellet resuspension, no precision pipetting, no humidity chamber, no temperature or humidity monitoring, and no vortexing except for nLR WBDPs.
FIGURE 2.
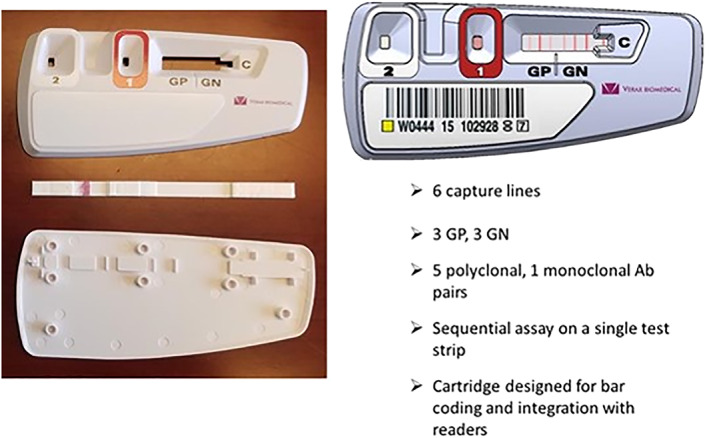
Layout of the PGDprime test device
1.1.2. Design factors for improved specificity
Immunoassays are known to be susceptible to interference from human endogenous antibodies that may interact nonspecifically with the antibodies used in the immunoassay. Human anti‐animal antibodies and heterophile antibodies are present in some human plasma. These can react with the Fc portion of an immobilized capture antibody and the Fc portion of the detector antibody, creating a false‐positive sandwich complex. Figure 3 depicts a true‐positive result and a false‐positive result due to bridging of the capture and detector antibodies by a heterophile agent via their Fc regions. With the original PGD test, this heterophile interference accounted for most of the 0.5% false‐positive rate observed for the method. This error mode was later corrected in PGD by removing the Fc fragment from one of the antibodies. 3
FIGURE 3.
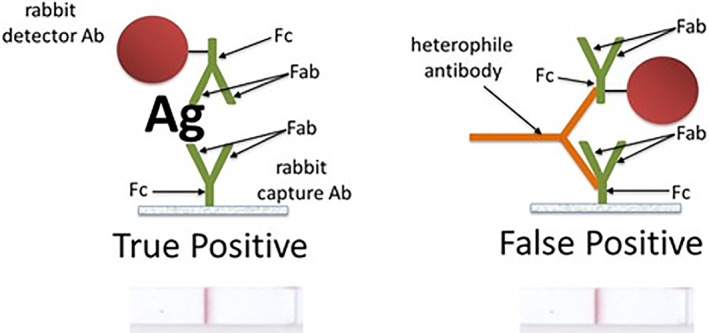
Heterophile antibody creates a false‐positive bridging complex between the capture and detector antibodies via their Fc regions. Removal of the Fc from one of the antibodies prevents this
To prevent the formation of these anomalous sandwich complexes, PGDprime uses F(ab')2 fragments of antibodies for six of seven detectors. F(ab')2 are antibody fragments resulting from the enzymatic cleavage of the Fc portion from the whole antibody. With the Fc no longer present on the detector antibody, false‐positive sandwich complexes created by heterophilic agents are efficiently avoided. For the seventh detector antibody, added animal immunoglobulin was found to be sufficient to block these interactions.
1.1.3. Assay control line
The control line in PGDprime is a true assay control line. It comprises streptavidin immobilized on the distal end of the membrane. Excess biotinylated detectors are captured by this line. Excess gold‐streptavidin particles are subsequently captured by the immobilized detectors at the control line. If the operator forgets a step in the workflow of the assay, the control line will not appear, thus serving as a true indicator of validity or error. In the older PGD test, the control was a liquid‐flow indicator, which, although an indirect indication of correct user manipulation, was not indicative of the success or failure of the immunoassay process.
2. MATERIALS AND METHODS
2.1. Assay components and procedure
The components required to run a single test include the following:
Reagent 1A: A sample pretreatment reagent to digest complexed bacteria and interferents.
Reagent 1B: A neutralizing agent containing detector antibodies.
Reagent 2: Chase buffer to release immobilized gold conjugate.
The PGDprime test device (Figure 2).
The test procedure is summarized as follows:
Obtain 150 μL of a PLT sample and transfer to a sample processing tube.
Add six drops of Reagent 1A, mix, and wait at least 2 minutes.
Add six drops of Reagent 1B and mix.
Transfer 50 μL of the pretreated sample to Well 1 of the test device.
Add six drops of Reagent 2 to Well 2 of the test device.
Read result in the results window after all traces of the conjugate have disappeared and a control line has formed.
2.2. Evaluation of Performance
The performance of PGDprime was evaluated using consecutive lots of test devices and reagents manufactured under cGMP. The following studies were conducted:
2.2.1. Specificity
The specificity of the updated test was evaluated at three sites. 4 A total of 3802 individual PLT samples of various types were tested with PGDprime using blinded aerobic and anaerobic culture as the predicate method. A negative result on PGDprime was classified as nonreactive. The current PGD confirmation protocol was followed when an initially reactive (IR) sample was encountered on PGDprime. IR samples were retested on two additional PGDprime devices. If at least one retest was also reactive, the final result was determined to be repeat reactive and the sample considered to be a bacteria positive specimen. Invalid test runs were also tracked.
The following PLT types were tested: LR WBDP, poststorage LR WBDPp, LRAP, nLR WBDP, poststorage nLR WBDPp, LRAP in PAS‐C, and prestorage pools of LR PLTs (PSP). Samples that were culture positive but PGDprime nonreactive were determined to be false negative. Samples that were culture negative but PGDprime repeat reactive were determined to be false positive.
2.2.2. Sensitivity
The sensitivity of PGDprime was compared to that of PGD in a limit of detection (LoD) equivalence study by testing the PGD claim bacteria at three levels. 5 The PGD claim bacteria are Bacillus cereus, Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus agalactiae, Clostridium perfringens, Klebsiella aerogenes, Pseudomonas aeruginosa, Escherichia coli, Serratia marcescens, and Klebsiella pneumoniae. For each bacterial claim strain, a three‐member panel was prepared.
Lower: 0.4 to 0.9 logs below the PGD LoD
Middle (LoD): within 0.5 logs of the PGD LoD
Upper: 0.6 to 1.0 logs above the PGD LoD
All concentrations are after a 1‐in‐21 dilution into a PLT matrix. Two negative panel members were also included in the blinded study. Each three‐member panel was diluted into 10 individual indate apheresis in plasma PLT samples and tested with three separate lots of PGDprime and one lot of PGD in a blinded study. PGDprime would therefore have 30 results to 10 with PGD.
In a subsequent study to confirm LoD in other PLT types, the middle (LoD) and negative panel members were diluted into at least 5 units each of the following PLT types:
PSP;
Poststorage pools of non‐LR random‐donor PLTs (nLR pools);
Apheresis PLTs in PAS.
PGD is not able to detect Streptococcus oralis. 6 In a separate study, the detection of S. oralis by the new test was evaluated by diluting the organism into PLTs and determining the colony‐forming units (CFU)/mL by dilution plate count of the highest dilution detected by PGDprime.
2.2.3. Breadth of detection
To assess the improvement in breadth of detection, several wild‐type Gram‐negative bacteria isolated from various contamination events since the initial release of PGD were inoculated at 1000 CFU/mL into individual apheresis PLTs samples that were then tested at 6‐hour intervals with PGD and PGDprime. 7 The times to initial detection for PGD and PGDprime were compared.
2.2.4. Reproducibility
The results of all of the tests using LoD panels performed during the sensitivity studies were analyzed to assess reproducibility. There were 42 test runs on each of three device lots for each LoD panel member and 52 test runs for negative samples on each of three device lots. Three reagent lots were rotated through the testing. Three operators were involved in testing.
2.2.5. Analytical and ultralow‐inoculum growth studies
Growth studies were conducted to evaluate the ability of the PGDprime test to detect bacteria initially present at low levels that culture may miss due to sampling errors. Detection by PGDprime was compared to that by PGD. 8 Initially, an analytical growth study was conducted comparing the time to detection by PGDprime and PGD of nine aerobic claim bacteria inoculated at low CFU/mL in apheresis in plasma PLT bags (≤21 CFU/mL).
In a follow‐on ultralow‐inoculum growth study, three bacteria, B. cereus, K. pneumoniae, and S. epidermidis, were inoculated individually at an initial population of not more than 188 CFU/bag (≤0.6 CFU/mL) into three different types of PLTs, apheresis in plasma, PAS‐C, and LR WBDPp. The time to detection by the two methods of these three representative bacteria in low CFU/bag levels were compared.
2.2.6. Interfering substances
The detection of 11 bacteria, B. cereus, C. perfringens, K. aerogenes, E. coli, K. pneumoniae, P. aeruginosa, S. marcescens, S. aureus, S. epidermidis, S. agalactiae, and S. oralis, and the accuracy of negative samples were evaluated in the presence of abnormal levels of the following potentially interfering substances and sample conditions. 9
Rheumatoid factor High cholesterol
HAMA High total protein
ANA Low total protein
ds‐DNA High and low pH
High IgA Red blood cells
High IgM White blood cells
High IgG PLT concentration (0.5×, 1×, 2×)
Lipemia PAS‐C content
For each species, a concentration of bacteria representing a lower midrange detection signal on the test was used. A minimum of 5 units or pools of each of the following sample types were tested:
Apheresis PLTs;
LR WBDPp or prestorage LR PLT pools;
nLR WBDPp.
2.2.7. User guardbands
The robustness of PGDprime to environmental extremes and to procedural errors that may be committed by users was tested by reconstituting the middle (LoD) level panel member associated with each of the PGD bacterial claim species in two apheresis in plasma PLT units and two nLR PLT pools. 10 A negative panel member was also reconstituted in the PLT units. The accuracy of test results was evaluated when these samples were tested using PGDprime under extremes of test environment and when deviations from test instructions were applied:
Humidity: 10%–90% RH;
Ambient temperature: 15–30°C;
Ambient airflows due to room and equipment ventilation;
Deviation of reagent and sample volumes added up to ±2 drops;
Sample pretreatment incubation time > 2 minutes.
3. RESULTS
3.1. Specificity
The specificity study evaluated the performance of PGDprime across seven PLT types and ages (Day 2‐Day 6) at three independent sites. Each sample was blindly tested and confirmed to be negative via aerobic and anaerobic plate culture. Table 1 summarizes the PGDprime results obtained at the three sites with 3802 samples.
TABLE 1.
Statistical analysis of PGDprime specificity study results
| PLT type | Results | All a | Specificity | ||||
|---|---|---|---|---|---|---|---|
| IR/rate | Repeat reactive/rate | Nonreactive | Indeterminate | Observed | Lower one‐sided 95% confidence limit | ||
| LR WBDP | 0/0% | 0/0% | 611 | 0 | 611 | 100% | 99.6% |
| LR WBDPp | 0/0% | 0/0% | 75 | 0 | 75 | 100% | 96.5% |
| LRAP | 1/0.06% | 0/0% | 1598 | 0 | 1599 b | 100% | 99.8% |
| nLR WBDP a | 1/0.2% | 0/0% | 501 | 1 | 503 | 99.8% | 99.1% |
| 0/0% | 502 | 100% | 99.5% | ||||
| nLR WBDPp | 1/1.5% | 0/0% | 64 | 0 | 65 | 100% | 96.0% |
| PAS | 2/0.7% | 0/0% | 295 | 0 | 297 | 100% | 99.1% |
| PSP | 0/0% | 0/0% | 650 | 0 | 650 | 100% | 99.6% |
| All a | 5/0.13% | 0/0% | 3794 | 1 | 3800 | 100% | 99.9% |
| 100% | 99.9% | ||||||
Reported both with (n = 503) and without (n = 502) the sample classified as Ind (indeterminate) because the IR result for that sample was retested with only a single additional test instead of two, making it impossible to interpret as nonreactive or repeat reactive. (Note: The single repeat retest was nonreactive.).
Two samples excluded due to repeat invalid results. These were the only invalid results in the study (0.05%).
Two samples were repeatedly invalid and were excluded from the statistical analyses. There were five IR samples (0.13%) but no repeat reactive results (confirmed false positives) in the population tested. One of these IR samples was classified as indeterminate after the study site failed to retest the sample twice per protocol. The single confirmatory repeat test run at the site gave a nonreactive (negative) result. Culture results for this sample were also negative. With this sample excluded as neither confirmed positive nor confirmed negative, the observed specificity was 100%.
3.2. Sensitivity
The results of the LoD equivalence study comparing the sensitivity of PGDprime with PGD are summarized in Table 2. PGDprime detected all of PGD's claim bacteria at the LoD (middle) level. With three of the 10 claim species, S. epidermidis, S. agalactiae, and S. marcescens, PGDprime consistently detected the strain at a level lower than the LoD of PGD. With an additional three organisms, B. cereus, S. aureus, and C. perfringens, PGDprime detected the lower level 73% to 97% of the time, implying that the true LoD of the updated test for each of these bacteria was indeed significantly lower than the PGD LoD but somewhat higher than the lower level tested. In comparison, PGD detected the lower level of these three bacteria 0% of the time.
TABLE 2.
Results of LoD equivalence study comparing detection of claim strains by PGD and PGDprime
| LoD comparison (reactive results/units tested) | ||||
|---|---|---|---|---|
| Bacteria | PGD LoD (CFU/mL) | Level (middle = LoD) | PGD | PGDprime (three lots) |
| B. cereus | Upper | 10/10 | 30/30 | |
| 1.2 × 104 | Middle | 10/10 | 30/30 | |
| Lower | 0/10 | 26/30 | ||
| S. aureus | Upper | 10/10 | 30/30 | |
| 8.2 × 103 | Middle | 10/10 | 30/30 | |
| Lower | 0/10 | 22/30 | ||
| S. epidermidis | Upper | 10/10 | 30/30 | |
| 9.2 × 103 | Middle | 10/10 | 30/30 | |
| Lower | 0/10 | 30/30 | ||
| S. agalactiae | Upper | 10/10 | 30/30 | |
| 5.5 × 104 | Middle | 10/10 | 30/30 | |
| Lower | 0/10 | 30/30 | ||
| C. perfringens | Upper | 10/10 | 30/30 | |
| 8.9 × 104 | Middle | 10/10 | 30/30 | |
| Lower | 1/10 | 29/30 | ||
| K. aerogenes | Upper | 10/10 | 30/30 | |
| 1.0 × 104 | Middle | 10/10 | 30/30 | |
| Lower | 0/10 | 2/30 | ||
| P. aeruginosa | Upper | 10/10 | 30/30 | |
| 8.2 × 103 | Middle | 10/10 | 30/30 | |
| Lower | 0/10 | 4/30 | ||
| E. coli | Upper | 10/10 | 30/30 | |
| 2.8 × 104 | Middle | 10/10 | 30/30 | |
| Lower | 0/10 | 2/30 | ||
| S. marcescens | Upper | 10/10 | 30/30 | |
| 8.6 × 105 | Middle | 10/10 | 30/30 | |
| Lower | 1/10 | 30/30 | ||
| K. pneumoniae | Upper | 10/10 | 30/30 | |
| 2.0 × 104 | Middle | 10/10 | 30/30 | |
| Lower | 0/10 | 0/30 | ||
| Negative | 0/20 | 0/60 a | ||
Two initial reactive results; no repeat‐reactive results.
In the subsequent study of LoD detection in three other PLT types, PGDprime also detected all of PGD's bacterial claim strains at the LoD level. Detection at all LoDs were observed with three lots of PGDprime in 10 individual units of PSP, six individual units of poststorage pools of non‐LR random donor PLTs (nLR pools) and in seven individual units of apheresis PLTs in PAS.
The lowest detectable concentration of S. oralis in the dilution study was 1.95 × 106 CFU/mL. The actual LoD would be somewhere between this level and the next higher dilution at 9.75 × 105 CFU/mL.
3.3. Breadth of detection
The time to detection by PGD and PGDprime of several wild‐type Gram‐negative isolates associated with PLT contamination are compared in Table 3. In all cases, PGDprime was able to detect bacterial growth from 6 to 30 hours earlier than PGD.
TABLE 3.
Comparison of time to detection by PGD and PGDprime of wild‐type Gram‐negative bacteria growing in PLTs (initial inoculum = 1000 CFU/mL)
| GN PLT isolate grown in apheresis PLTs | Time to detection (hours) | Difference (hours) | |
|---|---|---|---|
| PGD | PGDprime | ||
| Citrobacter koseri A0053 | 36 | 18 | 18 |
| E. coli 660366 | 42 | 12 | 30 |
| E. coli No.36 | 30 | 18 | 12 |
| E. coli No.50 | 30 | 18 | 12 |
| E. coli Grenoble | 24 | 12 | 12 |
| E. coli NBL‐4 | 30 | 24 | 6 |
| E. coli TX | 48 | 24 | 24 |
| K. pneumoniae No.65 | 24 | 12 | 12 |
| K. pneumoniae NBL‐1 | 30 | 18 | 12 |
| S. marcescens E5021556 | 42 | 30 | 12 |
| S. marcescens No.31 | 24 | 12 | 12 |
| P. aeruginosa NBL‐5 | 72 | 48 | 24 |
3.4. Reproducibility
The results of all of the tests using LoD panels performed during the sensitivity studies were analyzed to assess reproducibility. There were 42 test runs on each of three device lots for each LoD panel member and 52 test runs for negative samples on each of three device lots. Three reagent lots were rotated through the testing. Three operators were involved in testing.
All LoD samples were detected in each run. All negative samples tested nonreactive. There was no effect of operator, reagent lot, or device lot on the test results. Overall, the average intensity of results is slightly higher on one lot of the three tested, with no effect on specificity.
3.5. Analytical and ultra low inoculum growth studies
Growth studies are conducted to evaluate the ability of the PGDprime test to detect bacteria initially present at low levels that culture may miss due to sampling errors. Initially, an analytical growth study was conducted comparing the time to detection by PGDprime and PGD of nine aerobic claim bacteria present at low CFU/mL (≤21 CFU/mL) in PLT bags. Duplicates of three lots of PGDprime and one lot of PGD were used in the 12‐hour interval testing. Table 4 shows that both tests detected bacteria within a 12‐hour interval of each other.
TABLE 4.
Time to detection by PGD and PGDprime of bacteria inoculated at low CFU/mL in PLT units
| Bacteria | Initial CFU/mL in bag | Hours after inoculation to initial reactivity (no. detected/no. tested) | |
|---|---|---|---|
| PGD | PGDprime | ||
| B. cereus | 18.8 | 24 (2/2) | 24 (6/6) |
| S. aureus | 17.3 | 48 (2/2) | 48 (6/6) |
| S. epidermidis | 16.3 | 96 (2/2) | 84 (6/6) |
| S. agalactiae | 13.8 | 120 (2/2) | 108 (6/6) |
| K. aerogenes | 6.3 | 48 (2/2) | 60 (6/6) |
| 5.8 | 72 (2/2) | 72 (6/6) | |
| E. coli | 9.8 | 96 (2/2) | 96 (6/6) |
| K. pneumoniae | 6.5 | 36 (2/2) | 36 (6/6) |
| P. aeruginosa | 21 | 84 (2/2) | 84 (6/6) |
| S. marcescens | 3.8 | 48 (2/2) | 48 (6/6) |
In a follow‐on ultralow‐inoculum growth study, the time to detection by the two methods of three representative bacteria in low CFU/bag levels (≤ 188 CFU/bag) were compared in three PLT types. The results are presented in Table 5.
TABLE 5.
Time to detection by PGD and PGDprime of bacteria inoculated at ultralow CFU/bag in PLT units
| Bacteria | CFU/bag at inoculation | CFU/mL at inoculation | Time to detection (hours) | |
|---|---|---|---|---|
| PGD | PGDprime | |||
| Apheresis PLTs | ||||
| B. cereus | 117.3 | 0.514 | 24 | 24 |
| 11.73 | 0.051 | 36 | 36 | |
| K. pneumoniae | 1.45 | 0.007 | 36 | 36 |
| S. epidermidis | 17 | 0.075 | 96 | 96 |
| 162.8 | 0.74 | 96 | 96 | |
| PAS‐C PLTs | ||||
| B. cereus | 56.1 | 0.301 | 36 | 36 |
| K. pneumoniae | 25.1 | 0.078 | 84 | 36 |
| 188.4 | 0.78 | 60 | 36 | |
| S. epidermidis | 66.1 | 0.234 | 96 | 96 |
| LR WBDPp | ||||
| B. cereus | 8.1 | 0.029 | 48 | 36 |
| 75.4 | 0.29 | 48 | 36 | |
| K. pneumoniae | 24.6 | 0.099 | 96 | 48 |
| S. epidermidis | 65.5 | 0.23 | 96 | 96 |
These results show that in most but not all PLT bags inoculated with extremely low levels of bacteria, both PGDprime and PGD detected bacteria at the same time intervals. However, in several cases, PGDprime detected bacterial growth much earlier than PGD by 12 to 48 hours.
It has been demonstrated in various growth studies that the time to detection of any bacterial strain will vary from PLT to PLT depending on the presence of growth‐inhibiting factors such as endogenous antibodies and other sample‐specific conditions that may impair or enhance bacterial proliferation. 11
3.6. Interfering substances
With the exception of one elevated IgM sample in the presence of E. coli, there were no effects of interfering substances or conditions on performance of the PGD prime when testing 11 bacteria‐positive samples. One elevated IgM sample yielded a repeatable false‐negative result with E. coli while all other bacteria were detected in the presence of this IgM sample. One possible explanation is that the IgM sample contained a high titer of antibodies specific to E. coli.
Potential interferents were not provided as sterile materials from the sample supplier. Six HAMA samples and one ANA sample produced reactive results on single Gram‐negative detector/capture pairs, results consistent with the presence of bacterial antigens. Had the HAMA samples truly created a false‐positive reaction, the false positive would have been a Gram‐positive reaction since the only mouse antibody pair used detects Gram‐positive, not Gram‐negative, bacteria. One ds‐DNA sample and two elevated protein samples yielded signal on multiple detector/capture pairs, results consistent with nonspecific reactions. All other results from valid PGDprime assays yielded nonreactive results. Some high‐protein samples (>10 g/dL) did not flow, resulting in invalid test results.
3.7. User guardbands
PGDprime was tolerant of most expected user errors and environmental variations:
The test performs accurately at relative humidity between 10% and 90%, obviating the need for the humidity chamber required by PGD.
The test can be performed with valid results at temperatures as low as 15°C and as high as 30°C.
The test is not susceptible to local ambient airflows due to room and equipment ventilating fans.
Each of the test reagents is added in six‐drop increments. The test can tolerate a ± 2 drop error.
Sample pretreatment requires 2 minutes of exposure. Valid test results are obtained with up to 30 minutes of pretreatment.
No loss in accuracy was observed when these conditions were stacked or combined in multiple combinations.
No vigorous mixing of sample/reagent mixtures is required except for nLR WBDPs.
The test does not require a centrifuge or precision pipetting.
4. DISCUSSION
When comparing the older PGD rapid test with PGDprime, several improvements are notable. Ease of use has been enhanced by the elimination of centrifugation and precision pipetting. By so doing, the number of user manipulations has been reduced from seven to four steps.
Specificity has been significantly improved by using F(ab')2 fragments for detector reagents. Although five initial reactives out of 3800 samples were reported (0.13%), no repeat‐reactive results (confirmed false positives) were obtained in the study with the entire population of culture‐confirmed negative samples. The improved test shows earlier detection and lower LoDs of many claim bacteria tested. PGDprime detects other bacteria at the same levels as PGD. No bacteria tested has shown a poorer LoD with PGDprime compared to PGD.
The PGD rapid test has been used for many years as a safety measure for the detection of bacteria in PLTs within 24 hours of transfusion (or 4 hours for poststorage pools or individual WBDs). It has been proven to detect dangerously contaminated PLTs that are released as falsely negative by early culture methods due to sampling error when the bacterial contamination is still at a very low CFU/mL level. Comparable performance data for other 7‐day testing approaches using culture (safety measure on Day 4 or later or large‐volume delayed sampling) have either not been generated or published. 12 The updated PGDprime test represents improvement of the original PGD test in ease of use and time to detection of many bacteria, including wild‐type strains reported in recent years as PLT contaminants. Rapid testing as a safety measure permits dating extension to 7 days, which has been reported to save more money than it costs in many blood centers and transfusion services. 13
In an independent study, the PGD test had failed to detect the viridans group S. oralis at 2 × 107 CFU/mL in an LRAP unit. 6 PGDprime now detects this organism at a level between 1.95 × 106 CFU/mL and 9.75 × 105 CFU/mL. Jacobs and coworkers 14 have reported no morbidity associated with viridans group Streptococci at ≤5 × 106 CFU/mL.
In 2018, four contamination events with Acinetobacter calcoaceticus‐baumannii complex were reported resulting in patient morbidity and mortality. 15 In two morbidity cases, PGD had been used as a safety measure. Both PGD and PGDprime had not been designed to efficiently detect Acinetobacter spp. since these bacteria had not been reported heretofore as PLT contaminants. An updated version of PGDprime with enhanced Acinetobacter detection has now been optimized and is in validation studies in the US. 16
In conclusion, the Platelet PGD Test for the detection of bacteria in PLTs for transfusion has been updated using an innovative sequential lateral‐flow format that has enabled the simplification of workflow, the improvement of the breadth of bacterial strain detection, and in several cases, the improvement of sensitivity of detection. The specificity of the assay has also been enhanced via manipulation of antibody structure. The updated version, PGDprime, is a robust assay that can tolerate a wide range of potential sample interferents, user error modes, and environmental extremes. It may be used to extend the outdate of PLTs in storage containers cleared by the FDA for use to 7 days, thereby improving availability while reducing expenses associated with PLT discards.
CONFLICT OF INTEREST
All authors are employees of Verax Biomedical Incorporated.
Vallejo RP, Shinefeld L, LaVerda D, et al. Performance profile of an updated safety measure rapid assay for bacteria in platelets. Transfusion. 2020;60:2622–2632. 10.1111/trf.16000
REFERENCES
- 1. Bacterial Risk Control Strategies for Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion. Guidance for Industry Silver Spring, MD: U.S. Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research. 2019.
- 2. Lousararian A, Lawrence G, Shinefeld L, Lamkin M, et al. Next generation rapid test format for the detection of bacteria in platelets. Transfusion. 2015;55(3S):195A. [Google Scholar]
- 3. Best N, Lamkin M, Shinefied L. Reducing false‐positive reactions in a rapid test for bacteria in platelets. Transfusion. 2015;55(3S):193A. [Google Scholar]
- 4. Shinefeld L, Hornbaker N, Rasmusson P, et al. Validation of the specificity of the PGDprime test for bacteria in platelets with commercial scale lots. Transfusion. 2019;59(S3):40A. [Google Scholar]
- 5. Shinefeld L, LaVerda D, Best N, et al. Validation of the analytical sensitivity of commercial scale lots of the verax PGDprime test for bacteria in platelets. Transfusion. 2019;59(S3):155A. [Google Scholar]
- 6. Platelet PGD® Test Instructions for Use. P00583 2019/04 Rev J:7. Marlborough, MA: Verax Biomedical, 2019.
- 7. LaVerda D, Boudreau E, McKenzie N, et al. Improved detection of gram‐negative platelet isolates with next‐generation platelet PGD test. Transfusion. 2015;55(3S):185A. [Google Scholar]
- 8. LaVerda D, Lisitu J, Shinefeld L, et al. Ultra‐Low inoculum growth study to validate commercial scale lots of the PGDprime® rapid test for detection of bacteria in platelets. Transfusion. 2019;59(S3):65A–66A. [Google Scholar]
- 9. Shinefeld L, Best N, Williams M, et al. Robustness of the verax PGDprime test for bacteria in platelets to interfering substances in plasma. Transfusion. 2018;58(S2):239A–240A. [Google Scholar]
- 10. Shinefeld L, Best N, Williams M, et al. User guardband study to determine the robustness of the verax platelet PGDprime test for bacteria in platelets. Transfusion. 2018;58(S2):228A. [Google Scholar]
- 11. LaVerda D, Boudreau E, Lisitu J, et al. Growth model studies highlight the variability of bacterial growth in platelets. Transfusion. 2019;59(S3):75A. [Google Scholar]
- 12. Kundrapu S, Srivastava S, Good CE, et al. Bacterial contamination and septic transfusion reaction rates associated with platelet components before and after introduction of primary culture: experience at a US Academic Medical Center 1991 through 2017. Transfusion. 2020;60(5):974–985. [DOI] [PubMed] [Google Scholar]
- 13. Mintz PD, Sanders JR. Outdate reduction and cost savings with rapid testing for seven‐day platelet storage. Ann Clin Lab Sci. 2020;50(3):404–407. [PubMed] [Google Scholar]
- 14. Jacobs MR, Good CE, Lazarus HM, Yomtovian RA. Relationship between bacterial load, species virulence, and transfusion reaction with transfusion of bacterially contaminated platelets. Clin Infect Dis. 2008;46:1214–1220. [DOI] [PubMed] [Google Scholar]
- 15. Jones SA, Jones JM, et al. Sepsis attributed to bacterial contamination of platelets associated with a potential common source – multiple states. MMWR. 2019;68(23):519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. LaVerda D, Shinefeld L, Best N, et al. Updating the Platelet PGDprime Rapid Test® for bacteria in platelets to detect acinetobacter [abstract]. Transfusion. 2020;60. [Google Scholar]


