Summary
Associations between skin microbes or biomarkers and pathological conditions have been reported in the literature. However, there is a lack of clarity on the interaction between the coexistence of common skin microbes with skin physiology and subsequent development of clinical symptoms, and the role of biomarkers in mediating these changes before the development of skin disease. In this review, we aim to identify areas in which extensive research for the studied factors has already been conducted, and which research areas are under‐represented. The SciFinder database was searched for articles containing key words including specific skin microbes, biomarkers, skin physiology and diseases from the beginning of the SciFinder data record to 26 April 2016, and we included an additional relevant recent publication from our group. Among the 8000 + articles selected, the frequency of keyword pairs between two roles [microscopic markers (microflora or biomarkers) and reactions (skin physiology or clinical symptoms, or skin disease)] was investigated. Associated research between the individual factors such as skin microflora or biomarkers (chosen based on our earlier publication) and specific biophysical parameters, symptoms or skin disease was identified. The present research heatmap emphasizes the significance of a structured review of research on concerned factor associations to identify early/subclinical clues that can be used to prevent progression to overt skin disease with the help of precise skin care or early intervention, as indicated by skin microflora, biomarkers and an interactive skin biophysics profile. The findings provide a novel approach to explore such associations and may guide future research directed towards predicting disease from early/subclinical symptoms.
Short abstract
Click here for the corresponding questions to this CME article.
Introduction
The skin is the largest and most exposed protective organ of the human body, 1 which is inhabited by numerous residents (commensal and mutualistic microflora) and is constantly exposed to the external environment. Usually, these inhabitants remain largely in a state of homeostasis, contributing to the skin’s natural defence mechanisms. 2 , 3 These protective mechanisms form a dynamic skin barrier including, but not limited to, physical barriers [relatively acidic surface pH (4.5–5.5), lower temperature (29‒34 °C) compared with the internal body (37 °C), timely desquamation, presence of tight junction proteins] and chemical barriers [host defence molecules released by keratinocytes, e.g. antimicrobial peptides (AMPs)], which provide unfavourable growth conditions for pathogenic microbes to attack or disturb the homeostasis. 1 Further, immunological barriers are provided by a number of hematopoietic cells, including T cells, neutrophils, eosinophils, Langerhans/dendritic cells, natural killer cells and mast cells.
The integrity of the skin’s physical barrier depends on the biophysical properties of the skin, 2 , 3 , 4 which in turn is affected by multiple factors such as age, sex, skin site, seasonal variations and body temperature. 5 , 6 , 7 , 8 Any change in the skin’s natural defence mechanisms or biophysical parameters can influence microbial colonization and create an imbalance in the skin microflora, which serves as the first‐line defence and promotes crosstalk with other protective mechanisms, resulting in physiological changes in skin health.
Researchers have been striving for many years to elucidate the pattern of distribution of skin microflora, their association with other (pathogenic) microbes and skin disease, and factors that influence the relative abundance of micro‐organisms at different skin sites. The current major initiatives (e.g. by the National Institute of Health 9 ) are focused on identifying skin microflora and their relationship with skin disease. Some studies have also compared the skin microflora of healthy volunteers with that of patients with skin infections to determine the changes in the microflora during skin disease. 10 Recently, several studies have attempted to identify the relationship of commonly found skin microflora and/or biomarkers with skin physiology, clinical symptoms and skin disease. 2 , 3 , 4 Although individual associations have been reported, there is a lack of research on how coexistence of skin microflora affects skin physiology, what factors lead to the development of clinical symptoms and what is the role of biomarkers that mediate these changes before the advent of skin disease. These associations can not only serve as predictive or diagnostic markers, but may also empower dermatologists to comprehend the series of changes that occur along the development of skin disease and devise treatment strategies to combat the development of such disease. In a recent study, we established the microbial distribution and co‐occurrence of six commonly found microflora (Propionibacterium acnes, Staphylococcus aureus, Staphylococcus epidermidis, Lactobacillus spp., Pseudomonadaceae and Malassezia furfur) and their association with biophysical parameters (e.g. transepidermal water loss, pH, skin scaling and roughness, and sebum and hydration levels) and biomarkers [LL‐37, human β‐defensin (hBD)‐2, hBD‐3 and claudin‐1] in the skin of healthy Chinese women. 4
In this review, we aimed to leverage ‘big data’/text‐mining technology for literature mapping in a knowledge‐based structured manner, based on clinical research on skin microflora and skin biophysical parameters. We hoped that this would validate previous observations of our group 4 and help to establish a link between subclinical symptoms and their clinical manifestations for appropriate skin disease management. We mapped the literature in a structured manner to gain insights and identify potential opportunities or gaps in the current published literature (including our recent publication described above) in the field of dermatology and skin care research and development, based on an association between five factors [previously identified common skin microbes and biomarkers, 4 versus skin physiology (which can be precisely evaluated at normal and subclinical status) perceived or observed clinical symptoms on skin and dermatological disease based on clinical diagnosis] (Fig. 1).
Figure 1.
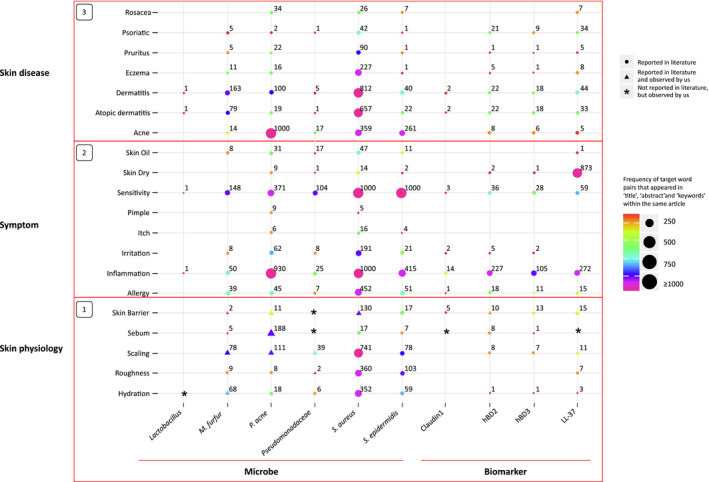
Patterns of reactions as a function of impulse from reported literature. The size and colour of particular dots/triangles represent the number of articles that studied an association between either skin microflora or biomarkers with a specific biophysical parameter, symptom or skin disease. The triangles show that the above parameters were observed in the literature and confirmed in studies from our group, while asterisks represents a gap in the literature that was previously identified and addressed in our recent publication. 9
Methods
A literature review was performed using the SciFinder electronic database, which included all articles from the beginning of the SciFinder data record to 26 April 2016. An additional relevant publication from our group in 2017 9 reported significant associations between subclinical signs and microscopic factors, and hence, this was included in the analysis to identify confirmed associations and less explored factor pairs that had been identified and addressed in our report.
The following keywords were selected: ‘S. aureus’, ‘S. epidermidis’, ‘Lactobacillus’, ‘M. furfur’, ‘P. acne’, ‘Pseudomonadaceae' (for microflora); ‘hBD‐2’, ‘hBD‐3’, ‘LL37’, ‘claudin‐1’ (for biomarkers); ‘skin barrier’, ‘roughness’, ‘scaling’, ‘hydration’, ‘sebum’ (for skin physiology); ‘pimple’, ‘allergy’, ‘irritation’, ‘sensitivity’, ‘itch’, ‘inflammation’, ‘skin dry’, ‘skin oil’ (for clinical symptoms or conditions) and ‘acne’, ‘psoriatic’, ‘atopic ‘dermatitis’, ‘rosacea’, ‘pruritus’, ‘dermatitis’ and ‘eczema’ (for skin disease). Only articles in English were selected. The frequency of co‐occurrence of keyword pairs connected with two roles [microscopic markers (microflora or biomarkers) and reactions (skin physiology or clinical symptoms, or skin disease)] in the selected articles was investigated.
Results
Using the selected keywords, over 8000 English‐language articles were identified. We included the reported literature describing an association between common skin conditions such as rosacea, psoriasis, pruritus, eczema, dermatitis, atopic dermatitis (AD) and acne, with either an imbalance in skin microflora or an immune response to external/internal stimuli. The total number of literature reports for the reactions as a function of impulse are represented in Fig. 1. The size and colour of the dots/triangles represent the number of articles that reported an association between either skin microflora (factor 1, marked on the x axis; for instance, lactobacillus) or biomarkers (factor 2, marked on the x axis; for instance, claudin‐1) and a specific biophysical parameter, symptom or skin disease (factors 3–5, represented as horizontal boxes stacked on the y axis).
Association of skin physiology with skin microflora and biomarkers
Figure 1 (Box 1) shows the number of articles that assessed different skin physiological changes observed with common skin microflora and the biomarkers expressed during skin disease. The size of the points shows that few of the microbe/biomarker–skin physiology combinations compared with other boxes. A large number of articles evaluated associations between S. aureus and either scaling, roughness or hydration. Microbe–pathological factor combinations such as M. furfur–skin barrier, Pseudomonadaceae–roughness/hydration and S. epidermis–sebum were co‐studied in at least a few articles, but such studies were rare for Lactobacillus and Pseudomonadaceae, as observed in a previous study from our group. 4 Overall, very few articles reported the association of the studied targeted biomarkers with biophysical parameters.
Association of skin microflora and biomarkers with clinical symptoms
Figure 1 (Box 2) shows the number of articles in which an association of clinical symptoms with common skin microflora and biomarkers were observed. A large number of articles evaluated associations between sensitivity and inflammation with five of the six micro‐organisms we assessed (exception was Lactobacillus), allergy with four micro‐organisms (exceptions were Lactobacillus and Pseudomonadaceae); and irritation with three micro‐organisms (exceptions were Lactobacillus, Pseudomonadaceae and M. furfur); their relation with skin physiological factors may be inferred from Box 1.
Similarly, associations were reported between dry skin and LL‐37, and inflammation and three AMPs, with a few indicating a relationship with factors such as skin barrier and scaling (Fig. 1, Box 1).
Intermediary changes from an imbalance in skin microflora and biomarkers to the development of skin diseases
As seen in Fig. 1 (Box 3) among the microbes and biomarkers co‐reported with rosacea/acne, associations were most common for P. acnes and mediated through LL37, indicating a possible mechanistic association between the three factors. Similarly, associations were observed between clinical conditions such as dermatitis/AD, eczema, pruritus and psoriasis and S. aureus; these associations were often mediated through LL37 and hBD2.
With the understanding of subclinical physiological changes in skin and appearance of symptoms during the development of skin diseases in the vertical boxes and association between micro‐organisms or biomarkers on the horizontal axis, an association frequency between the various micro‐organisms and skin physiology, symptoms and disease was observed: S. aureus > P. acne > S. epidermis > M. furfur > Pseudomonadaceae > Lactobacillus. Similarly, for the biomarkers, the reported association frequencies for the biomarkers with the same three factors (skin physiology, symptoms and diseases) are denoted vertically in Fig. 1 (Box 3): LL‐37 > hBD‐2 > hBD‐3 > claudin‐1.
Discussion
The present study reviewed the available published literature to identify reported associations between skin microbes or biomarkers and skin physiology, clinical symptoms and skin disease. Association between the studied microbes and pathological conditions such as scaling, skin barrier and sebum were observed in a number of publications. These physiological changes later develop severity and manifest as symptoms of particular clinical diseases. The mapping provided a systematic approach to review specific skin microbes related to disease and the pattern of their association (from a subclinical impact on the skin physiology to its manifestation into clinical symptoms) as the initial association develops into a skin disease. A holistic review of the three boxes (Fig. 1) for a particular microbe could provide pathological clues. The mapped association of M. furfur is an example; M. furfur is commonly associated with dermatitis/dandruff, and is correlated with skin barrier function, scaling and hydration. This association appears clinically as skin inflammation/sensitivity, and was confirmed in our recent publication. 4 Similarly, P. acne, which has been implicated in inflammation/skin sensitivity related to acne and dermatitis, can also be commonly detected on skin, and from a skin physiology point of view, is linked to sebum, scaling and oily skin. 4 The co‐occurrence of the studied microbes or biomarkers and some clinical symptoms or diseases was observed in a large number of reports. However, there were a few microbes (e.g. Lactobacillus 4 ) and biomarkers for which we observed gaps in the literature (including our recent publication) regarding their association with common skin physiology, clinical symptoms and skin diseases, suggesting a connection with skin benefits and potential skin care opportunities for future studies.
Although the studies evaluating associations between skin microflora and pathological parameters were relatively few, some associations reported in this mapping confirm the observations in an earlier report from our group. 4 Furthermore, few of these microbe–pathological parameter combinations, e.g. Lactobacillus and any of the skin physiology parameters, or Pseudomonadaceae and sedum/skin barrier, have not been reported in the historical literature.
Although understanding the initial events is crucial for early diagnosis or interventions in skin diseases, 11 , 12 , 13 , 14 eliminating the cycle of progression from changes in skin physiology to clinical symptoms can also provide clinical benefit. The current findings are of significance, as they may be helpful in understanding how microscopic skin microflora and biomarkers can indicate disease progression from changes in skin physiology to the development of clinical symptoms and ultimately the actual disease.
The present study has some limitations. First, the literature search was performed in April 2016, hence some recent articles (except for a recent article from our group published in 2017, 4 which was included) may have been excluded from the search. Secondly, only one database was used to search for the articles. Lastly, the associations of concerned factors were identified based on frequency of target word pairs that appeared in the title, abstract/summary and/or keywords within the same article, and did not include synonyms. Despite these limitations, a robust number of published articles that evaluated various combinations of the studied factors were screened and analysed, providing a valuable overview of the current research in this field.
Conclusion
This literature mapping visualizes the recent research in skin physiology, symptoms and disease associated with common skin microflora and biomarkers in a novel way, which may be applicable to other research from overall microflora (macroscopic) to specific microbial interaction (microscopic) analysis. Some of these associations, such as underlying changes in skin physiology with commonly found skin microbes and biomarkers, have been reported previously. 4
The review further identified areas where extensive literature on association between the studied factors (micro vs. macro association) existed and analysed these. These associations may help in predicting disease from the first appearance of symptoms or when these are still subclinical. Furthermore, knowledge of the microscopic skin microflora, biomarkers and associated skin biophysical profile may assist in preventing progression of the subclinical condition to the development of skin disease, with the help of precise skin care prevention or early intervention.
Moreover, the gaps in literature identified in the present study may guide future research opportunities.
Overall, this big data‐based approach of literature review and meta‐analysis could potentially guide the development of accurate skin‐disorder prevention solutions, help researchers and dermatologists efficiently connect research findings and opportunities from micro to macro, and shed light on early clinical diagnosis.
CPD questions
Learning objective
To demonstrate an overall understanding of how skin commensal/transit microbials associate with skin physiological/clinical conditions and relate to specific skin diseases to guide disease prevention and early intervention strategies.
Question 1
Which of the following protective mechanisms form the skin’s dynamic barrier function?
Chemical barrier.
Immunological barrier.
Microbial barrier.
Physical barrier.
All of the above.
Question 2
Which of the following is an antimicrobial peptide?
Human β‐defensin 2.
C‐peptide.
Claudin‐1.
Interleukin 1.
Procollagen III.
Question 3
Which of the following is a fungal skin opportunistic pathogen?
Candida pseudotropicalis.
Lactobacillus spp.
Malassezia furfur.
Propionibacterium acnes.
Staphylococcus aureus.
Question 4
Colonization with which of the following skin microbes is associated with disease progression in atopic dermatitis?
Propionibacterium acnes.
Roseomonas mucosa.
Staphylococcus aureus.
Staphylococcus hominis.
Staphylococcus epidermidis.
Question 5
Which of these microbe combinations has been studied and reported in the context of skin allergy?
Lactobacillus, Malassezia furfur and Propionibacterium acnes.
Lactobacillus, P. acnes, Staphylococcus aureus and Staphylococcus epidermidis.
Lactobacillus, M. furfur, S. aureus and S. epidermidis.
Lactobacillus, M. furfur, P. acnes and S. aureus.
M. furfur, P. acnes, S. aureus and S. epidermidis.
Instructions for answering questions
This learning activity is freely available online at http://www.wileyhealthlearning.com/ced
Users are encouraged to
Read the article in print or online, paying particular attention to the learning points and any author conflict of interest disclosures
Reflect on the article
Register or login online at http://www.wileyhealthlearning.com/ced and answer the CPD questions
Complete the required evaluation component of the activity
Once the test is passed, you will receive a certificate and the learning activity can be added to your RCP CPD diary as a self‐certified entry.
This activity will be available for CPD credit for 2 years following its publication date. At that time, it will be reviewed and potentially updated and extended for an additional period.
Acknowledgement
We thank Dr J. Ramanathan and Dr R. Bhandari (SIRO Clinpharm Pvt. Ltd, India) for providing writing assistance during the development of the manuscript. This study was supported by Johnson & Johnson (China) Ltd.
Learning points.
This literature mapping analysis provides an overview of research of skin physiology, symptoms and disease associated with common skin microbes and biomarkers.
Associations between the studied factors, such as underlying changes in skin physiology with commonly found skin microbes and biomarkers, may be useful for predicting disease from early/subclinical symptoms and for developing early intervention strategies.
The gaps in reported associations, such as between skin microbes and pathological parameters, identified in the present mapping may guide future research opportunities.
Conflict of interest: XL and LX are employees of Johnson & Johnson (China) Ltd, Shanghai, China. The other authors declare that they have no conflicts of interest.
References
- 1. Egert M, Simmering R, Riedel CU. The association of the skin microbiota with health, immunity, and disease. Clin Pharmacol Ther 2017; 102: 62–9. [DOI] [PubMed] [Google Scholar]
- 2. Del Rosso JQ. Repair and maintenance of the epidermal barrier in patients diagnosed with atopic dermatitis: an evaluation of the components of a body wash‐moisturizer skin care regimen directed at management of atopic skin. J Clin Aesthet Dermatol 2011; 4: 45–55. [PMC free article] [PubMed] [Google Scholar]
- 3. Thiboutot D, Del Rosso JQ. Acne vulgaris and the epidermal barrier: is acne vulgaris associated with inherent epidermal abnormalities that cause impairment of barrier functions? Do any topical acne therapies alter the structural and/or functional integrity of the epidermal barrier? J Clin Aesthet Dermatol 2013; 6: 18–24. [PMC free article] [PubMed] [Google Scholar]
- 4. Li X, Yuan C, Xing L, Humbert P. Topographical diversity of common skin microflora and its association with skin environment type: an observational study in Chinese women. Sci Rep 2017; 7: 18046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boer M, Duchnik E, Maleszka R, Marchlewicz M. Structural and biophysical characteristics of human skin in maintaining proper epidermal barrier function. Postepy Dermatol Alergol 2016; 33: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Darlenski R, Fluhr JW. Influence of skin type, race, sex, and anatomic location on epidermal barrier function. Clin Dermatol 2012; 30: 269–73. [DOI] [PubMed] [Google Scholar]
- 7. Darlenski R, Sassning S, Tsankov N, Fluhr JW. Non‐invasive in vivo methods for investigation of the skin barrier physical properties. Eur J Pharm Biopharm 2009; 72: 295–303. [DOI] [PubMed] [Google Scholar]
- 8. Ying S, Zeng DN, Chi L et al The influence of age and gender on skin‐associated microbial communities in urban and rural human populations. PLoS One 2015; 10: e0141842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peterson J, Garges S, Giovanni M et al The NIH Human Microbiome Project. Genome Res 2009; 19: 2317–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosenthal M, Goldberg D, Aiello A et al Skin microbiota: microbial community structure and its potential association with health and disease. Infect Genet Evol 2011; 11: 839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Addor FA, Aoki V. Skin barrier in atopic dermatitis. An Bras Dermatol 2010; 85: 184–94. [DOI] [PubMed] [Google Scholar]
- 12. Li X, Galzote C, Yan X et al Characterization of Chinese body skin through in vivo instrument assessments, visual evaluations, and questionnaire: influences of body area, inter‐generation, season, sex, and skin care habits. Skin Res Technol 2014; 20: 14–22. [DOI] [PubMed] [Google Scholar]
- 13. Marina ME, Roman II, Constantin AM et al VEGF involvement in psoriasis. Clujul Med 2015; 88: 247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberson ED, Bowcock AM. Psoriasis genetics: breaking the barrier. Trends Genet 2010; 26: 415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]


