Abstract
Molibresib (GSK525762), an orally bioavailable small molecule with 2 major equipotent active metabolites, is being developed for the treatment of cancers. Molibresib is a substrate of cytochrome P450 (CYP) 3A4 and P‐glycoprotein (P‐gp). To enable administering safe doses of molibresib to healthy volunteers, this 2‐part randomized, open‐label, crossover drug‐drug interaction trial was conducted as an adaptive design study using physiologically based pharmacokinetic (PBPK) modeling and simulation to predict the lowest doses of molibresib that could be safely administered alone (10 mg) or with itraconazole and rifampicin (strong inhibitors and inducers of CYP3A and P‐gp, respectively). PBPK simulation guided the molibresib dose (5 mg) to be administered along with itraconazole in part 1. Itraconazole increased total exposure (AUC) of molibresib by 4.15‐fold with a 66% increase in Cmax, whereas the total AUC and Cmax for the 2 major active metabolites of molibresib decreased by about 70% and 87%, respectively. A second PBPK simulation was conducted with part 1 data to also include the active metabolites to update the recommendation for the molibresib dose (20 mg) with rifampicin. With rifampicin, the AUC and Cmax of molibresib decreased by approximately 91% and 80%, respectively, whereas the AUC of the 2 active metabolites decreased to a lesser extent (8%), with a 2‐fold increase in Cmax. The results of this study confirmed the in vitro data that molibresib is a substrate for CYP3A4. The adaptive design, including Simcyp simulations, allowed evaluation of 2 drug interactions of an oncology drug in a single trial, thus minimizing time and exposures administered to healthy subjects.
Keywords: molibresib, GSK525762, itraconazole, rifampicin, Simcyp, drug‐drug interaction, pharmacokinetics
Bromodomains (BRDs) are small protein domains found in a variety of proteins that recognize and bind to acetylated histone tails. This binding affects chromatin structure and thereby epigenetically regulates controlled processes including gene transcription and mRNA elongation. 1 , 2 Inhibitors of the bromodomain extraterminal (BET) family of BRD proteins have been shown to have antitumor effects by blocking both c‐Myc and its downstream transcriptional function in leukemia and multiple myeloma studies. 3 , 4
Molibresib (GSK525762) is an orally bioavailable small molecule that is a potent inhibitor of the binding of BET proteins to acetylated histones and is being developed by GlaxoSmithKline for the treatment of various cancers. 1 , 5 The safety, tolerability, clinical activity, and pharmacokinetics (PK) of molibresib have been evaluated in 2 first‐time‐in‐human (FTIH) studies (study BET115521, NCT01587703, and study BET116183, NCT01943851) in patients with solid and hematologic malignancies over the dose range of 2 to 125 mg once daily. Following single and repeat administrations of oral doses, molibresib was rapidly absorbed and eliminated, with maximum concentration (Cmax) occurring within 2 hours after dosing and an average terminal‐phase half‐life of 3 to 7 hours. 6 Both Cmax and the area under the concentration curve (AUC) tended to increase in a dose‐proportional fashion, with a large overlap for individual AUC values at doses ≥30 mg. However, within‐subject dose proportionality over the dose range of 6 to 80 mg was evaluated and confirmed in a relative bioavailability study in cancer patients as part of FTIH study BET115521 using a stable isotope approach (G.M. Ferron‐Brady et al, unpublished data, 2020). In addition, in patients with acute myeloid leukemia who received molibresib concomitant with moderate or potent cytochrome P450 (CYP) 3A enzyme inhibitors (eg, fluconazole, voriconazole, or posaconazole) for prevention of fungal pneumonia, the average and the maximum molibresib exposure (AUC) was approximately 2 and 4.5 times higher, respectively, in these patients than those not receiving a CYP3A inhibitor. These observations provided early indications for the potential involvement of CYP3A in the metabolism of molibresib.
The precise in vivo metabolic liability for molibresib has not yet been assessed. Preliminary analysis of human plasma and urine samples from study BET115521 showed the presence of oxidative metabolites of molibresib. Two major metabolites (GSK3529246 [N‐desethyl], GSK3536835 [ethyl hydroxy]) as well as several minor metabolites have been identified in human plasma and urine. 6 These 2 major metabolites have demonstrated inhibitory activity against tumor cell lines with a similar potency to the parent molecule. 6 Urinary excretion of unchanged molibresib accounted for approximately 9.5% of the total dose administered. An in vitro CYP phenotyping study evaluated 7 CYP enzymes (CYP1A1, ‐2C9, ‐2C8, ‐2C19, ‐2D6, ‐2B6, and ‐3A4), and data suggested that CYP3A4 is the predominant enzyme responsible for the metabolism of molibresib and the generation of its 2 major metabolites. This was further supported by in vitro inhibition data, in which complete inhibition of molibresib metabolism was observed in the presence of azamulin (a potent and specific CYP3A4 inhibitor). Molibresib is a substrate for P‐glycoprotein (P‐gp) and breast cancer resistance protein transporters, but it does not inhibit these transporters (IC50 > 100 μM).
Taken together, the emerging clinical PK data, the in vitro CYP phenotyping study, and in vitro inhibition data indicate that CYP3A4 may be the major route of clearance for molibresib, and coadministration of drug therapies that modulate CYP3A4 (ie, CYP3A4 inhibitors and inducers) is likely to alter the exposure of molibresib. As cancer patients could be in need of receiving medications that would affect CYP3A and P‐gp activity, it was important to identify the impact of such therapies early in drug development, thus necessitating the conduct of a drug interaction study in a healthy volunteer population to minimize confounding factors.
Using an adaptive design approach to minimize exposure to healthy volunteers, the current study investigated the clinical drug interaction of molibresib, as the victim, with 2 strong perpetrators: itraconazole (CYP3A4 and P‐gp inhibitor) and rifampicin (CYP3A and P‐gp inducer). 7 , 8
Methods
This study (NCT02706535) was conducted and completed at a single center in the United States as per the ethical principles of “good clinical practice” and the Declaration of Helsinki after obtaining written informed consent from each subject. The protocol was approved by Aspire Investigational Review Board (Santee, California).
Molibresib Physiologically Based PK Model Development and Qualification
Physiologically based PK (PBPK) simulations were used to design the clinical study in the various phases, and the general workflow is shown in Figure 1. Molibresib pharmacokinetics were simulated using Simcyp ADME Simulator (version 14; Simcyp Ltd, Sheffield, UK); PBPK model parameters are summarized in Table 1. Molibresib rate of absorption (Ka), plasma protein binding, and blood: plasma ratios were fixed to their observed values. Molibresib volume of distribution was estimated using the parameter estimation module within version 14. Systemic clearance of molibresib was simulated using the enzyme kinetics elimination mode, and an initial estimate of molibresib intrinsic clearance was made based on clinically observed oral clearance values using the retrograde calculator in Simcyp. Renal clearance values were set to match the dose excreted unchanged in urine over 24 hours (∼10%). The final molibresib intrinsic clearance value was optimized to match the clinical pharmacokinetics of molibresib observed in clinical study BET115521. 6 A similar approach was used to build the PBPK model for metabolite GSK3529246, with model parameters summarized in Table 1.
Figure 1.
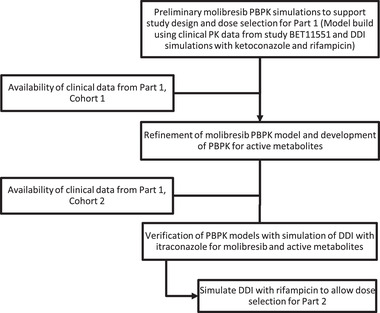
PBPK simulation workflow.
Table 1.
Preliminary Simcyp PBPK Model Parameters
| Parameter | Value for Molibresib | Value for GSK3529246 |
|---|---|---|
| Molecular weight | 423.9 | 395.84 |
| Log P | 2.38 | 1.6 |
| pKb | 7.49 | 7.36 |
| Compound type | Monoprotic base | Monoprotic base |
| Blood:plasma ratio | 0.9 | 0.9 |
| Fu plasma | 0.2 | 0.2 |
| Model type | First order | – |
| Fa a | 0.732 | – |
| Ka (1/h) a | 3.4 | – |
| Permeability (nm/s) | 84 | – |
| Volume of distribution method | Method 2 | Method 2 |
| Oral CL (L/h), CL/F a | ∼14 | 11 b |
| Renal contribution (%) a | 10.5 | – |
| CYP3A4 contribution to hepatic CL (%) a | 99 | – |
Log P, partition coefficient; pKb, negative logarithm of the base dissociation constant (Kb); Fu, fraction unbound; Fa, fraction absorbed; Ka, rate of absorption; CL, clearance.
Estimates were taken from the then‐ongoing first‐time in‐patient study BET115521.
CLiv predicted by Simcyp.
A retrospective simulation of the pharmacokinetic profile of molibresib was performed to evaluate whether Simcyp could accurately simulate the observed exposure after an oral dose of 60 or 100 mg. Simulations were performed as 10 trials of 10 individuals per trial, using the “healthy volunteer” population with a proportion of women of 0.5 and ages ranging from 20 to 50 years. The sampling interval of the simulation was every 0.05 hours. Simcyp‐predicted AUC and Cmax values were computed from the predicted concentrations. At the time of analysis, molibresib had not been dosed to healthy volunteers, and availability of a validated virtual cancer population was limited. Therefore, available data in patient subjects clear of comedications that are inhibitors of CYP3A4 were used as a surrogate for model building and PK comparison.
Molibresib PBPK Model Applications
Initial Molibresib Model Applications
Prior to conducting simulations with molibresib, PBPK model files for ketoconazole and rifampicin were verified by simulating the effect of these perpetrators on the exposure of the sensitive CYP3A4 index substrate midazolam. Dosing regimens for ketoconazole and rifampicin used a typical perpetrator study design and were simulated to steady state. 9 , 10 It is important to note that at the time of this assessment, utilization of ketoconazole as a clinical probe inhibitor was prohibited. However, as verification of the PBPK models for the newly recommended CYP3A4 probe inhibitor (itraconazole) was still underway, ketoconazole was used to allow prediction of the worst‐case scenario and recommend a safe as well as analytically feasible molibresib dose with itraconazole. Comparing the PBPK simulations with observed clinical data from the itraconazole drug‐drug interaction (DDI) study also confirmed the role of CYP3A in molibresib metabolism and clearance.
Simcyp simulations exploring the effect of ketoconazole and rifampicin on molibresib exposure were performed as 10 trials of 10 individuals per trial, with age ranging from 20 to 50 years. Ketoconazole was administered at 400 mg once daily on days 1 through 8, and molibresib was administered as a single oral dose on day 5. Rifampicin was administered at 600 mg once daily on days 1 through 7, and molibresib was administered as a single 20‐mg oral dose on day 7.
Refined Molibresib Model Applications
Prior to conducting simulations with molibresib, the PBPK model file for itraconazole was verified by simulating the effect of these perpetrators on the exposure of the sensitive CYP3A4 index substrate midazolam. Dosing regimens for itraconazole and midazolam used a typical perpetrator study design 11 and were simulated to steady state.
Simcyp simulations exploring the effect of itraconazole and rifampicin (previously verified) on molibresib exposure were performed as 10 trials of 10 individuals per trial, with age ranging from 20 to 50 years. Itraconazole was administered at 200 mg twice daily on day 1, followed by 200 mg once daily on days 2 through 7, and molibresib was administered as a single oral dose on day 5. Rifampicin was administered at 600 mg once daily on days 1 through 17, and molibresib was administered as a single 20‐mg oral dose on day 16.
Study Population and Design
Eligibility criteria included healthy female subjects of nonchildbearing potential (postmenopausal or premenopausal with documentation of tubal ligation, hysteroscopic tubal occlusion, hysterectomy, or bilateral oophorectomy) aged between 18 and 70 years with a body weight ≥ 45 kg and body mass index (BMI) within the range of 18.0 to 29.9 kg/m2 (inclusive) at the time of screening.
This study was a phase 1 single‐center, 2‐part, randomized, open‐label, crossover adaptive study, as illustrated in Figure 2. Part 1 of this study evaluated the PK, safety, and tolerability of molibresib when administered alone and when coadministered following repeat dosing of itraconazole in 2 cohorts. Part 2 (1 cohort) evaluated the PK, safety, and tolerability of molibresib when administered alone and when coadministered following repeat dosing of rifampicin.
Figure 2.
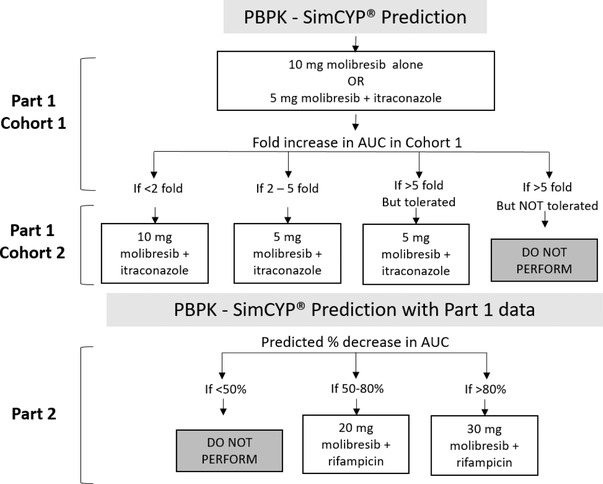
Adaptive study design.
In advance of the study, a mechanistic physiologically based pharmacokinetic model (PBPK), as part of the Simcyp population‐based PBPK simulator, was used to predict the Cmax and AUC with and without coadministration of a CYP3A4 inducer (rifampin) and inhibitor (ketoconazole). On the basis of safety data from BET115521, doses up to 30 mg were tolerated during the first week of daily administration, with only 1 incident of dry mouth, nausea, and international normalized ratio increase reported; only low‐grade (grades 1 to 2) adverse events limited to gastrointestinal (nausea, vomiting, dysgeusia, loss of appetite, dry mouth) events were reported at these doses with longer duration of treatment. 6 Predicted exposure for doses to be selected in this DDI study were to remain below the exposure observed for the 30‐mg dose. A single dose of molibresib 10 mg alone or 5 mg combined with itraconazole was selected as the initial doses in part 1 of the study, as these are the minimum doses needed to provide a plasma concentration‐time course that is sufficiently quantifiable for the objective of the study while minimizing exposure of healthy female volunteers to molibresib. A decision to proceed to part 2 was made based on review of safety and PK data from part 1 as well as updated predictions for rifampicin effect based on predictions from the Simcyp population‐based PBPK simulator.
Part 1
To mitigate the risk of a ≥4‐fold increase in exposure that was predicted using the preliminary ketoconazole DDI simulations, part 1 contained 2 cohorts, in which data from cohort 1 informed doses for cohort 2. In cohort 1 of part 1, a sentinel group of 3 subjects were dosed with molibresib 10 mg alone and molibresib 5 mg in combination with itraconazole. Following an interim review of safety data, the remaining 3 subjects were dosed at the initial doses. Subjects received a single dose of molibresib 10 mg on day 1, followed by a 48‐hour PK sampling. On day 3, subjects received itraconazole 200 mg twice daily, and from days 4 to 6, subjects received single doses of itraconazole 200 mg followed by coadministration with 5 mg molibresib on day 7 and a single dose of itraconazole 200 mg alone on days 8 and 9. PK samples were collected over 72 hours following administration on day 7. Cohort 2 of part 1 was designed to provide a more robust quantitative evaluation of the effect of itraconazole on molibresib after establishing tolerability and approximate estimates of fold change in molibresib exposure in cohort 1 of part 1. Cohort 2 of part 1 was a contingent design with 3 dosing options depending on the PK and tolerability data from cohort 1 of part 1, as illustrated in Figure 1.
On the basis of the 4‐fold increase in molibresib AUC observed in the first 6 subjects in cohort 1, subjects in cohort 2 (n = 7) were also dosed with 5 mg of molibresib with itraconazole on day 7.
Part 2
The decision to proceed to part 2 and the selected doses of molibresib were made based on review of the safety and PK data obtained from part 1 as well as updated predictions for rifampicin effect from a PBPK‐Simcyp population‐based simulation, as illustrated in Figure 1. Part 2 of the study investigating the effect of rifampicin used a dose of 10 mg of molibresib when administered alone and a dose of 20 mg of molibresib when administered after repeat administration of rifampicin.
On day 1, subjects received a single dose of 10 mg molibresib followed by a 48‐hour PK sampling. From days 3 to 17 inclusive, subjects received a single dose of 600 mg rifampicin. On day 18, subjects received a single dose of 20 mg molibresib (selected based on data from cohort 1) along with the dose of rifampicin. On day 19, subjects received a single dose of 600 mg rifampicin. PK samples were collected for 48 hours starting on day 18.
Pharmacokinetic Assessments
Blood samples were taken via an indwelling cannula (or by direct venipuncture), collected into a dipotassium ethylenediaminetetraacetic acid tube and kept at 4°C (refrigerated) or on water ice (not dry ice). Samples were centrifuged refrigerated at approximately 1500g for 10 minutes at approximately 4°C within 1 hour of blood collection. All resultant plasma was transferred into a labeled 2.0‐mL Matrix screw cap cryovial for PK analysis. The PK plasma samples were frozen and stored immediately at −70°C.
Analytical Method
Plasma samples were analyzed for molibresib and its 2 active metabolites, GSK3529246 and GSK3536835, by Covance Bioanalytical Services, LLC.
GSK3536835 was unstable in matrix and readily converted to GSK3529246 during all aspects of sample processing. Under basic conditions at ambient temperature, GSK3536835 converted to GSK3529246 with demonstrated stability following full conversion. Thus, GSK3536835 was converted to GSK3529246 during extraction and subsequently quantified as GSK3529246 in the assay. Quality control samples (QCs) containing GSK3536835 only were processed alongside QCs containing molibresib and GSK3529246 for each individual validation and sample analysis run to evaluate precision and accuracy and to ensure the conversion of GSK3536835 to GSK3529246 was complete. The concentrations of GSK3529246 and GSK3536835 were quantified as GSK3529246 in all QCs, and clinical samples and reported as the sum of GSK3529246 and GSK3536835 metabolite concentrations. To account for the mass difference between GSK3536835 (molecular weight [MW] of 439 g/mol) and GSK3529246 (MW of 395 g/mol), a conversion factor of 1.1 was applied to the QCs containing GSK3536835 only as calculated by the ratio of the MW of GSK3536835 to the MW of GSK3529246.
Prior to extraction, internal standard working solution containing stable isotope‐labeled internal standards [13C6]‑GSK525762 and [13C6]‑GSK3529246 were added to each standard, QC, or clinical sample. To convert GSK3536835 to GSK3529246 in clinical samples and GSK3536835‐only QCs, 10% ammonium hydroxide (aqueous) was added to all samples, which were subsequently incubated for 2 hours at ambient temperature prior to protein precipitation extraction.
Molibresib and its total metabolites were extracted from human plasma by protein precipitation using 5/95 (v/v) ammonium hydroxide/acetonitrile. Extracts were analyzed by ultrahigh‐pressure liquid chromatography (UPLC)‑tandem mass spectrometry using a TurboIonSpray interface and multiple reaction monitoring. The chromatography system comprised a Waters Acquity Classic UPLC fitted with a Waters 2.7 μm BEH C18 (2.1 × 50 mm) column heated to 55°C and used 0.1% ammonium hydroxide (aqueous) and 0.1% ammonium hydroxide in acetonitrile as the mobile phases. All samples were stored on the autosampler at ambient temperature.
The characteristic precursor [M+H]+ to product ions transitions m/z 424 to 269 for GSK525762, m/z 396 to 269 for GSK3529246, m/z 430 to 385 for [13C6]‐GSK525762, and 402 to 275 for [13C6]‐GSK3529246 were consistent with the structures of GSK525762, GSK3529246, and the internal standards, respectively. The ionization source temperature was set to 675°C.
At all validation sample concentrations examined, the bias was less than 15% (20% at the lower limit of quantitation [LLQ]). The maximum bias observed was −16.8% for GSK525762 and −14.5% for GSK3529246.
At all validation sample concentrations examined, the within‐ and between‐run precision values were less than or equal to 15% (20% at the LLQ) and therefore considered acceptable. The maximum within‐ and between‐run precision values observed were 11.1% and 15.5%, respectively, for GSK525762, and 11.8% and 8.1%, respectively, for GSK3529246.
As defined by the lower and upper validation sample concentrations possessing acceptable accuracy and precision, the validated range of the method for GSK525762 was 1 to 1000 ng/mL and for total metabolites as GSK3529246 was 1 to 1000 ng/mL.
Data Analysis
The primary objective was to describe the impact of coadministration of itraconazole and rifampicin on the PK of molibresib and its 2 major metabolites (GSK3529246 and GSK3536835, quantitatively reported as GSK3529246). The impact was also evaluated on the total active moiety computed as the sum of the molar concentrations of molibresib and its 2 major equipotent active metabolites.
The sample size for this study was not determined based on hypothesis testing consideration. Instead, the sample size was determined to ensure that the end points could be estimated with reasonable precision. In part 1 and part 2, the planned sample size was 12 to 15 subjects. Assuming a 20% within‐subject variability and 12 subjects per study part, it was estimated that the widths of the 90% confidence intervals (CIs) around the geometric mean ratios would be within approximately 16% of the point estimates. An estimation approach was used to assess the effect of repeat oral doses of itraconazole and rifampicin on the PK of the single oral dose of molibresib. An interim review was conducted in part 1 to review the preliminary safety and PK data from cohort 1 of part 1.
Plasma molibresib, GSK3529246, and total active moiety concentration‐time data were analyzed by noncompartmental analysis methods using Phoenix WinNonlin (Certara, St Louis, Missouri). Concentration‐time data were used to generate maximum observed plasma concentration (Cmax), time to Cmax (tmax), area under the plasma concentration‐time curve from time zero to the last quantifiable concentration (AUC0‐t), and area under the plasma concentration‐time curve from time zero extrapolated to infinity (AUC0‐∞), elimination rate constant (λz), and apparent terminal‐phase half‐life (t1/2) using noncompartmental methods and were listed for each subject and summarized by planned time and regimen. PK parameters were listed and summarized descriptively by treatment.
For each part, after loge‐transformation, the AUC and Cmax of molibresib, GSK3529246, and total active moiety dose‐normalized to a 10‐mg dose of molibresib with and without itraconazole (part 1) or rifampicin (part 2) and were analyzed separately by a mixed‐effects model with a fixed‐effect term for treatment and subject as a random effect. Point estimates and associated 90%CIs for the difference were constructed using the residual variance.
For final analyses, data were analyzed separately by part and summarized by treatment. Data from cohort 1 of part 1 and cohort 2 of part 1 were combined if the same regimen was administered.
Results
PBPK Simulations to Support Study Design
The Simcyp‐simulated and observed plasma concentration‐time curves for molibresib following oral administration of 60 and 100 mg are shown in Figure 3. Simulated parameters were within 2‐fold of those observed clinically at both doses (Table 2).
Figure 3.
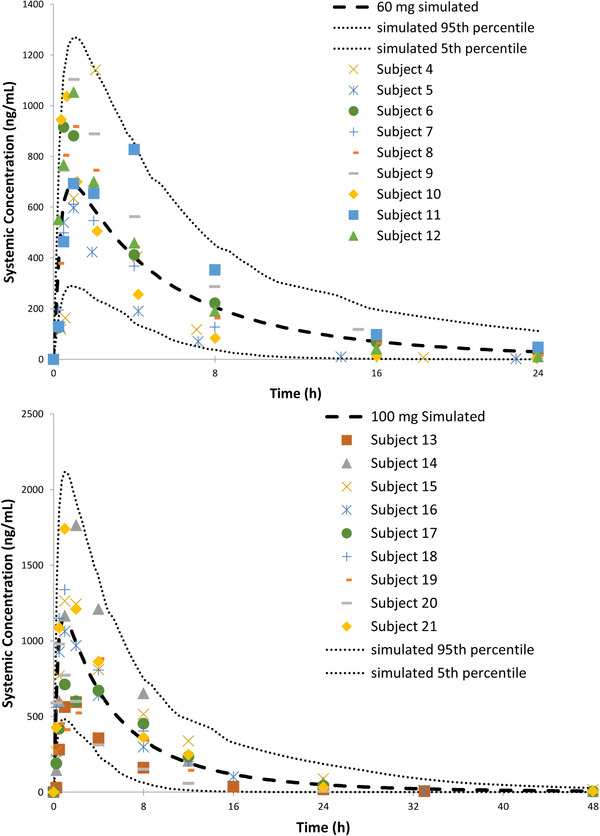
Observed and Simcyp‐simulated molibresib concentration‐time profile after a single 60‐ or 100‐mg dose (BET115521).
Table 2.
Summary of Simcyp‐Simulated and Observed Molibresib PK Parameters
| AUC0‐t (ng·h/mL), CV% | Cmax (ng/mL), CV% | |||
|---|---|---|---|---|
| Dose | Predicted | Observed a | Predicted | Observed a |
| 60 mg | 4271 (56) | 4255 (39.2) | 628.7 (46) | 889.5 (24.5) |
| 100 mg | 7119 (56) | 6958 (43.5) | 1048 (46) | 1081 (38.8) |
Cmax, maximum concentration; AUC0‐t, area under the curve from 0 to time t, t being 24 hours.
Piha‐Paul et al. 6
The Simcyp‐predicted versus observed AUC ratio for midazolam in the presence of ketoconazole and rifampicin were within 2‐fold of that observed clinically. Simulations were then conducted to predict the effect of the CYP3A4 inhibitor ketoconazole and inducer rifampicin on the exposure molibresib. The PBPK model predicted a moderate drug interaction following coadministration of itraconazole, with AUC and Cmax ratios of 4.7 and 1.29, respectively. Using the adaptive design study approach shown in Figure 3, a 5‐mg dose in combination with itraconazole was recommended for part 1. Utilizing the same PBPK model used for the itraconazole simulations, the predicted change in molibresib AUC and Cmax following repeat‐dose rifampicin was 0.24‐ and 0.61‐fold, respectively. Preliminary simulations are shown in Table 3.
Table 3.
Simcyp Preliminary Prediction for Ketoconazole and Rifampicin Effects on Molibresib PK Parameters
| Predicted ratio (5th and 95th percentiles) | ||
|---|---|---|
| Parameter | Ketoconazole | Rifampicin |
| Cmax | 1.29 (1.26‐1.32) | 0.61 (0.58‐0.64) |
| AUC | 4.7 (4.39‐5.03) | 0.24 (0.23‐0.26) |
Cmax, maximum concentration; AUC, area under the curve from 0 to infinity.
Subject Disposition and Demographics
A total of 13 female subjects participated in part 1 (n = 6 in cohort 1; n = 7 in cohort 2) and were included in the analysis of the PK data. One subject had mild elevations in liver enzymes that appeared on day 6, which was noted on day 7 after dosing. Because this event was considered related to itraconazole by the investigator, this subject did not receive itraconazole on days 8 and 9 as planned. In part 2, 15 female subjects were enrolled, and all but 1 subject completed the study as planned. One subject in part 2 was withdrawn postdose for a protocol deviation resulting in exclusion of the PK data from analysis. Subject demographics were similar across all study cohorts in both parts of the study (n = 28). Overall, 61% of the subjects were white, with a mean ± SD age of 51.7 ± 7.94 years and a mean ± SD BMI of 25.7 ± 2.79 kg/m2.
Itraconazole Drug‐Drug Interaction — Part 1
During the initial evaluation of itraconazole coadministration in cohort 1 of part 1, itraconazole was shown to increase Cmax of molibresib by 48% while increasing the total exposure to molibresib (AUC0‐∞) by 3.97‐fold. Conversely, itraconazole led to am 88% and 72% reduction in its active metabolites’ Cmax and AUC0‐∞, respectively. On the basis of this information, along with the safety information, subjects in cohort 1 of part 1 received the same doses of molibresib as the subjects in cohort 1 of part 1. Summary of PK parameters for molibresib, its metabolites, and active moiety are presented in Table 4.
Table 4.
Summary of PK Parameters of Molibresib, Its Two Active Metabolites (GSK3529246), and Total Active Moiety—Itraconazole Interaction (Part 1)
| Analyte | Cohort | Parameter | Molibresib 10 mg Alone | Molibresib 5 mg + Itraconazole |
|---|---|---|---|---|
| Molibresib |
Cohort 1 n = 6 |
Cmax (ng/mL) | 167 (27) | 124 (10) |
| AUC0‐∞ (ng·h/mL) | 672 (11) | 1330 (11) | ||
| Tmax (h) | 0.52 (0.52‐1.5) | 1.5 (0.52‐3.0) | ||
| t1/2 (h) | 7.7 (16) | 10.2 (18) | ||
|
Cohort 2 n = 7 |
Cmax (ng/mL) | 176 (27) | 161 (25) | |
| AUC0‐∞ (ng·h/mL) | 683 (38) | 1470 (14) | ||
| Tmax (h) | 0.52 (0.50‐0.58) | 0.52 (0.27‐0.53) | ||
| t1/2 (h) | 6.6 (42) | 12.5 (18) | ||
|
Overall n = 13 |
Cmax (ng/mL) | 172 (26) | 142 (23) | |
| AUC0‐∞ (ng·h/mL) | 678 (27) | 1410 (13) | ||
| Tmax (h) | 0.52 (0.50‐1.5) | 0.52 (0.27‐3.0) | ||
| t1/2 (h) | 7.07 (32) | 11.4 (20) | ||
| Molibresib 2 active metabolites (GSK3529246) |
Cohort 1 n = 6 |
Cmax (ng/mL) | 44.1 (10) | 2.65 (18) |
| AUC0‐∞ (ng·h/mL) | 492 (13) | 68.2 (33) | ||
| Tmax (h) | 1.5 (0.52‐3.0) | 6.0 (1.5‐8.0) | ||
| t1/2 (h) | 11.2 (18) | 13.7 (31) | ||
|
Cohort 2 n = 7 |
Cmax (ng/mL) | 43.8 (14) | 2.92 (21) | |
| AUC0‐∞ (ng·h/mL) | 551 (15) | 83.4 (20) | ||
| Tmax (h) | 1.5 (1.0‐4.0) | 8.0 (4.0‐8.1) | ||
| t1/2 (h) | 10.7 (25) | 16.2 (48) | ||
|
Overall n = 13 |
Cmax (ng/mL) | 43.9 (12) | 2.80 (20) | |
| AUC0‐∞ (ng·h/mL) | 523 (15) | 76.0 (28) | ||
| Tmax (h) | 1.5 (0.52‐4.0) | 8.0 (1.5‐8.1) | ||
| t1/2 (h) | 10.9 (21) | 15.0 (40) | ||
| Total active moiety |
Cohort 1 n = 6 |
Cmax (nmol/mL) | 478 (22) | 297 (9) |
| AUC0‐∞ (nmol·h/mL) | 2850 (9) | 3310 (12) | ||
| Tmax (h) | 0.52 (0.52‐1.5) | 1.5 (0.52‐3.0) | ||
| t1/2 (h) | 10.7 (27) | 10.0 (20) | ||
|
Cohort 2 n = 7 |
Cmax (nmol/mL) | 494 (24) | 382 (25) | |
| AUC0‐∞ (nmol·h/mL) | 3040 (25) | 3680 (13) | ||
| Tmax (h) | 0.52 (0.50‐0.58) | 0.52 (0.27‐0.53) | ||
| t1/2 (h) | 9.6 (26) | 11.7 (19) | ||
|
Overall n = 13 |
Cmax (nmol/mL) | 486 (23) | 340 (23) | |
| AUC0‐∞ (nmol·h/mL) | 2950 (19) | 3510 (13) | ||
| Tmax (h) | 0.52 (0.50‐1.5) | 0.52 (0.27‐3.0) | ||
| t1/2 (h) | 10.1 (26) | 10.9 (20) |
Cmax, maximum concentration; AUC0‐∞, area under the curve from 0 to infinity; tmax, time of occurrence of Cmax; t1/2, terminal phase elimination half‐life.
Cmax, AUC, and t1/2 are presented as geometric means (CVb%) with 3 significant digits for the geometric mean and 2 for the CVb%; Tmax is presented as median (range) with 2 significant digits.
GSK3529246 represents the sum of GSK3529246 and GSK3536835; GSK3536835 was converted to GSK3529246 prior to analysis. The total active moiety represents the sum of molibresib and its 2 active metabolites reported as GSK3529246.
As illustrated in Figure 4 and summarized in Table 5, in the presence of itraconazole (part 1), molibresib exposure was increased, as evidenced by an observed 4.15‐fold increase in AUC0‐∞ and a 66% increase in Cmax. Similarly, the exposure of the total active moiety molibresib was increased with a 2.38‐fold increase in AUC0‐∞ and a Cmax increase of 40%.
Figure 4.
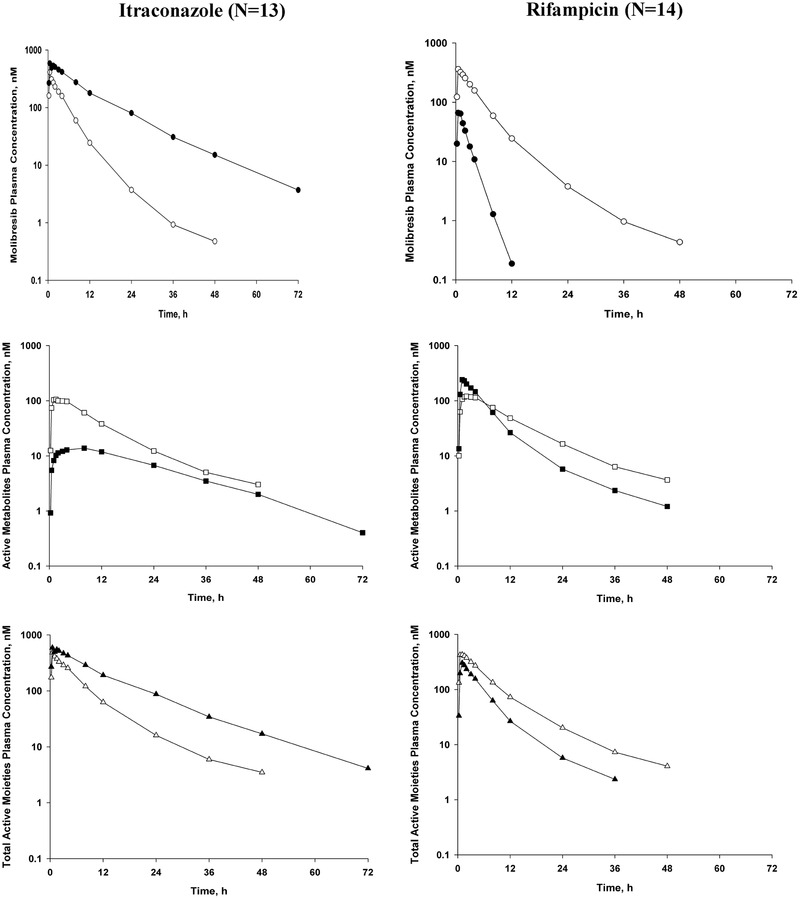
Dose‐normalized mean molibresib, active metabolite, and total active moiety concentration‐versus‐time plots when molibresib is administered alone (open symbols) or in combination (closed symbols).
Table 5.
Simcyp Prediction and Actual Observed Effect of Itraconazole on PK Parameters of Molibresib, Its Two Active Metabolites (GSK3529246), and Total Active Moiety—Part 1
| Molibresib | GSK3529246 | Total Active Moiety | |||
|---|---|---|---|---|---|
| Parameter | Observed Geometric Mean Ratio (90%CI) | Predicted Ratio (5th and 95th Percentiles) | Observed Geometric Mean Ratio (90%CI) | Predicted Ratio (5th and 95th Percentiles) | Observed Least‐Squares Mean Ratio (90%CI) |
| Cmax | 1.66 (1.43‐1.92) | 1.30 (1.14‐1.52) | 0.13 (0.11‐0.14) | 0.14 (0.03‐0.55) | 1.40 (1.22‐1.61) |
| AUC | 4.15 (3.77‐4.58) | 4.41 (2.95‐6.02) | 0.29 (0.26‐0.32) | 0.29 (0.09‐0.82) | 2.38 (2.24‐2.52) |
CI, confidence interval; Cmax, maximum concentration; AUC, area under the curve from 0 to infinity.
The predicted ratios are presented as geometric mean and 5th and 95th percentiles for 100 simulated subjects. The total active moiety represents the sum of molibresib and its 2 active metabolites reported as GSK3529246.
Simcyp Model Refinement
At the completion of part 1, the PBPK model was refined using the updated to reflect oral clearance and volume of distribution estimates (part 1, cohort 1) from a healthy volunteer population (previous estimates taken from cancer subjects). The model was subsequently verified using the exposure data from cohort 2 of part 1. In addition, a PBPK model was built for the active metabolite GSK3529246, prior to simulating the effect with itraconazole. Observed versus simulated plasma profiles for molibresib and its active metabolite are shown in Figure 5.
Figure 5.
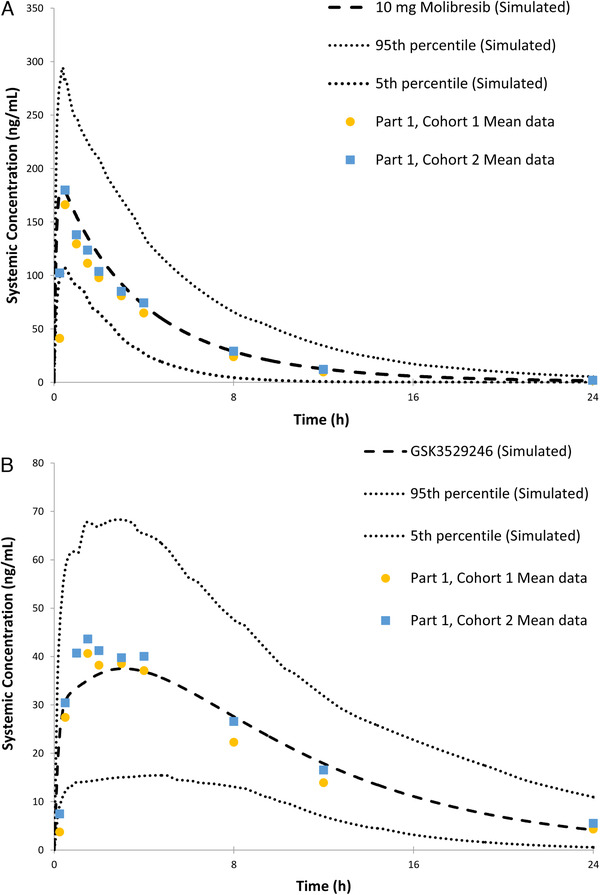
Observed and Simcyp‐simulated molibresib (A) and GSK3529246 (B) concentration‐time profiles after a single 10‐mg dose.
Simulations of molibresib and GSK3529246 AUC in the absence and presence of itraconazole were comparable to those observed in part 1 of this study, with the predicted mean geometric AUC ratio within 10% of the observed AUC ratio (Table 2). A slight underprediction of the effect of itraconazole on the Cmax of molibresib was noted. Molibresib was shown to be a P‐gp substrate in vitro; it could be postulated that this minor discrepancy seen in the predicted and observed Cmax ratio might be because of inhibition of P‐gp‐mediated transport of molibresib by itraconazole, an effect not incorporated in the Simcyp model. However, based on the predicted and observed Cmax ratio values, this contribution was deemed minimal and not clinically significant. In addition, the alteration of P‐gp, either via inhibition or induction, would have a limited impact on the formation of GSK3529246. In summary, Simcyp simulations were in agreement with the observed data confirming that the Simcyp model could be used to predict the effect of rifampicin on the PK of molibresib and GSK3529246.
Rifampicin Drug‐Drug Interaction—Part 2
Based on results from the itraconazole part of the study, including PK, safety, and tolerability data, and the refined prediction using the Simcyp model, the effect of rifampicin on molibresib AUC was predicted to be 0.21 (∼80%; Table 6). Accordingly, in part 2, a dose of 10 mg of molibresib was administered alone, and a dose of 20 mg of molibresib was administered after repeat administration of rifampicin.
Table 6.
Simcyp Prediction for Rifampicin Effect on PK Parameters of Molibresib and Its Two Active Metabolites (GSK3529246)
| Molibresib | GSK3529246 | |
|---|---|---|
| Parameter | Predicted Ratio (5th and 95th Percentiles) | Predicted Ratio (5th and 95th Percentiles) |
| Cmax | 0.51 (0.48, 0.54) | 1.13 (1.12, 1.14) |
| AUC | 0.21 (0.20, 0.23) | 1.98 (1.91, 2.06) |
Cmax, maximum concentration; AUC, area under the curve from 0 to infinity.
The predicted ratios are presented as geometric mean and 5th and 95th percentiles for 100 simulated subjects.
A dose‐normalized analysis of the observed data indicated that coadministration with rifampicin decreased the AUC0‐∞ of molibresib approximately 91% and decreased the total exposure of the 2 active metabolites by approximately 8% (Figure 4 and Table 7). Total exposure of the active moiety molibresib was decreased approximately 50% with coadministration of rifampicin. Coadministration with rifampicin decreased the Cmax of molibresib approximately 80% and increased the Cmax of the 2 active metabolites 104%. Peak exposure of the total active moiety molibresib was decreased approximately 33% with rifampicin coadministration. PK parameters in the presence and absence of rifampicin are summarized in Table 7. The PBPK model underpredicted the extent of rifampicin‐mediated CYP3A induction‐resultant decrease in molibresib exposure. The underprediction may be because of P‐gp induction, or it may be from the variability in the in vitro CYP3A induction parameters used to build the rifampicin PBPK model. Although rifampicin PBPK has also undergone substantial updates and successful applications over the last 5 years, 12 the induction parameters are further updated within the model, but the rifampicin model used for these molibresib simulations was those that were available for simulations about 5 years ago. Although out of the scope of the current article exercise, this may be a good case study to test the updated rifampicin models.
Table 7.
Summary of Observed PK Parameters of Molibresib, Its Two Active Metabolites (GSK3529246), and Total Active Moiety—Rifampicin Interaction (Part 2)
| Analyte | Parameter | Molibresib 10 mg Alone | Molibresib 20 mg + Rifampicin | Geometric Mean Ratio, Dosed Normalized (90%CI) |
|---|---|---|---|---|
| Molibresib (n = 14) | Cmax (ng/mL) | 158 (30) | 64.0 (59) | 0.202 (0.174‐0.235) |
| AUC0‐∞ (ng·h/mL) | 669 (36) | 115 (48) | 0.0864 (0.0744‐0.100) | |
| tmax (h) | 0.52 (0.27‐2.0) | 0.53 (0.50‐1.0) | – | |
| t1/2 (h) | 5.8 (42) | 1.3 (15) | – | |
| Molibresib 2 active metabolites, GSK3529246 (n = 14) | Cmax (ng/mL) | 49.0 (24) | 200 (24) | 2.04 (1.78‐2.32) |
| AUC0‐∞ (ng·h/mL) | 622 (26) | 1140 (26) | 0.917 (0.850‐0.989) | |
| tmax (h) | 2.0 (1.0‐4.1) | 1.0 (0.53‐1.5) | – | |
| t1/2 (h) | 10.0 (22) | 10.0 (24) | – | |
| Total active moiety (n = 14) | Cmax (nmol/L) | 468 (23) | 631 (27) | 0.673 (0.623‐0.727) |
| AUC0‐∞ (nmol·h/L) | 3190 (28) | 3180 (26) | 0.498 (0.480‐0.517) | |
| tmax (h) | 0.77 (0.52‐2.0) | 1.0 (0.50‐1.5) | – | |
| t1/2 (h) | 9.1 (25) | 10.2 (28) | – |
Cmax, maximum concentration; AUC0‐∞, area under the curve from 0 to infinity; tmax, time of occurrence of Cmax; t1/2, terminal phase elimination half‐life.
Cmax, AUC, and t1/2 are presented as geometric means (CVb%) with 3 significant digits for the geometric mean and 2 for the CVb%; Tmax is presented as median (range) with 2 significant digits.
GSK3529246 represents the sum of GSK3529246 and GSK3536835; GSK3536835 was converted to GSK3529246 prior to analysis.
Total active moiety represents the sum of molibresib and its active metabolite GSK3529246.
Safety
Administration of molibresib alone and when coadministered with itraconazole or rifampicin was safe and well tolerated in this study. The number of subjects with adverse events (AEs) included 8 (25%) in part 1 and 15 (100%) in part 2. Of these, 3 subjects reported AEs when molibresib was administered alone (1 [8%] in part 1, 2 [13%] in part 2). The interaction between molibresib and itraconazole was minimal, with 3 subjects (23%) reporting AEs, whereas no AEs were reported with coadministration of molibresib and rifampicin in part 2. The majority of the AEs in both parts of the study were reported during the days when either itraconazole or rifampicin was administered alone. All AEs were mild or moderate in severity; no serious AEs were reported during the study. In addition, there were no clinically relevant findings in vital signs, electrocardiograms, or clinical laboratory parameters.
Discussion
Conducting drug‐drug interactions (DDI) trials in healthy subjects is the preferred method for such studies because of the lack of confounding effects of other concomitant medications. However, because of the expected toxicity of administering most oncology products to healthy volunteers at therapeutic doses, the current trial used a more feasible adaptive design approach to administer the lowest possible doses of molibresib for a DDI study in healthy subjects. This adaptive design also utilized a Simcyp population‐based PBPK modeling simulation to predict the lowest doses of molibresib that could be administered with itraconazole and rifampicin, respectively, to minimize exposure in healthy volunteers, keeping the exposures in safe but at the same time analytically feasible as well. In preclinical findings, molibresib had adverse effects on the testes including sperm retention and disorganized spermatogenesis. In addition, there was a substantiated risk for adverse effects on embryofetal development and female fertility. Based on these observations, this study was conducted in female volunteers of nonchildbearing potential.
The interaction with itraconazole, a known strong CYP3A inhibitor and a P‐gp inhibitor, was evaluated in part 1 as a 2‐cohort study, in which preliminary data from the first cohort informed further dosing in the second cohort based on predefined criteria based on observed changes in AUC levels (Figure 2). The use of itraconazole was per the recommendations of the US Food and Drug Administration and the European Medicines Agency advising against the use of ketoconazole because of safety concerns 13 , 14 with a dosing schedule supported by the simulation work of Ke et al. 8 Overall, in the presence of itraconazole, total exposure (AUC) to molibresib increased by 4.15‐fold along with a 66% increase in Cmax, whereas the total AUC and Cmax for the 2 major active metabolites of molibresib decreased by approximately 70% and 87%, respectively. These results also showed that the predicted effects based on Simcyp modeling were similar to the observed effects of itraconazole on the PK of molibresib and its major metabolites. Prior to the start of this study, Simcyp simulation based on data from prior trials predicted an approximately 4.41‐fold increase in molibresib exposure, which was confirmed by the observed 4.15‐fold increase. The major changes were observed in the AUC and Cmax with minimal changes in the tmax for molibresib. This suggests that the DDI observed with itraconazole is primarily because of CYP3A inhibition with minimal effect from P‐gp, as itraconazole is also a known inhibitor of P‐gp and molibresib is a P‐gp substrate. The agreement between the PBPK predicted and clinically observed DDI also further engraves confidence in the CYP3A‐mediated metabolism assignment in the PBPK model.
In the presence of rifampicin, a strong CYP3A and P‐gp inducer, AUC and Cmax of molibresib decreased by approximately 91% and 80%, respectively. The total active metabolite exposure decreased to a lesser extent of 8% in the presence of rifampicin; this observation is not unexpected for a pathway with a high fm (fraction metabolized), as induction will not lead to a large fold‐increase in metabolite exposure (AUC), which is already high. For the total active moiety, coadministration with rifampicin resulted in a total exposure decrease of approximately 50%. The observed results for the rifampicin interaction showed a higher interaction than expected based on the Simcyp simulation. Possible explanations for this discrepancy could include p‐glycoprotein induction, which was not incorporated in the original model, or potential underestimation of first‐pass metabolism and hence overestimation of the Fg (fraction escaping gut metabolism) The other plausible reason for the underprediction may be the unavailability of a clinically representative in vitro CYP3A induction value calibration in the rifampicin PBPK model used at the time of these simulations. Previous rifampicin models have been shown to underestimate the CYP3A‐mediated interaction with drugs, as may be the case here. The rifampicin model has been updated over the years with successful applications in some recent case studies. 12
Overall, the results of this study confirmed the in vitro data that molibresib is a substrate for CYP3A4. The adaptive design of this study, including PBPK simulations, allowed for the investigation of 2 DDI studies in a single trial, thus minimizing the time for conducting such studies. In addition, with the utility of PBPK simulations, this study was conducted at very low doses with an oncology drug that is considered intolerable at high doses in healthy subjects. The PBPK simulations also allowed for selecting analytically feasible doses. The early availability of a PBPK model describing not only parent drug but also the active metabolites will provide support in predictions of DDIs with an array of cotherapies, common in oncology patients.
Conclusion
An adaptive design DDI study with itraconazole and rifampicin showed that CYP3A enzymes are playing a major role in the elimination of molibresib, as drug exposure was greatly increased with itraconazole (strong CYP3A inhibitor) and decreased with rifampicin (strong CYP3A inducer). The use of PBPK was key in supporting the selection of doses, and inclusion of the major active major metabolites will be valuable for the clinical development of this compound.
Conflicts of Interest
All authors are employees of GSK, hold company stock, and meet the criteria for authorship set forth by the International Committee for Medical Journal Editors. Editorial support (development of the first draft, assembling tables and figures, collating author comments, and referencing) was provided by Guissou Dabiri, PhD, and was funded by GSK.
Funding
Funding for this study was provided by GlaxoSmithKline (NCT02729038).
Data‐Sharing Statement
Anonymized individual participant data and study documents can be requested for further research from www.clinicalstudydatarequest.com.
Acknowledgment
The authors thank PAREXEL Baltimore for the clinical conduct of the study.
References
- 1. Nicodeme E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468(7327):1119‐1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. French CA, Ramirez CL, Kolmakova J, et al. BRD‐NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27(15):2237‐2242. [DOI] [PubMed] [Google Scholar]
- 3. Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c‐Myc. Cell. 2011;146(6):904‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mertz JA, Conery AR, Bryant BM, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669‐16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung CW, Coste H, White JH, et al. Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J Med Chem. 2011;54(11):3827‐3838. [DOI] [PubMed] [Google Scholar]
- 6. Piha‐Paul S, Hann C, French C et al. Phase I study of molibresib (GSK525762), a bromodomain and extra terminal protein (BET) inhibitor, in patients with advanced solid tumors including NUT carcinoma (NC). JNCI Cancer Spectrum. 2019;4(2):pkz093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ke AB, Zamek‐Gliszczynski MJ, Higgins JW, Hall SD. Itraconazole and clarithromycin as ketoconazole alternatives for clinical CYP3A inhibition studies. Clin Pharm Ther. 2014;95(5):473‐C>. [DOI] [PubMed] [Google Scholar]
- 8. Chen J, Raymond K. Roles of rifampicin in drug‐drug interactions: underlying molecular mechanisms involving the nuclear pregnane X receptor. Ann Clin Microbiol Antimimicrob. 2006;5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stoch SA, Friedman E, Maes A, et al. Effect of different durations of ketoconazole dosing on the single‐dose pharmacokinetics of midazolam: shortening the paradigm. J Clin Pharmacol. 2009;49(4):398‐406. [DOI] [PubMed] [Google Scholar]
- 10. Backmann JT, Olkkola KT, Neuvonen PJ. Rifampin drastically reduces plasma concentrations and effects of oral midazolam. Clin Pharmacol Ther. 1996;59(1):7‐13. [DOI] [PubMed] [Google Scholar]
- 11. Olkkola KT, Backman JT, Neuvonen PJ. Midazolam should be avoided in patients receiving the systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther. 1994;55(5):481‐485. [DOI] [PubMed] [Google Scholar]
- 12. Almond LM, Mukadam S, Gardner I, et al. Prediction of drug‐drug interactions arising from CYP3A induction using a physiologically based dynamic model [published correction appears in Drug Metab Dispos. 2016;44(6):877]. Drug Metab Dispos. 2016;44(6):821‐832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenblatt HK, Greenblatt DJ. Liver injury associated with ketoconazole: review of the published evidence. J Clin Pharmacol. 2014;54(12):1321‐1329. [DOI] [PubMed] [Google Scholar]
- 14. Liu L, Bello A, Dresser MJ, et al. Best practices for the use of itraconazole as a replacement for ketoconazole in drug‐drug interaction studies. J Clin Pharm. 2016;56(2):143‐151. [DOI] [PubMed] [Google Scholar]


