Summary
Synthetic biology can greatly aid the investigation of fundamental regulatory mechanisms and enable their direct deployment in the host organisms of choice. In the field of plant hypoxia physiology, a synthetic biology approach has recently been exploited to infer general properties of the plant oxygen sensing mechanism, by expression of plant‐specific components in yeast. Moreover, genetic sensors have been devised to report cellular oxygen levels or physiological parameters associated with hypoxia, and orthogonal switches have been introduced in plants to trigger oxygen‐specific responses. Upcoming applications are expected, such as genetic tailoring of oxygen‐responsive traits, engineering of plant hypoxic metabolism and oxygen delivery to hypoxic tissues, and expansion of the repertoire of genetically encoded oxygen sensors.
Keywords: flooding, genetically encoded sensors, hypoxia, metabolic engineering, plants, synthetic biology
Short abstract
See also the Editorial on this article by Sasidharan et al., 229: 5–7.
| Contents | ||
|---|---|---|
| Summary | 50 | |
| I. | Introduction | 50 |
| II. | In vivo biosensors for plant cellular hypoxia | 51 |
| III. | Synthetic biology strategies to improve plant performance during flooding | 53 |
| IV. | Synthetic oxygen‐dependent metabolism | 54 |
| V. | Conclusions | 55 |
| Acknowledgements |
55 |
|
| References | 55 | |
I. Introduction
Proposed in the beginning of the 20th century, synthetic biology has rapidly expanded from the early 2000s until now, as a result of the advancements and progressive affordability of DNA sequencing, synthesis and manipulation technologies. Nowadays, this research area combines approaches from biology, physics, mathematics and chemistry with the aim of modifying existing biological systems, or creating entirely new ones, to generate tools and knowledge for research or practical applications. To achieve such goals, synthetic biologists apply engineering principles (Andrianantoandro et al., 2006) in an iterative process that generates predictable and reliable models to be tested experimentally and thus gather information for future improved design (Cameron et al., 2014).
In the last decade, this framework has also been adopted in plant biology (Liu & Stewart, 2015). Continuous progress in the collection and functional characterization of DNA modules already enables the construction of synthetic genetic circuits in plant cells (Andres et al., 2019). Some bright examples, in which the aforementioned approaches have been used to ameliorate plant photosynthetic (South et al., 2019) and water‐use efficiency (Park et al., 2015; Papanatsiou et al., 2019), demonstrate that synthetic biology can provide unprecedented opportunities to assist and speed up the generation of crops with improved responses to the environment. Perhaps the most prolific field of application in plants is that of (genetically encoded) biosensors (Box 1), with a number of them engineered to date to respond to endogenous and environmental molecules or parameters. These endeavours have created new analytical tools to measure specific inputs, but have also led to discoveries concerning fundamental aspects of plant biology (Walia et al., 2018), demonstrating once more that, besides its technological value, synthetic biology can provide a new conceptual framework to expand the borders of scientific knowledge (de Lorenzo & Danchin, 2008). Here, we will outline how the plant community has taken advantage of the synthetic biology perspective to shed light on oxygen biology, and propose ideas for its next application towards improved plant performance under hypoxia‐related environmental stresses.
Box 1. General architecture of genetically encoded sensors.
1.
Biosensors are devices that incorporate a biological sensing element, able to reveal a biological analyte (e.g. a biomolecule) or its concentration, and convert the biological signal into a measurable output (Turner et al., 1987). Biosensors of a particular kind are the genetically encoded sensors, whose functions are covered by DNA‐encoded parts (e.g. proteins, peptides, RNA molecules, aptamers).
Genetically encoded sensors can be conceptually described as generated by combination of three kinds of functions: a sensory function, which has first‐hand interaction with the desired analyte or stimulus (biological sensory components may, for instance, use the analyte as a substrate for biochemical reactions, or undergo a spontaneous allosteric, conformational, or chemical change upon the interaction); an effector function that transduces the information; and an output function that produces a change in a measurable parameter as the biosensor readout. Functions are expressed by DNA modules (most frequently consisting of transcriptional units) of native or exogenous origin (i.e. introduced upon genetic transformation of the host organism), whose connection reconstitutes small genetic circuits. Individual functions can be executed by separate modules, constituted by one or more components, or aggregated. In this way, direct biosensors can be defined as those in which a single module is able to react to and report the status of the desired stimulus. Indirect biosensors, instead, require additional components, encoded either by separate modules or by the cellular machinery, to enable the production of the output (Wright & Nemhauser, 2019).
II. In vivo biosensors for plant cellular hypoxia
Oxygen levels in plant tissues display variations that depend on their organisation, distance from the body surface, and metabolic activity. This picture has been established through measurements of internal oxygen concentrations obtained with physical sensors. Optical sensors and, more recently, miniaturized Clarke‐type electrodes have been employed to reveal that in plant tissues oxygen can range from below the detection limit (0.07 kPa) to above 50 kPa (Pedersen et al., 2016). Oxygen‐sensitive foils have also been developed to visualize and quantify oxygen distribution in sectioned plant organs (Tschiersch et al., 2012). However, physical sensors are still constrained by their spatial resolution and by the unavoidable mechanical injury caused by piercing or slicing. Introduction of the thinnest probe available (as small as 5 μm in diameter; Schmidt et al., 2018) can still locally perturb cellular oxygen dynamics and biochemical responses.
When cell size resolution is sought, invasive detection methods are preferentially substituted with genetically encoded circuitries designed to report on O2 abundance or O2‐associated responses. Genetically encoded sensors have the potential to disclose phenomena that take place in undisturbed subcellular compartments and depict their dynamics over time, opposite to static single point surveys. Recently, knowledge gathered on the mechanisms of oxygen perception across life kingdoms has been deployed to design and test synthetic circuits that are able to reveal oxygen fluctuations in plant cells.
Transcriptional biosensors
Higher plants possess a direct oxygen‐sensing mechanism that relies on the conditional degradation of master activators of hypoxic gene expression, the ERF‐VII ethylene responsive factors. In the presence of oxygen, plant cysteine oxidase enzymes (PCOs) oxidize a conserved N‐terminal cysteine exposed by the ERF‐VII proteins, thereby targeting them to proteasomal degradation through the dedicated cysteine N‐degron pathway (van Dongen & Licausi, 2017). Conversely, inhibition of PCO activity under hypoxia makes the ERF‐VIIs stable in this condition. The endogenous plant oxygen‐sensing machinery has been exploited in Arabidopsis thaliana, to drive the expression of a synthetic promoter, derived from the DNA cis‐element recognized by the ERF‐VIIs (Gasch et al., 2016). Here, the transcriptional output module is coupled to native sensory and effector modules from Arabidopsis, respectively constituted by PCOs and the ERF‐VIIs (Fig. 1a). In Arabidopsis shoot apices, activation of the hypoxia‐responsive promoter element (HRPE) output occurs in cell layers where low oxygen concentrations have been measured with a Clark‐type electrode (Weits et al., 2019). Although use of output reporters based on hypoxia‐inducible promoters is not entirely novel, enhanced specificity is expected to be conferred by the absence of any additional cis‐element in the synthetic HRPE promoter beyond the one bound by the ERF‐VII. This should prevent the output module from responding to unrelated transcription factors, different from native hypoxia‐responsive promoters on which multiple signalling pathways can, in principle, converge. Dedicated comparisons with reporters based on native plant promoters will reveal whether the HRPE reporter is, in fact, characterized by more specific patterns of response to oxygen in a range of plant organs.
Figure 1.
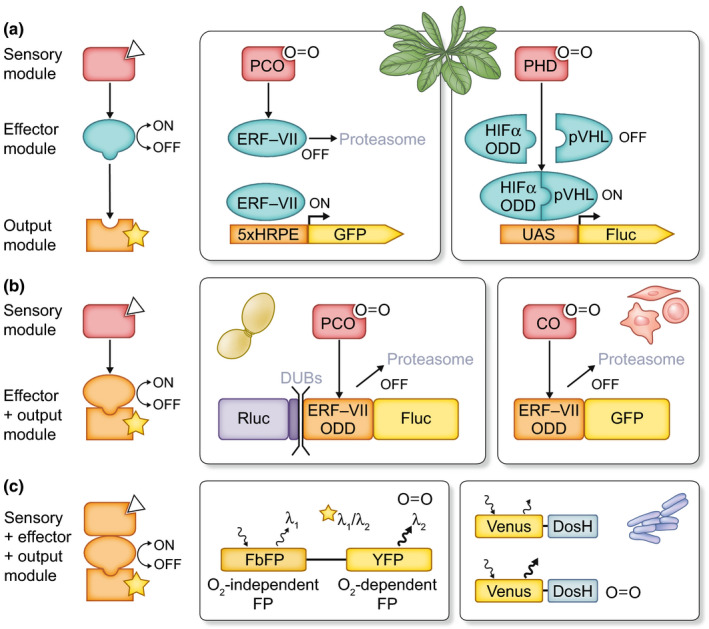
Synthetic strategies for the design of genetic biosensors of oxygen. Three types of biosensor designs are conceptualized on the left, based on the architecture of their constituents. In the schematics, a generic analyte, recognized by the sensory module, is represented by a triangle; a generic output is indicated by a star. Examples of oxygen biosensors of each kind, implemented in various hosts, are provided in the shaded boxes. Names in grey indicate endogenous components from the host cell. (a) Transcriptional (indirect) biosensors. Left panel: a synthetic 5 × hypoxia‐responsive promoter element‐green fluorescent protein (5xHRPE‐GFP) output module responded to the intracellular oxygen concentrations (Weits et al., 2019) by way of endogenous modules providing sensory (plant cysteine oxidases, PCO) and effector functions (ethylene‐responsive factors group VII (ERF‐VII) transcription factors), and other functions (Cys N‐degron pathway proteins and the proteasome system) needed to complete the genetic circuitry. Right panel: a molecular switch composed of three synthetic modules of fully exogenous origin was activated by oxygen in a context‐independent (orthogonal) fashion (Iacopino et al., 2019). A human prolyl hydroxylase domain (PHD) enzyme enables recognition of a hypoxia‐inducible factor’s α subunit (HIF1‐α) oxygen‐dependent degradation domain (ODD)‐based effector module by a von Hippel–Lindau protein (pVHL) β‐domain present on a second effector module. The interaction brings into contact two associated domains of the GAL4 transcription factor (not shown in the graphics) and enables the expression of the output module. (b) Degradation‐based (indirect) biosensors. Left panel: in yeast, ERF‐VII ODD served as effector domain incorporated in a ratiometric luminescent output module. Plant PCO was supplied as sensor (Puerta et al., 2019). Right panel: in human cells, ERF‐VII ODD‐containing output modules can work in a circuitry that provides an endogenous sensory function, thanks to human cysteine oxidases (COs) (Masson et al., 2019). (c) Direct biosensors. One example of a Foerster resonance energy transfer (FRET)‐based maturation biosensor is provided on the left (Potzkei et al., 2012; see main text for further detail). Right: a haem‐based biosensor design. When haem is bound to oxygen, a haem‐containing DosH unit is less efficient in quenching fluorescence of a Venus yellow fluorescent protein (YFP) linked to it (Nomata & Hisabori, 2018). Both designs were implemented in bacteria. DUBs, deubiquitinating enzymes; Fluc, firefly luciferase; Rluc, renilla luciferase; UAS, GAL4 upstream activating sequence.
The HRPE reporter may nonetheless retain residual responsivity to other inputs than the mere oxygen concentration: in particular, its output might be influenced by any O2‐independent factor impacting on ERF‐VII activity or abundance. Indeed, it should not be disregarded that indirect biosensors rely on cellular machinery components, whose status can substantially affect the output of the system (Wright & Nemhauser, 2019), and thus careful setup of the experimental controls is needed to avoid misinterpretations. In this regard, a desirable property of synthetic gene circuits and biological devices is the so‐called orthogonality (or context‐free behaviour), defined as their ability to work (nearly) uncoupled from all extant cellular processes that are not strictly required for the response of interest.
A recent study (Iacopino et al., 2019) attempted to attain a fully orthogonal sensor, by engineering human oxygen‐sensing components in Arabidopsis. Direct oxygen perception in humans revolves around the oxygen‐dependent degradation of Hypoxia Inducible Factor α (HIFα) transcription factors (Semenza, 2007), through a pathway that is structurally similar to the Cys N‐degron pathway in plants (Licausi et al., 2020). Minimum regulatory domains isolated from HIF‐1α and its cognate E3 ligase Von Hippel‐Lindau protein (pVHL) were used to build the effector modules of a transcriptional biosensor, by reconstruction of a GAL4‐based two‐hybrid system (Fig. 1a). The sensory module was constituted by the human oxygen sensor, prolyl hydroxylase domain 3 (PHD3); Schofield & Ratcliffe, 2004). In this strategy, incorporation of protein modules with no phylogenetic relationships to the plant proteome has proved successful to achieve high specificity for oxygen. When specifically tested, the effector modules showed no interaction with the extant functions of the cell, indicating that the biosensor was selective for PHD3 as a switch; in turn, this sensory module proved to be selective for oxygen as an input (Iacopino et al., 2019).
Degradation‐type biosensors
In the case of the ERF‐VII factors, the exposed cysteine acts as a regulatory target that, combined with proper structural features (yet to be generalized), turns the N‐terminal extremity of these proteins into an O2‐dependent protein degradation domain (ODDs). Originally described for HIF‐1α (Huang et al., 1998), ODDs link a protein’s half‐life to oxygen concentration, with the involvement of the ubiquitin proteasome system. ODDs are versatile units to be incorporated into oxygen biosensors. When ODD‐containing reporter proteins are expressed in cells where the cognate degradation machinery is present, indirect biosensors can be obtained in which effector and output modules are combined (Fig. 1b). In this way, the human HIF‐CODD peptide (C‐terminal ODD) fused to a fluorescent protein has been used to report hypoxia in Drosophila (Misra et al., 2017); moreover, HIF‐CODD‐based synthetic reporters have been successfully exploited as tracers in animals, where they could be delivered by injection (Iglesias et al., 2019). If not directly delivered to intact plant cells, whose low propensity to uptake exogenous compounds is probably a result of peculiar cell surface properties, such as the presence of a glycan‐rich cell wall (Cedeño et al., 2017), synthetic ODD fusion proteins can nonetheless be genetically encoded. Plant oxygen‐sensitive domains have been effectively exploited in ‘degradation‐type’ biosensors. The cysteine N‐degron from the barley ERF‐VII factor BERF1 has been associated with a visual reporter and expressed in barley, to obtain the only known oxygen reporter developed for a crop so far (Mendiondo et al., 2016). As in the case of plant HRPE‐based reporters, the endogenous partners of this indirect sensor in principle make it sensitive to additional stimuli to oxygen. In heterologous systems, in turn, cysteine N‐degrons from Arabidopsis have proved able to report oxygen in human cells (Masson et al., 2019) and yeast (Puerta et al., 2019) (Fig. 1b). Moreover, such an approach has made it possible to exclude the involvement of nitric oxide in PCO‐mediated cysteine oxidation (Puerta et al., 2019), as an advancement towards unravelling the hierarchical position of nitric oxide during ERF‐VII proteostasis (Gibbs et al., 2014).
Some of the developed oxygen biosensors are ratiometric (their readout is independent of probe concentration). In the degradation‐type sensor tested in yeast, this feature has been attained by translational fusion of an O2‐insensitive luminescent moiety to the output module (Fig. 1b). Otherwise, suitable fluorescent proteins have been paired to set up intrinsically ratiometric Foerster resonance energy transfer (FRET) sensors, where conformational changes triggered by interaction of HIF‐CODD and pVHL domains enabled FRET in an O2‐dependent fashion (Youssef et al., 2016).
Maturation‐type biosensors
All green fluorescent protein (GFP)‐like fluorescent proteins strictly require oxygen for the autocatalytic production of a functional chromophore from a nonfluorescent precursor: this property has been exploited to realize direct O2 biosensors that can be dubbed as ‘maturation type’ (Fig. 1c). They have so far been employed as quantitative reporters in bacteria or animals, but their evaluation in plants is yet to come. Combination of GFP‐like proteins with O2‐independent fluorophores based on flavin mononucleotides has generated a FRET biosensor in E. coli (Potzkei et al., 2012). Furthermore, a two‐color DsRed protein has been developed, whose properties of O2‐dependent maturation determined a quantitative shift in FRET emission from red to green that could be used to monitor hypoxia in Drosophila (Lidsky et al., 2018). Maturation‐type ratiometric biosensors may also be obtained using tandem fluorescent protein timers (tFT). Fusions between O2‐dependent and O2‐independent fluorescent proteins have been successfully used to spot hypoxia in human cells (Erapaneedi et al., 2016). The amenability of tFTs as in vivo sensors in plants has recently been demonstrated by coupling them to the N‐degron and auxin signalling pathways in Arabidopsis and tobacco (Zhang et al., 2019). Finally, haem‐binding protein domains can potentially serve as further direct sensors. In E. coli, oxygen‐dependent conformational change in the native haem‐binding domain DosH has been used to obtain a FRET probe that showed in vivo O2 sensitivity in the micromolar range (Nomata & Hisabori, 2018).
Acute hypoxia in cells displays physiological hallmarks, mostly connected to the impairment of mitochondrial respiration. Analytes different from O2 can therefore be deployed to investigate its onset and consequences. Indeed, genetic sensors have been designed in plants to detect relevant compounds and parameters that vary with O2 availability for metabolism, such as adenylates, cytosolic pH, free calcium, reactive oxygen species (ROS), redox state, and nicotinamide adenine dinucleotide oxidation. Five of the aforementioned reporters have recently been used to achieve parallel monitoring of different cytosolic parameters during hypoxia in Arabidopsis (Wagner et al., 2019).
III. Synthetic biology strategies to improve plant performance during flooding
Hypoxia is a component of the flooding stress, which arises as a result of water saturation in the soil (waterlogging) or complete submergence of the plant body (Sasidharan et al., 2017). Besides the direct inhibition of oxidative phosphorylation, and the consequent energy crisis, this environmental condition entails photosynthesis inhibition as a result of water turbidity, accumulation of gasotransmitters such as ethylene, nitric oxide and hydrogen sulphide, and increased availability of phytotoxic compounds (Bailey‐Serres & Colmer, 2014). Additionally, drought‐like stress is experienced when aerobic conditions are restored and plants need to rapidly readapt to the gaseous atmosphere (Yeung et al., 2019). Endeavours to improve crop tolerance to flooding have succeeded through transfer of single genes or loci, as a result of the practical and economical limitation in manipulating complex traits (Xu et al., 2006; Kretzschmar et al., 2015; Mendiondo et al., 2016). Moreover, as these loci are effective in conferring submergence resistance at specific developmental stages, a full‐spectrum resistance requires gene stacking, when they do not conflict with each other or impair yield (Lee et al., 2009).
Consequently, synthetic biology would prove advantageous to activate in a timely and tuneable manner some key metabolic or anatomical features that enhance tolerance (Fig. 2). Taking oxygen levels as bona fide proxy of the submergence status, molecular switches based on O2‐dependent reactions constitute a promising solution to toggle adaptive strategies, and possibly attune them to metabolic or developmental parameters. A number of reports proposed enhanced fermentation, inhibition of growth and enhanced ROS scavenging as means to improve plant tolerance (Ismond et al., 2003; Tesniere et al., 2006). While the efficacy of these strategies is based on large phenotypic surveys over natural variation within plant species, transcriptomic analyses and gene inactivation assays, an unsupervised attempt to generate plants with improved submergence tolerance has been successfully carried out by Vartapetian et al. (2014). Here, the authors selected hypoxia‐tolerant wheat and sugarcane calli by exposure to anoxia and, from these, regenerated plants with superior tolerance to waterlogging; unfortunately, the molecular determinants of this feature were not subsequently identified. Nowadays, however, the synthetic biology framework enables us to plan bolder endeavours. The oxygen‐sensing modules reviewed earlier could be linked to features that have been proposed as crucial for flooding and de‐submergence survival, such as promotion of water escape by elongation, energy‐saving by quiescence, prompt stomatal closure, and senescence retardation (Yeung et al., 2019). The availability of nonplant modules could provide the opportunity to achieve their orthogonal regulation. Moreover, extra copies of hormonal regulators (transcription factors or regulatory partners) could be made O2‐dependent by conjugation with ODDs. These strategies could involve ‘highjacking’ gibberellin or brassinosteroid signalling to control underwater growth, ethylene and cytokinin downstream targets to inhibit early senescence, and ABA perception to govern stomatal aperture during flooding and at de‐submergence (Fig. 2).
Figure 2.
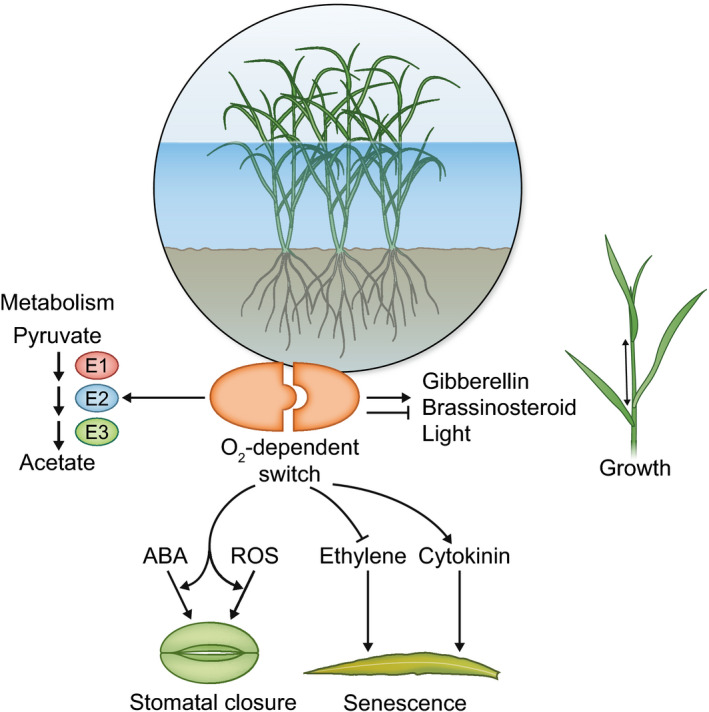
Proposed exploitation of synthetic biology to enhance flooding tolerance. Damage to plants and yield losses caused by flooding stress could be limited or overcome by exploiting (low) oxygen‐dependent switches. These can be adopted to (on the left) guide existing metabolic pathways or induce heterologous ones dedicated to pyruvate consumption and sustain glycolysis, such as acetic fermentation. Other applications (following towards to the right) entail the stimulation of abscisic acid (ABA) and reactive oxygen species (ROS)‐driven stomata closure in order to prevent hyperhydricity during flooding and dehydration during desubmergence, selective manipulation of hormonal control of premature senescence after reoxygenation and, finally, induction or repression of growth to establish escape or quiescence strategies, respectively.
IV. Synthetic oxygen‐dependent metabolism
A third area of application of synthetic biology in plant hypoxia research pertains to metabolic engineering. Oxygen is consumed by cytochrome c (COX) and alternative oxidases (AOX) in the mitochondrial electron transport chain, to drive ADP phosphorylation or prevent accumulation of over‐reduced electron carriers, respectively (Schertl & Braun, 2014). Exploitation of these enzymatic activities has been proposed in order to ensure nitrogenase protection in plant cells engineered to perform nitrogen fixation in a nonsymbiontic context. Indeed, the nitrogenase complex is extremely sensitive to oxygen and requires high energy expense for its functioning. In this light, specialized mitochondria would constitute a new site to host nitrogen fixation (Burén & Rubio, 2018). Expression and activity of COX and AOX should be attuned to those of nitrogenase components, low oxygen and high ATP levels, therefore requiring a synthetic coordinator able to integrate these signals and generate a robust output.
As ATP synthesis is inhibited when oxygen availability falls below COX affinity, engineering of metabolic flexibility to sustain chemical energy fixation under these circumstances would be beneficial for plant tissues. Alternative reactions tapping from the cellular pyruvate pool would also serve the purpose of avoiding accumulation of this metabolite, which is suspected to stimulate respiration and promote a dangerous anoxic state (Zabalza et al., 2008; Bui et al., 2019). Pathways directed at these aims could be borrowed from aerobic algae, fungi and prokaryotes that behave as facultative anaerobes. Examples of these pathways are the acetic fermentation pathways reported in Chlamydomonas reinhardtii (Yang et al., 2015), or parallel respiratory pathways that use alternative electron acceptors in the absence of oxygen (Lecomte et al., 2018). Attempting these strategies poses challenges akin to those of engineering autonomous nitrogen fixation, beginning from the need of coordinated expression of a number of proteins targeted to the same subcellular compartment. Progress towards this end has been made by the engineering of multicistronic giant genes, whose translation products are cleaved by the tobacco etch virus protease (TEVp) (Yang et al., 2018). Future investigations with this aim in mind will doubtlessly benefit from the design–build–test–learn approach typical of synthetic biology. The outcome of this research seems promising not only for application in whole plants but also to support metabolite production and biomass yield in large‐scale cell cultures.
V. Conclusions
Plant oxygen biologists have embraced synthetic biology principles to pursue some long‐standing goals in hypoxia research, such as the live detection of oxygen variations in a cell‐resolved fashion and the introduction of highly specific responses to enhance plant tolerance under low oxygen‐associated environmental stresses. Some significant limitations have to be overcome in the near future to make these novel strategies highly effective (Box 2). Above all, avoiding unintended interference by the synthetic switches with cell regulation or metabolism will be particularly crucial to attain high precision application of the strategies outlined here towards flooding‐induced metabolic control.
Box 2. Challenges in synthetic biology of plant hypoxia.
1.
This Insight describes a range of synthetic, genetically encoded devices devised to return oxygen‐specific outputs, such as the in vivo imaging of O2 gradients, the control O2‐dependent switching of development, the implementation of space‐resolved responses, or the improvement of plant hypoxic metabolism. In most instances, the functional space of these devices is yet to be explored in a systematic way. The ability to address the following aspects will be of paramount importance for the success of the strategies designed.
Functional standards. The range of device activity as a function of oxygen concentration needs to be defined in the host organism, at least in a controlled setup. For instance, oxygen biosensors remain, to a large extent, qualitative, whereas this standard has been set for different plant‐based biosensors.
High‐throughput procedures for device optimization. The possibility to iteratively test sequence variants of the modules directly in the plant host is crucial to the outcome of every synthetic biology strategy among those presented. Effective screening methods for a large number of variants in plant cells (e.g. isolated protoplasts) will aid in identifying the best combinations of sequence elements (e.g. promoters and coding sequences) that enable a balanced production of functional modules in aerobic and hypoxic conditions, according to the specific experimental design.
Orthogonality in plant cells. The interaction between the existing cellular context and the synthetic devices has to be evaluated case by case both from the input side (Is the device regulated by oxygen only? Is the input managed through the sensory module?) and the output side (Does the operating device impact on unintended downstream pathways?).
Acknowledgements
We thank Ole Pedersen for detailed information on oxygen measurements in plant tissues. We apologize to all colleagues whose contribution to the field could not be further highlighted on this occasion, owing to space constraints.
References
- Andres J, Blomeier T, Zurbriggen MD. 2019. Synthetic switches and regulatory circuits in plants. Plant Physiology 179: 862–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E, Basu S, Karig DK, Weiss R. 2006. Synthetic biology: new engineering rules for an emerging discipline. Molecular Systems Biology 2: 2006.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey‐Serres J, Colmer TD. 2014. Plant tolerance of flooding stress – recent advances. Plant, Cell & Environment 37: 2211–2215. [DOI] [PubMed] [Google Scholar]
- Bui LT, Novi G, Lombardi L, Iannuzzi C, Rossi J, Santaniello A, Mensuali A, Corbineau F, Giuntoli B, Perata P et al 2019. Conservation of ethanol fermentation and its regulation in land plants. Journal of Experimental Botany 70: 1815–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén S, Rubio LM. 2018. State of the art in eukaryotic nitrogenase engineering. FEMS Microbiology Letters 365: doi: 10.1093/femsle/fnx274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DE, Bashor CJ, Collins JJ. 2014. A brief history of synthetic biology. Nature Reviews in Microbiology 12: 381–390. [DOI] [PubMed] [Google Scholar]
- Cedeño C, Pauwels K, Tompa P. 2017. Protein delivery into plant cells: toward in vivo structural biology. Frontiers in Plant Science 8: 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Licausi F. 2017. Oxygen sensing and signaling. Annual Reviews in Plant Biology 66: 345–367. [DOI] [PubMed] [Google Scholar]
- Erapaneedi R, Belousov VV, Schäfers M, Kiefer F. 2016. A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO Journal 35: 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch P, Fundinger M, Müller JT, Lee T, Bailey‐Serres J, Mustroph A. 2016. Redundant ERF‐VII transcription factors bind to an evolutionarily conserved cis‐motif to regulate hypoxia‐responsive gene expression in Arabidopsis. The Plant Cell 28: 160–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Md Isa N, Movahedi M, Lozano‐Juste J, Mendiondo GM, Berckhan S, Marín‐de la Rosa N, Vicente Conde J, Sousa Correia C, Pearce SP et al 2014. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Molecular Cell 53: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Gu J, Schau M, Bunn HF. 1998. Regulation of hypoxia‐inducible factor 1alpha is mediated by an O2‐dependent degradation domain via the ubiquitin‐proteasome pathway. Proceedings of the National Academy of Sciences, USA 95: 7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacopino S, Jurinovich S, Cupellini L, Piccinini L, Cardarelli F, Perata P, Mennucci B, Giuntoli B, Licausi F. 2019. A synthetic oxygen sensor for plants based on animal hypoxia signaling. Plant Physiology 179: 986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias P, Penas C, Barral‐Cagiao L, Pazos E, Costoya JA. 2019. A bio‐inspired hypoxia sensor using HIF1a‐oxygen‐dependent degradation domain. Scientific Reports 9: 7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismond KP, Dolferus R, de Pauw M, Dennis ES, Good AG. 2003. Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiology 132: 1292–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T, Pelayo MAF, Trijatmiko KR, Gabunada LFM, Alam R, Jimenez R, Mendioro MS, Slamet‐Loedin IH, Sreenivasulu N, Bailey‐Serres J et al 2015. A trehalose‐6‐phosphate phosphatase enhances anaerobic germination tolerance in rice. Nature Plants 1: 1–5. [DOI] [PubMed] [Google Scholar]
- Lecomte SM, Achouak W, Abrouk D, Heulin T, Nesme X, Haichar FZ. 2018. Diversifying anaerobic respiration strategies to compete in the rhizosphere. Frontiers in Environmental Science. doi: 10.3389/fenvs.2018.00139. [DOI] [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, David Ho TH, Yu SM. 2009. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Science Signaling 2: ra61. [DOI] [PubMed] [Google Scholar]
- Licausi F, Giuntoli B, Perata P. 2020. Similar and yet different: oxygen sensing in animals and plants. Trends in Plant Science 25: 6–9. [DOI] [PubMed] [Google Scholar]
- Lidsky PV, Lukyanov KA, Misra T, Handke B, Mishin AS, Lehner CF. 2018. A genetically encoded fluorescent probe for imaging of oxygenation gradients in living Drosophila. Development 145: dev156257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Stewart CN Jr. 2015. Plant synthetic biology. Trends in Plant Science 20: 309–317. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V, Danchin A. 2008. Synthetic biology: discovering new worlds and new words. EMBO Reports 9: 822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson N, Keeley TP, Giuntoli B, White MD, Puerta ML, Perata P, Hopkinson RJ, Flashman E, Licausi F, Ratcliffe PJ. 2019. Conserved N‐terminal cysteine dioxygenases transduce responses to hypoxia in animals and plants. Science 365: 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiondo GM, Gibbs DJ, Szurman‐Zubrzycka M, Korn A, Marquez J, Szarejko I, Maluszynski M, King J, Axcell B, Smart K et al 2016. Enhanced waterlogging tolerance in barley by manipulation of expression of the N‐end rule pathway E3 ligase PROTEOLYSIS6. Plant Biotechnology Journal 14: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T, Baccino‐Calace M, Meyenhofer F, Rodriguez‐Crespo D, Akarsu H, Armenta‐Calderón R, Gorr TA, Frei C, Cantera R, Egger B et al 2017. A genetically encoded biosensor for visualising hypoxia responses in vivo . Biology Open 6: 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomata J, Hisabori T. 2018. Development of heme protein based oxygen sensing indicators. Scientific Reports 8: 11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanatsiou M, Petersen J, Henderson L, Wang Y, Christie JM, Blatt MR. 2019. Optogenetic manipulation of stomatal kinetics improves carbon assimilation, water use, and growth. Science 363: 1456–1459. [DOI] [PubMed] [Google Scholar]
- Park SY, Peterson FC, Mosquna A, Yao J, Volkman BF, Cutler SR. 2015. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature 520: 545–548. [DOI] [PubMed] [Google Scholar]
- Pedersen O, Colmer TD, Borum J, Zavala‐Perez A, Kendrick GA. 2016. Heat stress of two tropical seagrass species during low tides – impact on underwater net photosynthesis, dark respiration and diel in situ internal aeration. New Phytologist 210: 1207–1218. [DOI] [PubMed] [Google Scholar]
- Potzkei J, Kunze M, Drepper T, Gensch T, Jaeger KE, Büchs J. 2012. Real‐time determination of intracellular oxygen in bacteria using a genetically encoded FRET‐based biosensor. BMC Biology 10: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta ML, Shukla V, Dalle Carbonare L, Weits DA, Perata P, Licausi F, Giuntoli B. 2019. A ratiometric sensor based on plant N‐terminal degrons able to report oxygen dynamics in Saccharomyces cerevisiae . Journal of Molecular Biology 431: 2810–2820. [DOI] [PubMed] [Google Scholar]
- Sasidharan R, Bailey‐Serres J, Ashikari M, Atwell BJ, Colmer TD, Fagerstedt K, Fukao T, Geigenberger P, Hebelstrup KH, Hill RD et al 2017. Community recommendations on terminology and procedures used in flooding and low oxygen stress research. New Phytologist 214: 1403–1407. [DOI] [PubMed] [Google Scholar]
- Schertl P, Braun HP. 2014. Respiratory electron transfer pathways in plant mitochondria. Frontiers in Plant Science 5: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RR, Weits DA, Feulner CFJ, van Dongen JT. 2018. Oxygen sensing and integrative stress signaling in plants. Plant Physiology 176: 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield CJ, Ratcliffe PJ. 2004. Oxygen sensing by HIF hydroxylases. Nature Reviews in Molecular Cell Biology 5: 343–354. [DOI] [PubMed] [Google Scholar]
- Semenza GL. 2007. Hypoxia‐inducible factor 1 (HIF‐1) pathway. Sci STKE 407: cm8. [DOI] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR. 2019. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363: eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesniere C, Torregrosa L, Pradal M, Souquet J‐M, Gilles C, Dos Santos K, Chatelet P, Gunata Z. 2006. Effects of genetic manipulation of alcohol dehydrogenase levels on the response to stress and the synthesis of secondary metabolites in grapevine leaves. Journal of experimental botany 57: 91–99. [DOI] [PubMed] [Google Scholar]
- Tschiersch H, Liebsch G, Borisjuk L, Stangelmayer A, Rolletschek H. 2012. An imaging method for oxygen distribution, respiration and photosynthesis at a microscopic level of resolution. New Phytologist 196: 926–936. [DOI] [PubMed] [Google Scholar]
- Turner APF, Karube I, Wilson GS, eds. 1987. Biosensors: fundamentals and applications. Oxford, UK: Oxford University Press. [Google Scholar]
- Vartapetian BB, Dolgikh YI, Polyakova LI, Chichkova NV, Vartapetian AB. 2014. Biotechnological approaches to creation of hypoxia and anoxia tolerant plants. Acta Naturae 6: 19–30. [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Steinbeck J, Fuchs P, Lichtenauer S, Elsässer M, Schippers JHM, Nietzel T, Ruberti C, Van Aken O, Meyer AJ et al 2019. Multiparametric real‐time sensing of cytosolic physiology links hypoxia responses to mitochondrial electron transport. New Phytologist 224: 1668–1684. [DOI] [PubMed] [Google Scholar]
- Walia A, Waadt R, Jones AM. 2018. Genetically encoded biosensors in plants: pathways to discovery. Annual Review of Plant Biology 69: 497–524. [DOI] [PubMed] [Google Scholar]
- Weits DA, Kunkowska AB, Kamps NCW, Portz KMS, Packbier NK, Nemec Venza Z, Gaillochet C, Lohmann JU, Pedersen O, van Dongen JT et al 2019. An apical hypoxic niche sets the pace of shoot meristem activity. Nature 569: 714–717. [DOI] [PubMed] [Google Scholar]
- Wright RC, Nemhauser J. 2019. Plant synthetic biology: quantifying the “known unknowns” and discovering the “unknown unknowns”. Plant Physiology 179: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang‐Rodriguez R, Heuer S, Ismail AM, Bailey‐Serres J, Ronald PC, Mackill DJ. 2006. Sub1A is an ethylene‐response‐factor‐like gene that confers submergence tolerance to rice. Nature 442: 705–708. [DOI] [PubMed] [Google Scholar]
- Yang J, Xie X, Xiang N, Tian ZX, Dixon R, Wang YP. 2018. Polyprotein strategy for stoichiometric assembly of nitrogen fixation components for synthetic biology. Proceedings of the National Academy of Sciences, USA 115: E8509–E8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Catalanotti C, Wittkopp TM, Posewitz MC, Grossman AR. 2015. Algae after dark: Mechanisms to cope with anoxic/hypoxic conditions. The Plant Journal 82: 481–503. [DOI] [PubMed] [Google Scholar]
- Yeung E, Bailey‐Serres J, Sasidharan R. 2019. After the deluge: plant revival post‐flooding. Trends in Plant Science 24: 443–454. [DOI] [PubMed] [Google Scholar]
- Youssef S, Ren W, Ai HW. 2016. A genetically encoded FRET sensor for hypoxia and prolyl hydroxylases. ACS Chemical Biology 11: 2492–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalza A, van Dongen JT, Froehlich A, Oliver SN, Faix B, Gupta KJ, Schmalzlin E, Igal M, Orcaray L, Royuela M et al 2008. Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiology 149: 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Linster E, Gannon L, Leemhuis W, Rundle CA, Theodoulou FL, Wirtz M. 2019. Tandem fluorescent protein timers for noninvasive relative protein lifetime measurement in plants. Plant Physiology 180: 718–731. [DOI] [PMC free article] [PubMed] [Google Scholar]


