Abstract
Asymmetric cell division is one of the most elegant biological systems by which cells create daughter cells with different functions and increase cell diversity. In particular, PAR polarity in the cell membrane plays a critical role in regulating the whole process of asymmetric cell division. Numerous studies have been conducted to determine the underlying mechanism of PAR polarity formation using both experimental and theoretical approaches in the last 10 years. However, they have mostly focused on answering the fundamental question of how this exclusive polarity is established but the precise dynamics of polarity domain have been little notified. In this review, I focused on studies on the shape, length, and location of PAR polarity from a theoretical perspective that may be important for an integrated understanding of the entire process of asymmetric cell division.
Keywords: asymmetric cell division, cell polarity, pattern formation
Asymmetric cell division is one of the most elegant biological systems for cell diversity. The studies on the shape, length, and location of PAR polarity domain in C. elegans embryo are reviewed from a theoretical perspective.
1. POLARITY FORMATION IN ASYMMETRIC CELL DIVISION
The process of asymmetric cell division is among the most elegant biological systems by which cells create daughter cells with different functions and increase cell diversity (Campanale, Sun, & Montell, 2017; Knoblich, 2008; Rose & Gönczy, 2014). In a mother cell, various substances are asymmetrically distributed before cell division, and these substances are differentially transferred into each daughter cell. After then, the mother cell divides around the boundary of two exclusive polarities (Figure 1a). This leads to differences in gene expression between the two daughter cells, which exhibit different functions and sizes.
Figure 1.
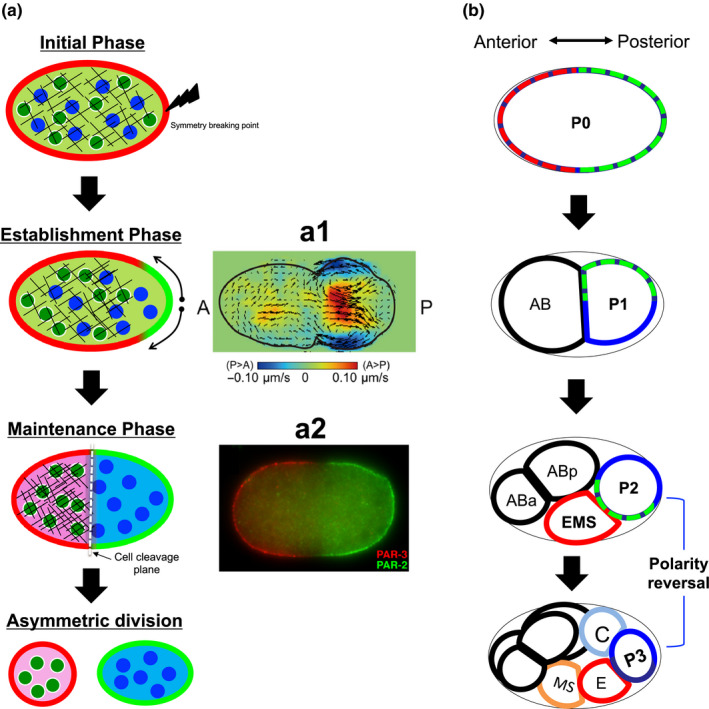
Asymmetric cell division in C. elegans. (a) Schematic diagram of asymmetric cell division in P0 cell stage. (a1) Flow velocity induced by acto‐myosin contraction (adapted from Niwayama et al., 2011) and (a2) Imaging data of PAR‐3(red) and PAR‐2(green) polarity (adapted from Nance & Zallen, 2011) (b) Sequence of asymmetric cell divisions and PAR‐2 polarity (Green dotted line).  , aPARs;
, aPARs;  , pPARs;
, pPARs;  , Actomyosin;
, Actomyosin;  , Mex‐5/6;
, Mex‐5/6;  , PIE‐1
, PIE‐1
In recent decades, polarity formation in asymmetric cell division has been extensively explored using Caenorhabditis elegans embryos as a biological model system. In a single fertilized egg cell (P0 cell) of C. elegans, the symmetry is disrupted at the pole where the posterior end is formed after sperm entry. Concurrently, the acto‐myosin network in the cell cortex begins contracting from the site of symmetry breakage and stops around the middle of the cell, which induces advective dynamics inside the cell (Figure 1a; Mayer, Depken, Bois, Julicher, & Grill, 2010; Niwayama, Shinohara, & Kimura, 2011; Goehring, Hoege, Grill, & Hyman, 2011a; Nishikawa, Naganathan, Jülicher, & Grill, 2017). Before sperm entry, PAR‐6, PAR‐3, and PKC‐3, known as posterior proteins (pPARs), are distributed homogeneously in the membrane and cytosol. Additionally, PAR‐2 and PAR‐1, known as anterior proteins (aPARs), are distributed homogeneously in the cytosol. When the symmetry breaks, these protein groups form exclusive polarity domains in the membrane (Figure 1a). Simultaneously, a similar exclusive polarity occurs for cytoplasmic proteins, which are regulated by the most upstream polarity of PARs (Cuenca, Schetter, Aceto, Kemphues, & Seydoux, 2002; Daniels, Perkins, Dobrowsky, Sun, & Wirtz, 2009; Schubert, Lin, Vries, Plasterk, & Priess, 2000; Wu et al., 2018). In C. elegans embryos, asymmetric cell division is not a single event but rather involves consecutive events. The polarity dynamics occur in a sequence (Figure 1b) and the PAR polarity is considered to be conserved in the daughter cells.
Numerous studies have been conducted to identify the underlying mechanism of PAR polarity formation using both experimental and theoretical approaches in the last 10 years (Hoege & Hyman, 2013; Lang & Munro, 2017; Zonies, Motegi, Hao, & Seydoux, 2010), and it is well known that the formation of exclusive PAR domains is based on mutual inhibition dynamics between anterior proteins and posterior proteins in which the two protein groups transfer each other from the membrane to cytosol (Hoege & Hyman, 2013; Figure 2a). Biologically, their networks are more complex (Lang & Munro, 2017), but the essential structures of the exclusive polarity domains have been predicted by mathematical modeling, including the mass conservation property and a bi‐stability structure based on the mutual inhibition dynamics (Goehring, Trong, et al., 2011b; Morita & Sakamoto, 2019; Seirin‐Lee, 2016b; Seirin‐Lee & Shibata, 2015; Trong, Nicola, Goehring, Kumar, & Grill, 2014). Furthermore, the effect of mechanical dynamics of advection transport induced by acto‐myosin contraction has been explored in detail with experiments, and it is shown to play a critical role in the establishment of PAR polarity (Goehring, Trong, et al., 2011b; Gross et al., 2019).
Figure 2.
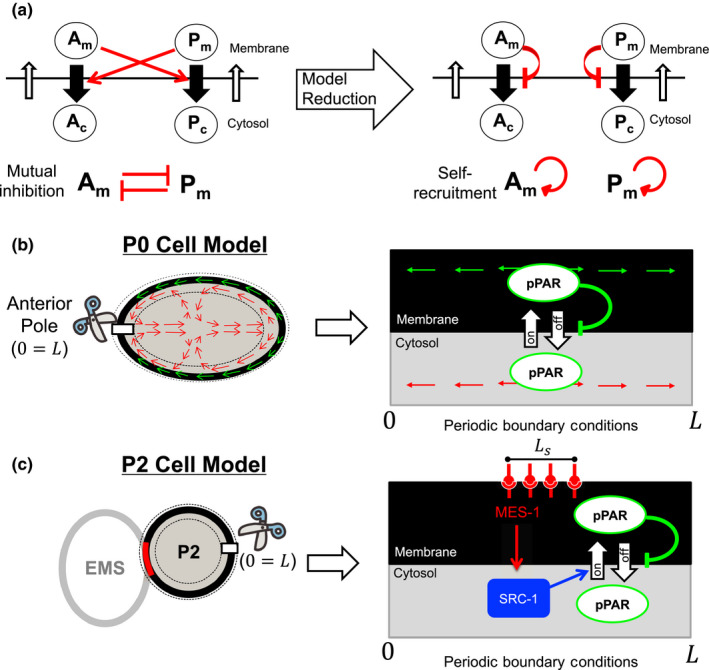
Mathematical models for P0 cell and P2 cell. (a) Concept diagram of self‐recruitment model. (b) P0 model diagram. The green and red arrows indicate the flow direction in membrane and cytosol, respectively, and the black dotted lines imply the region considered for the conceptional modeling. (c) P2 model diagram. Ls indicates the contact zone of EMS and P2 cells and the black dotted lines imply the region considered for the modeling
On the contrary, most studies on PAR polarity have focused on answering the fundamental question of how the exclusive polarity is established or how biochemical/biomechanical networks are involved in this polarity formation in the single mother cell stage (Lang & Munro, 2017). To understand the polarity dynamics from a global perspective such as the whole dynamics of asymmetric cell division, it is necessary to understand the robustness of polarity and capture more precise dynamics of polarity. However, such questions have been poorly understood. For example, how can cells always make a single rather than multiple peak domain per protein? In other words, how can the shape of the polarity domain be robustly determined as a single peak? how can the size of the polarity domain be robustly determined? how can the location of the polarity domain be robustly determined? Indeed, it has been reported that the formation of a single polarity domain per protein is crucial for the normal process of asymmetric division (Morton et al., 2002) and two exclusive domains segregated by PARs in the membrane are indispensable for polarizing cytoplasmic proteins (Cuenca et al., 2002; Wu et al., 2018). Furthermore, either multi‐mode polarization or abnormal length scale polarity domains of PARs in the membrane is typically accompanied by abnormal asymmetric cell division (Cuenca et al., 2002; Morton et al., 2002). In this review, I focused on suggesting hypotheses to answer the questions above from a theoretical perspective, particularly how polarity dynamics can differ between P0 cell in the single‐cell stage and P2 cell in the multi‐cell stage. The hypotheses by mathematical models might deliver biologically novel aspects and findings, which are not usually highlighted or might have been missed in biological studies.
2. PAR POLARITY MODELS FOR A SINGLE MOTHER CELL STAGE AND MULTIPLE DAUGHTER CELL STAGE
The conceptional models for PAR dynamics have been suggested by Seirin‐Lee and Shibata (2015) and Seirin‐Lee (2016b) for P0 cell case and P2 cell case, respectively (Figure 2b,c). The self‐recruitment PAR model (Figure 2a) suggested by Seirin‐Lee and Shibata (2015) shows that either aPAR or pPAR dynamics can be captured independently. The model for pPAR dynamics is based on the following three processes: (a) translocation between the membrane and cytosol by association and disassociation with anterior proteins, (b) diffusion in both the membrane and cytosol, and (c) advection by cortical and cytoplasmic flow (Figure 2b).
For the concentrations of posterior proteins in the membrane and cytosol, [Pm] and [Pc], respectively, the pPAR model is given as follows in a one‐dimensional circular space, [0,L]:
| (1) |
The second terms on the left‐hand side represent the advection term with the velocity of the cortical and cytosol flow, vm and vc, respectively. The first terms on the right‐hand side are diffusion coefficients, and the diffusion coefficient in the membrane is considered as smaller than that in the cytoplasm, i.e., Dm < Dc. The second and third terms on the right‐hand side are the association and dissociation reactions with the membrane concentration‐dependent on‐ and off‐rates. The non‐linear term of the off‐rate indicates that the location at which the concentration of posterior protein is low corresponds to that showing strong activity of an anterior protein to disassociate pPAR.
In P2 cells, an intracellular transducer SRC‐1 transmits a signal from the EMS cell (Arata, Lee, Goldstein, & Sawa, 2010). Through MES‐1 ligand‐receptor binding on the boundary of EMS and P2 cells, SRC‐1 in the cytosol is activated. It is known that SRC‐1 signaling regulates PAR‐2 in the cytosol to a certain degree, and the effect of SRC‐1 signaling on the on‐rate of PAR‐2 protein can be combined in a concentration‐dependent manner (Figure 2c). A previous study by Seirin‐Lee (2016b) suggested a positive correlation between SRC‐1 and the on‐rate of PAR‐2, which plays a key role in determining the polarity location of PAR‐2 in P2 cells. Denoting the width for the contact zone of P2 and EMS cells and the concentrations of SRC‐1 and MES‐1 by Ls, [Sc]and [Mm], respectively, the model suggested by Seirin‐Lee (2016b) is given as.
| (2) |
where µ2 and µ4 are the basal decay rates of MES‐1 and SRC‐1, respectively. χ(x) is given as.
where L 0 is the center of the contact boundary between P2 and EMS cells. Note that some quasi‐steady state assumptions in P2 cell model (2) can drive the following model.
where µ = εµ 1 µ 3/µ 2 µ 4. This model indicates that P2 cell situation is initially similar as the assumption of the spatial heterogeneity of the on‐rate of pPAR in P0 model (1).
In what follows, I will discuss how the shape, length, and location of PAR polarity domain can be determined robustly with the numerical results based on P0 cell model (1) and P2 cell model (2). The discussions also convey how the spatial heterogeneity of the on‐rate in polarity model makes the model dynamics differ mathematically from the basal model (1).
3. POLARITY SHAPES
How is the exclusive unimodal peak formed in PAR polarity? This is a very important question in asymmetric cell division because PAR polarity is the most upstream polarity that may be involved in not only downstream polarities of the cytoplasmic proteins but also the determination of spindle position for division (Cowan & Hyman, 2004), which consequently affects all properties of daughter cells such as normal differentiation, size determination, and function.
In P0 cell, the symmetry breaking position is determined very robustly at the pole of future posterior side (Figure 1a). It has been shown that microtubules help PAR‐2 to recruit in the invasion point of the membrane and may induce symmetry breaking (Motegi et al., 2011). However, in the mathematical model, a locally concentrated stimulus can show a unimodal polarity in some very specific perturbation type or parameter region (Figure 3a), and, in general, a mass conservation reaction‐diffusion system shows high sensitivity under the initial conditions including small random perturbations around a homogeneous steady state (spatially homogeneous initial conditions) (Figure 3b), in which patterns with multi‐wave modes emerge in the first stage and then the multi‐peaks merge into a unimodal peak but over a long time scale (Otsuji et al., 2007; Figure 3b, second panel). Such dynamics of reaction‐diffusion models fail to explain the unimodal polarity formation that occurs not only in the P0 cell but also after the P0 cell embryo stage such as in P2 cells (Rose & Gönczy, 2014).
Figure 3.
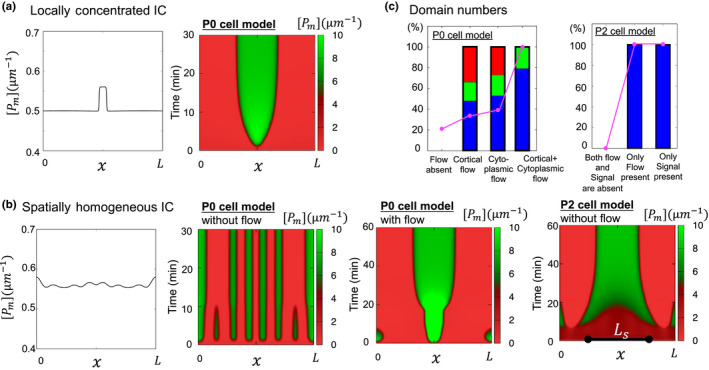
Polarity shapes. (a) Single mode polarity for locally concentrated initial conditions. (b) Polarity pattern for spatially homogeneous initial conditions. Ls is the extracellular signal length. (c) Effect of flow or extracellular signal for the number of polarity peak numbers. The red bar shows an increased peak number compared to the absence of flow. The green bar is the constant case and the blue bar is the decreased case of the peak number compared to the flow absent case. The pink linedots indicate the number of a single polarity domain. (a) and (b) were replotted with the same parameter suggested in Seirin‐Lee and Shibata (2015); Seirin‐Lee (2016b) and (c) was replotted with the same source data used in Seirin‐Lee and Shibata (2015); Seirin‐Lee (2016b).  , ratio of single domain number
, ratio of single domain number
How can we understand the robustness of unimodal polarity from a theoretical perspective? Numerical tests have suggested that the flows strongly affect the robustness of unimodal polarity (Figure 3b, third panel and Figure 3c, first panel). In contrast, similar numerical tests for the model of P2 cells showed that the extracellular signal is sufficient to increase the robustness of unimodal polarity even without flow (Figure 3b, third panel and Figure 3c, second panel). These two results indicate that the acto‐myosin contraction in a single mother cell stage plays an important role not only in symmetry breaking for the establishment phase (Gross et al., 2019) but also the unimodal shape of polarity. It is shown that in the zygote of a mutant spd‐5 of C. elegans, which lacked the cortical flow, the PAR‐2 domains is formed at either the anterior or posterior poles or both poles, indicating that the robustness of polarity formation may be increased by flow dynamics (Tsai & Ahringer, 2007). On the other hand, such acto‐myosin contraction may not be required for the robustness of polarity shape in the stage of multi‐daughter cells once the extracellular signaling is introduced between cells.
4. POLARITY LENGTH
Another notable feature of polarity formation in the C. elegans embryo is the robustness of the polarity domain size. The asymmetrically divided C. elegans embryo cells show a polarity pattern with a constant relative length scale of the posterior domain to the cell size (Arata et al., 2010). The abnormal length scale of the posterior domain in AP polarity formation is a common feature observed with an aberrant division (Cuenca et al., 2002; Morton et al., 2002; Rose & Kemphues, 1998). However, the underlying mechanism by which the length scale of polarity is determined robustly is still unclear in biology. Then, what possibilities can we suppose from the mathematical models?
It has been found that the length of the polarity domain is uniquely determined by the total mass of polarity protein regardless of the initial total mass in the membrane in P0 cell (Figure 4a). This also suggests that once a daughter cell that becomes half the size of a mother cell has half the total mass, the relative domain size of polarity can be conserved. In contrast, the length of polarity in the P2 cell case is highly affected by the width of the extracellular signal from neighboring cells, although it has the same total mass (Figure 4b). Notably, an increase in the amount of pPAR in the membrane caused by SRC‐1 is not the only determinant of the length of the domain. The non‐monotonicity of the pPAR domain length is essentially related to the shape of the pPAR distribution in the membrane, which is based on the shape of the SRC‐1 distribution (Figure 4b). Thus, the shape (distribution) of the extracellular signal can significantly affect polarity dynamics, and the time scale of polarity establishment can be significantly influenced by the extracellular signal (Kuwamura, Seirin‐Lee, & Ei, 2018).
Figure 4.
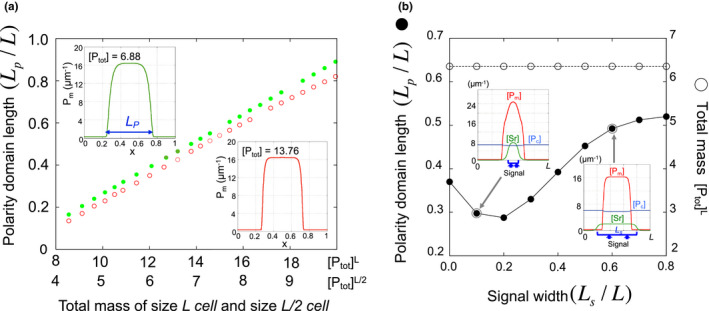
Polarity Length. (a) Relationship between total mass and polarity length with respect to cell size in P0 cell model (1) (adapted from Seirin‐Lee & Shibata, 2015). (b) Polarity length with respect to signal width and total mass in P2 cell model (2) (adapted from Seirin‐Lee, 2016b).  , cell perimeter = L/2;
, cell perimeter = L/2;  , cell perimeter = L
, cell perimeter = L
How the polarity length and concentration could be involved in asymmetric cell division? It has been shown that par‐2 mutation affects the orientation of the cleavage spindle (Cheng, Kirby, & Kemphues, 1995). Although the property of the PAR‐2 domain that directly regulates the division plan of germline precusors (or determination of spindle location) remains to be determined, the mathematical observations suggest that both the maximal concentration and length scale of the PAR‐2 domain potentially may regulate the spindle location. If the length scale of the polarity domain is a more critical factor than its concentration to determine the spindle location, then the length scale of the site of contact between mother cells would also be a critical factor in ensuring the robust size of daughter cells. Additionally, the total mass may play an important role in determining a spindle location. In contrast, if the spindle location is affected by the concentration of the polarity domain rather than the length scale and there exists a threshold concentration of the polarity domain that determines the spindle location, then the sizes of daughter cells are robustly determined without being affected by the length scale of the site of contact between mother cells.
5. POLARITY LOCATION
Where the polarity is formulated is another important question in the process of asymmetric cell division. The location of PARs polarity in the membrane determines the anterior and posterior axes in the P0 cell stage, which consequently determines the position of daughter cells that should interact with another specific daughter cell in the next stage such as P2 and after the P2 cell stages.
For this problem, mathematical approaches suggest that the symmetry of acto‐myosin contraction is important in the P0 cell because the asymmetry of flow velocity can result in a shift in the polarity domain location in the P0 cell stage (Figure 5a). Indeed, a recent study shows that the positioning of PAR‐2 polarity could be affected when flow‐induced perturbations are given (Mittasch et al., 2018). However, the flow may have the opposite effect on polarity reversal when it works together with extracellular signals. The parameter spaces for the case of flow showed a confined area for the polarity reversal (Figure 5b). Furthermore, the positioning of flow was highly sensitive to the directional relationship between the cortical and cytoplasmic flow, and a failed region of polarity reversal exists when the signal width is large, indicating that if the extracellular signal affects a wider region in the cell membrane, the flow effect is increased to strongly perturb the polarity domain in the membrane. Analytical study shows that an extracellular signal (although small) is sufficient to induce the positioning of the polarity domain robustly in the absence of flow (Kuwamura et al., 2018). In the presence of flow, the positioning of the polarity domain is sensitively determined by flow directions or the starting position of flow. Otherwise, the extracellular signal plays an important role in determining polarity positioning.
Figure 5.
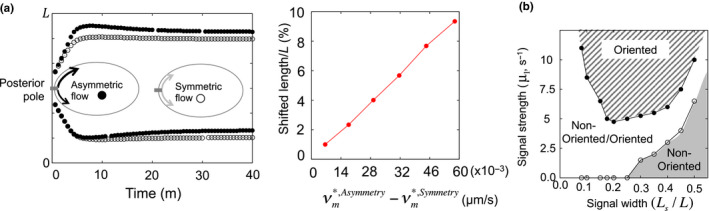
Polarity location. (a) Left panel shows the position of the boundary of polarity with respect to asymmetric/symmetric flow velocity for P0 cell model (1). Right panel shows the shifted distance of the polarity location from the symmetric velocity case. The figure was replotted with the same source data used in Seirin‐Lee and Shibata (2015). (b) Parameter space for the oriented/non‐oriented polarity location with respect to the width and strength of the extracellular signal for P2 cell model (2) (adopted from Seirin‐Lee, 2016b).  , sgn(vm) = sgn(vc);
, sgn(vm) = sgn(vc);  , sgn(vm) ≠ sgn(vc)
, sgn(vm) ≠ sgn(vc)
6. PROSPECTIVE FOR CELL GEOMETRY AND PAR POLARITY
Minimal mathematical models are among the standard approaches to capture an essential mechanism of a complex biological system, and simple mathematical models have led to numerous biological findings. In this review, I introduced hypotheses and findings based on such simple mathematical models described as a one‐dimensional space. On the other hand, the embryo of C. elegans has an elliptical form which is constrained by a solid eggshell (Figure 1a). It is reported that a geometrical characteristic may influence biochemical dynamics and consequently affect polarity dynamics (Dawes & Iron, 2013). Currently, with the dramatic development of computer sciences, multi‐dimensional modeling and numerical simulations have greatly advanced. Thus, extending a mathematical model in high‐dimensional space to reflect the cell geometry may give rise to additional biological and mathematical findings in polarity formation of asymmetric cell division (Aras, Zhou, Dawes, & Chou, 2018; Geβele, Halatek, Wurthner, & Frey, 2019; Seirin‐Lee, 2016a, 2017). Finally, it may be interesting to verify the hypotheses addressed in mathematical models in biology.
ACKNOWLEDGMENT
This work was partially supported by Grants‐in‐Aid for Scientific Research from JSPS (JP19H01805 and JP17KK0094) and the JST PRESTO program, Japan (JPMJPR16E2).
Seirin‐Lee S. Asymmetric cell division from a cell to cells: Shape, length, and location of polarity domain. Develop Growth Differ. 2020;62:188–195. 10.1111/dgd.12652
The copyrightline for this article was changed on 2 November 2020 after original online publication
REFERENCES
- Aras, B. S. , Zhou, Y. C. , Dawes, A. , & Chou, C.‐S. (2018). The importance of mechanical constraints for proper polarization and pseudo‐cleavage furrow generation in the early Caenorhabditis elegans embryo. PLOS Comutational Biology, 14(7), e1006294 10.1371/journal.pcbi.1006294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arata, Y. , Lee, J.‐Y. , Goldstein, B. , & Sawa, H. (2010). Extracellular control of PAR protein localization during asymmetric cell division in the C. elegans embryo. Development, 137, 3337–3345. 10.1242/dev.054742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanale, J. P. , Sun, T. Y. , & Montell, D. J. (2017). Development and dynamics of cell polarity at a glance. Journal of Cell Science, 130, 1201–1207. 10.1242/jcs.188599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, N. N. , Kirby, C. M. , & Kemphues, K. J. (1995). Control of cleavage spindle orientation in Caenorhabditis elegans: The role of the genes par‐2 and par‐3. Genetics, 139, 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan, C. R. , & Hyman, A. A. (2004). Asymmetric cell division in C. elegans: Cortical polarity and spindle positioning. Annual Review of Cell and Developmental Biology, 20, 427–453. [DOI] [PubMed] [Google Scholar]
- Cuenca, A. A. , Schetter, A. , Aceto, D. , Kemphues, K. , & Seydoux, G. (2002). Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development, 130, 1255–1265. 10.1242/dev.00284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, B. R. , Perkins, E. M. , Dobrowsky, T. M. , Sun, S. X. , & Wirtz, D. (2009). Asymmetric enrichment of pie‐1 in the Caenorhabditis elegans zygote mediated by binary counterdiffusion. Journal of Cell Biology, 184(4), 473–479. 10.1083/jcb.200809077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes, A. T. , & Iron, D. (2013). Cortical geometry may influence placement of interface between par protein domains in early Caenorhabditis elegans embryos. Journal of Theoretical Biology, 333, 27–37. 10.1016/j.jtbi.2013.04.024 [DOI] [PubMed] [Google Scholar]
- Geβele, R. , Halatek, J. , Wurthner, L. , & Frey, E. (2019). Geometric cues stabilise long‐axis polarisation of PAR protein patterns in C. elegans . bioRxiv 15, 10.1101/451880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring, N. W. , Hoege, C. , Grill, S. W. , & Hyman, A. A. (2011a). PAR proteins diffuse freely across the anterior‐posterior boundary in polarized C. elegans embryos. Journal of Cell Biology, 193(3), 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring, N. W. , Trong, P. K. , Bois, J. S. , Chowdhury, D. , Nicola, E. M. , Hyman, A. A. , & Grill, S. W. (2011b). Polarization of PAR proteins by advective triggering of a pattern‐forming system. Science, 334(6059), 1137–1141. [DOI] [PubMed] [Google Scholar]
- Gross, P. , Kumar, K. V. , Goehring, N. W. , Bois, J. S. , Hoege, C. , Julicher, F. , & Grill, S. W. (2019). Guiding self‐organized pattern formation in cell polarity establishment. Nature Physics, 15, 293–300. 10.1038/s41567-018-0358-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege, C. , & Hyman, A. A. (2013). Principles of PAR polarity in Caenorhabditis elegans embryos. Molecular Cell Biology, 14, 315–322. 10.1038/nrm3558 [DOI] [PubMed] [Google Scholar]
- Knoblich, J. A. (2008). Mechanisms of asymmetric stem cell division. Cell, 132, 583–597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Kuwamura, M. , Seirin‐Lee, S. , & Ei, S.‐I. (2018). Dynamics of localized unimodal patterns in reaction‐diffusion systems related to cell polarization by extracellular signaling. SIAM Journal on Applied Mathematics, 78(6), 3238–3257. [Google Scholar]
- Lang, C. F. , & Munro, E. (2017). The PAR proteins: From molecular circuits to dynamic self‐stabilizing cell polarity. The Company of Biologists, 144, 3405–3416. 10.1242/dev.139063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. , Depken, M. , Bois, J. S. , Julicher, F. , & Grill, S. W. (2010). Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature, 467, 617–621. 10.1038/nature09376 [DOI] [PubMed] [Google Scholar]
- Mittasch, M. , Gross, P. , Nestler, M. , Fritsch, A. W. , Iserman, C. , Kar, M. , … Kreysing, M. (2018). Non‐invasive perturbations of intracellular flow reveal physical principles of cell organization. Nature Cell Biology, 20, 344–351. 10.1038/s41556-017-0032-9 [DOI] [PubMed] [Google Scholar]
- Morita, Y. , & Sakamoto, K. (2019). Turing type instability in a diffusion model with mass transport on the boundary. Preprint. [Google Scholar]
- Morton, D. G. , Shakes, D. C. , Nugent, S. , Dichoso, D. , Wang, W. , Golden, A. , & Kemphues, K. J. (2002). The Caenorhabditis elegans par‐5 gene encodes a 14‐3‐3 protein required for cellular asymmetry in the early embryo. Developmental Biology, 241, 47–58. 10.1006/dbio.2001.0489 [DOI] [PubMed] [Google Scholar]
- Motegi, F. , Zonies, S. , Hao, Y. , Cuenca, A. A. , Griffin, E. , & Seydoux, G. (2011). Microtubules induce self‐organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nature Cell Biology, 13(11), 1361–1367. 10.1038/ncb2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance, J. , & Zallen, J. A. (2011). Elaborating polarity: PAR proteins and the cytoskeleton. Development, 138, 799–809. 10.1242/dev.053538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, M. , Naganathan, S. R. , Jülicher, F. , & Grill, S. W. (2017). Symmetry breaking in a bulk surface reaction diffusion model for signalling networks. Elife, 6, e19595.28117665 [Google Scholar]
- Niwayama, R. , Shinohara, K. , & Kimura, A. (2011). Hydrodynamic property of the cytoplasm is sufficient to mediate cytoplasmic streaming in the Caenorhabiditis elegans embryo. Proceedings of the National Academy of Sciences, 108(29), 11900–11905. 10.1073/pnas.1101853108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuji, M. , Ishihara, S. , Co, C. , Kaibuchi, K. , Mochizuki, A. , & Kuroda, S. (2007). A mass conserved reaction‐diffusion system captures properties of cell polarity. PLoS Computational Biology, 3(6), e108 10.1371/journal.pcbi.0030108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, L. S. , & Gönczy, P. (2014). Polarity establishment, asymmetric division and segregation of fate determinants in early C. elegans embryo. WormBook. [DOI] [PubMed] [Google Scholar]
- Rose, L. S. , & Kemphues, K. J. (1998). Early patterning of the C. elegans embryo. Annual Review of Genetics, 32, 521–545. [DOI] [PubMed] [Google Scholar]
- Schubert, C. M. , Lin, R. , de Vries, C. J. , Plasterk, R. H. A. , & Priess, J. R. (2000). MEX‐5 and MEX‐6 unction to establish soma/germline asymmetry in early C. elegans embryo. Molecular Cell, 5, 671–682. [DOI] [PubMed] [Google Scholar]
- Seirin‐Lee, S. (2016a). Lateral inhibition‐induced pattern formation controlled by the size and geometry of the cell. Journal of Theoretical Biology, 404, 51–65. 10.1016/j.jtbi.2016.05.025 [DOI] [PubMed] [Google Scholar]
- Seirin‐Lee, S. (2016b). Positioning of polarity formation by extracellular signaling during asymmetric cell division. Journal of Theoretical Biology, 400, 52–64. 10.1016/j.jtbi.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Seirin‐Lee, S. (2017). The role of domain in pattern formation. Development, Growth and Differentiation, 59, 396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seirin‐Lee, S. , & Shibata, T. (2015). Self‐organization and advective transport in the cell polarity formation for asymmetric cell division. Journal of Theoretical Biology, 382, 1–14. 10.1016/j.jtbi.2015.06.032 [DOI] [PubMed] [Google Scholar]
- Trong, P. K. , Nicola, E. M. , Goehring, N. W. , Kumar, K. V. , & Grill, S. W. (2014). Parameter‐space topology of models for cell polarity. New Journal of Physics, 16, 065009 10.1088/1367-2630/16/6/065009 [DOI] [Google Scholar]
- Tsai, M.‐C. , & Ahringer, J. (2007). Microtubules are involved in anterior‐posterior axis formation in C. elegans embryos. Journal of Cell Biology: Report, 179, 397–402. 10.1083/jcb.200708101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Han, B. , Li, Y. , Munro, E. , Odde, D. J. , & Griffin, E. E. (2018). Rapid diffusion‐state switching underlies stable cytoplasmic gradients in Caenorhabditis elegans zygote . Proceedings of the National Academy of Sciences of the United States of America, 115(36), E8440–E8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonies, S. , Motegi, F. , Hao, Y. , & Seydoux, G. (2010). Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR‐2. Development, 137, 1669–1677. 10.1242/dev.045823 [DOI] [PMC free article] [PubMed] [Google Scholar]


