Characterization of the human placental macrophages, Hofbauer cells, and their role in maternal–fetal interface
Abstract
In this issue of JEM, Thomas et al. (https://doi.org/10.1084/jem.20200891) provide elegant technological and conceptual advances that further our understanding of the immune cells enriched at the maternal–fetal interface. Using new isolation strategies to better separate maternal- and fetal-derived cells, the authors identify previously undefined maternal-derived immune cells associated with the fetal-derived placenta and provide an in-depth analysis of the markers and characteristics of placental Hofbauer cells.
Immune cells of fetal and maternal origin are abundant at the maternal–fetal interface throughout pregnancy. This interface is composed of the fetal-derived placenta and the maternal-derived decidua, which must balance the immunological complexities of pregnancy without harming the developing fetus or maternal host. Given the complex immunological crosstalk at the maternal–fetal interface, it may not be surprising that both the placenta and decidua are enriched in immune cells, although the function of many of these cells remains largely unknown, particularly across gestation in humans. The maternal decidua contains ∼40% immune cells, including uterine natural killer (uNK) cells, innate lymphoid cells, and macrophages, which change in abundance throughout gestation (Moffett and Colucci, 2014; Vento-Tormo et al., 2018). In the first trimester, uNKs represent >70% of all immune cells in the human decidua but decline in abundance as pregnancy progresses (reviewed in Moffett and Colucci, 2014; Yang et al., 2019). These uNKs play an essential role in the maintenance of healthy pregnancy, due in part to their ability to secrete factors that promote fetal growth (Fu et al., 2017). In the fetal-derived placenta, macrophages termed Hofbauer cells (HBCs) are the most prominent immune cells localized to placental villi and may play a role in the establishment and/or maintenance of pregnancy.
Insights from Christina Megli and Carolyn B. Coyne
The placenta begins to form within ∼5 d after conception and is derived from the trophectoderm that surrounds the blastocyst. Throughout the first trimester, the human placenta develops into a highly branched villous-like tree structure containing distinct types of trophoblasts, fibroblasts, developing fetal capillaries, and HBCs. HBCs have been identified as early as 18 d after fertilization, a stage well before the connection of the placenta to the fetal circulation (Castellucci et al., 1987; Boyd and Hamilton, 1970). HBCs are important in placental development, and abnormalities in their function or polarization have been implicated in a variety of pregnancy pathologies (reviewed in Reyes and Golos, 2018). Despite their abundance in placental villi, many of the functional and phenotypic characteristics of HBCs remain poorly defined, due largely to difficulties in both segregating these cells and maintaining their viability in order to characterize their phenotype and function. In a recent study published in JEM, Thomas et al. (2020) develop a novel approach to characterize placental HBCs and further apply this approach to profile placenta-associated maternal monocyte/macrophage (PAMM) immune cell subsets (Thomas et al., 2020). This work provides highly impactful insights into the unique properties of HBCs and PAMMs and innovative tools to profile the characteristics of these cells.
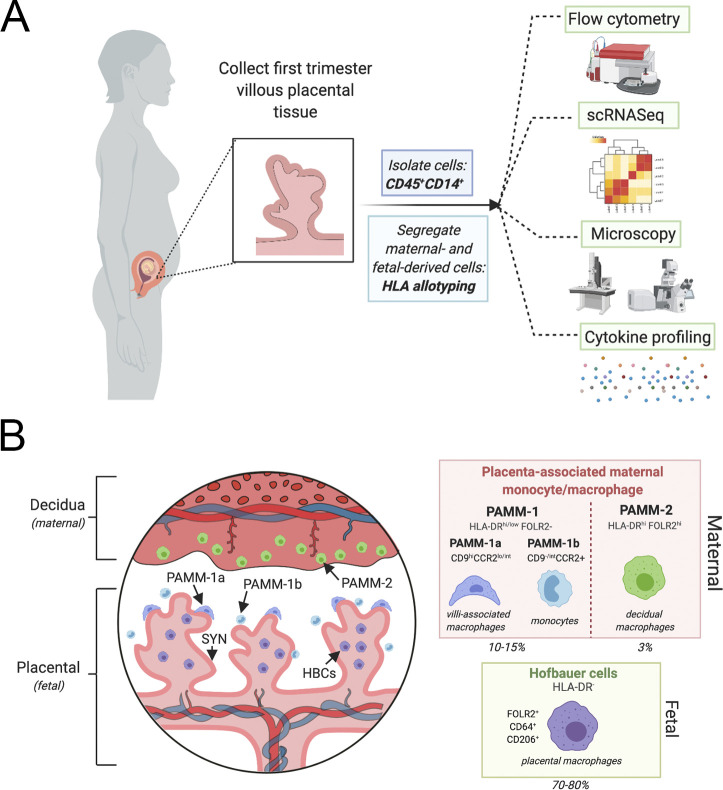
Within the context of immunity, defining how resident immune cells function in their native tissue environment allows for a better understanding of how tissues function in a complex environment. Thomas et al. (2020) report a new protocol for the identification and purification of HBCs directly from placental tissue collected in the first trimester. Moreover, the use of HLA allotyping to differentiate between maternal and fetal tissues demonstrated a substantial technological improvement from previously reported studies because it allows for retention of cell viability and additional ability for functional characterization of both fetal- and maternal-derived immune cells (see figure, panel A). When the authors applied this technique to first trimester placental tissue, they uncovered several novel findings relevant to understanding placentation, early pregnancy, and immune signaling at the maternal–fetal interface. First, they found that maternal-derived macrophages make up a surprisingly high number of total cells in the early placenta. Given that these cells were identified before the formation of maternal spiral arteries and delivery of maternal blood to the surfaces of placental villi, these findings suggest that these cells play direct roles in early placentation. With this initial establishment of differential cell populations, the authors then applied transcriptomic profiling to identify and establish HBC-specific markers. This further facilitated the identification and segregation of maternal- and fetal-derived macrophage populations independent of HLA matching, further improving on their initial isolation techniques. The authors demonstrate clear and consistent separation of cells using CD45, CD14, HLA-DR, and FOLR2 surface expression to allow for sorting and segregation of differential maternal and fetal macrophage populations while retaining cell viability. This is a substantial development in the field, and this methodological breakthrough has implications in the study of immunobiology at the maternal–fetal interface. The authors then go on to further characterize both PAMMs and HBCs.
Thomas et al. (2020) identified two PAMM subsets in the fetal placenta based on CCR9 and FOLR2 staining (see figure, panel B). They found that PAMM-2 represented decidual macrophages whereas PAMM-1 was a separate subtype that could be separated into both macrophage and monocyte subtypes based on cell marker, cell phenotype, and transcriptional analysis. The monocytes (termed PAMM-1b) were transcriptionally distinct from circulating maternal cells and represented a newly defined cell population at the maternal–fetal interface that has the capability to regulate the immune response at this unique niche. In contrast, PAMM-1 macrophages (termed PAMM-1a) have direct contact with the syncytiotrophoblast, the outer fused trophoblast layer covering the surface of placental villi, suggesting that these cells have the potential to regulate placental development and/or function, particularly given that these cells are transcriptionally active in cell regeneration and repair pathways. Remarkably, the authors then performed microscopy and found that PAMM-1a were directly adhered to the syncytiotrophoblast layer and were enriched in lipid droplets. These findings suggest that these cells are important in facilitating the survival and integrity of the multinucleated syncytiotrophoblast by assisting in cellular maintenance and/or other functions. Luminex profiling demonstrated that the PAMM-1a subsets secreted IL-1β and IL-6, consistent with a transition to active macrophages. These novel findings demonstrate that maternal monocytes/macrophages develop functional niches at the maternal–fetal interface and suggest that these cells directly interact with and regulate the fetal-derived placenta.
Isolation and characterization of placental associated macrophages and monocytes. (A) Cells were isolated from first trimester placental explants. CD14/CD45+ cells stained for HLA allotype to distinguish between maternal- and fetal-derived cells were sorted through a flow cytometer and characterized by transcriptional profile, microscopic evaluation, and staining as well as cytokine profiling to identify and characterize the distinct maternal and fetal macrophages in the placenta. (B) Left: The maternal–fetal interface is composed of the maternal-derived decidua and fetal-derived placenta. In the first trimester, chorionic villi are immature and are covered by a contiguous layer called the syncytiotrophoblast (dark pink, SYN). Thomas et al. (2020) define the characteristics of both maternal- and fetal-derived macrophages, termed PAMMs and HBCs. PAMM-1 (dark blue and light blue) and PAMM-2 (green) cells are identified by their HLA-DR, FOLR2 surface expression. PAMM-1 subtypes are distinct monocytes (PAMM-1b, light blue) and macrophages (PAMM-1a, dark blue). PAMM-1b macrophages are in direct contact with the SYN layer. The fetal-derived HBCs are in direct contact with the stroma of the chorionic villi and make up the majority of CD14+ cells in the placenta (70–80%), whereas PAMM-1 and PAMM-2 cells make up ∼10% and 3%, respectively. These cells are able to be identified by HLA-DR, FOLR2, CD64, and CD206 and are functionally and transcriptionally distinct from the maternal macrophage subsets. This figure was constructed using BioRender.
The authors then compared the transcriptional profile of the newly isolated HBCs to previously characterized single-cell RNA sequencing datasets and demonstrated that these cells are similar to “primitive macrophages” that are derived from monocytes, including by being mitotically active and not yet polarized. Luminex profiling demonstrated that these cells secreted both inflammatory (e.g., IL-8 and CCL2-4), extracellular matrix modeling (e.g., TIMP-1 and MMP-9) and pro-angiogenic cytokines (e.g., VEGF-A and FGF) and point to a functional role of these cells in stroma development within chorionic villi. The authors then compare these data to single-cell RNA sequencing data to identify cellular subtypes that express receptors for these cytokines, which suggests potential for endothelial, fibroblast, and cytotrophoblast interactions.
Lastly, Thomas et al. (2020) determine whether HBCs can be stimulated by various ligands representing infectious agents including viruses and bacteria. Given that some pathogens, such as Zika virus (Zimmerman et al., 2018; Jurado et al., 2016), replicate in HBCs as a mechanism to reach the intraamniotic cavity, these studies are critical to further understand the role of HBCs in placental defenses. These studies revealed that HBCs express various TLRs, and synthetic ligands of these receptors induced robust signaling. Interestingly, stimulating TLR2/6 heterodimers with FSL-1 induced differential responses in PAMMs and HBCs, further pointing to divergent roles for these cells in placental function. Consistent with this, HBCs exhibited increased phagocytotic activity and equivalent microbicidal activity as maternal placental macrophages.
In summary, this study provides elegant technological and conceptual advances that further our understanding of the immune cells enriched at the maternal–fetal interface. Notably, the technological innovations in this manuscript allow for the ability to segregate and functionally characterize the contributions of both maternal and fetal immune cells to placental function and antimicrobial defenses. These fundamental advances will undoubtedly lead to insights into the complex immunological network at the maternal–fetal interface and the impact of microbial infections on this network.
Acknowledgments
C. Megli is supported by the National Institute of Child Health and Human Development Research Scientist Development Program (K12 HD000849), and C.B. Coyne is supported by the National Institutes of Health (AI081759, AI150151, AI145828, and AI145296).
References
- Boyd, J.D., and Hamilton W.J.. 1970. Stroma of Villi. In The Human Placenta. Heffer, Cambridge. [Google Scholar]
- Castellucci, M., et al. 1987. Placenta. 10.1016/0143-4004(87)90040-3 [DOI] [Google Scholar]
- Fu, B., et al. 2017. Immunity. 10.1016/j.immuni.2017.11.018 [DOI] [Google Scholar]
- Jurado, K.A., et al. 2016. JCI Insight. 10.1172/jci.insight.88461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett, A., and Colucci F.. 2014. J. Clin. Invest. 10.1172/JCI68107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, L., and Golos T.G.. 2018. Front. Immunol. 10.3389/fimmu.2018.02628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.R., et al. 2020. J. Exp. Med. 10.1084/jem.20200891 [DOI] [Google Scholar]
- Vento-Tormo, R., et al. 2018. Nature. 10.1038/s41586-018-0698-6 [DOI] [Google Scholar]
- Yang, F., et al. 2019. Front. Immunol. 10.3389/fimmu.2019.02317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, M.G., et al. 2018. Cell Host Microbe. 10.1016/j.chom.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]