Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread rapidly around the world, posing a major threat to human health and the economy. Currently, long-term data on viral shedding and the serum antibody responses in COVID-19 patients are still limited. Herein, we report the clinical features, viral RNA loads, and serum antibody levels in a cohort of 112 COVID-19 patients admitted to the Honghu People’s Hospital, Hubei Province, China. Overall, 5.36% (6/112) of patients showed persistent viral RNA shedding (> 45 days). The peak viral load was higher in the severe disease group than in the mild group (median cycle threshold value, 36.4 versus 31.5; P = 0.002). For most patients the disappearance of IgM antibodies occurred approximately 4–6 weeks after symptoms onset, while IgG persisted for over 194 days after the onset of symptoms, although patients showed a 46% reduction in antibodies titres against SARS-CoV-2 nucleocapsid protein compared with the acute phase. We also studied 18 asymptomatic individuals with RT-qPCR confirmed SARS-CoV-2 infection together with 17 symptomatic patients, and the asymptomatic individuals were the close contacts of these symptomatic cases. Delayed IgG seroconversion and lower IgM seropositive rates were observed in asymptomatic individuals. These data indicate that higher viral loads and stronger antibody responses are related to more severe disease status in patients with SARS-CoV-2 infection, and the antibodies persisted in the recovered patient for more than 6 months so that the vaccine may provide protection against SARS-CoV-2 infection.
Electronic supplementary material
The online version of this article (10.1007/s12250-020-00329-9) contains supplementary material, which is available to authorized users.
Keywords: Coronavirus disease 2019 (COVID-19), Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Viral shedding, Antibody response, Disease severity
Introduction
Since the outbreak in Wuhan, Hubei Province, China in December 2019, Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread rapidly around the world, resulting in a very high number of infections and deaths, threatening public security, and placing a heavy burden on global healthcare systems and economy (Guan et al. 2020; Organization 2020b; Zheng 2020). Although it is less virulent than Middle East respiratory syndrome coronavirus (MERS-CoV) and severe acute respiratory syndrome coronavirus (SARS-CoV), a greater number of infections and deaths have been caused by SARS-CoV-2 because of its higher transmissibility (Cheng et al. 2007; Chan et al. 2015; Li Q et al. 2020). Unfortunately, to date, there are no specific antiviral drugs or licensed vaccines for COVID-19, and isolation is the most effective way to control the spread of infection (Skurnik et al. 2020). Therefore, serial data on viral RNA dynamics and antibody responses in COVID-19 patients are urgently needed to guide the management of the pandemic and the development of antiviral therapies and vaccines.
Although there has been research on the kinetics of viral shedding and antibody responses during SARS-CoV-2 infection, the data are limited by the short monitoring period (around 40 days) (Long et al. 2020a, b; Okba et al. 2020; To et al. 2020; Zhao et al. 2020). As such, long-term changes in antibody profiles in cases of SARS-CoV-2 infection and how long these antibody responses will maintained remain unknown. In order to gain a greater understanding of viral shedding and the immune response in humans following SARS-CoV-2 infection, here we described the long-term profile of viral nucleic acid shedding in respiratory tract and the longitudinal antibody response against the nucleocapsid protein (NP) and the surface spike protein receptor-binding domain (RBD) of SARS-CoV-2 in 112 patients admitted to the Honghu People’s Hospital, Hubei Province, China. Besides, 18 asymptomatic individuals together with 17 symptomatic COVID-19 patients were also included in our study, and the asymptomatic individuals were the close contacts of these 17 symptomatic cases.
Materials and Methods
Patients
A total of 112 patients who were diagnosed by real-time quantitative PCR (RT-qPCR) testing between January and March 2020 and admitted to Honghu People’s Hospital, Hubei Province were included in this retrospective cohort study. These patients were followed up biweekly until 80-days post onset. Throat swabs and serum samples were collected for viral load monitoring and measurement of antibody levels, respectively. Demographic information and clinical examination results were obtained from the electronic medical records. The diagnosis of COVID-19 and evaluation of disease severity is based on the 6th edition of the New Coronavirus Pneumonia Prevention and Control Program released by the National Health Commission of the People’s Republic of China (2020). As described, SARS-CoV-2 infection was diagnosed based primarily on clinical manifestations, chest computerized tomography, and detection of viral nucleic acid in respiratory tract swab samples by RT-q PCR, which is currently the gold standard diagnostic test. Patients presenting with fever, mild clinical symptoms, or mild pneumonia by chest imaging were defined as mild cases, while severe cases were those with respiration rate ≥ 30 times/min, blood oxygen saturation ≤ 93% in the resting state, arterial partial pressure of oxygen/fraction of inspiration oxygen ≤ 300 mmHg, or chest imaging showing > 50% lesion progression within 24–48 h. The date of symptoms onset was set as the day that fever, cough, chills, dizziness, headache, or fatigue appeared.
In late July, we conducted another round of follow-up in which 54 convalescent patients participated. The follow-up time ranged from 158 to 194 days post onset of symptoms. Serum samples were collected for antibodies measurement.
Additionally, 18 individuals who were confirmed to be infected with SARS-CoV-2 by RT-qPCR but did not show any signs or symptoms and had normal chest imaging were included in the analysis together with 17 symptomatic COVID-19 patients, and the asymptomatic individuals were the close contacts of these 17 symptomatic cases.
SARS-CoV-2 RNA Measurement
SARS-CoV-2 RNA load in throat swabs was measured by RT-qPCR using primers and probes targeting the NP and the open reading frame1b genes of SARS-CoV-2 according to the World Health Organization (WHO) guidelines (Organization 2020a). Specimens with cycle threshold values (Ct) < 40 were considered positive; Ct > 40 was assigned a value of 40. The diagnostic criteria were according to the recommendations of the National Institute for Viral Disease Control and Prevention in China (China. 2020).
Serology Testing
IgM and IgG antibodies against SARS-CoV-2 NP and RBD were detected using enzyme-linked immunosorbent assay (ELISA) kits (Guangzhou Darui Biotechnology Co., Guangzhou, China) according to the manufacturer’s instructions. Briefly, these assays employed the quantitative sandwich enzyme immunoassay technique. Serum samples were diluted at 1:100 and added to 96-well polystyrene microplates coated with recombinant SARS-CoV-2 protein antigens, followed by secondary antibody labelled with the conjugate. The optical density (OD) at 450 nm of each well was measured to quantify the amount of antibody in the serum sample. Positivity was determined based on the manufacturer’s recommended cutoff values of 0.4 for IgM and 0.3 for IgG. The total binding IgG and IgM antibodies against SARS-CoV-2 NP in serum were measured by a chemiluminescence microparticle immunoassay (CIMA). Antibody levels were expressed as the ratio of the chemiluminescence signal over the cut-off value (S/CO). A S/CO value of > 1.0 was considered positive.
Statistical Analysis
Prism v8.0 (GraphPad, La Jolla, CA, USA) and SPSS v26.0 (SPPS Inc, Chicago, IL, USA) were used for statistical analyses. Statistical comparisons between groups were performed using the Mann–Whitney U test for continuous variables, and the χ2 test for categorical variables. Correlations were assessed using a Spearman’s Rank correlation coefficient (r). All tests were 2-sided, and P values < 0.05 were considered statistically significant.
Results
Demographic and Clinical Characteristics of the Study Population
Of the 112 patients confirmed as positive for SARS-CoV-2 infection between January and March 2020, 87 (77.7%) were mild cases and 25 (22.3%) were severe. The demographic and clinical characteristics of each group are summarized in Table 1. The median age of the severe group was older than the mild group (median age, 65 vs 50 years, P < 0.001), and showed a tendency for higher male-to-female ratio (68.0% vs 51.7%, P = 0.176). Fever (76.8%) was the most common symptom among the 112 patients, followed by dry cough (54.5%), fatigue (33.0%), and shortness of breath (19.6%). Significantly higher proportions of patients in the severe disease group experienced fatigue (52.0% vs 27.6%, P = 0.030), chill (24.0% vs 5.8%, P = 0.015), and palpitation (20.0% vs 3.5%, P = 0.013), and showed greater abnormalities in biochemical indices at the time of admission than the milder patients group, including higher levels of C-reactive protein (51.57 vs 6.27 mg/L, P < 0.001), d-dimer (1.09 vs 0.7 mg/L, P = 0.033), and lactate dehydrogenase (306 vs 226.5 U/L, P = 0.003). Leukocyte and neutrophil counts were higher in the severe group than in the mild group (7.26 × 109 vs 5.48 × 109/L, P = 0.016 and 5.72 × 109 vs 3.33 × 109/L, P < 0.001, respectively), while the lymphocyte count was lower (0.93 × 109 vs 1.35 × 109/L, P = 0.003) in patients with more severe disease.
Table 1.
Clinical characteristics of COVID-19 patients.
| Total (n = 112) | Mild (n = 87) | Severe (n = 25) | P value | |
|---|---|---|---|---|
| Age, years | 54 (40.25–66) | 50 (38–64) | 65 (52–75.5) | < 0.001* |
| Male | 62 (55.36%) | 45 (51.72%) | 17 (68.00%) | 0.176 |
| Onset of symptoms to | ||||
| Hospital admission, days | 7 (2.25–13) | 9 (3–13) | 6 (1–8) | 0.064 |
| RNA confirmation, days | 7 (3–12) | 9 (3–14) | 5 (2–7.5) | 0.004* |
| Signs and symptoms | ||||
| Fever (temperature ≥ 37.3 °C) | 86 (76.79%) | 66 (75.86%) | 20 (80.00%) | 0.792 |
| Chest tightness | 17 (15.18%) | 11 (12.64%) | 6 (24.00%) | 0.205 |
| Night sweats | 2 (1.79%) | 1 (1.15%) | 1 (4.00%) | 0.398 |
| Shortness of breath | 22 (19.64%) | 15 (17.24%) | 7 (28.00%) | 0.259 |
| Chill | 11 (9.82%) | 5 (5.75%) | 6 (24.00%) | 0.015* |
| Fatigue | 37 (33.04%) | 24 (27.59%) | 13 (52.00%) | 0.030* |
| Dry cough | 61 (54.46%) | 50 (57.47%) | 11 (44.00%) | 0.261 |
| Vomiting | 3 (2.68%) | 2 (2.30%) | 1 (4.00%) | 0.535 |
| Anorexia | 17 (15.18%) | 10 (11.49%) | 7 (28.00%) | 0.058 |
| Palpitation | 8 (7.14%) | 3 (3.45%) | 5 (20.00%) | 0.013* |
| Myalgia | 3 (2.68%) | 1 (1.15%) | 2 (8.00%) | 0.125 |
| Dyspnoea | 15 (13.39%) | 11 (12.64%) | 4 (16.00%) | 0.740 |
| Expectoration | 16 (14.29%) | 13 (14.94%) | 3 (12.00%) | 1.000 |
| Pharyngalgia | 9 (8.04%) | 9 (10.34%) | 0 (0.00%) | 0.204 |
| Diarrhoea | 8 (7.14%) | 7 (8.05%) | 1 (4.00%) | 0.681 |
| Nausea | 7 (6.25%) | 6 (6.90%) | 1 (4.00%) | 1.000 |
| Dizziness | 5 (4.46%) | 2 (2.30%) | 3 (12.00%) | 0.073 |
| Headache | 3 (2.68%) | 1 (1.15%) | 2 (8.00%) | 0.125 |
| Serologic test on admission (normal range) | ||||
| C-reactive protein, mg/L (< 8.2) | 9.83 (2.03–47.71) | 6.27 (1.45–22.62) | 51.57 (23.99–113.50) | < 0.001* |
| d-Dimer, mg/L (< 0.05) | 0.87 (0.36–1.57) | 0.7 (0.31–1.46) | 1.09 (0.82–2.38) | 0.033* |
| Lactate dehydrogenase, U/L (318–618) | 232 (199–288) | 226.5 (190–258) | 306 (205–371) | 0.003* |
| White blood cell count, × 109/L (4–10) | 5.65 (4.67–7.42) | 5.48 (4.47–6.81) | 7.26 (5.01–9.66) | 0.016* |
| Lymphocyte count, × 109/L (0.8–4) | 1.27 (0.9–1.61) | 1.35 (1.09–1.71) | 0.93 (0.73–1.42) | 0.003* |
| Neutrophil count, × 109/L (2–7) | 3.8 (2.85–5.23) | 3.33 (2.75–4.83) | 5.72 (3.76–7.87) | < 0.001* |
| Antibodies against NP, S/CO | 443.09 (181.77–793.02) | 400.12 (172.95–732.51) | 548.26 (392.21–1031.98) | 0.044* |
| Peak viral load (Ct value) | 35.63 (30.87–37) | 36.39 (31.62–37) | 31.48 (24.03–36.64) | 0.002* |
| SARS-CoV-2 RNA persistence, days | 15 (9–27) | 17.5 (10–27) | 12 (7.5–32.5) | 0.455 |
Data are presented as median (interquartile range) and n (%); *P < 0.05 (Student’s t test, χ2 test, or Mann–Whitney U test); Ct, cycle threshold; NP, nucleocapsid protein; S/CO, signal-to-cutoff.
SARS-CoV-2 RNA Dynamics
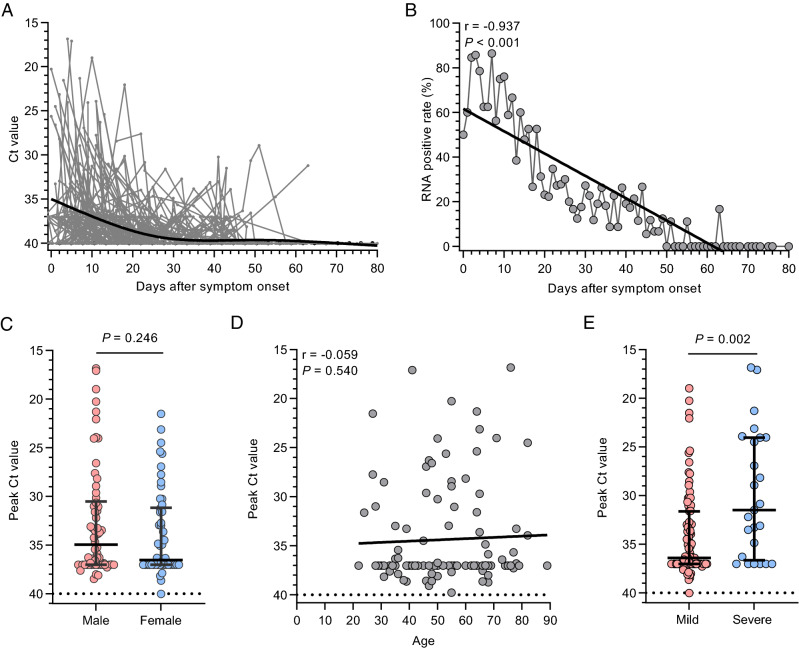
We collected 1065 throat swabs from 112 COVID-19 patients for viral load monitoring (mean of 9.5 throat specimens per patient). The median of viral RNA following-up time was 57 days (interquartile range [IQR], 49–63.75 days), and the longest was 80 days. The temporal profile of the viral load for all patients is shown (Fig. 1A, 1B). The peak viral load in throat swabs was significantly higher in the severe group than in the mild group but did not differ by sex and showed no correlation with age (Fig. 1C–1E). The median duration of viral shedding was 15 days (IQR, 9–27 days). In five patients (4.5%; four mild and one severe case), viral RNA had turn negative for two consecutive tests and then recurrence occurred 45 days after the initial symptom onset. In another patient with mild disease, viral RNA was detected by RT-qPCR up to 45 days after symptom onset.
Fig. 1.
SARS-CoV-2 RNA profiles in throat swabs of COVID-19 patients. A Temporal profile of viral load (inversely related to cycle threshold [Ct] value) in all patients (n = 112). Each line represents an individual patient. The thick line shows the trend in viral load using smoothing splines. B SARS-CoV-2 RNA positive rate at different time intervals. C Comparison of peak viral load according to sex. D Relationship between age and peak viral load. E Comparison of peak viral load between mild and severe patients. Results are shown as median and interquartile range; P values were derived from Mann–Whitney U test, or Spearman's test; r values were derived from Spearman's test.
Antibody Response Against SARS-CoV-2 NP and RBD
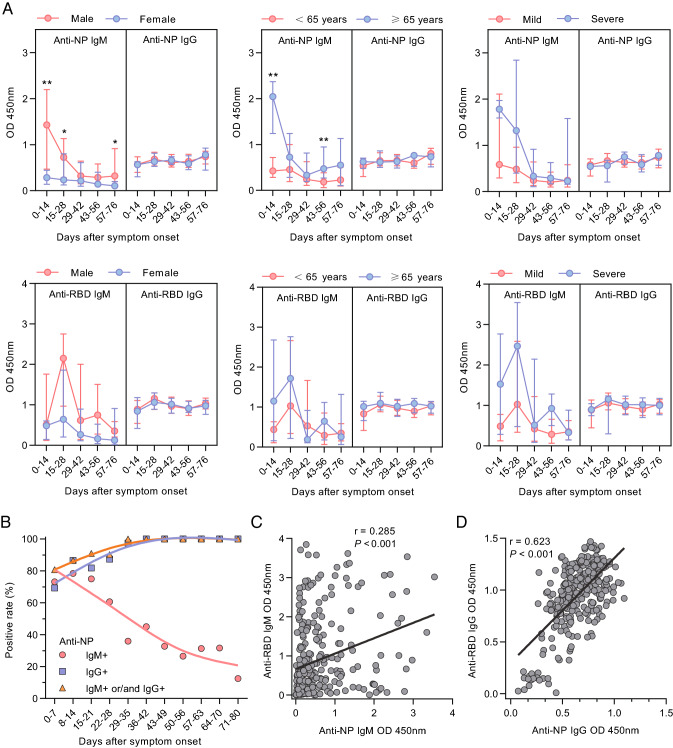
Longitudinal antibody responses were evaluated in our cohort. A total of 474 serum samples were obtained from the 112 patients (mean of 4.23 serum specimens per patient). These patients were followed up biweekly up to 80-days post disease onset. IgM and IgG antibodies against SARS-CoV-2 NP were detected by ELISA. To further understand the correlation between antibody responses to SARS-CoV-2 and clinical parameters, the antibody responses were stratified according to sex, age, and disease severity (Fig. 2A). Overall, the levels of IgM antibodies varied in the two groups, while there was no significant difference in IgG levels when patients were stratified according to sex, disease severity, and age. Anti-NP IgM levels peaked at the first 2 weeks after symptom onset and decreased steadily thereafter, and were undetectable in most patients after 4–6 weeks. Compared to females, male patients had significantly higher serum anti-NP IgM level; and the anti-NP IgM level was higher in older patients and the severe patients group compared to the younger group and the mild disease group, respectively, but the differences were not statistically significant. Anti-RBD IgM level reached its peak at 3–4 weeks post disease onset, which occurred later than anti-NP IgM level.
Fig. 2.
Profiles of antibodies against SARS-CoV-2 NP and RBD in COVID-19 patients (n = 112). A Antibodies against SARS-CoV-2 NP and RBD in patients according to sex, age, and disease severity. Results are shown as median and interquartile range; P values were derived from the Mann–Whitney U test. B Seropositive rates of IgM and IgG against SARS-CoV-2 NP at different intervals. C, D Relationship between levels of antibodies against SARS-CoV-2 NP (C) and RBD (D) measured by ELISA. The P and r values were derived from Spearman’s test. *P < 0.05; **P < 0.01.
We then evaluated the seropositive rates of anti-NP IgM and IgG antibodies based on the number of days after symptom onset (Fig. 2B; Supplementary Table S1). The positivity rate for IgM peaked in the second week with 86.49% and declined steadily afterward. The seroconversion rate of anti-NP IgG was 69.2% in the first week after symptom onset, reached 100% in the fifth week, and then remained 100% up to 80 days post disease onset.
Next, we estimated Spearman’s correlation among the antibody titres against NP and RBD (Fig. 2C, 2D). ELISA OD values for anti-RBD IgM showed a weak but significant positive correlation with anti-NP IgM levels (r = 0.285; P < 0.001; Fig. 2C), and anti-RBD IgG levels showed a significant positive association with anti-NP IgG levels (r = 0.623; P < 0.001; Fig. 2D). While there was no correlation between the persistence of SARS-CoV-2 RNA and antibodies levels (Supplement Fig. S1).
Follow-Up Study on Convalescent Patients
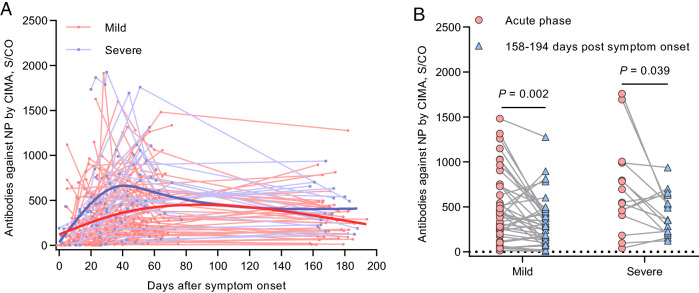
In late July, we conducted another round of follow-up in which included 54 recovered patients. The follow-up time ranged from 158 to 194 post onset of symptoms. All the samples collected in July tested positive (S/CO > 1) for antibodies against NP; the temporal profile is shown in Fig. 3A. At 158–194 days post onset of symptoms, the antibody levels decreased approximately 46.17% overall (median, 151.61 S/CO, IQR, 287.11–453.01 S/CO; P = 0.128; Fig. 3B) compared with those at the acute phase (the period when the viral RNA can be found in a respiratory specimen). Moreover, the median percentage of decrease was 46.31% (IQR 5.37%–68.96%) for antibodies against NP in the mild group, whereas it was 46.02% (IQR 11.01%–65.75%) in the severe group.
Fig. 3.
Profile of antibodies against SARS-CoV-2 NP measured by CIMA. A Temporal pattern of antibodies to NP in all patients (n = 112). Each line represents an individual patient. The thick line shows the trend using smoothing splines. B Dynamic changes in antibodies against NP in the acute phase (the period when the viral RNA can be found in a respiratory specimen) and 158–194 days post symptoms onset (n = 54). The P and r values were derived from Spearman’s test.
Comparison Between Asymptomatic Individuals and the related Symptomatic Cases
We compared SARS-CoV-2 RNA dynamics and antibody responses between 18 asymptomatic individuals and 17 symptomatic COVID-19 patients, and the asymptomatic individuals were the close contacts of these symptomatic cases. The clinical characteristics of these individuals are summarized in Table 2. The symptomatic group tended to have a higher median age and included more males than the asymptomatic group.
Table 2.
Characteristics of asymptomatic individuals and the related symptomatic cases.
| Asymptomatic (n = 18) | Symptomatic (n = 17) | P value | |
|---|---|---|---|
| Age, years | 38 (23–55.75) | 52 (37–63) | 0.06 |
| Male | 7 (38.89%) | 11 (64.71%) | 0.181 |
| IgM and IgG against NP peak titre, S/CO | 60.79 (9.99–194.54) | 302.44 (169.29–567.85) | < 0.001* |
| Peak viral load (Ct value) | 35.26 (32.1–37) | 34.3 (29.82–37) | 0.725 |
| RNA persistence, days | 2.5 (1–8.25) | 4 (1–20.5) | 0.508 |
Data are presented as median (interquartile range) and n (%); *P < 0.05 (χ2 test or Mann–Whitney U test); Ct, cycle threshold; NP, nucleocapsid protein; S/CO, signal-to-cutoff.
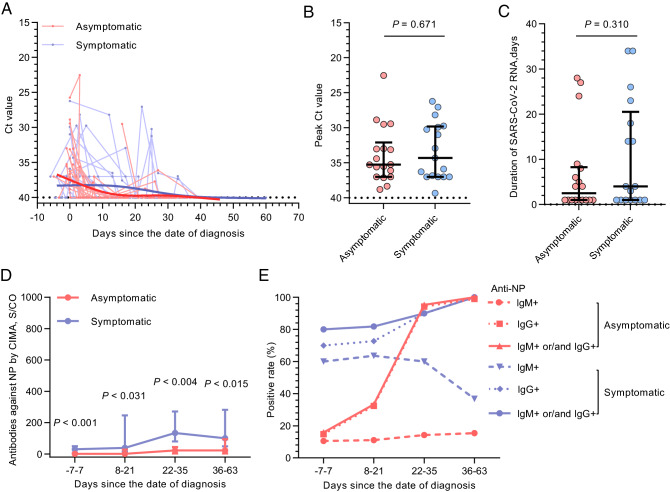
In total, 276 throat swabs (127 from 18 asymptomatic individuals and 149 from 17 symptomatic cases) were collected for viral RNA load monitoring. There were no statistically significant differences in the peak Ct value and the persistence of viral RNA between the asymptomatic individuals and the symptomatic cases (Fig. 4A–4C). A further 116 serum samples (2–5 per patient) were obtained at different time points and tested for antibodies against NP. The date of diagnosis was set as the starting point to compare the antibody dynamics between the asymptomatic individuals and symptomatic cases. The temporal profile of antibodies is shown in Supplementary Figure S2. Antibody levels against NP were significantly lower in the asymptomatic group (Fig. 4D). The serologic conversion of IgG occurred earlier in the symptomatic cases than in the asymptomatic carriers, and the IgM seroconversion rate in the asymptomatic group was much lower (16.7% vs 70.6%, P = 0.002; Fig. 4E).
Fig. 4.
SARS-CoV-2 RNA and antibody dynamics in asymptomatic patients (n = 18) and symptomatic cases (n = 17). A Temporal profile of viral RNA load (inversely related to cycle threshold [Ct] value). Each line represents an individual patient. The thick line shows the trend in viral load using smoothing splines. B, C Peak viral RNA load (B) and persistence of viral RNA (C) in asymptomatic patients and symptomatic cases. D Antibodies against NP measured by CIMA. E Seropositivity of antibodies against NP at different intervals measured by ELISA. Results are shown as median and interquartile range; P values were derived from the Mann–Whitney U test.
Discussion
This study illustrates the kinetics of SARS-CoV-2 RNA levels and the antibody responses to SARS-CoV-2 NP and RBD proteins in COVID-19 patients with different illness severities, including mild, severe, and asymptomatic. Compared to the peak viral load of SARS-CoV occurring at approximately 10 days and that of MERS-CoV occurring in the second week after symptom onset (Peiris et al. 2003a; Oh et al. 2015; Oh et al. 2016), in the present study, peak viral loads in throat swabs were in most cases already detectable at the initial presentation of symptoms and decreased during hospitalization. This finding is consistent with previous reports (Huang et al. 2020; To et al. 2020) and supports the view that peak transmissibility occurs at or before symptom onset (He et al. 2020; Rothe et al. 2020). Thus, strategies used to control the spread of SARS-CoV may not be applicable to SARS-CoV-2, which require stricter measures based on the viral dynamics. An association between higher viral load and worse outcome has been reported for SARS-CoV and MERS-CoV (Peiris et al. 2003b; Hung et al. 2004; Oh et al. 2016). Similarly, we observed much higher peak viral RNA loads and stronger antibody responses in patients with severe COVID-19 symptoms compared to patients with mild disease. The correlation of antibody responses with disease severity is still unclear. Higher viral load may lead to more severe disease severity and may stimulate the production of antibodies. In turn, a robust antibody response may result in tissue injury and drive the symptoms of COVID-19.
A few studies have reported prolonged shedding of SARS-CoV-2 RNA (Li N et al. 2020; Liu et al. 2020; Xiao et al. 2020; Yang et al. 2020). Notably, in our study, 5.36% (6/112) of patients showed prolonged viral shedding (> 45 days in throat swabs) and five patients among them had two successive swabs that were negative but experienced recurrence of COVID-19, although this may have been due to false-negative RT-qPCR results rather than actual recurrence or reinfection (Xiao et al. 2020). Another patient tested positive for SARS-CoV-2 RNA > 45 days post-symptom onset. However, antibody responses in these six patients did not differ from those in other patients. These data indicate that self-quarantine and regular monitoring of viral RNA levels are necessary in patients who have recovered from COVID-19, even after they have had 2 consecutive negative nucleic acid tests and met the discharge criteria.
All patients in this study achieved antibody seroconversion within a month post symptom onset. IgM is usually produced before IgG during viral infections (Aljumaili et al. 2014; Singh et al. 2015; Samanta and Willis 2016). This was also true in the case of SARS-CoV-2 infection: the median time for seroconversion was 17 days post symptom onset for anti-NP IgM and 20 days after the onset for anti-NP IgG. However, this finding contradicts earlier observations that the IgG response occurred simultaneously with or slightly earlier than the IgM response (To et al. 2020; Zhao et al. 2020). Some factors might have contributed to the variation in antibody response in different studies, including the frequency of specimen collection and different test kits with diverse specificity and sensitivity. Previous studies investigating the kinetics of antibodies to SARS-CoV-2 are limited to around 40 days after the onset of symptoms (Long et al. 2020a, 2020b; Okba et al. 2020; To et al. 2020; Zhao et al. 2020). The antibody response in patients following MERS-CoV infection could last more than 34 months after onset (Payne et al. 2016), and antibodies could still be detected up to 36 months post infection in 89% of individuals with SARS-CoV infection (Cao et al. 2007).
In this study, we described for the first time the 6-month longitudinal dynamics of the antibody response to SARS-CoV-2 in a cohort of COVID-19 patients. Our data showed that antibodies against SARS-CoV-2 NP can persist more than 194 days post illness onset, but most patients showed a variable degree of reduction in antibody levels. Although the antibody level was higher in patients with more severe symptoms in the acute phase, the degree of reduction was around 46% in the late convalescent stage (158–194 days after the onset) in both mild and severe patients. Whether the magnitude of antibody levels in the convalescent sera could prevent re-infection of SARS-CoV-2 is unknown and needs further research. Moreover, due to the decreasing trend observed in present study, antibodies to SARS-CoV-2 may disappear over a period of 18 months, suggesting the necessity of vaccine development.
The number of cases of asymptomatic SARS-CoV-2 infection is increasing rapidly (Chang et al. 2020; Li R et al. 2020; Team 2020). Asymptomatic individuals may be unaware of their infection status and therefore unknowingly transmit the virus to others if they are not isolated for medical observation. We monitored changes in viral RNA loads in throat swabs and serum antibody levels in several asymptomatic cases and symptomatic cases. In the former group, SARS-CoV-2 RNA persisted for a shorter time compared to the latter group and the anti-NP antibody levels were lower. Delayed IgG seroconversion and a lower IgM seropositive rates were observed in asymptomatic individuals, suggesting that asymptomatic individuals had a weaker immune response to SARS-CoV-2 infection and the antibody levels had a great potential to predict the progression of COVID-19.
Our study had some limitations. Firstly, we reported the viral RNA cycle threshold value, which does not distinguish between active and dead virus. Secondly, the time points of the sample collection were irregular and mainly relied on physician’s judgement, with some specimens collected after long intervals. Finally, given the small sample size, our results must be interpreted with caution and validated in a larger cohort.
In summary, we have determined the dynamics of the antibody response to SARS-CoV-2 up to 6 months after the onset of COVID-19 at a population level. IgM was detected earlier than IgG, and disappeared in most cases at around 4–6 weeks after symptoms onset, while IgG could last for more than 194 days after the onset, but patients showed approximately 46% reduction in antibody levels compared with the acute phase. These data imply that the antibody may persist for quite a long time and the vaccine could provide protection against SARS-CoV-2 infection. Delayed IgG seroconversion and a lower IgM seropositive rate were observed in asymptomatic individuals, indicating that asymptomatic individuals had a weak immune response to SARS-COV-2 infection.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to the patients enrolled in this study and to the nurses that assisted in the patient management and the collection of blood samples. This work was partly supported by grants from the National Science and Technology Major Project of China (2018ZX10301202-003) and the Collaboration and Innovation Health Care Major Project of Guangzhou (201803040013).
Author Contributions
XZ and JH conceived and designed the study. QL, Z Lu and YJ had roles in the patient recruitment, sample preparation and clinical data collection. Y Wang, Z Liu and HL performed the viral RNA and antibody detection experiments, data collection and analysis. SL, YL and XH had roles in statistical analysis. XZ and SL wrote the manuscript. JS, Y Wu and XX contributed to data interpretation and critical revision of the manuscript. All authors reviewed and approved the final version of the manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
Additional informed consent was obtained from all patients for which identifying information is included in this article.
Footnotes
Xiaoyong Zhang and Suwen Lu have contributed equally to this work.
References
- Aljumaili ZK, Alsamarai AM, Najem WS. Cytomegalovirus seroprevalence in women with bad obstetric history in Kirkuk, Iraq. J Infect Publ Health. 2014;7:277–288. doi: 10.1016/j.jiph.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007;357:1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- Chan JFW, Lau SKP, To KKW, Cheng VCC, Woo PCY, Yuen KY. Middle east respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Xu H, Rebaza A, Sharma L, Dela Cruz CS. Protecting health-care workers from subclinical coronavirus infection. Lancet Resp Med. 2020;8:e13. doi: 10.1016/S2213-2600(20)30066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng VCC, Lau SKP, Woo PCY, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for C (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720 [DOI] [PMC free article] [PubMed]
- He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Chen Y, Liao B, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung GM. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Huang JT, Ran RX, Lv ZH, Feng LN, Ran CY, Tong YQ, Li D, Su HW, Zhu CL, Qiu SL, Yang J, Xiao MY, Liu MJ, Yang YT, Liu SM, Li Y. Chronological changes of viral shedding in adult inpatients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung IF, Cheng VC, Wu AK, Tang BS, Chan KH, Chu CM, Wong MM, Hui WT, Poon LL, Tse DM, Chan KS, Woo PC, Lau SK, Peiris JS, Yuen KY. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550–1557. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wang X, Lv T. Prolonged SARS-CoV-2 RNA shedding: not a rare phenomenon. J Med Virol. 2020 doi: 10.1002/jmv.25952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WD, Chang SY, Wang JT, Tsai MJ, Hung CC, Hsu CL, Chang SC. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF, Wang DQ, Hu Y, Ren JH, Tang N, Xu YY, Yu LH, Mo Z, Gong F, Zhang XL, Tian WG, Hu L, Zhang XX, Xiang JL, Du HX, Liu HW, Lang CH, Luo XH, Wu SB, Cui XP, Zhou Z, Zhu MM, Wang J, Xue CJ, Li XF, Wang L, Li ZJ, Wang K, Niu CC, Yang QJ, Tang XJ, Zhang Y, Liu XM, Li JJ, Zhang DC, Zhang F, Liu P, Yuan J, Li Q, Hu JL, Chen J, Huang AL. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, Hu JL, Xu W, Zhang Y, Lv FJ, Su K, Zhang F, Gong J, Wu B, Liu XM, Li JJ, Qiu JF, Chen J, Huang AL. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020 doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- National Health Commission of the People’s Republic of China (2020) Guideline for Diagnosis and Treatment of SARS-CoV-2 (the sixth edition)
- Oh MD, Choe PG, Oh HS, Park WB, Lee SM, Park J, Lee SK, Song JS, Kim NJ. Middle east respiratory syndrome coronavirus superspreading event involving 81 persons, Korea 2015. J Korean Med Sci. 2015;30:1701–1705. doi: 10.3346/jkms.2015.30.11.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MD, Park WB, Choe PG, Choi SJ, Kim JI, Chae J, Park SS, Kim EC, Oh HS, Kim EJ, Nam EY, Na SH, Kim DK, Lee SM, Song KH, Bang JH, Kim ES, Kim HB, Park SW, Kim NJ. Viral load kinetics of MERS coronavirus infection. N Engl J Med. 2016;375:1303–1305. doi: 10.1056/NEJMc1511695. [DOI] [PubMed] [Google Scholar]
- Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken C, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization WH (2020a) Coronavirus disease (COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans
- Organization WH (2020b) Coronavirus disease (COVID-2019) situation reports
- Payne DC, Iblan I, Rha B, Alqasrawi S, Haddadin A, Al Nsour M, Alsanouri T, Ali SS, Harcourt J, Miao C, Tamin A, Gerber SI, Haynes LM, Al Abdallat MM. Persistence of antibodies against middle east respiratory syndrome coronavirus. Emerg Infect Dis. 2016;22:1824–1826. doi: 10.3201/eid2210.160706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, Law KI, Tang BS, Hon TY, Chan CS, Chan KH, Ng JS, Zheng BJ, Ng WL, Lai RW, Guan Y, Yuen KY, Group HUSS (2003a) Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361: 1767–1772 [DOI] [PMC free article] [PubMed]
- Peiris JS, Yuen KY, Osterhaus AD, Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M, Drosten C, Vollmar P, Zwirglmaier K, Zange S, Wolfel R, Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D, Willis E. Focal seizure associated with human parvovirus B19 infection in a non-encephalopathic child. World J Pediatr. 2016;12:118–120. doi: 10.1007/s12519-015-0060-0. [DOI] [PubMed] [Google Scholar]
- Singh L, Mishra S, Prasanna S, Cariappa MP. Seroprevalence of TORCH infections in antenatal and HIV positive patient populations. Med J Armed Forces India. 2015;71:135–138. doi: 10.1016/j.mjafi.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik D, Beaujouan L, Brun-Ney D, Lefevre-Utile A, Basmaci R, Soussan-Banini V, Guedj R, Rybak A, Gajdos V, Filser M, Yang DD, Ouldali N, Angoulvant F. COVID-19 pandemic: Impact caused by school closure and national lockdown on pediatric visits and admissions for viral and non-viral infections, a time series analysis. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team TNCPERE (2020) The epidemiological characteristics of an outbreak of 2019 novel coronavirus disease (COVID-19)—China [PMC free article] [PubMed]
- To KK-W, Tsang OT-Y, Leung W-S, Tam AR, Wu T-C, Lung DC, Yip CC-Y, Cai J-P, Chan JM-C, Chik TS-H, Lau DP-L, Choi CY-C, Chen L-L, Chan W-M, Chan K-H, Ip JD, Ng AC-K, Poon RW-S, Luo C-T, Cheng VC-C, Chan JF-W, Hung IF-N, Chen Z, Chen H, Yuen K-Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao AT, Tong YX, Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020 doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JR, Deng DT, Wu N, Yang B, Li HJ, Pan XB. Persistent viral RNA positivity during the recovery period of a patient with SARS-CoV-2 infection. J Med Virol. 2020 doi: 10.1002/jmv.25940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.