Abstract
Three dimensional (3D) bioprinting has recently advanced as an important tool to produce viable constructs that can be used for regenerative purposes or tissue models. To develop biomimetic and sustainable 3D constructs, several important processing aspects need to be considered, among which crosslinking is most important for achieving desirable biomechanical stability of printed structures which is reflected on subsequent behavior and use of these constructs. In this review, crosslinking methods used in 3D bioprinting studies are reviewed, parameters that affecting bioink chemistry discussed, and the potential towards improving crosslinking outcomes and construct performance are highlighted. Furthermore, current challenges and future prospects are discussed. Due to the direct connection between crosslinking methods and properties of 3D bioprinted structures, this review can provide basis for developing necessary modifications to design and manufacturing process of advanced tissue-like constructs, in future.
Keywords: crosslinking strategies, bioink, hydrogel-cell interactions, 3D bioprinting, tissue engineering
Graphical Abstract
When designing 3D bioprinting system, there is a need to choose an appropriate crosslinking approach for the desired applications. Appropriate crosslinking allows the printability of bioinks and ensures cytocompatibility, stability and sustainability of resulting living constructs. In this review, crosslinking techniques that have been employed in 3D bioprinting studies are critically reviewed, parameters that affect bioink hydrogel chemistry discussed and the potential towards improving crosslinking outcomes and construct performance are highlighted.
1. Introduction
Adult human organs have very limited capacity for regeneration, and their damage due to injury, disease or surgery leads to loss of function [1]. This requires treatment, which is usually achieved by tissue or organ transplantation [2]. Because of a limited supply of organs, many lives are lost while waiting for donated organs to become available [3]. With the recent advent of three-dimensional (3D) bioprinting [4, 5, 6], it is becoming more evident that tissue constructs can be printed ex vivo or even in situ [6, 7]. Despite remarkable research efforts on 3D bioprinting, several challenges that need to be resolved, to further advance research in this area. One of the most important challenges is related to the materials being printed (inks) [8]. Suitable inks should possess certain characteristics, such as good biocompatibility, structural stability, and sufficient mechanical and rheological properties [9]. To attain these properties, researchers have developed various materials, including hydrogels [10]. Hydrogels exhibit numerous attractive features, such as highly hydrated environment that mimics extracellular matrix (ECM), making them an ideal carrier for encapsulating cells [11]. Despite recent progress in using hydrogels for 3D bioprinting, many challenges need to be addressed in order to produce biomimetic and biologically inspired tissue constructs [12].
During the process of 3D bioprinting, a polymer solution is transformed into a 3D structure via crosslinking. Crosslinking is a key procedure that significantly influences the mechanical and physicochemical characteristics of the bioprinted constructs and the cellular behavior of loaded live cells. The most commonly used 3D bioprinting techniques are extrusion, injection, laser-assisted bioprinting, and stereolithography. The characteristics of these technologies are reviewed in depth elsewhere [13, 14, 15]. Extrusion among them has been shown to affect the rheological behavior of bioinks [16]. Hence, the effect of crosslinking reactions is more pronounced on the extrusion-based bioprinting methods. Extrusion-based printers divided into piston-driven, pneumatic, and screw-driven dispensing. There are also some advanced extrusion-based cell printing methods such as bath-Assisted approach, aerosol spraying, and light/thermo-assisted approach. Depending on the nature of the polymeric backbone and their functional groups, hydrogel bioinks (cell laden inks) could be crosslinked by using various methods that can be chemical, physical, enzymatic or a combination of them (Figure 1). Network formation in a chemically crosslinked hydrogel bioink occurs by non-reversible covalent bonding between polymeric chains, usually by adding a chemical crosslinkers (e.g. sodium bicarbonate [17], thrombin [18]) or through various chemical reactions including Schiff base chemistry [19], azide-alkyne cycloaddition [20], hydrazide-aldehyde coupling [21], thiolene coupling [22], enzymatic crosslinking [23], or various ultraviolet (UV) [24], visible [25], or near infrared light [26]. These hydrogels are usually strong enough to provide proper shape stability, but crosslinking kinetics should be precisely controlled to avoid any printer nozzle blockage. Physical crosslinking pathways, which are used for 3D bioprinting of hydrogels take place through the formation of non-covalent bonds, such as H-bonds [27, 28], hydrophobic interactions [29], electrostatic attraction [30], and ionic crosslinking [31]. These hydrogels are usually mechanically weak but provide a more cell-friendly environment than chemically crosslinked hydrogel. To overcome this limitation, nanofillers or chemical functionalities can be introduced to improve the stability of the bioprinted constructs [32]. Not only the type of crosslinking is important for 3D bioprinting but also the density of formed crosslinks plays an important role. There is a balance to strike between the degree of crosslinking. Reducing the degree of crosslinking may allow faster flow of the bioink, and increasing it may lead to a stiffer structure that may hamper printability.
Figure 1.
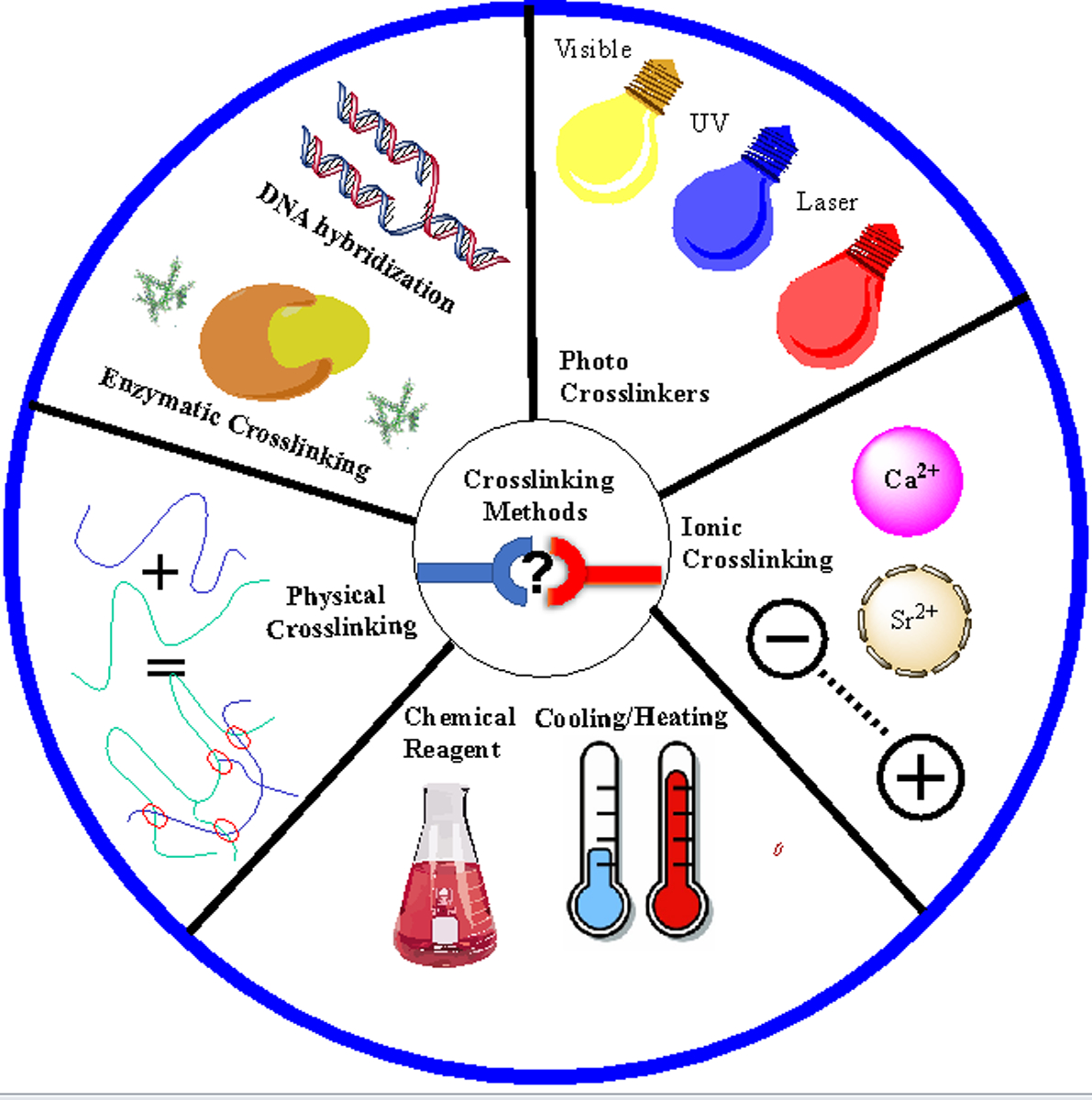
Various crosslinking methods that have been used for three-dimensional (3D) bioprinting of hydrogels.
Although various aspects of 3D bioprinting have been described in excellent reviews [33, 34], a proper systematic review of the crosslinking pathways used in 3D bioprinting including their effects on bioprinted constructs is lacking. In this review, crosslinking techniques that have been employed in 3D bioprinting studies are critically reviewed, parameters that affect bioink hydrogel chemistry are discussed and the potential towards improving crosslinking outcomes and construct performance are highlighted. In addition, the application of various techniques to develop advanced and biomimetic 3D constructs for regenerative therapeutics are presented. Finally, current challenges and future prospects are discussed. With the trend towards developing more biomimetic 3D printed constructs such as vascularized [35, 36], and four-dimensional (4D) constructs [5], it is becoming necessary to deeply study various crosslinking techniques and its effects that would undoubtedly result in developing more heterogeneous 3D bioprinted constructs that can mimic more closely native tissues and organs.
2. Crosslinking Methods
2.1. Physical Crosslinking
2.1.1. Ionic Interactions
- With the Addition of Metal ions (Metal coordination)
Ionic interaction is one of the most common methods used for crosslinking of hydrogels that are used in 3D bioprinting. It usually involves the addition of multivalent cations to the polymer solution to induce gelation. In this rapid crosslinking method, hydrogels can be formed under mild conditions, at room temperature and physiological pH, making it an attractive method for crosslinking. However, it has some drawbacks, such as mechanical weakness, poor stacking ability, and possibility of the release of metal ions into the body after implantation. Ionic interaction is mostly used as a crosslinking strategy for 3D bioprinting of sodium alginate (Table 1). Alginates are algae-derived anionic polysaccharides made up of linked β-d-mannuronate (M-blocks) and α-l-guluronate (G-blocks) residues [37, 38]. Carboxylic groups of adjacent polymer chains are capable of binding with multivalent cations Mn+ as shown in Figure 2A, resulting in an ionically crosslinked gel network via metal coordination. Beside alginate, other carboxylic containing polysaccharides such as gellan gum are also crosslinkable through metal ions addition (Figure 2B).
Table 1.
Summary of three dimensional (3D) bioprinting where ionic crosslinking, was employed showing materials used, type of biomolecules and target tissues. Various biomaterials have been used alone or in combination, such as collagen (Col), gelatin (gel), gelatin methacryloyl (GelMA) dentin (D), extracellular matrix (ECM), alginate (Alg), hyaluronic acid (HA), gellan (G), gellan gum (GG), cellulose (C) nanocellulose (NC), methylcellulose (MC), carboxymethylcellulose (CMC), silk fibroin (SF), chitosan (Ch), poly (vinyl alcohol) (PVA), polycaprolactone (PCL) or biosilica (BS). Cells used included human mesenchymal stem cells (HMSCs), rat mesenchymal stem cell (RMSCs), epidermal stem cells (ESCs), infrapatellar stem cells (ISCs), human immortalised keratinocyte cells (HIKCs), human induced pluripotent stem cells (HIPSCs), human umbilical vein endothelial cells (HUVECs), human coronary artery endothelial cells (HCAECs), rat heart endothelial cells (RHECs), porcine aortic valve interstitial cells (VICs), human fetal lung cells (HFUCs), human aortic root smooth muscle cells (SMCs), myoblasts (MBCs), corneal keratocytes cells (CKCs), human newborn foreskin cells (HNFCs), cartilage progenitor cells (CPCs), chondrocytes (CCs), neurolemmocytes (NUCs), osteoblast precursor cells (OPCs), osteosarcoma cells (OCs), and pancreatic endothelial cells (PECs).
| Biomaterial | Crosslinking/Metal Ion | Printing Technique | Cell type | Target tissue | Ref. |
|---|---|---|---|---|---|
| Alg | Ionic-CaCl2 | extrusion | HMSCs | Not specified | [44] |
| Alg | Ionic-CaCl2 | extrusion | HCAECs | Cardiac tissue | [47] |
| Alg | Ionic-CaCl2 | extrusion | HMSCs | Arbitrary tissue | [175] |
| Alg | Ionic-CaCl2 | extrusion | HIPSCs | Liver tissue | [60] |
| Alg | Ionic-CaCl2 | extrusion piston-driven | OCs | Not specified | [176] |
| Alg | Ionic-CaCl2 | Laser-assisted bioprinting | HIKCs | Skin Tissue | [177] |
| Alg | Ionic-CaCl2 | Laser-assisted bioprinting | OCs | Not specified | [178] |
| Alg | Ionic-CaCl2 | extrusion | Fibroblasts | Soft tissue | [179] |
| Alg | Ionic-CaCl2 | Laser-assisted bioprinting | Fibroblasts | Not specified | [45] |
| Alg | Ionic-CaCl2 | Inkjet bioprinting | OCs | Bone tissue | [180] |
| Alg | Ionic-CaCl2 | Inkjet bioprinting (piezo) | HeLa | Not specified | [181] |
| Alg | Ionic-CaCl2 | extrusion | CPCs | Vessel | [182] |
| Alg | Ionic-CaCl2 | extrusion | RHECs | Not specified | [43] |
| Alg | Ionic-CaCl2 | extrusion piston-driven | HMSCs | Human cartilage | [183] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion | ESCs | Not specified | [61] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion | HMSCs | Not specified | [54] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion | HFUC | Lung | [184] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | Inkjet bioprinting | ESCs | Not specified | [62] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion | Fibroblasts | Not specified | [185] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion | ESCs | Skin tissue | [186] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | Laser-assisted bioprinting | ESCs | Not specified | [187] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion | Fibroblasts | Not specified | [188] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion | OCs | Not specified | [189] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion piston-driven | VICs / SMCs | Aortic valve Conduits | [59] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion | MBCs | Not specified | [9] |
| Alg/gel | Thermal Pre-gelation + Ionic-CaCl2 | extrusion | MBCs | Soft tissue | [46] |
| Alg/GG | Ionic CaCl2 | extrusion | Rat MSCs | Not specified | [190] |
| Alg /gel/Col | Ionic-CaCl2 | extrusion piston-driven | CKCs | Not specified | [191] |
| Alg/MC | Ionic-CaCl2 | extrusion | HMSCs | Not specified | [53, 192] |
| Alg/CMC | Hydrogen bonds Pre-gelation + Ionic-CaCl2 | extrusion | HPCCs | Not specified | [52] |
| Alg/C | Ionic-CaCl2 | extrusion | HMSCs | Human Cartilage | [193] |
| Alg/NC | Ionic-CaCl2 | extrusion piston-driven | CPCs | Human Cartilage | [194] |
| Alg/D | Ionic-CaCl2 | extrusion | OCs | Dentin matrix | [195] |
| Alg/PVA | Ionic-CaCl2 | extrusion | OPCs | Bone tissue | [64] |
| Alg/ECM | Ionic-CaCl2 | extrusion | ISCs | Meniscal tissue | [196] |
| Alg/SF | Ionic-CaCl2 + (HRP) | extrusion piston-driven | Fibroblasts | Not specified | [197] |
| Alg/PCL | Ionic-CaCl2 | extrusion | Fibroblasts | Human Cartilage | [65] |
| Alg/PCL | Ionic-CaCl2 | extrusion | CCs | Cartilage tissue | [48] |
| Alg/BS | Ionic-CaCl2 | extrusion | OCs | Bone tissue | [198] |
| Alg/GelMA | Ionic-CaCl2 | extrusion | CKCs | Corneal stroma | [199] |
| Alg/GelMA | Ionic-CaCl2 | Inkjet bioprinting | HNFs | Not specified | [200] |
| Alg/HA | Ionic-CaCl2 | extrusion | NUs | Nerve tissue | [49] |
| Alg/C | Ionic-CaCl2 | extrusion | CCs | Human Cartilage | [63] |
| Alg/ GG | Ionic-SrCl2 | Inkjet bioprinting | CCs | Human Cartilage | [39] |
| Alg/MC | Ionic-SrCl2 | extrusion piston-driven | PECs | Liver Tissue | [201] |
| Peptide-ModifiedGG | Ionic-CaCl2 | extrusion | NUs | Brain tissue | [68] |
| Peptide-ModifiedGG | Ionic-CaCl2 | Inkjet bioprinting | MBCs | Not specified | [69] |
| Ch-HAp | Ionic- CaSO4 | extrusion | MSCs | Bone tissue | [42] |
Figure 2.
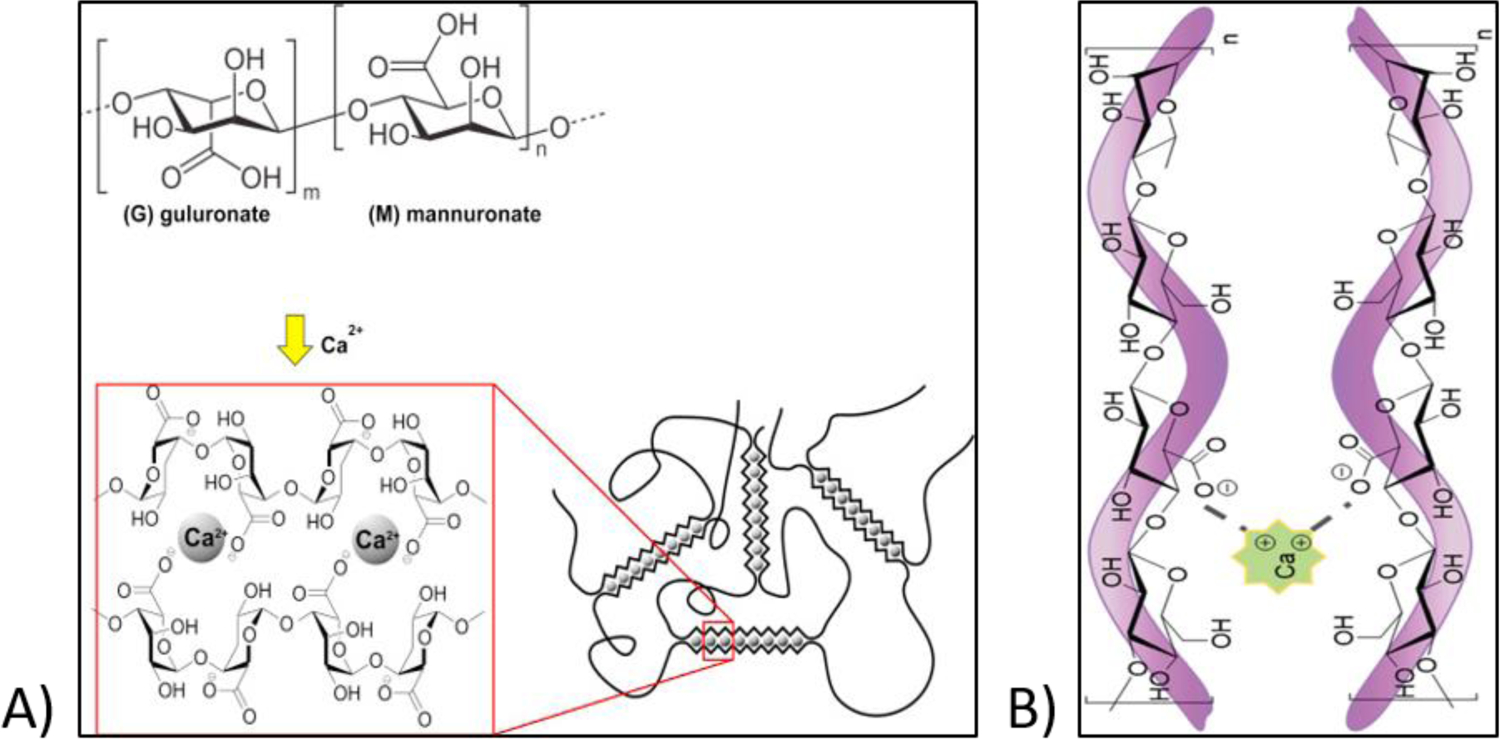
A) Chemical structure of sodium alginate and its network formation in the presence of calcium chloride. Reproduced from Bruchet and Melman [37], with permission from Elsevier, 2015. Network formation of gellan gum in the presence of calcium chloride. Reproduced from Valot et al. [168], with permission from the Royal Society of Chemistry, 2019.
The concentration of polymer solution and M-blocks/G-blocks ratio influences physiochemical and mechanical properties of alginate gels. Hydrogels with higher G-content were found to be mechanically stronger. The type of ionic crosslinker also has a significant influence on material printability. Several multivalent cations have been used for crosslinking of alginate system including calcium, barium, zinc, ferric and strontium [39, 40]. Among the tested metal ions, the biological properties of 3D bioprinted constructs were more stable when calcium was used as a crosslinking agent [40]. Therefore, alginate-based bioinks that are crosslinked by using calcium were utilized for 3D bioprinting of various and complex tissues structures (Figure 3). Among the different water soluble calcium salts, such as calcium chloride (CaCl2), calcium sulphate (CaSO4) and calcium carbonate (CaCO3) [31, 39, 41], CaCl2 is the most commonly used ionic crosslinker due to its higher solubility in aqueous media, which results in rapid gelation. However, this rapid gelation may also cause poor stability of the final 3D printed structure. This can take place because of faster deposition of calcium ions outside as compared to inside of the gel [42]. Therefore, the addition of ions to the polymer solution should be performed in a controllable manner. Sun et al. [43] showed that the concentration of ionic crosslinker must be carefully selected during the 3D bioprinting process. They studied the 3D bioprinting of different concentrations of sodium alginate/CaCl2 solutions and they found that the suitable fabrication parameters with high cell viability (83%) are in the range of 1.5% and 0.5% w/v for polymer solution and CaCl2 respectively. It was also found that required amount of crosslinker is also dependent on the molecular weight (MW) of alginate. To study this effect, Kelly et al. [44], studied the printability of alginates having different molecular weights in the presence of different concentration of ionic crosslinkers. The lowest spreading ratio, regardless of the type of crosslinker, for the alginate with high MW (75,000 g/mol) was found to be at 25:9 (alginate to crosslinker). By reducing the ratios, bioink viscosity increased significantly and undesirably affected the printing process (Figure 4). However, a bioink that was made by using low MW Alginate (28,000 g/mol) with crosslinking ratio of 4:3 (Alginate to crosslinker) exhibited the lowest spreading ratio. Furthermore, when CaSO4 was used as a crosslinker, the 3D-bioprinted structure was significantly stiffer than structures produced by using other ionic crosslinker.
Figure 3.
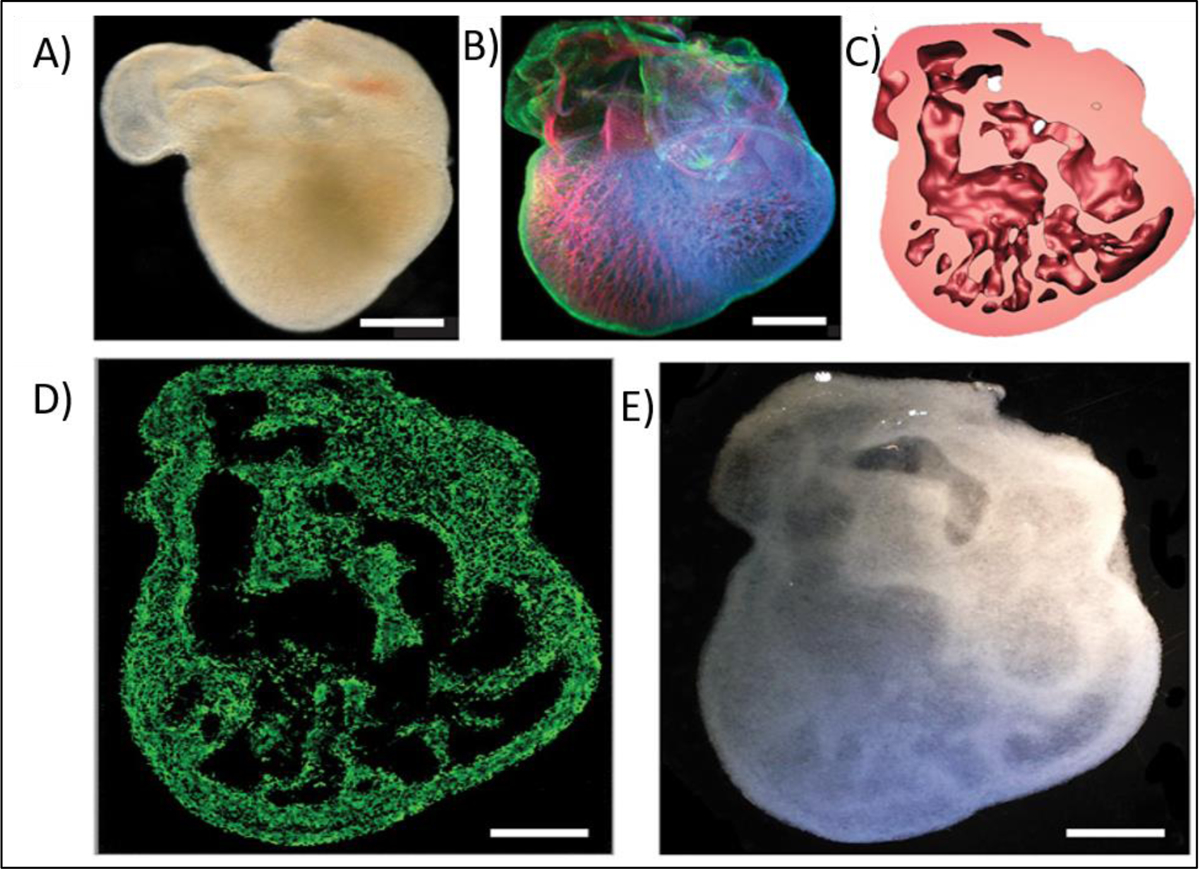
Three-dimensionally (3D) bioprinted of various complex structures based on 3d bioprinting of Alginate crosslinked by CaCl2; A) Explanted embryonic chick heart; B) stained embryonic chick heart after 5 days, C) 3D CAD model of the embryonic heart; D) Florescence microscopy of 3D printed heart; E) Dark field image of 3D printed heart; (scale bars 1 mm). Reproduced from Hinton et al. [169], with permission from American Association of the Advancement of Science, 2015.
Figure 4.
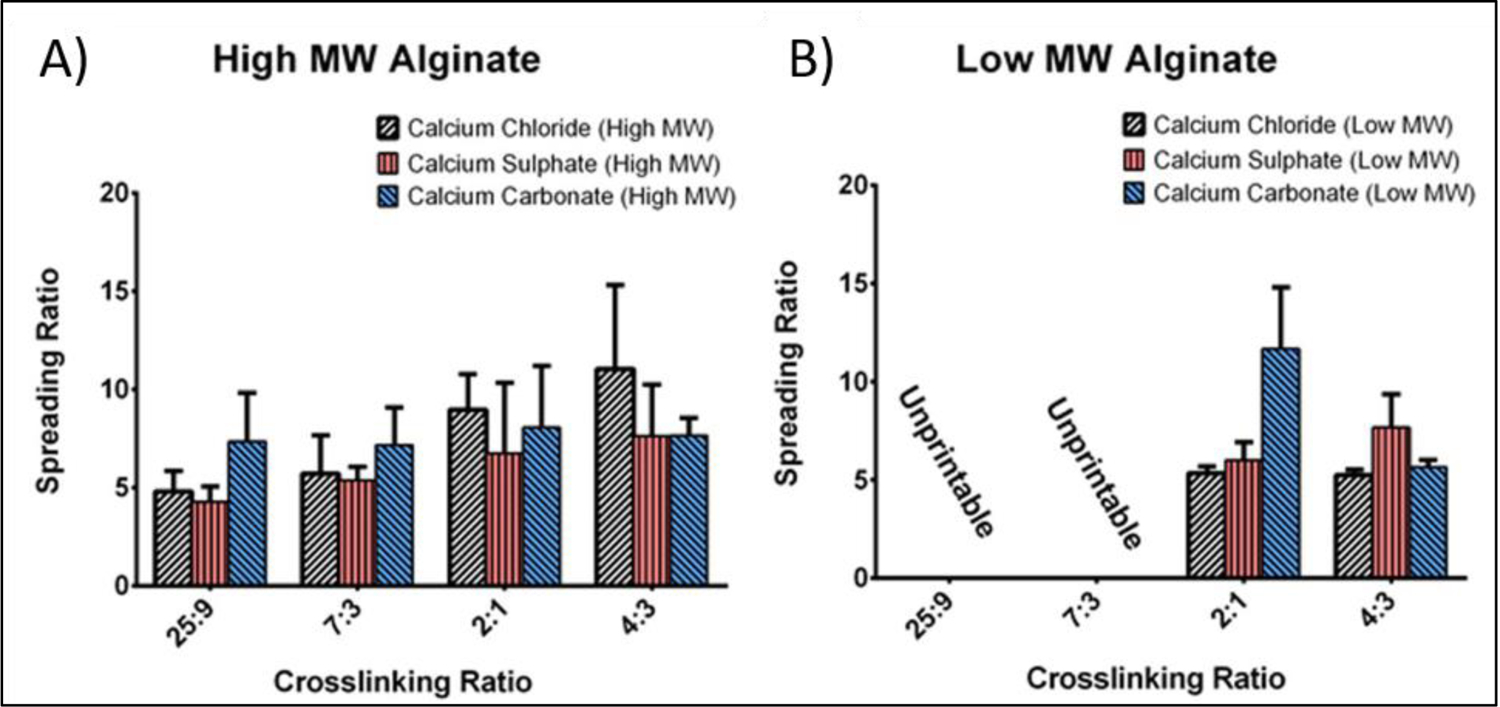
Spreading Ratio of (A) high molecular weight (MW) and (B) low MW sodium alginate with the crosslinking ratios of 25:9, 7:3, 2:1 and 4:3 (polymer to crosslinker) using calcium chloride (CaCl2), calcium sulphate (CaSO4) and calcium carbonate (CaCO3) as the crosslinkers. Reproduced from Freeman et al. [44], with permission from Nature Publishing Group, 2017.
The influence of ionic crosslinkers on cell damage/survival during and after the 3D bioprinting process is another challenging issue. Although 3D bioprinting using ionic crosslinking can avoid cell exposure to harsh chemical or thermal conditions, it can still expose the cells to a non-physiological environment, resulting in reduction in the viability of cells in the 3D bioprinted constructs. Various cell types such as fibroblasts [45], myoblasts [46], endothelial cells [47], chondrocytes [48], and Schwann cells [49] have been embedded in alginate solution and 3D bioprinted. Previous studies showed that the viability and proliferation capacity of cell-containing alginate-based hydrogels vary with changing the concentration of ionic crosslinker and crosslinking time. It was also found that excessive amounts of Ca2+ can be toxic to cells in the 3D bioprinted structure [42]. High concentration of calcium was found to damage cell membrane leading to disturbed state of cell electrolytes [50].
Providing an environment suitable for cell growth is difficult to achieve with the use of pure alginate. This is mainly due to its nonfouling characteristics, which limit cell adhesion and cell metabolism [51]. Another problem with the use of pure alginate hydrogel is the lack of appropriate porosities [52, 53]. To solve these problems, researchers suggested that the blending of non-ionic crosslinkable polymers with alginate may help the formation of more uniform gels having larger pore size, making overall gel properties more favorable for incorporation of living cells [52]. Lode and co-workers [53], compared the 3D bioprinting process of pure alginate (3 wt%) with alginate-methylcellulose (MC) blends (alginate:MC ratio of 1:3) using CaCl2 as a crosslinker. In this design, MC is not crosslinkable with metal ions, hence the MC polymers can be washed out over time from 3D bioprinted construct to obtain a more porous structure (sacrificial ink). SEM images showed that 3D printed pure alginate had a smooth surface with minimal porosity. In contrast, the 3D bioprinted alginate/MC blends had high porosity. The suitability of the alginate/MC blend for embedding mesenchymal stem cells (MSCs) was also evaluated and results showed the presence of high percentage of living cells in the alginate/MC blends.
The way ionic crosslinker is added to bioinks and the exposure time of polymeric solution to metal ions could also have a significant impact on bioink printability and cell viability. There are different methods to add ionic crosslinkers to the ion-crosslinkable polymers such as by direct printing into ionic solutions (Bath-assisted printing), spraying a mist of metal ions on the printing nozzle (Aerosol Spraying) and pre-crosslinking [54]. Bath-assisted printing or direct printing of polymer solution into a bath containing crosslinking agent (e.g. metal ions) allowing rapid gelation that supports the shape stability after the printing process. In this printing technique, it is important to use an optimum concentration of metal ions for the rapid gelation of the bioinks, while the cell viability maintained. The alginate-based bioink can provide excellent cell viability when the calcium concentration of the bath is >100 mM [55]. This technique was found to be able to print complex constructs, but, there are various challenges particularly in the preparation of suspension medium and the required extraction steps that put significant burdens on the manufacturing process [13, 56]. Crosslinking can be also achieved via spraying an aerosolized crosslinking agent on the printing nozzle. Based on this technique, Ahn et al. [57] successfully 3D bioprinted a preosteoblasts (MC3T3-E1) loaded 3.5% (w/w) alginate solution that were crosslinking via spraying 2% (w/w) calcium chloride solution. The cell viability remained 84% after 3D bioprinting process, indicating that the crosslinking method and printing technology did not alter the cell viability. Freeform deposition and repeatability of this technique have been mentioned as an advantage and challenge of this technique respectively [58]. Pre-crosslinking of alginate is also another sterategy that found to be beneficial for cell viability, shape fidelity and structure of the resulting 3D bioprinted products [59]. Pre-crosslinked alginate can be formed by mixing with low concentrations of ionic crosslinker prior to passing them through the nozzle of the printer. In this approach, the hydrogel will usually be fully crosslinked with metal ions after the bioinks are extruded, hence reduce required extrusion forces and increase cell survival during the printing process. Shu et al. [60] reported that mixing of alginate at the concentration of 10% (w/v) with 80 mM CaCl2 at a volume ratio of 1:1 is a suitable concentration for producing partially crosslinked alginate hydrogel. Lower ratios may cause difficulty in printing and negative effect on cell survival. Pre-crosslinking can also be achieved by integrating (blending) thermosensitive polymers into the bioink or changing the ionic strength throughout printing process [61]. Chung et al. [62] compared the 3D bioprinting process of alginates that were either blended or not blended with thermosensitive gelatin, and they used CaCl2 as a crosslinker. They found that alginate-gelatin blend (7.5%–0.75% w/v gelatin-alginate) hydrogel had better mechanical properties than non-blended alginate hydrogel. Hence, blended alginate protected embedded myoblasts from shear forces induced by the printing process. Thermal gelation of gelatin prior to ionic crosslinking of alginate can also provide a long-term stability of the printed constructs. The schematic illustration of combining the reversible thermal crosslinking behavior of gelatin with ionic crosslinking of alginate is shown in Figure 5. Ionic interaction crosslinking is also used for solidifying alginate based blends with other polymers such as hyaluronic acid [49], cellulose [52, 63], polyvinyl alcohol [64], and polycaprolactone [65]. Except alginate containing bioinks, other polysaccharides such as gellan gum are also crosslinkable through metal ions addition while the amount of knowledge on these bioinks are limited. Gellan gum (GG) is a water-soluble and negatively charged polysaccharides that has a tendency to crosslink in presence of cations (K+, Na+, Ca2+, and Mg2+). The mechanical properties of GG based hydrogels are greatly affected by the polymer concentration as well as the ionic content. Pure GG polymeric networks are usually mechanically weak. Hence, various chemical modifications on the GG backbone are made to address the mechanical weakness [66]. For example, Khademhosseini et al. [67] found that methacrylation of GG can improve the magnitude of compressive stress at failure to up to 60 MPa. Ferris et al. [68–70] used peptide modified gellan gum crosslinked using CaCl2 and suggested that this material can provide similar levels of mechanical strength as human soft tissues such as muscle, liver and cartilage, revealing that there is room to further investigate these sugar-based polymers as bioinks.
Figure 5.
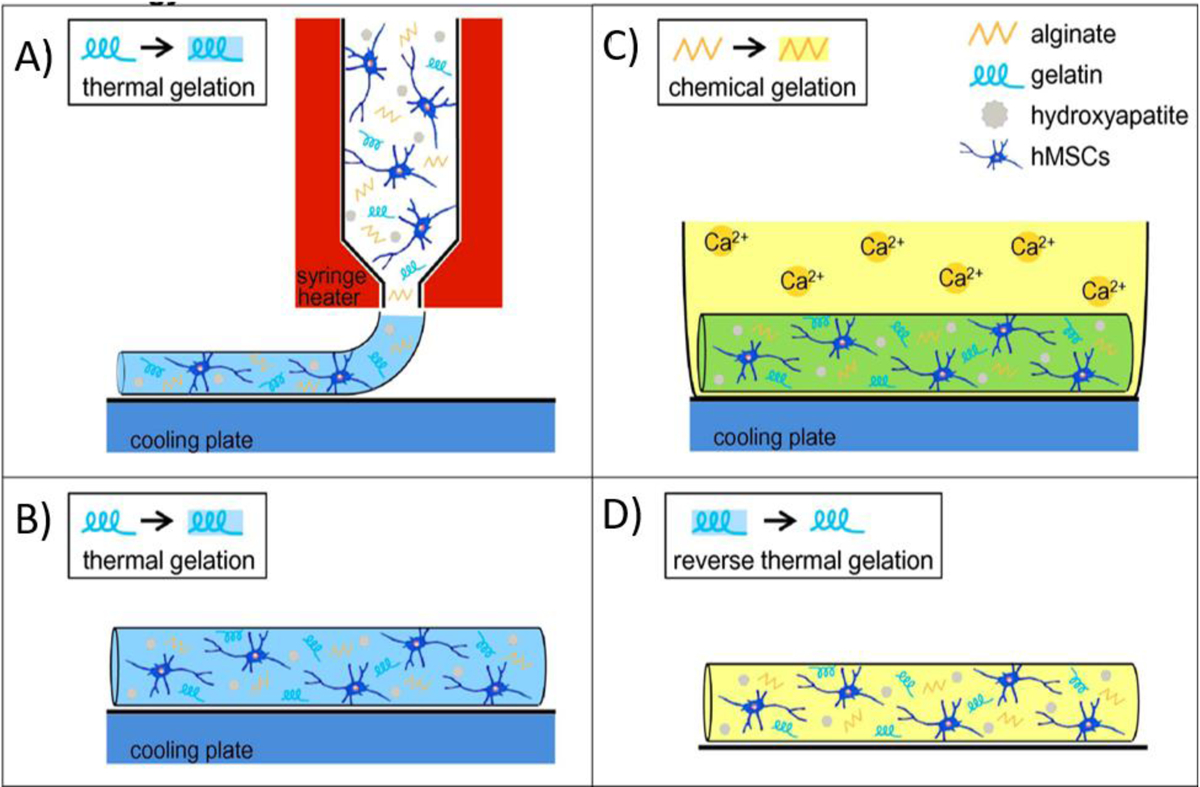
Schematic demonstration of combining thermo-gelation of gelatin (A, B) with ionic crosslinking of alginate (C, D) that improves the long-term stability of the 3d bioprinted construct. Reproduced from Wust et al. [170], with permission from Elsevier, 2014.
- Without the Addition of Metal Ions (Electrostatic)
Ionic crosslinking could also be achieved in the absence of potentially cytotoxic free metal ions and via electrostatic binding of ionic groups that exist in the backbone of polymer chains, which makes this strategy more cell friendly. This strategy has been used for 3D bioprinting of various ionic charge-containing hydrogels (Table 2). In terms of ionic charges, hydrogels can be divided into three groups, anionic hydrogels that contain negatively charged moieties (e.g., alginate, kappa (κ)-carrageenan and xanthan); cationic hydrogels that contain positively charged moieties (e.g., gelatin or chitosan), and neutral/zwitterionic hydrogels that contain equal numbers of positive and negative moieties (e.g., dextran, sulfobetaine and carboxybetaine) [71]. In this crosslinking approach, two opposite charge hydrogels can be printed to yield an electrostatic interaction network without the need for the addition of free metal ions. Electrostatic crosslinking is reversible in nature, which confers desirable characteristics to the extrusion-based 3D bioprinting. Similar to some dynamic covalent and physical bonds, most of the bioinks that are crosslinked via electrostatic interactions have a shear-thinning property (decrease in viscosity with increasing shear rate) during printing process followed by a quick rebuilding of internal structure to form a thixotropic hydrogel after printing. Hence, with the use of this method, hydrogel can be extruded smoothly through the nozzle of the printer with minimal mechanical stresses imposed on the cells. However, mixing of polycationic inks with polyanionic inks could lead to inhomogeneous gelation, because of the possibility of stronger electrostatic interactions that can take place at the interface of the two types of ink. Therefore, the internal crosslinking density of the 3D bioprinted material formed by this crosslinking method is usually low. To solve this problem and for maintaining the structure of 3D printed constructs for a long time, the final constructs are usually exposed to a second crosslinking process. Li et al. [30] used three anionic inks such as alginate, xanthan, and κ-carrageenan (Kca) and three cationic inks such as chitosan, gelatin, and gelatin methacryloyl (GelMA) to investigate the effect of using different charged bioinks on printability and cell viability. It was found that the Kca (2 wt%) and GelMA (10 wt%) hydrogels are the best mixture for 3D bioprinting of desired constructs. In this design, a strong electrostatic interaction formed between negatively charged sulfonic acid group of Kca and positively charged arginine and lysine residues of GelMA. During the printing process, crosslinks between polymer chains were broken by shear stress induced by pressing the bioink through the printer nozzle, which resulted in a decrease in the viscosity of bioinks. However, after printing, the hydrogel could rebuild broken crosslinks. Furthermore, authors showed that high cell viability (>96%) can be obtained, because of shear thinning/thickening behavior of bioinks throughout the printing process.
Table 2.
Summary of three dimensional (3D) bioprinting where electrostatic interaction crosslinking, was employed showing materials used, type of biomolecules and target tissues. Various materials have been used alone or in combination with each other, such as gelatin (gel), gelatin methacryloyl (GelMA), hyaluronic acid (HA), alginate (Alg), chitosan (Ch), methylcellulose (MC), silk fibroin (SF), polyethylene oxide (PEO), polyethylene oxide (PEO), maleic anhydride (MA), κ-carrageenan (Kca) and xanthan (Xa). Cells used included human mesenchymal stem cells (HMSCs), hepatic cells (HCs), murine preosteoblasts (MPs) and myoblasts (MBCs).
| Biomaterial | Crosslinking Method | Printing Technique | Cell Type | Target tissue | Ref. |
|---|---|---|---|---|---|
| PEO/MA | Electrostatic Interaction | extrusion | HCs | Not specified | [202] |
| PEG/Clay | Electrostatic Interaction | extrusion | MPs | Not specified | [75] |
| gel / Kca | Electrostatic Interaction | extrusion piston-driven | MBCs | Not specified | [203] |
| [Alg, Xa, Kca] / [cH, gel, GelMA] | Electrostatic Interaction | extrusion | MMCs | Not specified | [30] |
| GelMA/ Kca /Clay | Electrostatic Interaction/ UV Crosslinking | extrusion | MPs | Not specified | [204] |
| Alg/Ch | Electrostatic Interaction/ Metal ion crosslinking | extrusion | HMSCs | Not specified | [73] |
| HA | Electrostatic Interaction | Laser-assisted bioprinting | HMSCs | Corneal Structures | [205] |
| Peptide/Keratin | Electrostatic Interaction/Hydrophobic | Inkjet bioprinting | Fibroblasts | Not specified | [206] |
| SF/ Clay/ PEG | Electrostatic Interaction | extrusion | HMSCs | Not specified | [76] |
| gel/Ch | Electrostatic Interaction | extrusion | Fibroblasts | Skin | [72] |
| Alg /MC/Clay | Electrostatic Interaction | extrusion | HMSCs | Bone tissue | [207] |
In another study, gelatin–chitosan blend was utilized for the 3D bioprinting of skin [72]. Carboxylate groups gelatin exhibits negative charge when the pH of the medium is above 4.7. Therefore, the positively charged ammonium ions of chitosan could interact with carboxylate groups of gelatin resulting in the formation of an electrostatic crosslinking or vice versa (Figure 6 A, B). This method demonstrated a good spatial control over deposition of bioink at specific regions for skin bioprinting. In the study by Liu et al. [73], 10 wt% chitosan was mixed with 7 wt% alginate to print 3D hybrid constructs, which were then compared with alginate constructs that were crosslinked via CaCl2 (0.1 mol/L). To improve the viscosity of the bioink, chitosan powders were first mixed in an alginate solution, and then alginate-chitosan mixtures were treated with HCl (0.5 mol/L). By adding HCl, the amino group of chitosan was protonated to induce electrostatic crosslinking of the alginate and chitosan. The physicochemical properties and shape fidelity of the 3D printed hydrogels were found to be controllable by changing the chitosan content. The compression strength of 3D printed constructs that contained 7% chitosan and 10% alginate (w/v) was the strongest with the value of 1.5 MPa. Moreover, it was shown that, when alginate was crosslinked with Ca2+, the hydrogels were not stable in vivo due to exchange reactions taking place with monovalent cations that exist naturally in the body (e.g. sodium ions). In contrast, the addition of chitosan to alginate has significantly improved in vivo stability of the printed constructs due to enhanced entanglement between their polymeric chains. Due to the possible toxicity of HCl on cell activity [74], cells (Human adipose-derived stem cells) were seeded to hydrogels after the printing process. It should be mentioned that prior to the cell culture, the hydrogels were freeze-dried and then immersed in the cell culture medium for a day. By using this method, the cells adhered and proliferated to the 3D printed constructs. The formation of electrostatically crosslinked system between charged fillers and charged bioink is another method of enhancing the mechanical properties of 3D bioprinted constructs. Gaharwar et al. [75] used silicate nanoparticles to stabilize electrostatically crosslinked Kca/ GelMA bioink. These disc-shaped nanoparticles (mainly composed of MgO sheets sandwiched between SiO2 sheets) have dual ionic character of opposite signs on the faces (negative) and on the rims (positive) that allow reversible electrostatic associations between GelMA and Kca with faces and edges respectively. This additional electrostatic crosslinking mechanism of particle to polymer interactions resulted in the improvement of shear thinning characteristics of the material during bioprinting, and mechanical properties of the resulting constructs. Kca and GelMA individually showed a shear thinning behavior in response to a steady flow rate. However, with the addition of silicate nanoparticles, the size of the plug flow region was increased. The increase in plug flow region protected the cells from shear stress and thereby, 3D bioprinted preosteoblasts were found to better maintain their viability. In another study, Kaplan and colleagues [76] used silicate nanoparticles (2.5% w/v) for in situ crosslinking of silk fibroin (SF) conjugated polyethylene glycol (PEG) (6% w/v). By changing the pH value of SF solution to below and above isoelectric point (PI=3.8–3.9), SF could interact with the surface of these nanoparticles through an electrostatic interaction or ion-dipole bonding (Figure 6C). The conjugation of PEG could facilitate fibroin crystallization and improve thixotropic property of the final product. It was found that human skeletal muscle myoblasts have high cell viability (90%) after being loaded and printed using this bioink due to the possible positive influence of silicate particles on cellular metabolism.
Figure 6.
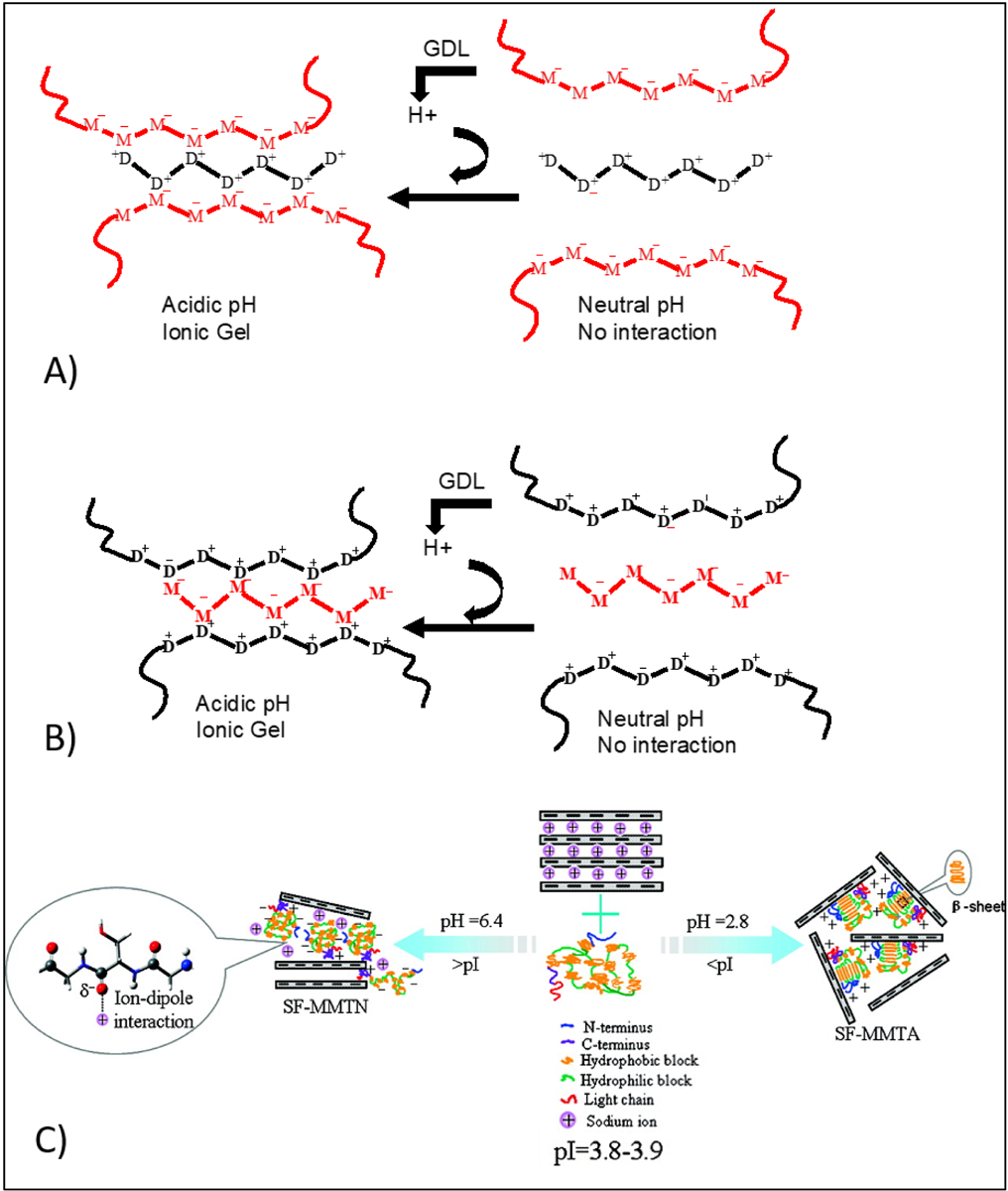
Gelation mechanism between sodium alginate and chitosan at acidic pH; Gelatin and chitosan can be used with various chain length ranging from oligomer to high molecular weight polymeric chains in this method of crosslinking. A) Alginate used as the main polymeric chain and interacted with chitosan oligomers, B) Chitosan used as the main polymeric chain and interacted with alginate oligomers. Reproduced from Khong et al. [171], with permission from American Chemical Society, 2013. C) Schematic illustration of interactions between silk fibroin and clay (MMT) through ion-dipole bonding and electrostatic attraction at above and below the isoelectric point Reproduced from Dang et al. [172], with permission from American Chemical Society, 2010.
2.1.2. Other Non-Covalent Interactions
Hydrogen bonds or H-bonds are intermolecular bonding interaction between the hydrogen atom and an electronegative atom [77]. They play an important role in the formation of various inorganic (e.g. water) and organic (e.g. DNA) molecules. H-bonds can be usually formed between two hydroxyl groups, or carboxyl and amide groups. They have a low binding energy (4 to 60 kJ/mol), but multiple hydrogen bonds are relatively stronger to preserve the polymer network. H-bonds have not been used alone for 3D bioprinting, however, there are some reports on using this strategy in 3D Printing. Wang et al. [78] reported a copolymer of N-acryloyl glycinamide and 1-vinyl-1,2,4- triazole as inks for extrusion-based printing. This system formed a crosslink via the h-bonding between dual amide motifs in the side chain of N-acryloyl glycinamide. The ink was extruded and printed structure maintained sufficient mechanical integrity. Beside H-bonds, polymeric bioinks can also be physically crosslinked via other non-covalent bonds such as hydrophobic interactions [79], π–π stacking [80], dipole-dipole interactions, host–guest recognition and β-sheet mediated crosslinking [16]. In addition to these, self-assembling peptides and peptide–DNA conjugation are other emerging candidates for crosslinking design. The list of these non-covalent bonds that used for 3D bioprinting can be seen at Table 3. Physically crosslinked bioinks are very attractive for use in extrusion 3D bioprinting as they could be extruded under applied shear force with a minimum impact on the cell viability. However, most of the 3D bioprinted constructs employing physical crosslinking methods are mechanically weak and they are prone to fracture. Therefore, this crosslinking strategy is used mainly for 3D bioprinting of super soft structures, trying to mimic tissues such as those of the brain or lung. Tan et al. [81] reported that super soft tissue-phantoms can be cryogenically 3D bioprinted using physical crosslinking method. They used a bioink containing phytagel and poly (vinyl alcohol) (PVA), which is crosslinked via hydrogen bonds after a freeze-thaw cycle. This freeze/thaw cycle induces the hydrogen bonds formation between the hydroxyl groups of phytagel and PVA and causes rapid gelation. Moreover, they showed that human fibroblasts had a good viability of 80% after the crosslinking process. However, it should be noted that the freeze/thaw induce crosslinking method cannot be applied to all cell types as each type of cell has its own protocol for freezing and some cannot yet be preserved by freezing [82].
Table 3.
Summary of three dimensional (3D) bioprinting where non-covalent crosslinking other than electrostatic interaction, was employed showing materials used, type of biomolecules and targeted tissues. Various biomaterials have been used alone or in combination, such as alginate (Alg), prolinated alginate (P-Alg), polycaprolactone (PCL), glycinamide (Gly), Silica nanoparticle (SN), silk fibroin (SF), polyethylene oxide (PEO), polyvinyl alcohol (PVA), polyethylene glycol (PEG). Cells used included human mesenchymal stem cells (HMSCs), human induced pluripotent stem cells (HIPSCs), chondrocytes cells (CCs) murine preosteoblasts (MPs), and epidermal stem cells (ESCs).
| Biomaterial | Crosslinking Method | Printing Technique | Cell Type | Targeted tissue | Ref. |
|---|---|---|---|---|---|
| Gly/Clay | Hydrogen bonding | extrusion | MPs | Bone Regeneration | [27] |
| PVA/ Phytagel | Hydrogen bonding | extrusion | ESCs | Soft Tissue Phantoms | [81] |
| P-Alg | Hydrogen binding + Ionic-Cacl2 | extrusion | HIPSCs | Not specified | [208] |
| SN-PCL | Hydrogen binding + π–π stacking | Inkjet bioprinting | CCs | Not specified | [209] |
| PCL-Pyrimidinone | Hydrogen binding + UV crosslinking | stereolithography | Not specified | Not specified | [210] |
| SF/PEG | Hydrophobic interactions | extrusion | HMSCs | Not specified | [29] |
| Polypeptide/DNA | DNA hybridization | extrusion | HIPSCs | Not specified | [211] |
| Polypeptide/DNA | DNA hybridization | extrusion | HIPSCs | Not specified | [83] |
DNA-based materials can also be physically crosslinked to form hydrogels through peptide–DNA conjugation and hybridization. DNA hybridization is a technique in which DNA solution is stressed by temperature or other stimuli, breaking the weak hydrogen bonds between matching bases in the DNA strands. When the solution is relaxed, complimentary sequences can bond back together again, and the double-helix form will reappear. The first study on 3D bioprinting of DNA based bioinks was performed by Li et al. [83] who found that mixing of polypeptide containing bioink (5 wt%) and double-stranded DNA containing bioink (5 wt%) leads to rapid gelation due to the hybridization of DNA (Figure 7). Using this method of crosslinking, hydrogels were formed under physiological conditions with cell viability of more than 98%. Host-guest interaction is another type of supramolecular binding that used for crosslinking. Host-guest interactions represent specific non-covalent interactions that are based on selective inclusion complexation between macrocyclic hosts, such as cucurbit[n]urils (CB), cyclodextrins (CD), crown ethers, and smaller guest molecules such alcohols, acids, amines, amino acids or less polar molecules such as alkyls, cycloalkanes, aromatic molecules. Host-guest interactions is used in combination photocrosslinking for 3D bioprinting of methacrylated hyaluronic acid (M-HA) functionalized with β-cyclodextrin and adamantine to fabricate a fibroblast laden grid-like construct [84]. In this dual crosslinking design, M-HA is photocrosslinkable and β-cyclodextrin (host) can bind to adamantine (guest) via host-guest assembly (Figure 8). By using either photocrosslinking or host-guest assembly the final structure was found to be unstable over time. However, combining these two crosslinking methods provided a more cell-friendly environment in which loaded fibroblasts demonstrated high viability for more than 30 days in culture. Silk based bioink has also been physically crosslinked for 3D bioprinting, by using pH or sonication induced formation of β-sheet crystallization [85]. However, β-sheet mediated crosslinking, like many other types of non-covalent crosslinking methods, still requires to have additional crosslinking to improve the mechanical properties of the resulting structures.
Figure 7.
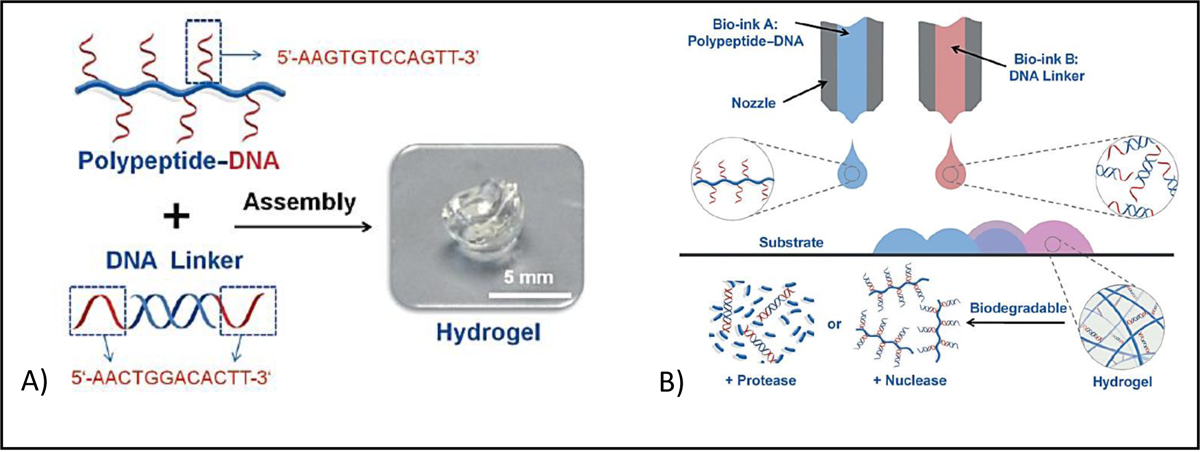
A) Gel formation of polypeptide–DNA upon mixing of polymer solution (bioink A -blue) with DNA linker (bioink B - red). B) Hybridization mediated crosslinking that used for 3D bioprinting. Reproduced from [83], with permission from Wiley, 2015
Figure 8.
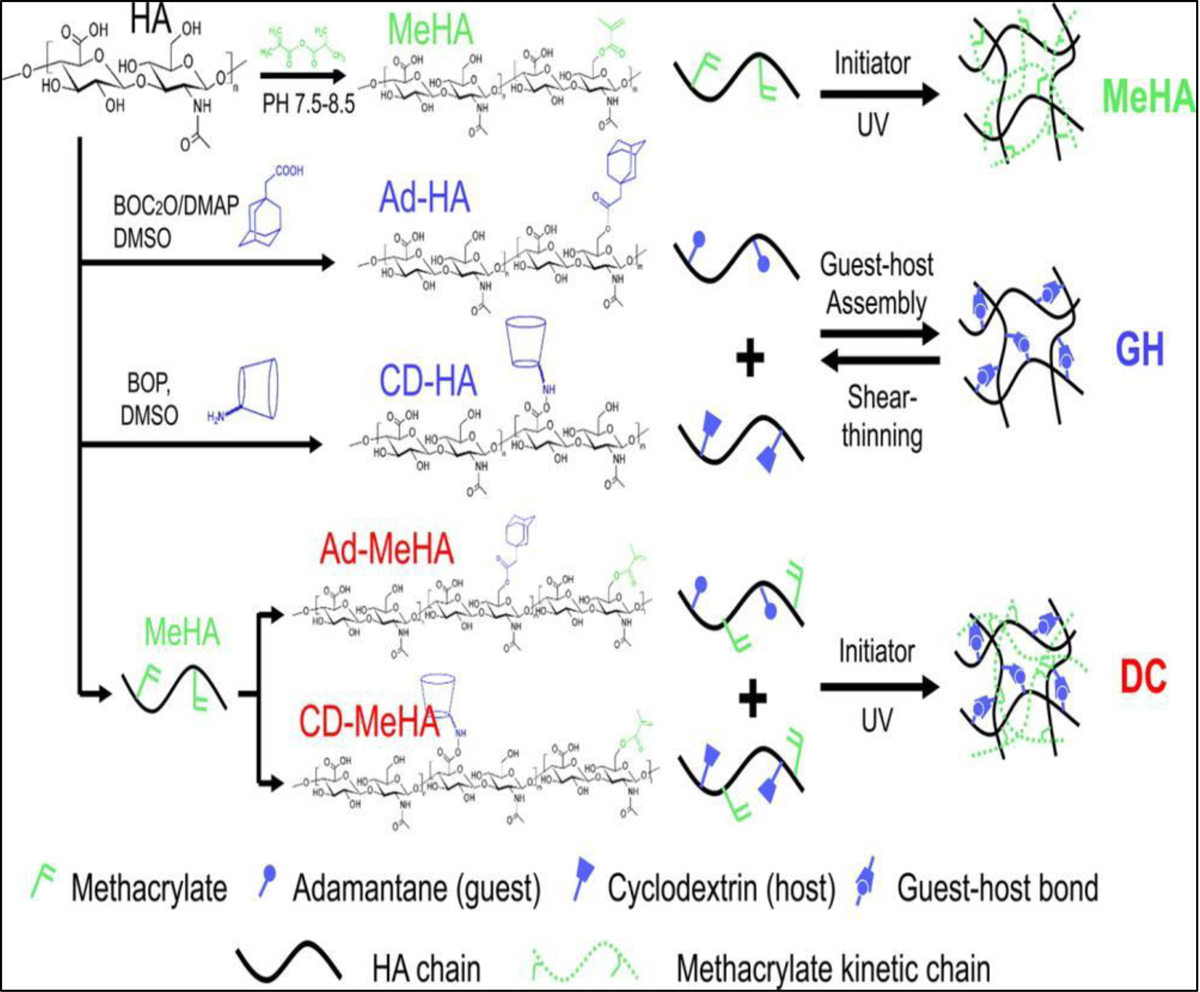
Dual crosslinking mechanism formed by ultraviolet (UV) crosslinking of methacrylated hyaluronic acid and hose-guest assembly of Cyclodextrin and Adamantane functional groups. Reproduced from [84], with permission from American Chemical Society, 2016.
2.2. Chemical crosslinking methods
2.2.1. Photocrosslinking
Photocrosslinking has special importance in 3D printing applications, as many 3D printing industries have been using this crosslinking method to fabricate their products (Table 4). This is because of its facile operation, spatiotemporal, and remote control. Photocrosslinking is a cost-effective method as it can be performed under room temperature, and consumption of energy is comparatively less in comparison with other techniques. In addition to extrusion, there are a several 3D bioprinting techniques such as stereolithography [86], digital light processing [15], laser‐assisted methods [87], and volumetric bioprinting [88] have been employed for converting photocurable bioinks to 3D architectures. The principle of all these methods is based on using photocurable bioinks (in the presence of photoinitiators) that can be crosslinked through either Chain-growth, Step-growth mechanisms or Redox based reactions.
Table 4.
Summary of three dimensional (3D) bioprinting where photocrosslinking was employed showing materials used, type of biomolecules and target tissues. Various biomaterials have been used alone or in combination, such as collagen (Col), gelatin (gel), gelatin methacryloyl (GelMA), dopamine functionalized gelatin methacryloyl (D-GelMA) fibrinogen (Fib), extracellular matrix (ECM), hyaluronic acid (HA), methacrylated-HA (M-HA), alginate (Alg), silk fibroin (SF), cellulose nanofibrils (CNF), chitosan (Ch), gellan (G), gellan gum (GG), methylcellulose (MC), soybean oil epoxidized acrylate (SOEA), galactoglucomannan methacrylates (GGM), polycaprolactone (PCL), polylactide (PLA), methacrylated κ-carrageenan (M-kca), poly(ethylene glycol) (PEG), PEG dimethacrylate (PEGDA), poly (vinyl alcohol) (PVA), polyethylene oxide (PEO), Pluronic F127 (Plu), biosilica (BS), hydroxyapatite (HAp) and carbon nanotubes (CNTs). Cells used included human mesenchymal stem cells (HMSCs), bone marrow derived stem cells (BMSCs), adipose tissue derived mesenchymal stem cells (ADMSCs), epidermal stem cells (ESCs), human induced pluripotent stem cells (HIPSCs), human umbilical vein endothelial cells (HUVECs), human coronary artery endothelial cells (HCAECs), porcine aortic valve interstitial cells (VICs), human aortic root smooth muscle cells (SMCs), myoblasts (MBCs), osteosarcoma cells (OCs), cartilage progenitor cells (CPCs), human articular chondrocytes (HACs), periodontal ligament cells (PLCs), hepatic cellss (HCs), neural progenitor cells (NPCs), pancreatic endothelial cells (PECs), human kidney cells (HKCs) and breast cancer cells (BCCs).
| Biomaterial | Crosslinking Method | Printing Technique | Cell Type | Target tissue | Ref. |
|---|---|---|---|---|---|
| GelMA | UV crosslinking/(Irgacure 2959) | extrusion piston-driven | Fibroblasts | Not specified | [96] |
| GelMA | UV crosslinking/(Irgacure 2959) | extrusion | HCs | Liver Tissue | [91] |
| GelMA | UV crosslinking/(Irgacure 2959) | extrusion | HMSCs | Not specified | [99] |
| GelMA | UV crosslinking/(Irgacure 2959) | extrusion | HMSCs | Trophoblast Migration in Preeclampsia | [212] |
| GelMA | UV crosslinking/(Irgacure 2959) | extrusion | HCs | Not specified | [212] |
| GelMA | UV crosslinking/(Irgacure 2959) | extrusion | CPCs | Cartilage regeneration | [213] |
| GelMA | UV crosslinking/(LAP) | extrusion | HMSCs | 3D in vitro models | [214] |
| GelMA | UV crosslinking/(eosin Y) | extrusion | HIPSCs | Human Cardiac Tissues | [215] |
| GelMA | UV crosslinking/(Irgacure 2959) | extrusion | OCs | Not specified | [216] |
| GelMa | UV crosslinking/(LAP) | Stereolithography (microscale continuous optical bioprinting) | HUVECs | Vascular tissue | [98] |
| GelMA | UV crosslinking/(Irgacure 2959) | extrusion | BMSCs | Not specified | [217] |
| GelMA/PLA | UV crosslinking/(Irgacure 2959) | extrusion | HUVECs | Bonetissue | [92] |
| GelMA/PEGDA | UV crosslinking/(LAP) | extrusion | HUVECs | Not specified | [218] |
| GelMA/PEGDA | UV crosslinking(Irgacure 2959) | Stereolithography | HMSCs | Cartilage tissue | [114] |
| GelMA/Silicate | UV crosslinking/(Irgacure 2959) | extrusion | HUVECs/ HMSCs | Bone tissue | [93] |
| GelMA/Alg | UV crosslinking/(Irgacure 2959) + Ionic-Cacl2 | extrusion | HUVECs | Heart tissue | [36, 113] |
| GelMA/Alg/PEGDA | UV crosslinking/(Irgacure 2959) + Ionic-Cacl2 | extrusion | HUVECs/HMSCs | Perfusable vascular constructs | [219] |
| GelMA/PEGDA | Photo/Lap | Stereolithography (microscale continuous projection printing) | NPCs | Spinal cord | [220] |
| GelMA/M-HA | UV crosslinking/(LAP) | extrusion | VICs | Human HeartValve | [115] |
| GelMA/M-HA | UV crosslinking/(Irgacure 2959) + Ionic-Cacl2 | extrusion | BMSCs | Neocartilage formation | [221] |
| GelMA/M-HA | UV crosslinking/(Irgacure 2959) | Stereolithography | CPCs | Adiposetissue | [222] |
| GelMA/Plu | UV crosslinking/(Irgacure 2959) + Cooling | extrusion | ESCs | Skin tissue | [117] |
| GelMA/Plu | UV crosslinking/(Irgacure 2959) | extrusion | HMSCs | Bone tissue engineering | [223] |
| GelMA/SF | Visible-light crosslinking/(Eosin Y) | extrusion | Fibroblasts | Not specified | [110] |
| GelMA/HAp | UV crosslinking/(Irgacure 2959) | extrusion piston-driven | BMCs | Breast cancer Metastasis | [120] |
| GelMA/G | UV crosslinking/(Irgacure 2959) | extrusion | HMSCs | Cartilage tissue | [224] |
| GelMA/Col | UV crosslinking/(Irgacure 2959) | extrusion | HUVECs | Promotion of angiogenesis | [118] |
| GelMA/Alg | UV crosslinking/(Irgacure 2959) +Ionic-Cacl2 | extrusion | HMSCs | Cartilage tissue | [225] |
| GelMA/Alg | UV crosslinking/(LAP) | extrusion | PECs | Liver tissue | [226] |
| GelMA/Alg | UV crosslinking/(Irgacure 2959) | extrusion | OCs | Bone tissue | [227] |
| Gelatin/HA/ECM | UV crosslinking/(Irgacure 2959) | extrusion | HKCs | Kidney tissue | [125] |
| GelMA/Alg/PEGMA | UV crosslinking/(Irgacure 2959) + Ionic-Cacl2 | extrusion | HMSCs | Bone tissue | [228] |
| GelMA/PEGDA | UV crosslinking/(Irgacure 2959) | extrusion | PLCs | Ligament | [229] |
| GelMA/PEGDA | UV crosslinking/(LAP) | extrusion | Fibroblasts | Not specified | [230] |
| GelMA/Gold nanorod | UV crosslinking/(Irgacure 2959) | extrusion | HCAECs | Cardiac tissue | [90] |
| GelMA/PEGDA | UV crosslinking/(Irgacure 2959) | extrusion | PLCs | Repair ofAlveolar Bone Defect | [231] |
| GelMA/CNT | UV crosslinking/(Irgacure 2959) | extrusion | HUVECs | Myocardial tissue | [119] |
| Dopamine-GelMA | UV crosslinking/(Irgacure 2959) | Stereolithography | Neural stem cells | Nerve tissue | [232] |
| GelMa/M-HA | UV crosslinking/(LAP) | extrusion | ADMSCs | Cartilage | [111] |
| PEGDA | Visible-light crosslinking/(VA-086) | Stereolithography | BCCs | Not specified | [102] |
| PEG/PCL | Visible-light crosslinking/(LAP) | extrusion | SMCs | Vascular tissue | [25] |
| PEGDA /Clay | UV crosslinking/(Irgacure 1173) | extrusion | OCs | Not specified | [100] |
| SOEA | UV crosslinking/(Irgacure 819) | extrusion | HMSCs | Not specified | [101] |
| M-kca | UV crosslinking/(Irgacure 2959) + Ionic-KCl/ | extrusion | HMSCs | Not specified | [233] |
| PEGDA | UV crosslinking/(Irgacure 2959) | extrusion | HACs | Cartilage tissue | [234] |
| PEGDA | UV crosslinking/(Irgacure 2959) | Inkjet bioprinting | HACs | Cartilage tissue | [235] |
| HA /dextran | UV crosslinking/(Irgacure 2959) | piston-driven | HACs | Not specified | [236] |
| PEGDA/HAp | UV crosslinking/(Irgacure 2959) | extrusion | HMSCs | Bone tissue | [237] |
| M-HA/A-Plu | UV crosslinking/(LAP) | extrusion | HACs | Not specified | [238] |
| M-Alg/M-HA | Visible-light crosslinking/(Eosin Y) | extrusion | BMCs | Not specified | [239] |
| M-SF | UV crosslinking/(LAP) | Stereolithography | Fibroblasts | Cartilage tissue | [240] |
| SF/gel | Visible-light crosslinking/Ruthenium | extrusion | HACs | Not specified | [143] |
| GGM /CNF | UV crosslinking/(Irgacure 2959) | extrusion | PECs | Liver tissue | [123] |
| M-HA | UV crosslinking/(Irgacure 2959) | extrusion | Fibroblasts | Not specified | [89] |
| M-HA | UV crosslinking/(Irgacure 2959) | extrusion | HMSCs | Not specified | [126] |
| Tyramine-HA | Visible-light crosslinking/Rose bengal | Laser-Assisted bioprinting | HMSCs | Not specified | [130] |
| M-HA/HAp | UV crosslinking/(Irgacure 2959) | extrusion | ADMSCs | Multizonal grafting | [241] |
| PEGDA/Fib/Alg | UV crosslinking/(Irgacure 2959) + Ionic-CaCl2 | extrusion | MBCs | Skeletal Muscle | [242] |
| PEGDA/Alg/clay | UV crosslinking/(Irgacure 2959) | extrusion | Fibroblasts | Not specified | [243] |
| PLA /GG- PEGDA | UV crosslinking/(Irgacure 2959) | extrusion | BMCs | Intervertebral disc (IVD) repair | [24] |
| A-PG/ T-HA | Thiol-ene coupling | extrusion | HMSCs | Cartilage | [128] |
| A-gel/ T-gel | Thiol-ene coupling | extrusion | CCs | Not specified | [22] |
| E-Alg | Thiol-ene coupling + Ionic-CaCl2 | extrusion | Fibroblasts | Not specified | [129] |
- Chain-Growth Crosslinking
The chain-growth polymerization such as free-radical polymerizations of (Meth)acrylate-based monomers is the most frequently used method in 3D bioprinting of photocrosslinkable constructs. In this crosslinking process, photo-radiation produces free radicals by dissociating photoinitiators that exist in the bioink. Produced radicals react with functional groups of the polymers and bind them together to create 3D network structures. This crosslinking strategy involves mostly the formation of irreversible bonds between two polymer chains. Therefore, for extrusion based 3D bioprinting, it would be better to crosslink these bioinks at the nozzle outlet (in situ) or immediately after extrusion rather than pre-crosslinking (Figure 9) [89].
Figure 9.
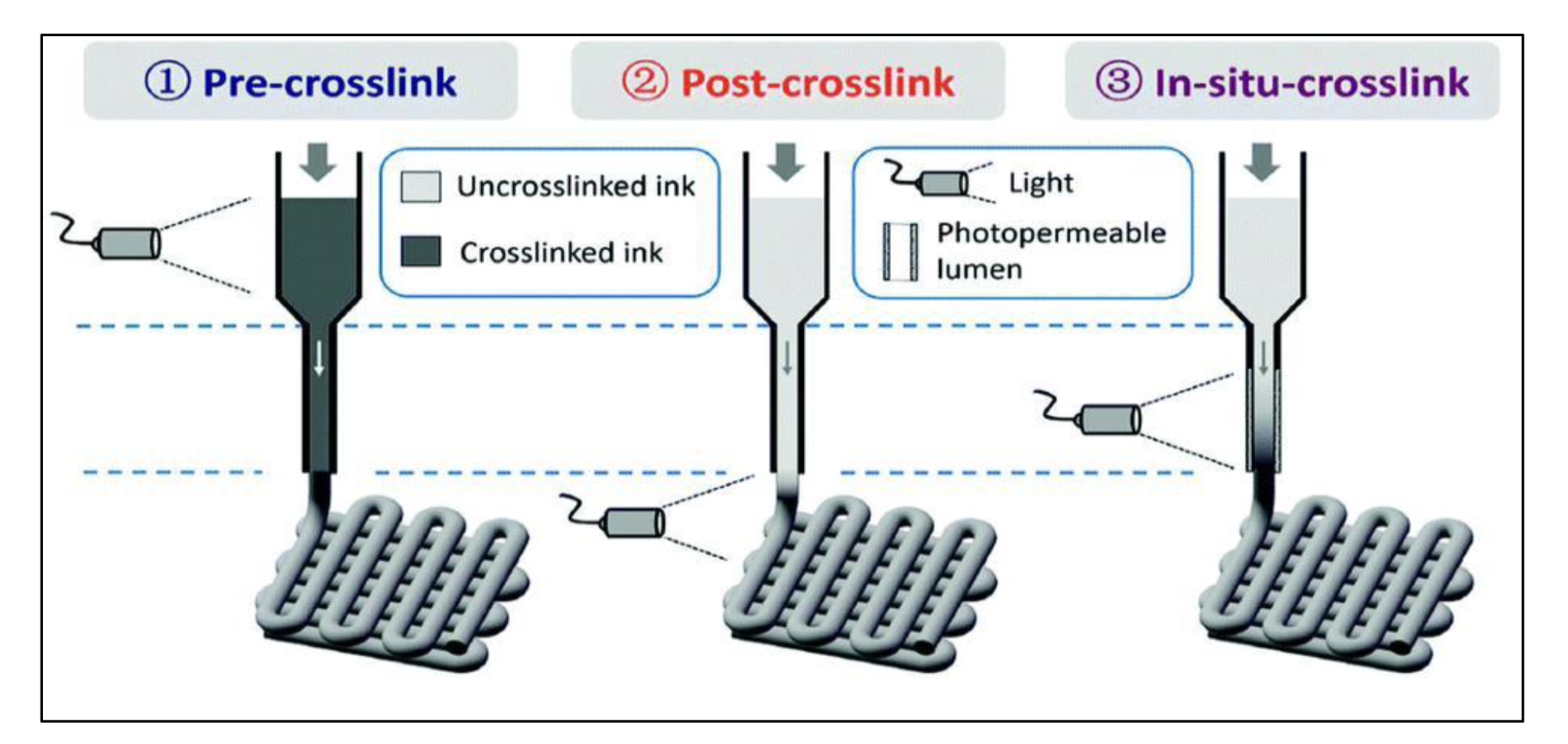
Pre-crosslinking, post-crosslinking and in situ crosslinking methods that used for three dimensional (3D) bioprinting of photo-crosslinkable bioinks. Reproduced from Ouyang et al. [89], with permission from Wiley, 2017.
Photocrosslinking takes place by light radiation either in the form of ultraviolet (UV), laser or visible light. Among the various lights, UV (320−365 nm) is the most commonly used one. However, despite the widespread use of UV light-based crosslinking in 3D bioprinting, UV light has potential biological risks. The use of UV light for photocrosslinking may damage cells in the printed constructs and can potentially be harmful to the operators. In this regard, the use of visible light is considered advantageous because the wavelengths of visible range are not harmful to cells. In photocrosslinking, all the crosslinking parameters should be optimized for the intended 3D bioprinted structures. Such parameters include light exposure time, intensity, and photoinitiator type and concentration that can influence the stiffness and cellular behaviors of the 3D bioprinted constructs. GelMA is one of the most commonly used photocrosslinkable biomaterials in 3D bioprinting and our group has pioneered the use of GelMA for 3D bioprinting of various tissues such as heart [36, 90], liver [91], bone [92–94] and blood vessels [95] (Figure 10). GelMA is an inexpensive protein-based polymer which can be synthesized by reacting acid or alkaline treated gelatin with methacrylic anhydride. GelMA is capable of being crosslinked in the presence of a water soluble photoinitiator and exposure to light (Figure 11). The methacrylamide and methacrylate side groups on GelMA chains form covalent bonds after the generation of free radicals by photoinitiator, to produce a network of gelatin chains bound by short polymethacryloyl chains. Mechanical properties and cell viability in GelMA based bioinks are highly depended on its degree of methacrylation and its concentration. The proportion of the methacryloyl substituent groups in the GelMA, can affect the crosslinking density of the 3D-bioprinted constructs. The crosslink density will decrease with the reduction in the amount of pendant methacrylate groups. Lower crosslink density leads to relatively greater swellability, larger pore size in forming GelMA hydrogels and provide a more suitable environment for incorporating biomolecules. However, bioinks with low-crosslinking density suffer from poor processability. Therefore, printability and biological functionality need to be balanced with choosing proper polymer concentration and degree of methacryloyl modification in GelMA-based constructs. Khademhosseini et al. showed that GelMA hydrogels can be successfully 3D bioprinted at concentrations ranging from 7 to 15% [96]. The UV exposure time for crosslinking was found to affect the elastic modulus, printability and cell viability. Among tested concentrations, 15% GelMA hydrogels had the highest elastic modulus (20 kPa) and 10% GelMA had the best fibroblast cell viability (75%). However, this concentration range can be changed in the presence of viscosity enhancers such as gellan gum. The effect of polymer concentration on printability of GelMA containing gellan gum bioink was studied by Zhuang et al. [97] and it was found that the concentration of GelMA in the bioink can be reduced to 5% by blending with 0.5% gellan gum without negative effect on printability (Figure 12).
Figure 10.
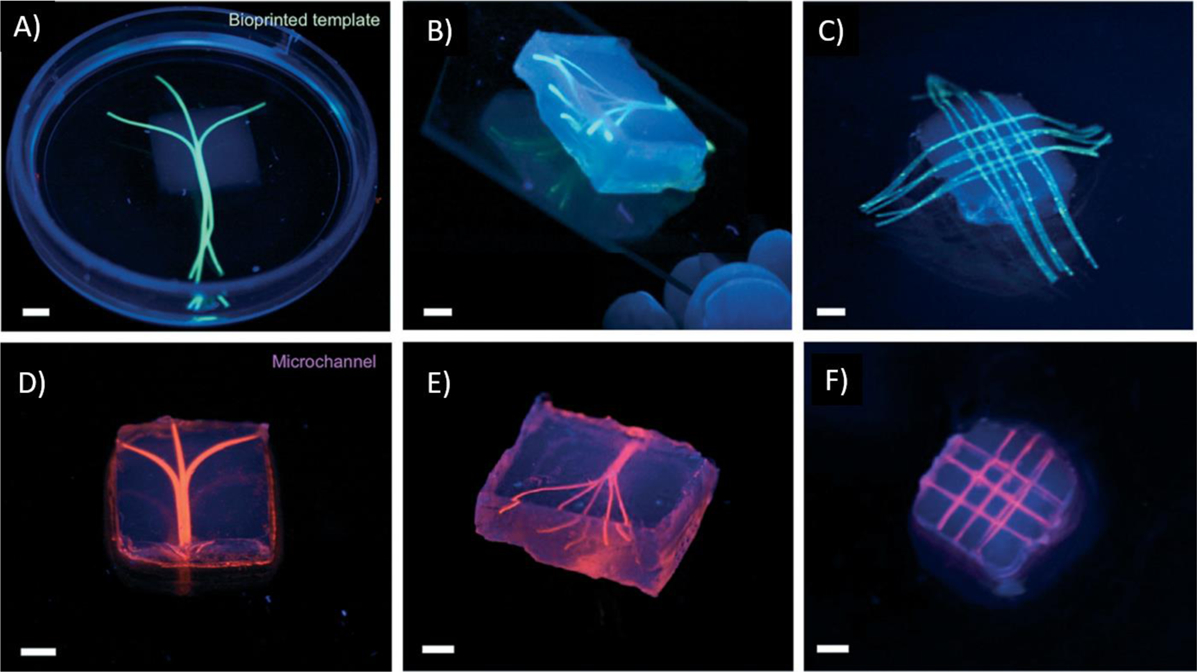
The 3D bioprinted vascular channels formed by gelatin methacryloyl (GelMA) based bioink; A, B, C) before and D, E, F) after perfusion of fluorescent suspension (pink), (scale bars 3 mm), Reproduced from Bertassoni et al. [173], with permission from Royal Society of Chemistry, 2014.
Figure 11.
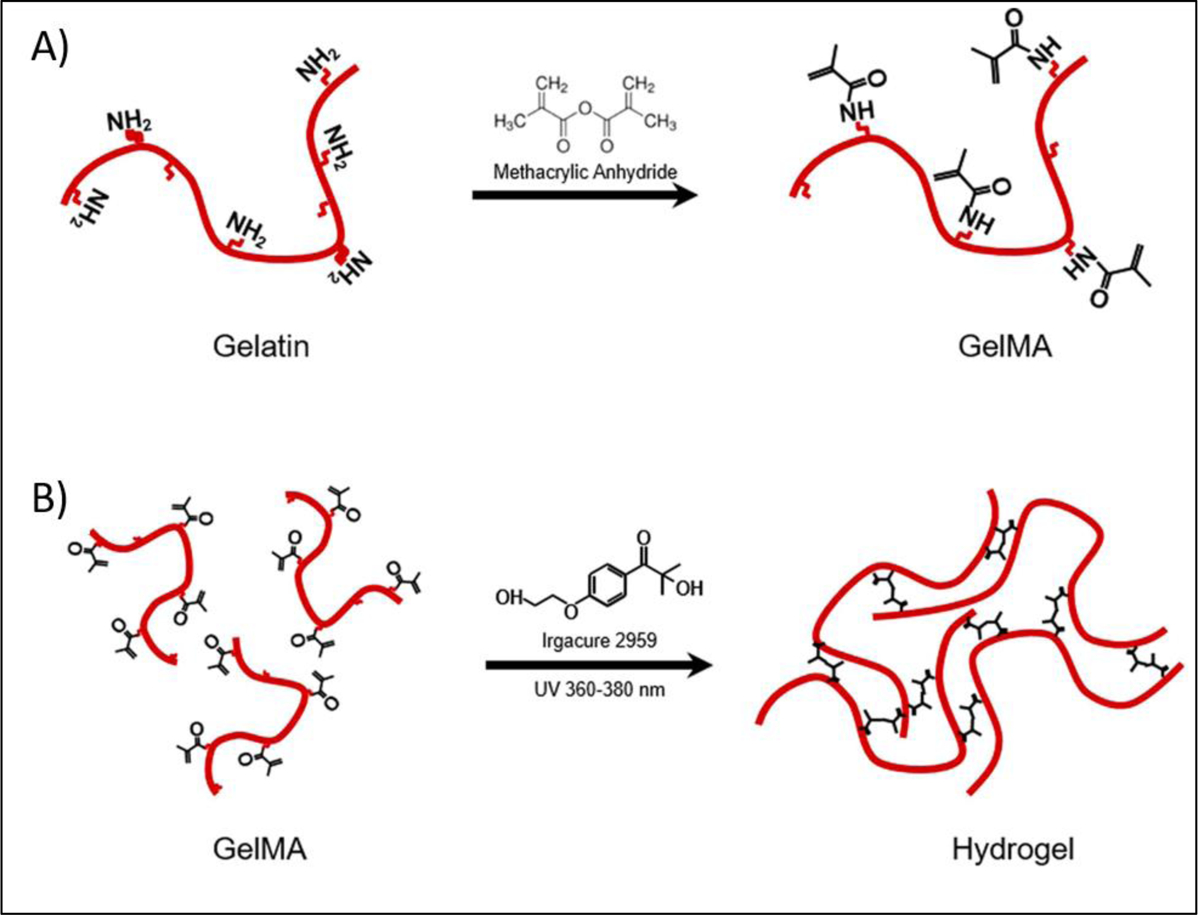
A) Synthesis of gelatin methacryloyl (GelMA) by reaction between gelatin with methacrylate anhydride at 50°C and B) GelMA network formation with free radicals generated by light irradiation in the presence of an initiator. Reproduced from Yoon et al. [174], with permission from Plos One, 2016.
Figure 12.
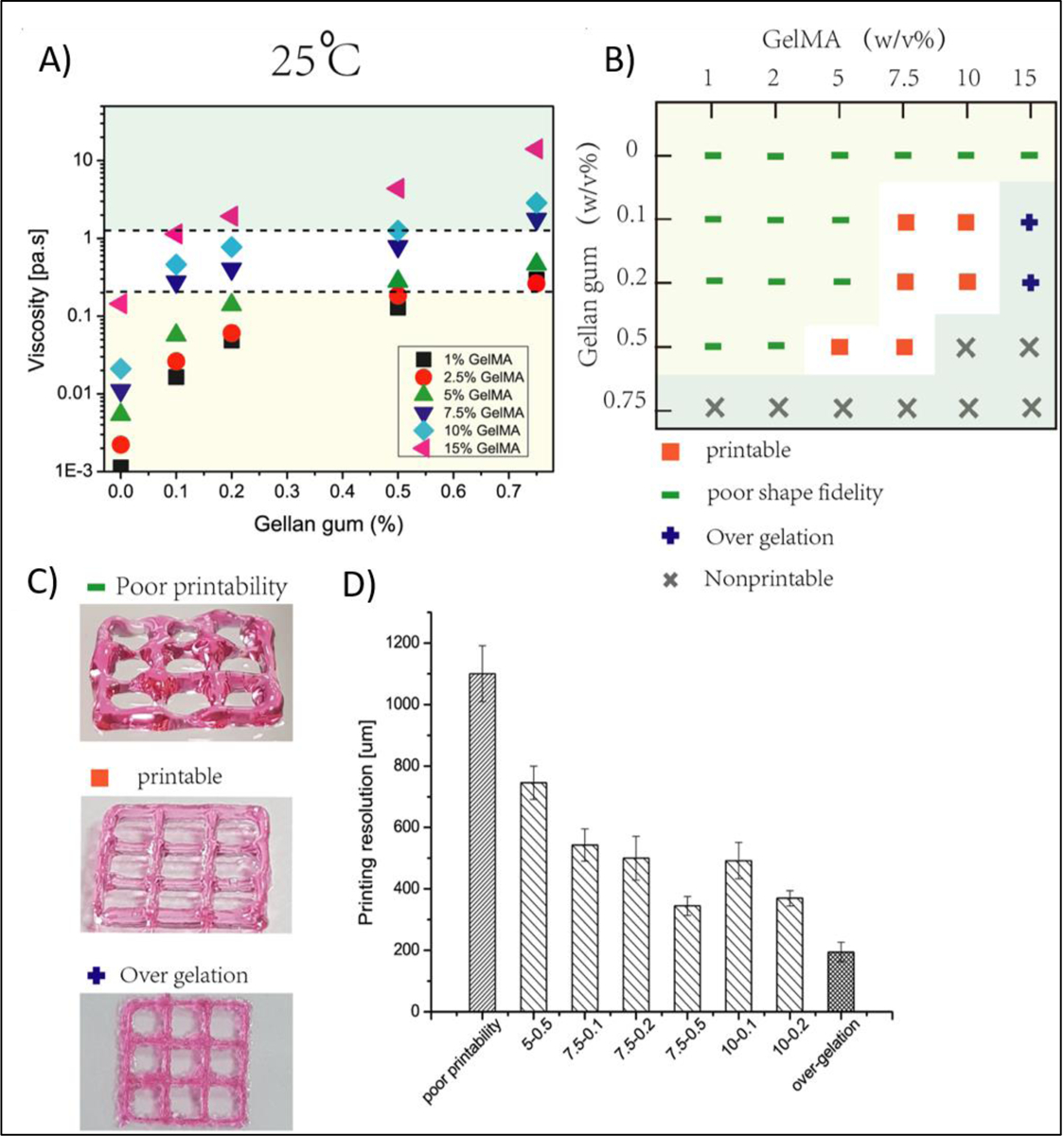
A) The effect of polymer concentration on viscosity of gelatin methacryloyl (GelMA)-gellan gum based bioinks; B) The concentration windows of GelMA and gellan gum for 3D bioprinting application and C) Photographs of printed structures and their printability resolution table. Reproduced from Zhauang et al.[97], Plos One, 2019.
Various photoinitiators such as lithium-acyl phosphinate (LAP) [98], Irgacure 2959 [99], Irgacure 1173 [100], Irgacure 819 [101], VA086 [102], camphorquinone [103], fluorescein [104], riboflavin [105], ruthenium (Ru)/sodium persulfate (SPS) [106–108], Rose Bengal [109], and eosin Y [110], have been used for photocrosslinking of bioinks (Figure 13). Duchi et al. [111] studied the crosslinking capacity of three different photoinitiators including LAP, Irgacure 2959 and VA086 at the constant concentration of 0.1% w/v. LAP was found to provide more adequate cell (Adipose-derived mesenchymal stem/stromal cell) viability (90%) in the final 3D bioprinted constructs. They have found that reduction in photoinitiator concentration and light intensity can further enhance cell viability. Similarly, Fairbanks et al. [112] demonstrated high levels of fibroblast cell viability (96%) when using LAP with low light intensity of 10 mW/cm2. However, it should be noted that when the concentration of initiator is decreased, time required for crosslinking needs to be increased, which is not desirable. The use of visible light for crosslinking is a better alternative to the use of UV, and it would provide a safer environment for operator and for cells. Among the photoinitiators, Eosin Y is a visible light initiator which has been mostly used in combination with other co-photoinitiators such as triethanolamine (TEA) and 1-vinyl-2-pyrrolidinone (VP). Eosin Y is a xanthene dye, and it can trigger the photocrosslinking reaction under visible light exposure (395–405 nm) and maintain high cell viability during 3D bioprinting process. To find the optimal concentration of this visible light reactive photoinitiator, Hyun et al. [110] tested different concentrations of eosin Y from 0–3 (mM), and found that fibroblasts encapsulated in GelMA hydrogel had more than 90% viability when the concentration of the photoinitiator was less than 1 mM. To enhance the processability and crosslinking of GelMA-based bioinks, GelMA was also blended with various polymers such as alginate [113], polylactide (PLA) [92], poly (ethylene glycol) diacrylate (PEGDA) [114], methacrylated HA [115], polyisocyanide [116], pluronic [117], collagen [118] and with various fillers such as gold nanorods (GNR) [90], carbon nanotubes (CNT) [119], silicate nanoplatelets [93], SF particles [110] and nanocrystalline hydroxyapatite [120]. Recently, the use of ruthenium based (ruthenium/sodium persulfate) visible-light initiator system has also gained a lot of attention in the 3D bioprinting of hydrogels [22, 106, 107, 121]. In this method, visible light would oxidize Ru2+ to Ru3+ in presence of a co-initiator such as persulfate salt. Then, co-initiator dissociates to form the persulfate anion and radicals that are able to initiate the polymerization. Woodfield et al. [122] 3D bioprinted a hydrogel made of 10 wt % GelMA and 0.6 wt % collagen in presence of various concentrations of Ru/SPS (0.2/2, 0.5/5, and 1/10 mM/mM) and different intensities of visible light (3, 30, 50 mW/cm2). The results have also compared to the UV crosslinking based system using I2959 (0.05, 0.125, 0,25, and 0.5 wt%). Increasing the concentration of photoinitiators and light intensities reduced the oxygen inhibition effect for both systems. However, the Ru/SPS system exhibited better biocompatibility with more than 85% of cell viability after 21 days.
Figure 13.
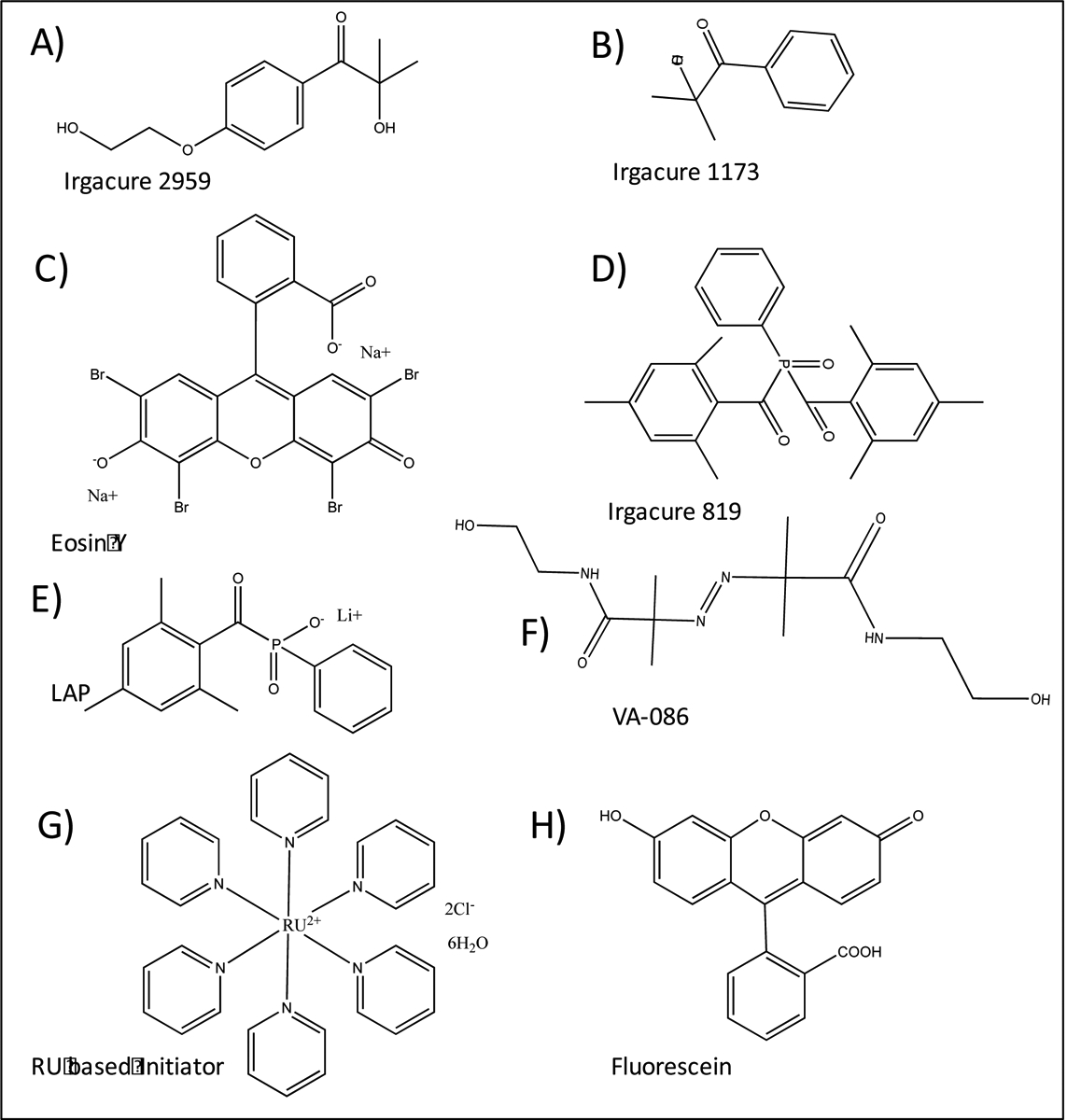
Chemical structures of A) Irgacure 2959, B) 1173, C) 819, D) Eosin Y, E) lithium-acyl phosphinate (LAP), F) 2,2’-Azobis[2-methyl-N-(2-hydroxyethyl) propionamide] (VA-086), G) tris(2,2‐bipyridyl) dichlororuthenium(II) hexahydrate and H) Fluorescein that commonly used as photoinitiators in three-dimensional (3D) bioprinting.
Apart from GelMA, other photocrosslinkable bioinks such as PEGDA [102], galactoglucomannan methacrylates [123], methacrylated HA [124], and decellularized extracellular matrix (dECM) have also been used for the 3D bioprinting. Among these, ECM-based bioink has recently got a considerable attention as it can provide a tissue-specific microenvironment for loaded cells. There are usually three steps to fabricate a photocrosslinkable ECM-based bioinks. These include, decellularization, solubilization, and methacrylation (Figure 14 A and B). For example, Ali et al. [125] synthesized a photocrosslinkable kidney ECM‐derived bioink and used for 3D bioprinting of renal constructs. They found that increased concentration of ECM in the bioink led to increased viscosity and stiffness of the bioink before and after crosslinking, respectively (Figure 14 C, D). They also demonstrated that with the use of this bioink, desirable shape can be printed, and cells viability maintained.
Figure 14.
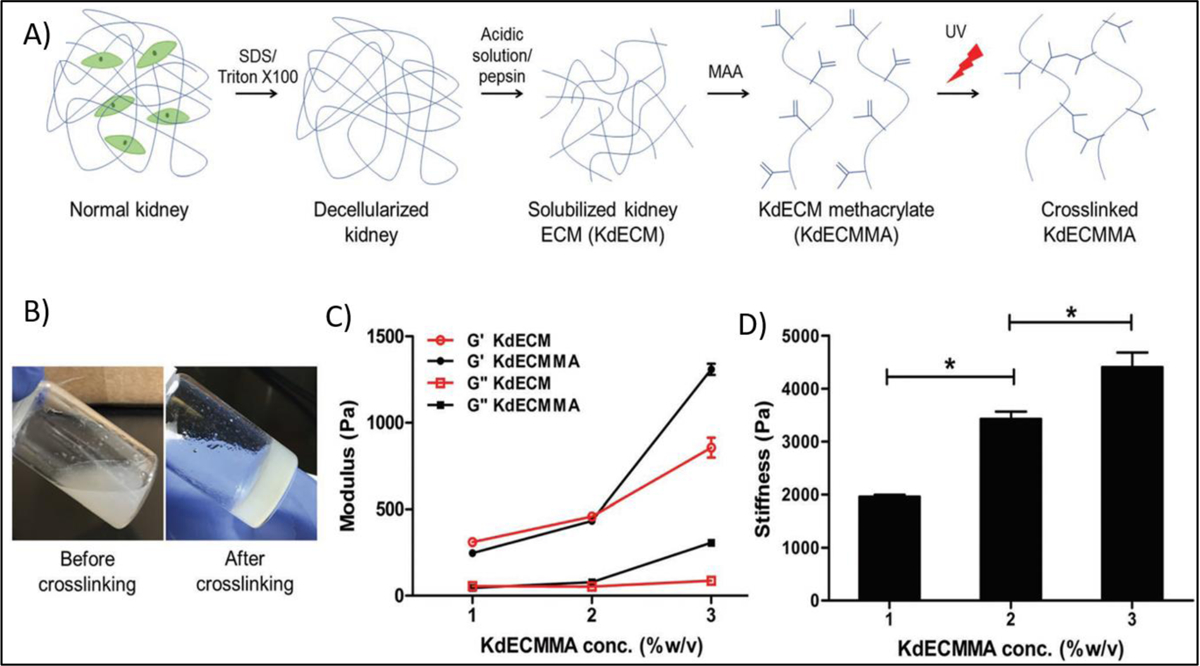
A) Preparation steps of a photo-crosslinkable kidney-specific extracellular matrix based bioink, B) Photographs of polymer solution before and after ultraviolet (UV) irradiation, C) Rheological behavior and D) Stiffness of bioinks at different polymer concentration Reproduced from Ali et al. [125], with permission from Wiley, 2019.
Poldervaat et al. [126] synthesized methacrylated HA in order to develop a bioink that undergoes fast photocrosslinking immediately after printing. The effects of different polymer concentrations [1–3% (w/v)] on mechanical and cell viability were studied. Mechanical properties were enhanced by increasing polymer concentration. Storage and elastic modulus for 3% (w/v) were determined to be 2.6 ± 0.1 and 10.6 ± 0.1 kPa respectively. After 21 days of culture, the viability of human bone marrow derived mesenchymal stromal cells within the 3D constructs containing 2% (w/v) methacrylated HA had the highest viability (64.4%) in comparison to other concentration. However, the highest amount of osteogenic differentiation was seen in the 2.5% (w/v) bioink. Authors concluded that the best suitable bioink for the 3D bioprinting of bone constructs is the 2.5% MeHA polymer. Although the free-radical polymerizations are highly used in the 3D bioprinting of photocrosslinkable constructs, but they have also some drawbacks, including the oxygen inhibition, shrinkage of printed structures, leakage of the unreacted functional groups, and the generation of inhomogeneities in the network of 3D bioprinted constructs.
- Step-Growth Crosslinking
Recently, bioorthogonal click reactions such as thiol–ene click chemistry have raised considerable attention as an alternative crosslinking mechanism to chain-growth polymerization counterpart. These reactions can proceed via Michael-addition reactions or a step-growth polymerization under light irradiation (mostly UV or visible light). This crosslinking method contain three steps of initiation, propagation and termination as shown in Figure 15A. Thiol is a −SH containing molecule which is capable of binding to unsaturated bonds such as carbon–carbon double bonds (“enes”), triple bond (“yne”) and epoxy groups to give various alkyl sulfides crosslinking. Among these, only thiol–ene reaction has been used in 3D bioprinting application, even though the important roles of thiol-yne and thiol-epoxy crosslinking have been widely emphasized in polymer science. Thiol–ene click chemistry is based on dimerization of thiols with reactive −ene groups which can form homogeneous hydrogel networks. Common −ene groups used in this crosslinking methods are acrylate, methacrylate, norbornene, vinyl ether, vinyl ester, alkene and malemide. Various polymers either contain −ene groups in their chemical structures or can be functionalized with them that makes this strategy applicable to a wide range of polymers. The use of thiol–ene can be an efficient method in minimizing polymerization shrinkage due to its rapid reaction kinetics. For example, the required time for the crosslinking of PEG norbornene via thiol–ene chemistry are reported to be only a few seconds (1 to 3 seconds). This reaction can provide temporal and spatial control of the crosslinking chemistry, which is not possible otherwise by using many crosslinking methods. Furthermore, using this method, many thiol-containing biomolecules can be bind to the bioink and be loaded to the structure of the final 3D bioprinted constructs. However, this approach has disadvantage of having a relatively short shelf-life and poor thermal storage stability due to the possible oxidation of disulfide bond [127]. Various synthetic and natural polymers such as polyethylene glycol (PEG), gelatin, and hyaluronic acid (HA), have been conjugated with either −SH or “ene” groups and used as a thiol-ene photocrosslinkable bioink in 3D bioprinting. Blunk et al. [128] used thiolated hyaluronic acid (HA-SH) as a bioink that was crosslinked with allyl-functionalized poly(glycidol)s (P(AGE-co-G)) for 3D bioprinting. In this study, 10 wt. % of total polymer concentration was used as the optimal concentration and gelation occurred by a UV-induced thiol-ene coupling between the thiol groups of HA and allyl groups of modified PEGs. It was found that this bioink is capable of accommodating human and equine MSCs over 21 days in vitro, and crosslinking method does not have negative effects on cell viability. In another study, dithiol PEG crosslinker was used with norbornene functionalized alginate to provide a rapid UV-induced thiol–ene crosslinking reaction (Figure 16) [129]. Different concentrations of thiolated PEG as a crosslinker having different molecular weights were used for bioprinting. Results showed that by increasing molecular weight from 1500 to 5000 Da, mesh size and swelling behavior of the hydrogel were increased. A more compact network was formed by using a 4-arm thiolated PEG instead of bifunctional crosslinker which decreased the swelling behavior of the hydrogel.
Figure 15.
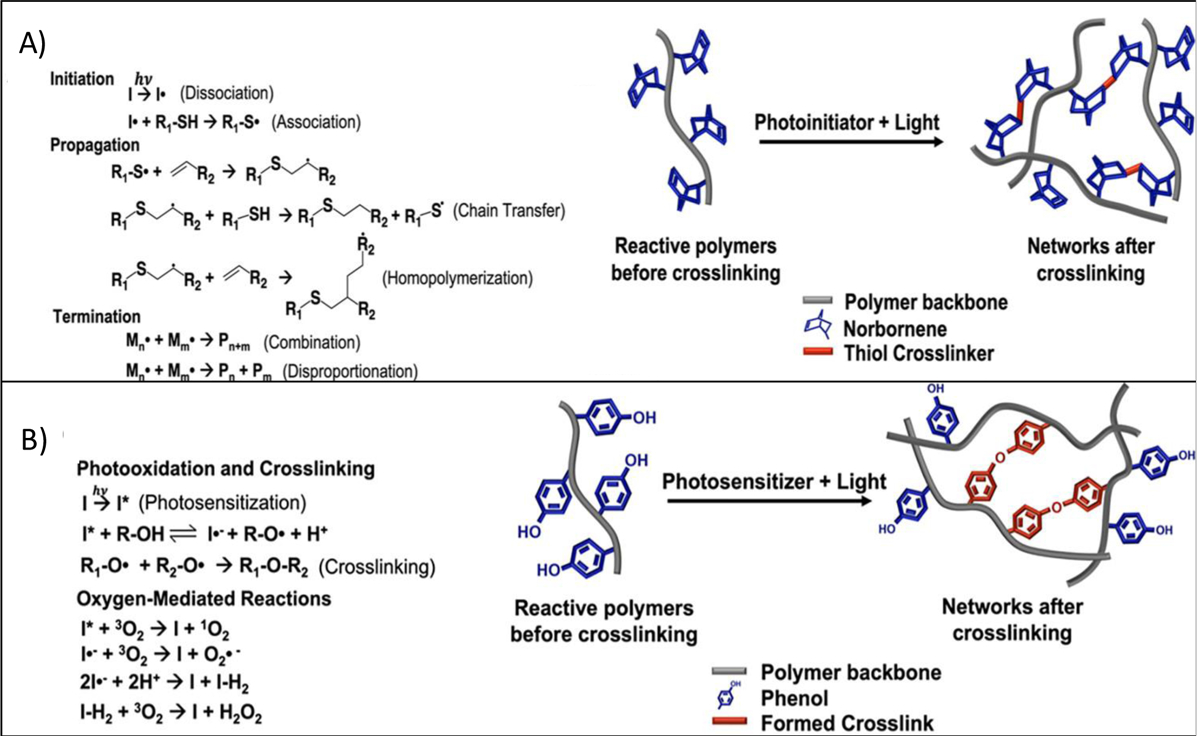
General mechanisem of the photocrosslinking through A) step growth polymerization and B) Photo mediated redox reactions. Reproduced from Woodfield et al.[34], with permission from American Chemical Society, 2020.
Figure 16.
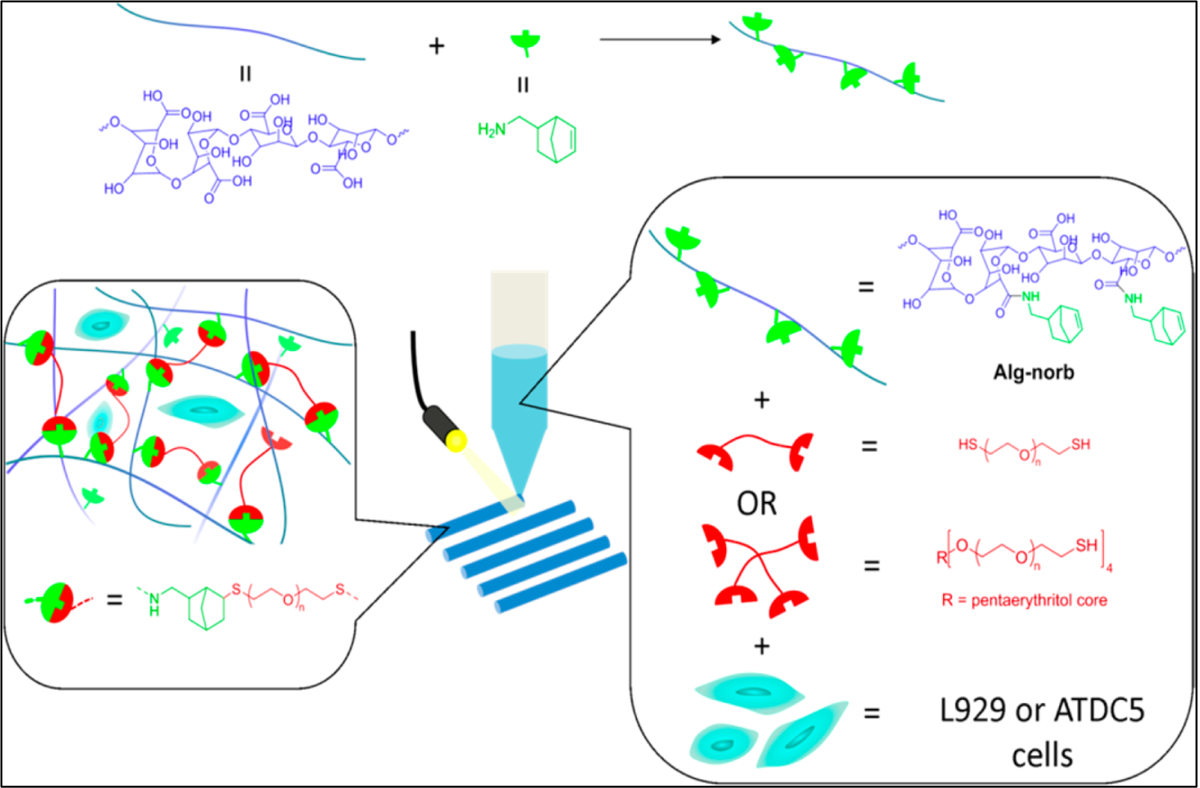
Schematic illustration of crosslinking mechanism that used for three dimensional (3D) bioprinting of photoactive Alginate based bioink. Thiol containing molecules and Norbornene functionalized alginate (Alg-norb) can form thiol–ene crosslinking reaction under ultraviolet (UV) exposure with minimal effect on cell viability. Reproduced from Ooi et al. [129], with permission from American Chemical Society, 2017.
- Redox Based Crosslinking
Another photo mediated crosslinking strategy involves the use of redox reactions. Redox responsive materials such as phenol containing polymers can be crosslinked through photoxidation of reactive groups. In this crosslinking method, the photolysis of oxygen produces singlet form of oxygen and radicals in the presence of photosensitizer. The formed radical converts phenolic compounds (e.g. tyrosine) into a different form of radical (e.g. tyrosyl radicals) that could rapidly binds the paired reactive groups together to form 3D network structures. The general mechanism of these reactions is shown in Figure 15B. This strategy is barely investigated for the 3D bioprinting of phenolic functionalized polymer so far, but it holds great potential for further investigation due to its rapid network formation. Eglin and colleagues [130], photocrosslinked a bioink containing hyaluronic acid functionalized with tyramine (HA-Tyr, 7.8% modification, 3.5 wt %) in presence of the different amount of eosin Y (0.02 wt %) and rose bengal (0.05 wt %) via laser-assisted printing method. The gelation time was reported to be less than 30 and 10 seconds for bioink containing rose bengal and eosin Y respectively. Eosin Y containing bioink exhibited a faster gelation time (<10 sec), compared to rose bengal with more than 95% hMSCs viability. In another study, Lim et al. [107] synthesized a bioink based on 2% silk fibrolin with and without 0.5 wt% gelatin that rapidly gelify (<1 min) by using tyrosine-tyrosine (di-tyrosine) binding in the presence of Ru/SPS photosensitizer and under visible light. Bioink containing gelatin showed better cell viability (>75%) compared to pure silk fibroin. Unlike enzymatic crosslinking that would be discussed in Section 5, the resulting hydrogel also showed a stable mechanical property over time.
2.2.2. Thermal Crosslinking
Thermal crosslinking via heating or cooling of polymer solution is one of the simplest methods of crosslinking and it can easily be applied to those polymers that can sustain heat or cold during the process of 3D printing. Some examples of thermo-crosslinkable based bioinks are presented in Table 5. However, gelation time in thermal crosslinking is longer than other crosslinking methods. In addition, in thermal crosslinking method, the degree of crosslinking cannot be precisely controlled. Furthermore, thermal energy generated from heating modules can adversely affect cells present in the bioink. Agarose, which is an uncharged polysaccharide, is crosslinked through the use thermal method. At high temperature (above 40 °C), agarose chains have a random coil conformation but cooling the polymer solution (to approx. 32 °C) induces a change in the conformation from random-coil to helical structure and subsequent formation of a 3D network. The rate of this transition increases at higher concentrations of polymer solution. Kelly et al. [131] used MSC-laden agarose–as a bioink. MSCs were combined with melted agarose and the suspension was allowed to cool and crosslink for 30 min. Agarose bioinks were found to support the development of a hyaline-like cartilaginous tissues. In another research, low melting point agarose (1.5 w/v%) was combined with MSCs and used to produce a bioink [132]. The bioink was kept at 37 °C and then submerged in a hydrophobic perfluorotributylamine solution at room temperature to achieve gelation. The results showed that perfluorotributylamine can provide both mechanical and biological support during printing (~80% cell viability in day 1). Shastri et al. [133] used carboxylated agarose (CA) as a bioink that was thermally crosslinked and then compared its performance with native agarose (NA) (Figure 17). Agarose was used in different concentrations (0.5–2% w/v) for 3D bioprinting. The optimal concentration for achieving the highest modulus was found to be 2%, which is in agreement with other studies [131]. In addition, there was an independent effect of concentration on the hysteresis and thermal transitions and strong dependence on the viscosity of NA bioink. Carboxylation of agarose led to reduction in the number of helical structures that can be involved in crosslinking points, and therefore, mechanical properties, hysteresis and thermal transitions were reduced by increasing carboxylation. It was suggested that that the carboxylation is an effective process for adjusting the crosslinking density and elastic modulus of agarose based bioinks.
Table 5.
Summary of three-dimensional (3D) bioprinting where temperature (heating or cooling) crosslinking, was employed showing materials used, type of biomolecules and targeted tissues. Various biomaterials have been used alone or in combination, such as collagen (Col), thiolated gelatin (T-gel), fibrinogen (Fib), alginate (Alg), agarose (Ag), carboxylated agarose (C-Ag), κ-carrageenan (Kca), polylactide (PLA), polyurethane (PU), and peptidesamphiphiles (PPS). Cells used included human mesenchymal stem cells (HMSCs), bone marrow derived stem cells (BMSCs), epidermal stem cell (ESCs), embryonic cells (ECs), human umbilical vein endothelial cells (HUVECs), murine preosteoblasts (MPs), chondrocytes cells (CCs), myoblasts cells (MBCs), neural progenitor cells (NPCs), Hela cells (HCs), cholangiocytes (CECs), and osteosarcoma cells (OCs).
| Biomaterial | Crosslinking Method | Printing Technique | Cell Type | Target tissue | Ref. |
|---|---|---|---|---|---|
| Ag | Temperature-induced gelation | extrusion piston-driven | HMSCs | Hyaline cartilage | [131,132] |
| C-Ag | Temperature-induced gelation | extrusion | HMSCs | Not specified | [133] |
| MC | Temperature-induced Gelation | extrusion | Fibroblasts | cell-sheet engineering | [135] |
| MC/HA | Temperature-induced Gelation | extrusion | HMSCs | Not specified | [134] |
| PU | Temperature-induced Gelation | extrusion | NPCs | Nerve tissue | [244] |
| Col | Temperature-induced Gelation | extrusion | CCs | Cartilage | [245] |
| Col | Temperature-induced Gelation | Inkjet bioprinting | ECs | cardiac constructs | [246] |
| PCL/PLA | Temperature-induced Gelation | extrusion | NPCs | Not specified | [141] |
| Col | Temperature-induced Gelation | extrusion | Fibroblasts | Not specified | [136] |
| GelMA | Temperature-induced Gelation | extrusion | HUVECs | Not specified | [247] |
| GelMA | Temperature-induced Gelation + UV Crosslinking | extrusion piston-driven | CCs | Cartilage | [248] |
| Poloxamer | Temperature-induced Gelation + UV | extrusion | CCs | Not specified | [137] |
| HA/Col | Temperature-induced Gelation + Ionic-CaCl2 | extrusion | OCs | Not specified | [139] |
| Gel/Alg/Fib | Temperature-induced gelation +Ionic-CaCl2 | extrusion | HCs | Tumor model | [249] |
| Plu/Alg | Temperature-induced Gelation +Ionic-CaCl2 | extrusion | MBCs | Not specified | [250] |
| Kca /Silicates | Temperature-induced Gelation +Ionic-KCL | extrusion | MPs | Not specified | [140] |
| T-gel/PPS | Temperature-induced Gelation +Ionic-CaCl2 | extrusion | CECs | Cartilage | [251] |
Figure 17.
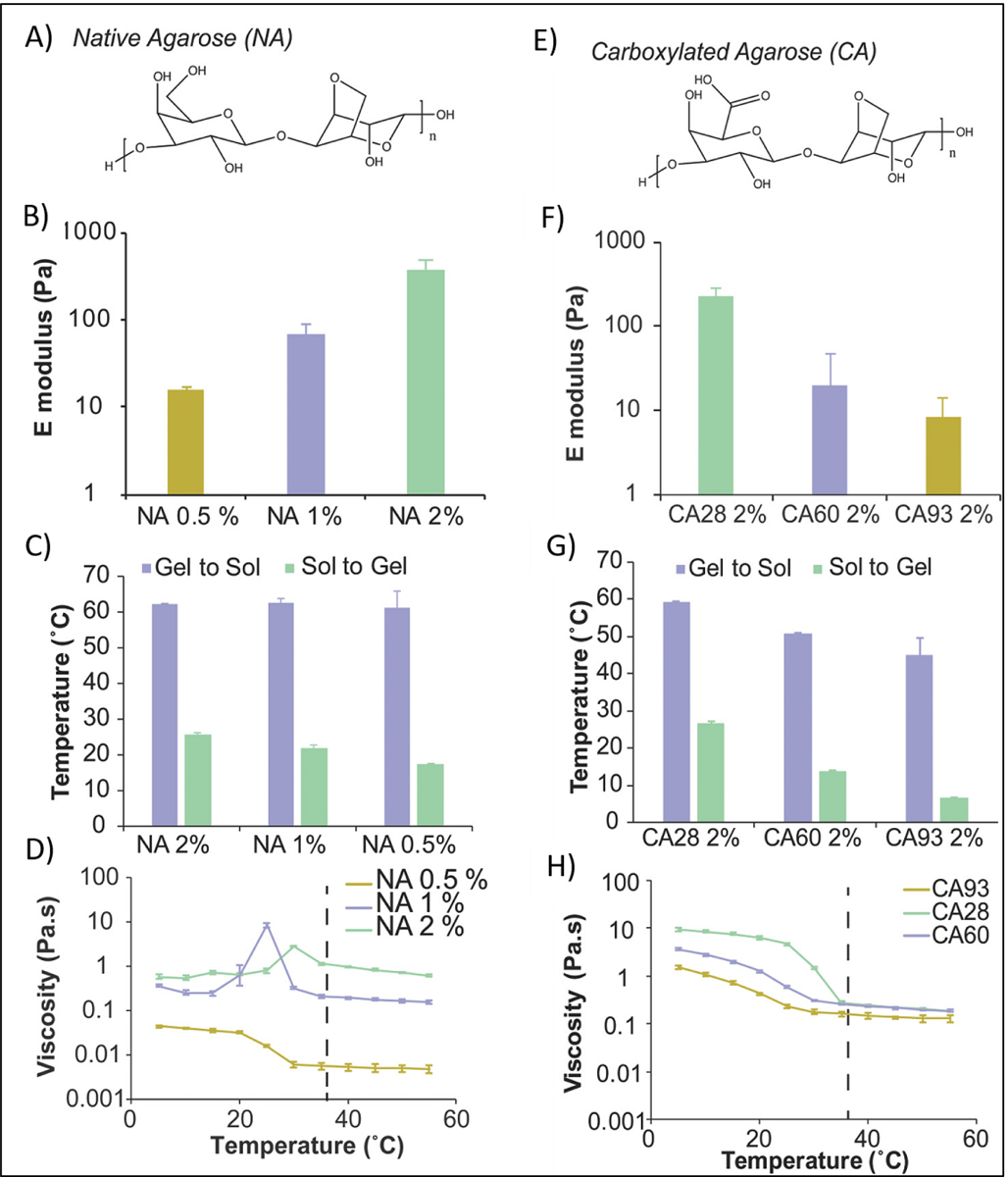
A) Native Agarose (NA) structure. B) Effect of NA concentration on Elastic modulus in compression (E, Young’s modulus). C) Tsol‐gel and Tgel‐sol for NA at different concentration. D) Viscosity of NA bioinks as a function of the temperature at different concentration. E) Carboxylated Agarose (CA) structure. F) Effect of CA concentration on Elastic modulus in compression (E, Young’s modulus). G) Tsol‐gel and Tgel‐sol for CA at different percentage of carboxylation. H) Viscosity of CA bioinks as a function of the temperature at different concentration. Reproduced from Forget et al. [133], with permission from Wiley, 2017.
MC is another biocompatible polymer that has been thermally crosslinked and used for 3D bioprinting. At room temperature, MC bioinks are in the form of a viscus solution due to the hydrogen bonding between the hydroxyl groups of the MC chains with the water molecules. By increasing temperature (37°C), the solution becomes a gel, because the formation of intermolecular hydrophobic interactions between the hydrophobic groups of MCs (–OCH3). This transition was found to be reversible, due to the absence of covalent bonds. Several studies have examined the printability of MC-based hydrogels using an extrusion printing technique [134, 135]. Collagen [136], poloxamers [137], N-isopropylacrylamine based copolymers [138], HA [139], Kca [140], poly (ε-caprolactone) poly (L-lactide) diol [141], are other thermo-crosslinkable polymeric based bioinks, which have been used for 3D bioprinting applications.
2.2.3. Chemical reactions
Network formation in chemically crosslinked hydrogel bioinks can take place by covalent bonding between polymeric chains through various chemical reactions including Schiff base coupling, hydrazide-aldehyde coupling, Diels–Alder linkage and azide-alkyne cycloaddition. These reactions can be usually triggered in the presence of light or heat [142]. Formed crosslinking points are usually strong enough to provide good mechanical properties, but crosslinking kinetics should be precisely controlled to avoid any printer nozzle blockage or cell toxicity.
- Schiff Base Chemistry
Schiff’s base crosslinking involves the reaction between polymers containing alcohol or amine groups with aldehydes/ketones to obtain a 3D network. This reaction usually occurs under mild conditions and has been shown to have adjustable rates in a time dependent manner. The formation of this crosslinking method was found to be enhanced at high pH. However, to be stable, they are required to be reduced by reductive amination. The by-product of this reaction is water. When the water molecule is not removed, the hydrolysis can occur, therefore, a dynamic equilibrium is possible by adjusting reaction conditions. To form a polymeric network based on Schiff base chemistry various amino-rich polymers, such as chitosan and polyacrylamide, with other aldehyde-functionalized polymers, such as oxidized alginate [143], dextran [144], and hyaluronic acid [145] have been used. Zhu et al. [19] investigated a hydrogel formed via Schiff-base crosslinking with taking the advantage of phase separation between oxidized dextran and gelatin. Figure 18 shows the design process of prepared hydrogel that involved the nucleophilic reaction of aldehydes on the oxidized dextran and addition of amines nucleophiles to the gelatin. In this design, the amino groups of gelatin chains were regulated the interface crosslinking between the dextran-rich phase and the gelatin-rich phase through adjusting the pH values. In addition, they reported that loaded fibroblasts had a high viability of 90%, following the printing process.
Figure 18.
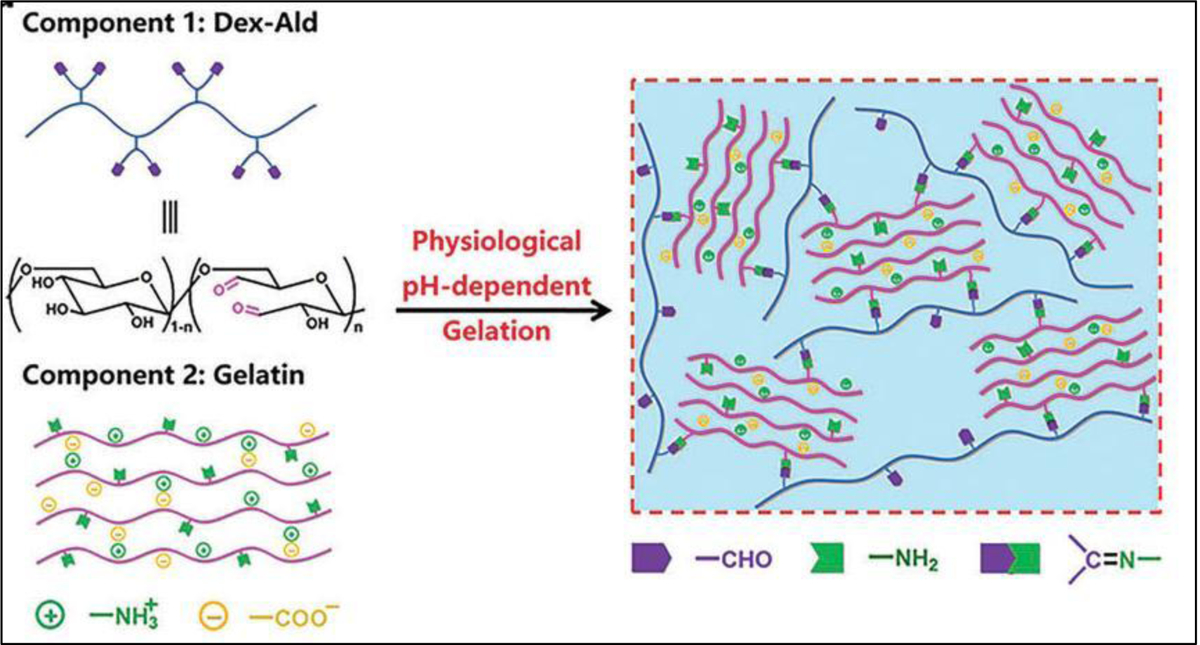
The chemical structure of crosslinking strategy that has been used between gelatin and oxidized dextran (Dex-Ald) that involved the nucleophilic reaction of aldehydes on the oxidized dextran and addition of amines nucleophiles to the gelatin, which causes crosslinking in a pH dependent manner. Reproduced from Reproduced from Du et al. [19], with permission from Royal Society of Chemistry, 2019.
- Hydrazide-Aldehyde Coupling
Aldehyde-containing macromolecules can react with hydrazide compounds without the need for photo-irradiation to form another form of Schiff bases, a dehydration reaction yielding a hydrazone linkages. The crosslinking formed by hydrazone bond is reported to be more stable than the crosslinking formed by aldehyde-amine bond. Fukuda et al. [21] used this in situ crosslinking strategy by mixing gelatin hydrazide (2.5 w/v %) with aldehyde-functionalized carboxymethylcellulose (2 w/v %) (Figure 19). When endothelial cells were encapsulated in the bioink, cell viability did not drop significantly and remained more than 80% after the crosslinking process, indicating the biocompatibility of hydrogels and the safety of crosslinked method.
Figure 19.
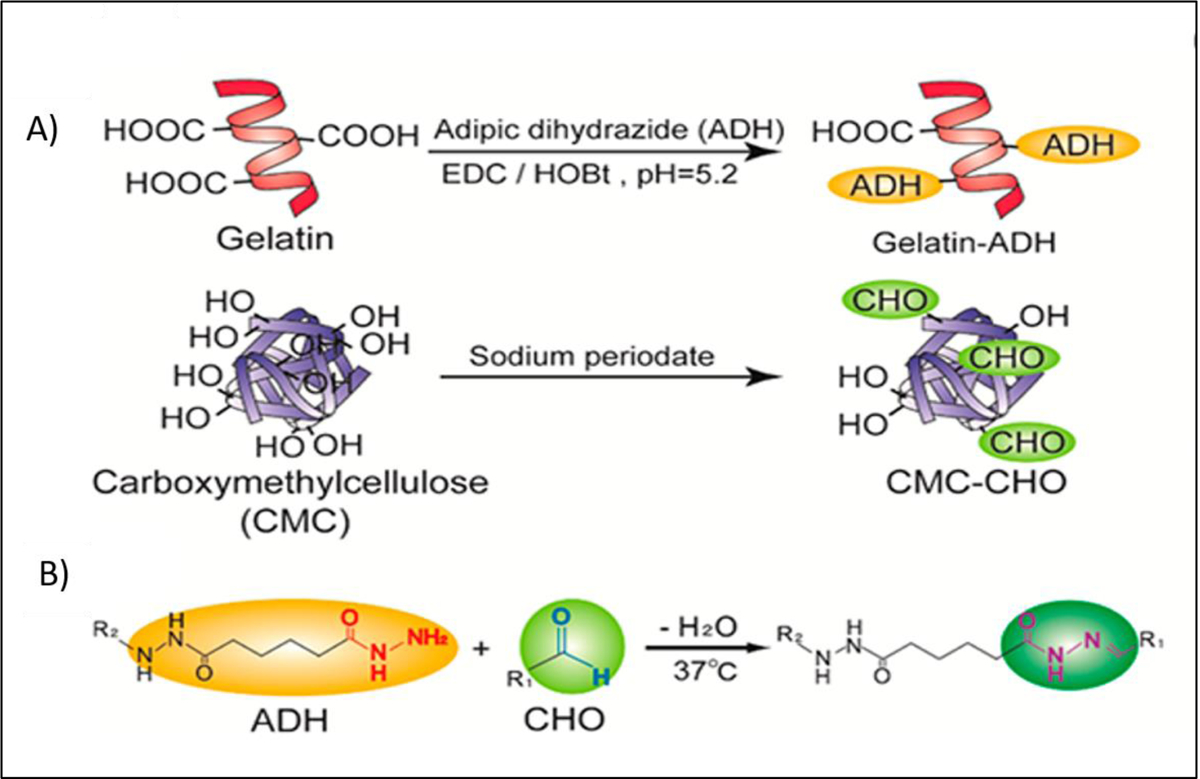
A) Functionalization of gelatin with hydrazide groups by reaction between gelatin and adipic dihydrozide in the presence of 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and hydroxybenzotriazole (HOBt) and functionalization of carboxymethylcellulose (CMC) with aldehyde groups by reaction between CMC and Sodium periodate, B) Network formation through hydrazide-aldehyde coupling. Reproduced from Kageyama et al. [21], with permission from American Chemical Society, 2017.
- Acylhydrazone
Acylhydrazone crosslinking can be formed by mixing of hydrazide and the carbonyl groups containing materials. Kim et al. [146] mixed chitosan modified adipic acid dihydrazide (ADH) (1 wt%) with partially oxidized hyaluronan (OHA) (2 wt%) to form a crosslink between the carbonyl group of OHA and hydrazide groups of ADH (Figure 20). The fabricated 3D bioprinted hydrogels showed a rapid self-healing behavior due to the fast healing capability of used crosslinking methods, including acylhydrazone bonds obtained via the reaction between ADH and OHA and imine bonds formed by Schiff base chemistry. This crosslinking strategy also showed a minimal effect on chondrogenic differentiation of mouse teratocarcinoma-derived cells (80% viability).
Figure 20.
(A) Chemical structures of oxidized hyaluronate (OHA), Glycol Chitosan (GC), and Adipic dihydrozide (ADH). (B) Gel formation upon mixing of OHA with GC+ADH polymer solutions. (C) Schematic illustration for the produced network. Reproduced from Kim et al. [146], with permission from American Chemical Society, 2019.
- Diels–Alder Linkage
Diels–Alder linkage is a click reaction (cycloaddition) between a conjugated diene and a substituted alkene or alkyne. This reaction involves overlapping of highest occupied molecular orbital (HOMO) containing 4π electrons of the diene with the lowest unoccupied molecular orbital (LUMO) containing 2π electrons of dienophile to form a cyclo-addition product. The driving force of the reaction is the formation of new σ-bonds, which are energetically more stable than the π-bonds. This crosslinking method can be thermally reversible, which has been applied to design of various smart self-healing materials. For example, Furan and Maleimide containing molecules can reversibly bind to each other to form a 3D network in a temperature-responsive manner (Figure 21) [147]. However, this method is barely used for 3D bioprinting due to the sensitivity of cells to relatively high temperature that should be used in this crosslinking strategy. Among a few research studies using Diels–Alder crosslinking, Zhang and his coworkers [148], used a Diels–Alder linkage between tetrazine and trans-cyclooctene to crosslink bioinks containing bone morphogenetic protein-2 (BMP2) and bone marrow derived MSCs (BMSCs). HA (2% (w/v)) and 4-arm-PEG (3% (w/v)) were functionalized with pendant tetrazine and trans-cyclooctene respectively. Then polymer solutions loaded with cells were gellified upon mixing at room temperature. The results of this work demonstrated that synthesized materials and crosslinking strategy could enhance the differentiation and proliferation of the cells and can be potentially used as a promising strategy for 3D bioprinting of living tissue constructs.
Figure 21.
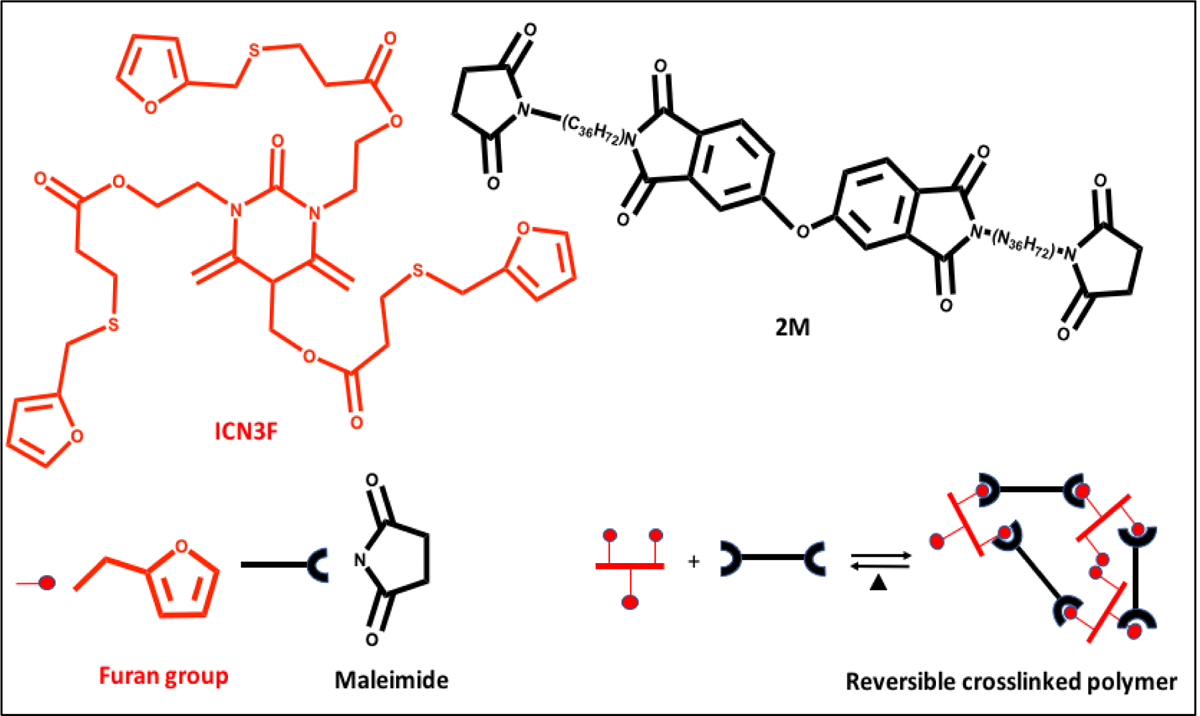
Chemical structures of bismaleimide (2M), and furan containing monomer (ICNF3) that can form a reversible covalent binding via Diels–Alder chemistry. Reproduced from Yang et al. [147], with permission from Wiley, 2017.
- Azide-Alkyne Cycloaddition
Azide and internal alkyne can react with each other to form azide-alkyne cycloaddition which can be used for the formation of hydrogel in the presence of copper based catalyst. This method is not suitable for cell encapsulation, due to the cytotoxicity of copper. However, catalyst‐free bio‐orthogonal crosslinking methods such as strain‐promoted azide‐alkyne cycloaddition eliminate the risk of cytotoxicity and allow biomaterial cell encapsulation, and thus these methods are anticipated to be useful in 3D bioprinting [20].
2.2.4. Chemical crosslinker
- Zero-Length and Non-zero-Length Crosslinker
Chemical crosslinker can be classified based on their contribution to the final structure of the hydrogel into two groups, 1) zero-length crosslinker, and 2) non-zero-length crosslinker. Zero-length crosslinkers covalently link carboxylic acid group with amine residues without contributing any part of the crosslinker molecules to the final structure of hydrogel. The most common example of this type of crosslinking involves the use of carbodiimide linkers such as 1-ethyl-3-(3-dimethyl aminopropyl) carbodiimide (EDC) which reacts with the carboxylic acid and converts them into O-acylisourea groups. In combination with photocrosslinking, EDC (15 Mm) has been used in 3D bioprinting of Schwann cell-laden alginate/HA and human dermal fibroblasts-laden PEG based bioinks [149]. Wang et al. studied how different concentrations of EDC (10,30, 50, and 100 mmol/L) influence the mechanical properties and cellular responses in Schwann cells loaded alginate/HA based hydrogels [150]. They have demonstrated that the degree of crosslinking and cell viability are highly dependent on the EDC concentrations. The non-zero-length crosslinker covalently binds two amino acid residues by an esterification reaction and crosslinker molecule or part of the molecule becomes part of the final materials after the crosslinking process. The network formation of a non-zero-length crosslinked hydrogel can also be achieved by the addition of synthetic or natural crosslinker agents to the polymer solution.
Several synthetic non-zero-length crosslinkers have been used for gelation of various polymeric systems, such as glyceraldehyde [151], sodium bicarbonate [17], formaldehyde [152], and glutaraldehyde (GA) [153]. However, they have been barely used in 3D bioprinting due to their high cytotoxicity, which can cause inflammation and calcification when implanted in vivo [154]. On the other hand, natural non-zero-length crosslinkers, for example genipin, have shown to be more suitable options for the 3D bioprinting. Genipin is a natural crosslinking agent isolated from the fruit of the gardenia plant and could spontaneously react with polymers containing the primary amine such as collagen, gelatin, or chitosan. Although genipin has been numerously applied for crosslinking of gelatin, its exact crosslinking mechanism is still under discussion. Rose et al. [154] proposed two steps mechanism for crosslinking of gelatin using genipin. In the first step a nucleophilic attack by primary amine to the C3 carbon of genipin results in the opening of the dihydropyran ring and the formation of a tertiary amine (Figure 22A). In the second step, oxygen in the genipin can be replaced with the tertiary nitrogen to from a second fragment of gelatin (Figure 22B). These two reactions are independent and they result in gelatin crosslinking. Kim et al. [155] developed crosslinking ability of the collagen bioink (3, 5 and 7 wt%) using genipin (0.5, 1, and 3 mM) as a crosslinker to print highly porous structure encapsulating osteoblast-like cells and human adipose stem cells. In their work, it was found that increasing crosslinking time and concentration of genipin improved the qualitative stiffness of the mesh structure of hydrogels. One-hour crosslinking time of 5 wt% polymer solution with 1 mM concentration of genipin was reported as an optimal condition. Higher concentrations of genipin and collagen or longer crosslinking time caused significant cell damage.
Figure 22.
Crosslinking mechanism of gelatin-genipin systems: (A) Michael addition used as a primary reaction; and (B) nucleophilic substitution of free lysine amine molecules into genipin activated ester used as a secondary reaction. Reproduced from Rose et al. [154], with permission from MDPI, 2014.
3. Enzymatic Reaction
Enzymes have also been used for crosslinking of bioinks. Enzymes can be employed as catalysts to promote the formation of covalent bonds between protein-based polymers. Enzymatic crosslinking is an attractive method of crosslinking for use in 3D bioprinting, due to the mildness of the enzymatic reactions which could avoid cell death. Various enzymes have been used for crosslinking of hydrogels such as transglutaminase, phosphopantetheinyl transferase, lysyl oxidase, plasma amine oxidase, peroxidases and horseradish peroxidase mimetic enzymes [156]. However, only few of them have been applied in 3D bioprinting and most often in combination with other crosslinking procedures (Table 6). Tan et al. [157] developed a GelMA hydrogel (10 wt%) based on partial enzymatic crosslinking triggered by microbial transglutaminase. Microbial transglutaminase induces crosslinking between the ε-amino group of lysine and carboxylic acid groups of glutamic acids (Figure 23A). This crosslinking reaction can lead to the formation of stable 3D polymeric networks, without the need to add any other reagent. It was found that the crosslinking reaction could influence the rheological properties and thermal stability of GelMA. Proposed bioinks displayed good printability when they were in extrusion-based printing. Zhang group [158] constructed a living skin tissue by using cell-laden GelMA and collagen bioink which was first enzymatically crosslinked with tyrosinase (Ty) and then photocrosslinked to improve the mechanical properties of bioink. Ty is an enzyme that in the presence of oxygen, catalyzes the oxidation of phenols to catechol and quinone groups which can to react with amine containing polymers (Figure 23B). Human melanocytes, keratinocytes and fibroblasts were incorporated in the bioinks. High cell viability of over 90% was observed for the three cell lines. Evaluation in vivo has further proved that cell-laden ty-doped bioinks could be used to form an epidermis and dermis.
Table 6.
Summary of three dimensional (3D) bioprinting where chemical reaction crosslinking, was employed showing materials used, type of biomolecules and target tissues. Various biomaterials have been used alone or in combination, such as collagen (Col), gelatin (gel), alkylated gel (A-gel), thiolated gel (T-gel), hydrazide modified gel (H-gel), aldehyde modified gel (AD-gel), gel methacryloyl (GelMA), fibrinogen (Fib), extracellular matrix (ECM), hyaluronic acid (HA), thiolated HA (T-HA), oxidized HA (O-HA), hydrazide-modified HA (H-HA), aldehyde-modified HA (AD-HA), Ene-functionalized alginate (E-Alg), silk fibroin (SF), chitosan (Ch), oxidized dextran (OD), polylysine (Pys), poly(vinyl alcohol) (PVA), polylactide (PLA), alkylated polyglycide (A-PGA), and polyvinylpyrrolidone (PVP). Cells used included human mesenchymal stem cells (HMSCs), human adipose tissue derived stem cells (HASCs), epidermal stem cells (ESCs), human umbilical vein endothelial cells (HUVECs), rat skin fibroblasts (RSFs), human dermal fibroblasts (HDFs), chondrocytes (CCs), cartilage progenitor cells (CPCs), hepatocellular carcinoma cells (HCCs), osteosarcoma cells (OCs), glioma stem cells (GSCs), and clonal mouse embryonic cells (CMECs).
| Biomaterial | Crosslinking Method | Printing Technique | Cell Type | Targeted tissue | Ref. |
|---|---|---|---|---|---|
| H-gel/ AD-gel | Hydrazide Aldehyde coupling | extrusion | HUVECs | Not specified | [21] |
| H-HA/ AD-HA | Hydrazide Aldehyde coupling + UV-Crosslinking | Extrusion piston-driven | Fibroblasts | Not specified | [252] |
| gel/OD | Schiff base chemistry | extrusion piston-driven | HDFs | Skin | [19] |
| ECM | EDC coupling | extrusion | RSFs | Skin | [253] |
| Elastin | Azide-alkyne cycloaddition | extrusion | HMSCs | Not specified | [20] |
| O-HA/Ch | acylhydrazone | extrusion | CKCs | Cartilage | [146] |
| gel/Fib/HA | Thrombin | extrusion | MBCs | Not specified | [254] |
| gel/Alg/Fib | Thrombin / Ionic CaCl2 | extrusion | GSCs | Brain tissue | [255] |
| Fib/gel/Alg | Thrombin / Ionic CaCl2 | extrusion | HMSCs | Bone tissue | [256] |
| GelMA | Transglutaminase/ UV | extrusion | HMSCs | Not specified | [157] |
| Fib/Col | Thrombin / transglutaminase/Ionic-CaCl2 | Inkjet bioprinting | HUVECs | Vascular tissue | [257] |
| SF | Enzymatic-crosslink | extrusion | HASCs | Not specified | [23] |
| PVA/PLA | Thrombin / transglutaminase | extrusion | HCCs | Not specified | [18] |
| Col | Vitamin B2 | extrusion | CPCs | Not specified | [258] |
| Col | Genipin | extrusion | OCs / HASCs | Not specified | [155] |
| gel/Pys | Genipin | extrusion | HMSCs / HUVECs | Vascular tissue | [259] |
| Col/PVP | Sodium bicarbonate | extrusion | Fibroblasts | Not specified | [17] |
| Alg | Horseradish peroxidase | Inkjet bioprinting | CMECs | Not specified | [260] |
| Alg/SF | Horseradish peroxidase + Ionic-CaCl2 | extrusion | Fibroblasts | Not specified | [197] |
Figure 23.
The enzymatic crosslinking mediated by A) transglutaminase, B) tyrosinase and C) peroxides. Reproduced from Teixeira et al. [156], with permission from Elsevier, 2012.
The use of horseradish peroxidase (HRP) as a catalyst in the crosslinking reaction is another enzymatic crosslinking process. In the presence of hydrogen peroxide, HRP catalyzes the coupling of aniline, phenol and their derivative (Figure 23C). Reis et al. [23] developed a fast-setting silk-based (silk-fibrolin) hydrogel (5 wt%), which was enzymatically crosslinked by using HRP as enzyme (0.84 mg/ml) and hydrogen peroxide (0.36 wt%) as substrate. The addition of enzyme to silk solution induces a change in conformation from a random coil into a β-sheet conformation, that could also cause a change in mechanical properties over time. The mechanical properties of the synthesized hydrogel are tunable by changing the concentration of HRP and H2O2. In vitro cellular assays demonstrated that obtained constructs can support HASCs proliferation and viability for up to seven days in culture.
4. Challenges and Future Perspectives
Three-dimensional bioprinting requires careful selection of crosslinking technique as it plays an important role in balancing the mechanical and chemical properties of the printed constructs, resulting in induce a positive cellular response. Although various crosslinking methods have been applied for 3D bioprinting of hydrogels, there are still many challenges which face the utilization of these methods. For ideal 3D bioprinting, utilized bioink should have sufficient crosslinking density to provide enough mechanical strength to first printed layers and support subsequent deposition of printed layers without leading to the collapse or impairment of the resulting structure. There is a relation between the 3D bioprinting technologies and crosslinking mechanisms. Unfortunately, the printing technologies aspects are often not described in detail in many research articles which makes it difficult to compare and draw a conclusion on the relation between the 3D bioprinting technologies and crosslinking mechanisms. There is a balance between degree of crosslinking and resulting structure. Low degree of crosslinking results in spreading of the structure, while high degree can prevent printability. Crosslinking method also raises challenges relating to cell viability issues. Some researchers have suggested promising strategies by performing various mild crosslinking strategies [159]. However, further research and development is required.
Photocrosslinking is the most widely used approach for developing 3D bioprinted constructs. One of the favorable advantages of the photo-mediated crosslinking such as UV crosslinking is the strong sterilization process, as the microorganisms are inactivated by UV irradiation due to the damage of nucleic acids [160]. Therefore, this crosslinking method can potentially overcome the sterilization issue associated with the 3D bioprinted constructs which is one of the important challenges facing clinical translation of 3D bioprinting. On the other hand, exposure to light irradiation or to free radicals generated from photoinitiators, can also increase the risk of phototoxic damage to cells and influence the functionality of the printed constructs [161]. These effects may become more significant at high-density cell containing bioinks due to the light absorption capability of cell components (such as DNA). Therefore, the crosslinking density, structural integrity, and cell viability need to finely be balanced in this method of crosslinking.
Bioinks formed by physical crosslinking methods such as hydrogen bonding, hydrophobic interaction, π–π stacking, dipole-dipole interactions, host–guest recognition and β-sheet mediated crosslinking are of huge interest for use in cell encapsulation and entrapment, especially when 3D bioprinting process is conducted under mild conditions [162]. However, the dynamic and reversible nature of crosslinking formed by noncovalent interactions results in poor mechanical stability. Crosslinking of bioinks with stronger non-covalent bonds such as coordination complexes or stronger supramolecular bindings may potentially overcome this problem. Non-covalent crosslinking can be also utilized for fabricating self-healing materials, which are in early stages of application in 3D bioprinting [146]. To date, only a few self-healing 3D bioprinted constructs have been fabricated, probably because of the complexity of their design and processing [163]. Therefore, further research and development of novel materials and crosslinking strategies in the future is essential to advance this part of the field. Recently 3D printing of shape memory constructs known as a 4D printing [5], has also gained attention. The used crosslinking methods to fabricate the 4D bioprinted structures could significantly influence the 4D effect. This is more important for thermally induced actuation, as it could affect transition temperature of the final structure.
The addition of chemical crosslinkers is another highly versatile method to improve hydrogel mechanical properties. The chemical crosslinkers are often toxic compounds and they result in unwanted reactions with biological moieties present on the surface of cells, with the risk of cytotoxicity and other side effects. However, there are also examples of some chemical crosslinkers that are less cytotoxic. But some cautions should be taken prior to using the chemical crosslinkers. For example, the concentration of crosslinker highly influences the mechanical properties and cellular responses. Moreover, in this crosslinking method, polymeric network forms by irreversible covalent bonds, hence, the shape of constructs can be irreversibly fixed upon mixing of polymer solution with the crosslinkers. Therefore, the mixing step is quite crucial to ensure shape fidelity. Ionic interaction is another common method of crosslinking that used in 3D bioprinting. Although 3D bioprinting through ionic crosslinking can help to avoid cell exposure to harsh chemical conditions, cells can still suffer from extensive membrane damage in the presence of metal ions [164]. Furthermore, ionic crosslinking can be accompanied with a non-uniform network formation due to the faster deposition of ions on the surface of bioinks that can affect swelling behaviors.
To date, there are still various methods of crosslinking that have not been investigated for 3D bioprinting. For instance, in coordination chemistry many ligands-based crosslinking strategies have not been applied to the field. Some of these ligands such as catechol or terpyridine are expected to provide strong interactions with metal ions that can be utilized for the crosslinking of bioinks. In photocrosslinking, various lasers with different wavelengths can be still utilized not only to crosslink the bioinks but also to pattern the cells into the bioinks and final structures [165]. Furthermore, polymers containing cinnamic, coumarin and anthracene groups have been reported to be able to form a reversible photocrosslinking, however, they have never been used for 3D bioprinting. Among the chemical reactions induce crosslinking, various reactions such as thiol-yne and thiol-epoxy, azide-alkyne cycloaddition have not been studied so far, even though their important roles has been recognized in polymer science [166]. Similarly, non-enzymatic crosslinking or sugar induced-crosslinking is never investigated in this field.
While in most of the 3D bioprinting systems the bioinks crosslinked either immediately prior or after printing, in situ crosslinking is barley studied. In situ crosslinking is expected to improve the crosslinking uniformity throughout the final structure. Moreover, to date, only a few studies have been focused on computational modeling of bioink crosslinking for bioprinting applications [167]. Mathematical modeling can be used to better understand of effect of crosslinking on the final state of both 3D and 4D bioprinted constructs. Modeling of crosslinking process for bioprinting applications can significantly reduce trial-and-error of experimental tests, and hence, it can facilitate the optimization of bioinks and lower the cost of 3D bioprinting process for further applications and translation of this technology to the clinic.
5. Conclusion
The field of 3D bioprinting has been advancing tremendously in the last few years. When designing 3D bioprinting system, there is a need to choose an appropriate crosslinking approach for the desired applications. Appropriate crosslinking allows the printability of bioinks and ensures cytocompatibility, stability and sustainability of resulting living constructs. While there are several crosslinking techniques that are limited by the type of material used in the bioink and levels/concentrations that can applied yet preserve cell viability, function. Although various methods of crosslinking methods have been developed and used in 3D bioprinting, challenges related to present methods still exist and warrant further development of current and new methods. This will help advancing the field of 3D bioprinting further and expedite its applications and clinical translation.
Acknowledgements
((The authors acknowledge that they have no competing interests. The authors also acknowledge funding from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2019-07204) to XYW, and National Institutes of Health (EB021857, AR066193, AR057837, CA214411, HL137193, EB024403, EB023052, EB022403 and R01EB021857).”))
Biographies
Bio:

Amin GhavamiNejad is a postdoctoral scholar and seminar coordinator at Leslie dan faculty of pharmacy, University of Toronto (Canada). He was previously a research assistant professor in Bionanosystem Engineering department at Chonbuk National university (South Korea). He has an extensive research experience in synthesizing polymers for various biomedical and pharmaceutical applications. He is one of the recipient of the Banting & Best Diabetes Centre Fellowship in Diabetes Care and has also obtained and supervised several projects funded by various federal and provincial funding agencies in South Korea and Canada.

Nureddin Ashammakhi is an associate director of the Center for Minimally Invasive Therapeutics at UCLA, leading translational research in regenerative therapy. He has extensive experience with biodegradable implants, drug release, and nanofiber-based scaffolds. Currently, he is working on 3D bioprinting and organ-on-a-chip models for regenerative and personalized medicine. He was previously a professor of Biomaterials Technology in Tampere University of Technology, Finland, Chair of Regenerative Medicine in Keele University, UK, and Adjunct Professor in Oulu University, Finland, before he joined University of California-Los Angeles first as a visiting professor (Scholar) and then as an adjunct professor.

Ali Khademhosseini is the Levi Knight Professor of Bioengineering, Chemical Engineering and Radiology at UCLA. He is the Founding Director of the Center for Minimally Invasive Therapeutics at UCLA. Previously, he was a professor of Medicine at Harvard Medical School. He is recognized as a leader in combining micro- and nanoengineering approaches with advanced biomaterials for regenerative medicine applications.
References
- [1].Langer R, Vacanti JP, Science 1993, 260, 920; F. Triolo, B. Gridelli, Cell Transplant 2006, 15, S3. [DOI] [PubMed] [Google Scholar]
- [2].Linden PK, Crit Care Clin 2009, 25, 165. [DOI] [PubMed] [Google Scholar]
- [3].Li PKT, Chu KH, Chow KM, Lau MF, Leung CB, Kwan BCH, Tong YF, Szeto CC, Ng MMM, Nephrology 2012, 17, 514. [DOI] [PubMed] [Google Scholar]
- [4].Murphy SV, Atala A, Nat Biotechnol 2014, 32, 773; S. Vijayavenkataraman, W. C. Yan, W. F. Lu, C. H. Wang, J. Y. H. Fuh, Adv Drug Deliver Rev 2018, 132, 296; N. Ashammakhi, S. Ahadian, M. A. Darabi, M. El Tahchi, J. Lee, K. Suthiwanich, A. Sheikhi, M. R. Dokmeci, R. Oklu, A. Khademhosseini, Advanced Materials 2019, 31. [DOI] [PubMed] [Google Scholar]
- [5].Ashammakhi N, Ahadian S, Fan ZJ, Suthiwanich K, Lorestani F, Orive G, Ostrovidov S, Khademhosseini A, Biotechnol J 2018, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ashammakhi N, Ahadian S, Pountos I, Hu SK, Tellisi N, Bandaru P, Ostrovidov S, Dokmeci MR, Khademhosseini A, Biomedical Microdevices 2019, 21. [DOI] [PubMed] [Google Scholar]
- [7].Di Bella C, O’Connell C, Blanchard R, Duchi S, Ryan S, Yue Z, Onofrillo C, Wallace G, Choong P, Tissue Eng Pt A 2017, 23, S9. [Google Scholar]
- [8].Gungor-Ozkerim PS, Inci I, Zhang YS, Khademhosseini A, Dokmeci MR, Biomater Sci-Uk 2018, 6, 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chung JHY, Naficy S, Yue ZL, Kapsa R, Quigley A, Moulton SE, Wallace GG, Biomater Sci-Uk 2013, 1, 763. [DOI] [PubMed] [Google Scholar]
- [10].Donderwinkel I, van Hest JCM, Cameron NR, Polym Chem-Uk 2017, 8, 4451. [Google Scholar]
- [11].Stanton MM, Samitier J, Sanchez S, Lab Chip 2015, 15, 3111. [DOI] [PubMed] [Google Scholar]
- [12].Zhang B, Luo YC, Ma L, Gao L, Li YT, Xue Q, Yang HY, Cui ZF, Bio-Des Manuf 2018, 1, 2. [Google Scholar]
- [13].Gao G, Kim BS, Jang J, Cho DW, Acs Biomater Sci Eng 2019, 5, 1150. [DOI] [PubMed] [Google Scholar]
- [14].Jungst T, Smolan W, Schacht K, Scheibel T, Groll J, Chem Rev 2016, 116, 1496; B. K. Gu, D. J. Choi, S. J. Park, Y. J. Kim, C. H. Kim, Adv Exp Med Biol 2018, 1078, 15; E. S. Bishop, S. Mostafa, M. Pakvasa, H. H. Luu, M. J. Lee, J. M. Wolf, G. A. Ameer, T. C. He, R. R. Reid, Genes Dis 2017, 4, 185. [DOI] [PubMed] [Google Scholar]
- [15].Lim KS, Levato R, Costa PF, Castilho MD, Alcala-Orozco CR, van Dorenmalen KMA, Melchels FPW, Gawlitta D, Hooper GJ, Malda J, Woodfield TBF, Biofabrication 2018, 10. [DOI] [PubMed] [Google Scholar]
- [16].Heidarian P, Kouzani AZ, Kaynak A, Paulino M, Nasri-Nasrabadi B, Acs Biomater Sci Eng 2019, 5, 2688. [DOI] [PubMed] [Google Scholar]
- [17].Ng WL, Goh MH, Yeong WY, Naing MW, Biomater Sci-Uk 2018, 6, 562. [DOI] [PubMed] [Google Scholar]
- [18].Pimentel CR, Ko SK, Caviglia C, Wolff A, Emneus J, Keller SS, Dufva M, Acta Biomater 2018, 65, 174. [DOI] [PubMed] [Google Scholar]
- [19].Du ZM, Li NF, Hua YJ, Shi Y, Bao CY, Zhang HB, Yang Y, Lin QN, Zhu LY, Chem Commun 2017, 53, 13023. [DOI] [PubMed] [Google Scholar]
- [20].Madl CM, Katz LM, Heilshorn SC, Adv Funct Mater 2016, 26, 3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kageyama T, Osaki T, Enomoto J, Myasnikova D, Nittami T, Hozumi T, Ito T, Fukuda J, Acs Biomater Sci Eng 2016, 2, 1059. [DOI] [PubMed] [Google Scholar]
- [22].Bertlein S, Brown G, Lim KS, Jungst T, Boeck T, Blunk T, Tessmar J, Hooper GJ, Woodfield TBF, Groll J, Adv Mater 2017, 29. [DOI] [PubMed] [Google Scholar]
- [23].Costa JB, Silva-Correia J, Oliveira JM, Reis RL, Adv Healthc Mater 2017, 6. [DOI] [PubMed] [Google Scholar]
- [24].Hu D, Wu DW, Huang L, Jiao YP, Li LH, Lu L, Zhou CR, Mater Lett 2018, 223, 219. [Google Scholar]
- [25].Xu CC, Lee WH, Dai GH, Hong Y, Acs Appl Mater Inter 2018, 10, 9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dinh ND, Luo RC, Christine MTA, Lin WN, Shih WC, Goh JCH, Chen CH, Small 2017, 13. [DOI] [PubMed] [Google Scholar]
- [27].Zhai XY, Ma YF, Hou CY, Gao F, Zhang YY, Ruan CS, Pan HB, Lu WW, Liu WG, Acs Biomater Sci Eng 2017, 3, 1109. [DOI] [PubMed] [Google Scholar]
- [28].GhavamiNejad A, Hashmi S, Vatankhah-Varnoosfaderani M, Stadler FJ, Rheol Acta 2016, 55, 163. [Google Scholar]
- [29].Zheng ZZ, Wu JB, Liu M, Wang H, Li CM, Rodriguez MJ, Li G, Wang XQ, Kaplan DL, Adv Healthc Mater 2018, 7. [DOI] [PubMed] [Google Scholar]
- [30].Li HJ, Tan YJ, Liu SJ, Li L, Acs Appl Mater Inter 2018, 10, 11164. [DOI] [PubMed] [Google Scholar]
- [31].Morch YA, Donati I, Strand BL, Skjak-Braek G, Biomacromolecules 2006, 7, 1471. [DOI] [PubMed] [Google Scholar]
- [32].Hribar KC, Choi YS, Ondeck M, Engler AJ, Chen SC, Adv Funct Mater 2014, 24, 4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mandrycky C, Wang ZJ, Kim K, Kim DH, Biotechnol Adv 2016, 34, 422; S. C. Ligon, R. Liska, J. Stampfl, M. Gurr, R. Mulhaupt, Chem Rev 2017, 117, 10212; S. A. N.Ashammakhi, .Xu H.Montazerian, H.Ko, R.Nasir, N.Barrosab, A.Khademhossein, Materials Today Bio 2019, 1; S. Ostrovidov, S. Salehi, M. Costantini, K. Suthiwanich, M. Ebrahimi, R. B. Sadeghian, T. Fujie, X. T. Shi, S. Cannata, C. Gargioli, A. Tamayol, M. R. Dokmeci, G. Orive, W. Swieszkowski, A. Khademhosseini, Small 2019, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Khoon JHG Lim S, Cui Xiaolin, Lindberg Gabriella C. J., Burdick Jason A.,, Woodfield a. T. B. F., Chem Rev 2020. [DOI] [PubMed] [Google Scholar]
- [35].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA, P Natl Acad Sci USA 2016, 113, 3179; A. N. Shahabipour F, Oskuee RK, Bonakdar S, Hoffman T, Shokrgozar MA, Khademhosseini A, Translational Research 2019, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A, Biomaterials 2016, 110, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bruchet M, Melman A, Carbohyd Polym 2015, 131, 57. [DOI] [PubMed] [Google Scholar]
- [38].Du L, GhavamiNejad A, Yan ZC, Biswas CS, Stadler FJ, Carbohyd Polym 2018, 199, 58. [DOI] [PubMed] [Google Scholar]
- [39].Kesti M, Eberhardt C, Pagliccia G, Kenkel D, Grande D, Boss A, Zenobi-Wong M, Adv Funct Mater 2015, 25, 7406. [Google Scholar]
- [40].Sarker M, Izadifar M, Schreyer D, Chen XB, J Biomat Sci-Polym E 2018, 29, 1126. [DOI] [PubMed] [Google Scholar]
- [41].Tabriz AG, Hermida MA, Leslie NR, Shu WM, Biofabrication 2015, 7. [DOI] [PubMed] [Google Scholar]
- [42].Demirtas TT, Irmak G, Gumusderelioglu M, Biofabrication 2017, 9. [DOI] [PubMed] [Google Scholar]
- [43].Khalil S, Sun W, J Biomech Eng-T Asme 2009, 131. [DOI] [PubMed] [Google Scholar]
- [44].Freeman FE, Kelly DJ, Sci Rep-Uk 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Xiong RT, Zhang ZY, Chai WX, Huang Y, Chrisey DB, Biofabrication 2015, 7. [DOI] [PubMed] [Google Scholar]
- [46].Zhang T, Yan KRC, Ouyang LL, Sun W, Biofabrication 2013, 5. [DOI] [PubMed] [Google Scholar]
- [47].Izadifar M, Babyn P, Kelly ME, Chapman D, Chen XB, Tissue Eng Part C-Me 2017, 23, 548. [DOI] [PubMed] [Google Scholar]
- [48].Kundu J, Shim JH, Jang J, Kim SW, Cho DW, J Tissue Eng Regen M 2015, 9, 1286. [DOI] [PubMed] [Google Scholar]
- [49].Ning LQ, Sun HY, Lelong T, Guilloteau R, Zhu N, Schreyer DJ, Chen XB, Biofabrication 2018, 10. [DOI] [PubMed] [Google Scholar]
- [50].Farber JL, Environ Health Persp 1990, 84, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Spring Ser Biomat S 2018, 11, 1; K. B. Fonseca, F. R. Maia, F. A. Cruz, D. Andrade, M. A. Juliano, P. L. Granja, C. C. Barrias, Soft Matter 2013, 9, 3283; F. You, X. Wu, X. B. Chen, Int J Polym Mater Po 2017, 66, 299; Y. Sapir, O. Kryukov, S. Cohen, Biomaterials 2011, 32, 1838; J. C. Sun, H. P. Tan, Materials 2013, 6, 1285. [Google Scholar]
- [52].Habib A, Sathish V, Mallik S, Khoda B, Materials 2018, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schutz K, Placht AM, Paul B, Bruggemeier S, Gelinsky M, Lode A, J Tissue Eng Regen M 2017, 11, 1574. [DOI] [PubMed] [Google Scholar]
- [54].Paxton N, Smolan W, Bock T, Melchels F, Groll J, Jungst T, Biofabrication 2017, 9. [DOI] [PubMed] [Google Scholar]
- [55].Gao G, Lee JH, Jang J, Lee DH, Kong JS, Kim BS, Choi YJ, Jang WB, Hong YJ, Kwon SM, Cho DW, Adv Funct Mater 2017, 27. [Google Scholar]
- [56].Andrew McCormack CBH, Nicholas R Ferry Leslie Melchels PW, Trends in Biotechnology 2020. [DOI] [PubMed] [Google Scholar]
- [57].Ahn S, Lee H, Bonassar LJ, Kim G, Biomacromolecules 2012, 13, 2997. [DOI] [PubMed] [Google Scholar]
- [58].Wilkinson NJ, Smith MAA, Kay RW, Harris RA, Int J Adv Manuf Tech 2019, 105, 4599. [Google Scholar]
- [59].Duan B, Hockaday LA, Kang KH, Butcher JT, J Biomed Mater Res A 2013, 101, 1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Faulkner-Jones A, Fyfe C, Cornelissen DJ, Gardner J, King J, Courtney A, Shu WM, Biofabrication 2015, 7. [DOI] [PubMed] [Google Scholar]
- [61].Li Z, Huang S, Liu YF, Yao B, Hu T, Shi HG, Xie JF, Fu XB, Sci Rep-Uk 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ding HZ, Tourlomousis F, Chang RC, Biomed Phys Eng Expr 2017, 3. [Google Scholar]
- [63].Markstedt K, Mantas A, Tournier I, Avila HM, Hagg D, Gatenholm P, Biomacromolecules 2015, 16, 1489. [DOI] [PubMed] [Google Scholar]
- [64].Bendtsen ST, Wei M, J Biomed Mater Res A 2017, 105, 3262. [DOI] [PubMed] [Google Scholar]
- [65].Izadifar Z, Chang TJ, Kulyk W, Chen XB, Eames BF, Tissue Eng Part C-Me 2016, 22, 173. [DOI] [PubMed] [Google Scholar]
- [66].Muthukumar T, Song JE, Khang G, Molecules 2019, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Coutinho DF, Sant SV, Shin H, Oliveira JT, Gomes ME, Neves NM, Khademhosseini A, Reis RL, Biomaterials 2010, 31, 7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lozano R, Stevens L, Thompson BC, Gilmore KJ, Gorkin R, Stewart EM, Panhuis MIH, Romero-Ortega M, Wallace GG, Biomaterials 2015, 67, 264. [DOI] [PubMed] [Google Scholar]
- [69].Ferris CJ, Stevens LR, Gilmore KJ, Mume E, Greguric I, Kirchmajer DM, Wallace GG, Panhuis MIH, J Mater Chem B 2015, 3, 1106. [DOI] [PubMed] [Google Scholar]
- [70].Ferris CJ, Gilmore KJ, Wallace GG, Panhuis MIH, Soft Matter 2013, 9, 3705. [Google Scholar]
- [71].GhavamiNejad A, Lu B, Giacca A, Wu XY, Nanoscale 2019, 11, 10167; A. GhavamiNejad, C. H. Park, C. S. Kim, Biomacromolecules 2016, 17, 1213. [DOI] [PubMed] [Google Scholar]
- [72].Ng WL, Yeong WY, Naing MW, Int J Bioprinting 2016, 2, 53. [Google Scholar]
- [73].Liu QQ, Li QT, Xu S, Zheng QJ, Cao XD, Polymers-Basel 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Croce N, Bernardini S, Di Cecca S, Caltagirone C, Angelucci F, J Neurosci Meth 2013, 217, 26. [DOI] [PubMed] [Google Scholar]
- [75].Peak CW, Stein J, Gold KA, Gaharwar AK, Langmuir 2018, 34, 917. [DOI] [PubMed] [Google Scholar]
- [76].Rodriguez MJ, Dixon TA, Cohen E, Huang WW, Omenetto FG, Kaplan DL, Acta Biomater 2018, 71, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Vatankhah-Varnoosfaderani M, Hashmi S, Stadler FJ, Macromol Rapid Comm 2015, 36, 447. [DOI] [PubMed] [Google Scholar]
- [78].Wang HB, Zhu H, Fu WG, Zhang YY, Xu B, Gao F, Cao ZQ, Liu WG, Macromol Rapid Comm 2017, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hashmi S, Vatankhah-Varnoosfaderani M, GhavamiNejad A, Obiweluozor FO, Du B, Stadler FJ, Rheol Acta 2015, 54, 501. [Google Scholar]
- [80].GhavamiNejad A, Hashmi S, Joh HI, Lee S, Lee YS, Vatankhah-Varnoosfaderani M, Stadler FJ, Phys Chem Chem Phys 2014, 16, 8675. [DOI] [PubMed] [Google Scholar]
- [81].Tan ZC, Parisi C, Di Silvio L, Dini D, Forte AE, Sci Rep-Uk 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Akiyama Y, Shinose M, Watanabe H, Yamada S, Kanda Y, P Natl Acad Sci USA 2019, 116, 7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Li C, Faulkner-Jones A, Dun AR, Jin J, Chen P, Xing YZ, Yang ZQ, Li ZB, Shu WM, Liu DS, Duncan RR, Angew Chem Int Edit 2015, 54, 3957. [DOI] [PubMed] [Google Scholar]
- [84].Ouyang LL, Highley CB, Rodell CB, Sun W, Burdick JA, Acs Biomater Sci Eng 2016, 2, 1743. [DOI] [PubMed] [Google Scholar]
- [85].Wang QS, Han GC, Yan SQ, Zhang Q, Materials 2019, 12. [Google Scholar]
- [86].Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, Johansson F, Randles A, Rosenkrantz JE, Louis-Rosenberg JD, Galie PA, Stevens KR, Miller JS, Science 2019, 364, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Guillotin B, Ali M, Ducom A, Catros S, Keriquel V, Souquet A, Remy M, Fricain JC, Guillemot F, Biofabrication: Micro- and Nano-Fabrication, Printing, Patterning, and Assemblies 2013, 95. [Google Scholar]
- [88].Bernal PN, Delrot P, Loterie D, Li Y, Malda J, Moser C, Levato R, Adv Mater 2019, 31. [DOI] [PubMed] [Google Scholar]
- [89].Ouyang LL, Highley CB, Sun W, Burdick JA, Adv Mater 2017, 29. [DOI] [PubMed] [Google Scholar]
- [90].Zhu K, Shin SR, van Kempen T, Li YC, Ponraj V, Nasajpour A, Mandla S, Hu N, Liu X, Leijten J, Lin YD, Hussain MA, Zhang YS, Tamayol A, Khademhosseini A, Adv Funct Mater 2017, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, Lang Q, Zhang YS, Shin SR, Calzone G, Annabi N, Shupe TD, Bishop CE, Atala A, Dokmeci MR, Khademhosseini A, Biofabrication 2016, 8. [DOI] [PubMed] [Google Scholar]
- [92].Cui HT, Zhu W, Nowicki M, Zhou X, Khademhosseini A, Zhang LG, Adv Healthc Mater 2016, 5, 2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Byambaa B, Annabi N, Yue K, Trujillo-de Santiago G, Alvarez MM, Jia WT, Kazemzadeh-Narbat M, Shin SR, Tamayol A, Khademhosseini A, Adv Healthc Mater 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ashammakhi N, Hasan A, Kaarela O, Byambaa B, Sheikhi A, Gaharwar AK, Khademhosseini A, Adv Healthc Mater 2019, 8. [DOI] [PubMed] [Google Scholar]
- [95].Richards D, Jia J, Yost M, Markwald R, Mei Y, Ann Biomed Eng 2017, 45, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bertassoni LE, Cardoso JC, Manoharan V, Cristino AL, Bhise NS, Araujo WA, Zorlutuna P, Vrana NE, Ghaemmaghami AM, Dokmeci MR, Khademhosseini A, Biofabrication 2014, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhuang P, Ng WL, An J, Chua CK, Tan LP, Plos One 2019, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhu W, Qu X, Zhu J, Ma XY, Patel S, Liu J, Wang PR, Lai CSE, Gou ML, Xu Y, Zhang K, Chen SC, Biomaterials 2017, 124, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Gurkan UA, El Assal R, Yildiz SE, Sung Y, Trachtenberg AJ, Kuo WP, Demirci U, Mol Pharmaceut 2014, 11, 2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zhai XY, Ruan CS, Ma YF, Cheng DL, Wu MM, Liu WG, Zhao XL, Pan HB, Lu WWJ, Adv Sci 2018, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Miao SD, Zhu W, Castro NJ, Nowicki M, Zhou X, Cui HT, Fisher JP, Zhang LG, Sci Rep-Uk 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Wang ZJ, Jin X, Tian ZL, Menard F, Holzman JF, Kim K, Adv Healthc Mater 2018, 7. [DOI] [PubMed] [Google Scholar]
- [103].Hu JL, Hou YP, Park H, Choi B, Hou SY, Chung A, Lee M, Acta Biomater 2012, 8, 1730. [DOI] [PubMed] [Google Scholar]
- [104].Al Mousawi A, Lara DM, Noirbent G, Dumur F, Toufaily J, Hamieh T, Buo TT, Goubard F, Graff B, Gigmes D, Fouassier JP, Lalevee J, Macromolecules 2017, 50, 4913. [Google Scholar]
- [105].Serna JA, Florez SL, Talero VA, Briceno JC, Munoz-Camargo C, Cruz JC, Polymers-Basel 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Sakai S, Kamei H, Mori T, Hotta T, Ohi H, Nakahata M, Taya M, Biomacromolecules 2018, 19, 672. [DOI] [PubMed] [Google Scholar]
- [107].Cui XL, Soliman BG, Alcala-Orozco CR, Li J, Vis MAM, Santos M, Wise SG, Levato R, Malda J, Woodfield TBF, Rnjak-Kovacina J, Lim KS, Adv Healthc Mater 2020, 9. [DOI] [PubMed] [Google Scholar]
- [108].Lim KS, Klotz BJ, Lindberg GCJ, Melchels FPW, Hooper GJ, Malda J, Gawlitta D, Woodfield TBF, Macromol Biosci 2019, 19. [DOI] [PubMed] [Google Scholar]
- [109].Mazaki T, Shiozaki Y, Yamane K, Yoshida A, Nakamura M, Yoshida Y, Zhou D, Kitajima T, Tanaka M, Ito Y, Ozaki T, Matsukawa A, Sci Rep-Uk 2014, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Na K, Shin S, Lee H, Shin D, Baek J, Kwak H, Park M, Shin J, Hyun J, J Ind Eng Chem 2018, 61, 340. [Google Scholar]
- [111].Duchi S, Onofrillo C, O’Connell CD, Blanchard R, Augustine C, Quigley AF, Kapsa RMI, Pivonka P, Wallace G, Di Bella C, Choong PFM, Sci Rep-Uk 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS, Biomaterials 2009, 30, 6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Colosi C, Shin SR, Manoharan V, Massa S, Costantini M, Barbetta A, Dokmeci MR, Dentini M, Khademhosseini A, Adv Mater 2016, 28, 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Zhu W, Cui HT, Boualam B, Masood F, Flynn E, Rao RD, Zhang ZY, Zhang LG, Nanotechnology 2018, 29. [DOI] [PubMed] [Google Scholar]
- [115].van der Valk DC, van der Ven CFT, Blaser MC, Grolman JM, Wu PJ, Fenton OS, Lee LH, Tibbitt MW, Andresen JL, Wen JR, Ha AH, Buffolo F, van Mil A, Bouten CVC, Body SC, Mooney DJ, Sluijter JPG, Aikawa M, Hjortnaes J, Langer R, Aikawa E, Nanomaterials-Basel 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Celikkin N, Padial JS, Costantini M, Hendrikse H, Cohn R, Wilson CJ, Rowan AE, Swieszkowski W, Polymers-Basel 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA, Adv Mater 2014, 26, 3124. [DOI] [PubMed] [Google Scholar]
- [118].Stratesteffen H, Kopf M, Kreimendahl F, Blaeser A, Jockenhoevel S, Fischer H, Biofabrication 2017, 9. [DOI] [PubMed] [Google Scholar]
- [119].Izadifar M, Chapman D, Babyn P, Chen XB, Kelly ME, Tissue Eng Part C-Me 2018, 24, 74. [DOI] [PubMed] [Google Scholar]
- [120].Zhou X, Zhu W, Nowicki M, Miao S, Cui HT, Holmes B, Glazer RI, Zhang LG, Acs Appl Mater Inter 2016, 8, 30017. [DOI] [PubMed] [Google Scholar]
- [121].Taya SSHOTHHKM, Biopolymers.
- [122].Lim KS, Schon BS, Mekhileri NV, Brown GCJ, Chia CM, Prabakar S, Hooper GJ, Woodfield TBF, Acs Biomater Sci Eng 2016, 2, 1752. [DOI] [PubMed] [Google Scholar]
- [123].Xu WY, Zhang X, Yang PR, Langvik O, Wang XJ, Zhang YC, Cheng F, Osterberg M, Willfor S, Xu CL, Acs Appl Mater Inter 2019, 11, 12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Kesti M, Muller M, Becher J, Schnabelrauch M, D’Este M, Eglin D, Zenobi-Wong M, Acta Biomater 2015, 11, 162. [DOI] [PubMed] [Google Scholar]
- [125].Ali M, Kumar PRA, Yoo JJ, Zahran F, Atala A, Lee SJ, Adv Healthc Mater 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Poldervaart MT, Goversen B, de Ruijter M, Abbadessa A, Melchels FPW, Oner FC, Dhert WJA, Vermonden T, Alblas J, Plos One 2017, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Bagheri A, Jin JY, Acs Appl Polym Mater 2019, 1, 593; L. Chen, Q. Y. Wu, G. Wei, R. Liu, Z. Q. Li, J Mater Chem C 2018, 6. [Google Scholar]
- [128].Stichler S, Bock T, Paxton N, Bertlein S, Levato R, Schill V, Smolan W, Malda J, Tessmar J, Blunk T, Groll J, Biofabrication 2017, 9. [DOI] [PubMed] [Google Scholar]
- [129].Ooi HW, Mota C, ten Cate AT, Calore A, Moroni L, Baker MB, Biomacromolecules 2018, 19, 3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Loebel C, Broguiere N, Alini M, Zenobi-Wong M, Eglin D, Biomacromolecules 2015, 16, 2624. [DOI] [PubMed] [Google Scholar]
- [131].Daly AC, Critchley SE, Rencsok EM, Kelly DJ, Biofabrication 2016, 8. [DOI] [PubMed] [Google Scholar]
- [132].Campos DFD, Blaeser A, Weber M, Jakel J, Neuss S, Jahnen-Dechent W, Fischer H, Biofabrication 2013, 5. [DOI] [PubMed] [Google Scholar]
- [133].Forget A, Blaeser A, Miessmer F, Kopf M, Campos DFD, Voelcker NH, Blencowe A, Fischer H, Shastri VP, Adv Healthc Mater 2017, 6. [DOI] [PubMed] [Google Scholar]
- [134].Law N, Doney B, Glover H, Qin YH, Aman ZM, Sercombe TB, Liew LJ, Dilley RJ, Doyle BJ, J Mech Behav Biomed 2018, 77, 389. [DOI] [PubMed] [Google Scholar]
- [135].Cochis A, Bonetti L, Sorrentino R, Negrini NC, Grassi F, Leigheb M, Rimondini L, Fare S, Materials 2018, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Ahn G, Min KH, Kim C, Lee JS, Kang D, Won JY, Cho DW, Kim JY, Jin S, Yun WS, Shim JH, Sci Rep-Uk 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Melchels FPW, Blokzijl MM, Levato R, Peiffer QC, de Ruijter M, Hennink WE, Vermonden T, Malda J, Biofabrication 2016, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Han D, Lu ZC, Chester SA, Lee H, Sci Rep-Uk 2018, 8; J. N. Tang, X. L. Cui, T. G. Caranasos, M. T. Hensley, A. C. Vandergriff, Y. Hartanto, D. L. Shen, H. Zhang, J. Y. Zhang, K. Cheng, Acs Nano 2017, 11, 9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Park JY, Choi JC, Shim JH, Lee JS, Park H, Kim SW, Doh J, Cho DW, Biofabrication 2014, 6. [DOI] [PubMed] [Google Scholar]
- [140].Wilson SA, Cross LM, Peak CW, Gaharwar AK, Acs Appl Mater Inter 2017, 9, 43449. [DOI] [PubMed] [Google Scholar]
- [141].Lin HH, Hsieh FY, Tseng CS, Hsu SH, J Mater Chem B 2016, 4, 6694. [DOI] [PubMed] [Google Scholar]
- [142].Hui Xie K-K, Wang Yu-Zhong, Progress in Polymer science 2019, 95, 32. [Google Scholar]
- [143].Wei Z, Yang JH, Liu ZQ, Xu F, Zhou JX, Zrínyi M, Osada Y, Chen YM, Advanced Functional Materials 2015, 25, 1352. [Google Scholar]
- [144].Guo B, Qu J, Zhao X, Zhang M, Acta Biomaterialia 2019, 84, 180. [DOI] [PubMed] [Google Scholar]
- [145].Chen YC, Su WY, Yang SH, Gefen A, Lin FH, Acta Biomater 2013, 9, 5181. [DOI] [PubMed] [Google Scholar]
- [146].Kim SW, Kim DY, Roh HH, Kim HS, Lee JW, Lee KY, Biomacromolecules 2019, 20, 1860. [DOI] [PubMed] [Google Scholar]
- [147].Yang K, Grant JC, Lamey P, Joshi-Imre A, Lund BR, Smaldone RA, Voit W, Adv Funct Mater 2017, 27. [Google Scholar]
- [148].Zhang YJ, Chen H, Zhang TT, Zan Y, Ni TY, Liu M, Pei RJ, Biomater Sci-Uk 2018, 6, 2578. [DOI] [PubMed] [Google Scholar]
- [149].Rutz AL, Hyland KE, Jakus AE, Burghardt WR, Shah RN, Adv Mater 2015, 27, 1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Wang MD, Zhai P, Schreyer DJ, Zheng RS, Sun XD, Cui FZ, Chen XB, Front Mater Sci 2013, 7, 269. [Google Scholar]
- [151].Sisson K, Zhang C, Farach-Carson MC, Chase DB, Rabolt JF, Biomacromolecules 2009, 10, 1675. [DOI] [PubMed] [Google Scholar]
- [152].Usta M, Piech DL, MacCrone RK, Hillig WB, Biomaterials 2003, 24, 165. [DOI] [PubMed] [Google Scholar]
- [153].Talebian A, Kordestani SS, Rashidi A, Dadashian F, Montazer M, Kem Ind 2007, 56, 537. [Google Scholar]
- [154].Rose JB, Pacelli S, El Haj AJ, Dua HS, Hopkinson A, White LJ, Rose FRAJ, Materials 2014, 7, 3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Kim YB, Lee HJ, Kim GH, Acs Appl Mater Inter 2016, 8, 32230. [DOI] [PubMed] [Google Scholar]
- [156].Teixeira LSM, Feijen J, van Blitterswijk CA, Dijkstra PJ, Karperien M, Biomaterials 2012, 33, 1281. [DOI] [PubMed] [Google Scholar]
- [157].Zhou MM, Leet BL, Tan LP, Int J Bioprinting 2017, 3, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Shi Y, Xing TL, Zhang HB, Yin RX, Yang SM, Wei J, Zhang WJ, Biomed Mater 2018, 13. [DOI] [PubMed] [Google Scholar]
- [159].Hu WK, Wang ZJ, Xiao Y, Zhang SM, Wang JL, Biomater Sci-Uk 2019, 7, 843. [DOI] [PubMed] [Google Scholar]
- [160].Smith KC, Radiat Res 1966, S, 54. [PubMed] [Google Scholar]
- [161].Kazemzadeh-Narbat M, Rouwkema J, Annabi N, Cheng H, Ghaderi M, Cha BH, Aparnathi M, Khalilpour A, Byambaa B, Jabbari E, Tamayol A, Khademhosseini A, Adv Healthc Mater 2017, 6; R. F. Pereira, P. J. Bartolo, J Appl Polym Sci 2015, 132. [DOI] [PubMed] [Google Scholar]
- [162].Liaw CY, Guvendiren M, Biofabrication 2017, 9. [DOI] [PubMed] [Google Scholar]
- [163].Wang YN, Adokoh CK, Narain R, Expert Opin Drug Del 2018, 15, 77. [DOI] [PubMed] [Google Scholar]
- [164].Angele-Martinez C, Goodman C, Brumaghim J, Metallomics 2014, 6, 1358. [DOI] [PubMed] [Google Scholar]
- [165].Xiong RT, Chai WX, Huang Y, Lab Chip 2019, 19, 1644. [DOI] [PubMed] [Google Scholar]
- [166].Hoyle CE, Lee TY, Roper T, J Polym Sci Pol Chem 2004, 42, 5301. [Google Scholar]
- [167].Valero C, Amaveda H, Mora M, Garcia-Aznar JM, Plos One 2018, 13; A. Hajikhani, F. Scocozza, M. Conti, M. Marino, F. Auricchio, P. Wriggers, 0, 0391398819856024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Valot L, Martinez J, Mehdi A, Subra G, Chem Soc Rev 2019, 48, 4049. [DOI] [PubMed] [Google Scholar]
- [169].Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue HJ, Ramadan MH, Hudson AR, Feinberg AW, Sci Adv 2015, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Wust S, Godla ME, Muller R, Hofmann S, Acta Biomater 2014, 10, 630. [DOI] [PubMed] [Google Scholar]
- [171].Khong TT, Aarstad OA, Skjak-Braek G, Draget KI, Varum KM, Biomacromolecules 2013, 14, 2765. [DOI] [PubMed] [Google Scholar]
- [172].Qinqin Dang SL, Yu Shen, Pingchuan Sun, Yuan Zhi, Biomacromolecules 2010, 11, 1796. [DOI] [PubMed] [Google Scholar]
- [173].Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang YZ, Khademhosseini A, Lab Chip 2014, 14, 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Yoon HJ, Shin SR, Cha JM, Lee SH, Kim JH, Do JT, Song H, Bae H, Plos One 2016, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Blaeser A, Campos DFD, Puster U, Richtering W, Stevens MM, Fischer H, Adv Healthc Mater 2016, 5, 326. [DOI] [PubMed] [Google Scholar]
- [176].Fedorovich NE, Schuurman W, Wijnberg HM, Prins HJ, van Weeren PR, Malda J, Alblas J, Dhert WJA, Tissue Eng Part C-Me 2012, 18, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Koch L, Deiwick A, Schlie S, Michael S, Gruene M, Coger V, Zychlinski D, Schambach A, Reimers K, Vogt PM, Chichkov B, Biotechnol Bioeng 2012, 109, 1855. [DOI] [PubMed] [Google Scholar]
- [178].Catros S, Guillemot F, Nandakumar A, Ziane S, Moroni L, Habibovic P, van Blitterswijk C, Rousseau B, Chassande O, Amedee J, Fricain JC, Tissue Eng Part C-Me 2012, 18, 62. [DOI] [PubMed] [Google Scholar]
- [179].Park J, Lee SJ, Chung S, Lee JH, Kim WD, Lee JY, Park SA, Mat Sci Eng C-Mater 2017, 71, 678. [DOI] [PubMed] [Google Scholar]
- [180].Loozen LD, Wegman F, Oner FC, Dhert WJA, Alblas J, J Mater Chem B 2013, 1, 6619. [DOI] [PubMed] [Google Scholar]
- [181].Arai K, Iwanaga S, Toda H, Genci C, Nishiyama Y, Nakamura M, Biofabrication 2011, 3. [DOI] [PubMed] [Google Scholar]
- [182].Yu Y, Zhang YH, Martin JA, Ozbolat IT, J Biomech Eng-T Asme 2013, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Rathan S, Dejob L, Schipani R, Haffner B, Mobius ME, Kelly DJ, Adv Healthc Mater 2019, 8. [DOI] [PubMed] [Google Scholar]
- [184].Jiang T, Munguia-Lopez JG, Flores-Torres S, Grant J, Vijayakumar S, De Leon-Rodriguez A, Kinsella JM, Sci Rep-Uk 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].He Y, Yang FF, Zhao HM, Gao Q, Xia B, Fu JZ, Sci Rep-Uk 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Huang S, Yao B, Xie JF, Fu XB, Acta Biomater 2016, 32, 170. [DOI] [PubMed] [Google Scholar]
- [187].Ouyang LL, Yao R, Zhao Y, Sun W, Biofabrication 2016, 8. [DOI] [PubMed] [Google Scholar]
- [188].Jin YF, Compaan A, Bhattacharjee T, Huang Y, Biofabrication 2016, 8. [DOI] [PubMed] [Google Scholar]
- [189].Wang XH, Tolba E, Schroder HC, Neufurth M, Feng QL, Diehl-Seifert B, Muller WEG, Plos One 2014, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Ahlfeld T, Schuster FP, Forster Y, Quade M, Akkineni AR, Rentsch C, Rammelt S, Gelinsky M, Lode A, Adv Healthc Mater 2019, 8. [DOI] [PubMed] [Google Scholar]
- [191].Wu ZJ, Su X, Xu YY, Kong B, Sun W, Mi SL, Sci Rep-Uk 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Ahlfeld T, Doberenz F, Kilian D, Vater C, Korn P, Lauer G, Lode A, Gelinsky M, Biofabrication 2018, 10. [DOI] [PubMed] [Google Scholar]
- [193].Apelgren P, Amoroso M, Lindahl A, Brantsing C, Rotter N, Gatenholm P, Kolby L, Plos One 2017, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Thayer PS, Orrhult LS, Martinez H, Jove-J Vis Exp 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Athirasala A, Tahayeri A, Thrivikraman G, Franca CM, Monteiro N, Tran V, Ferracane J, Bertassoni LE, Biofabrication 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Romanazzo S, Vedicherla S, Moran C, Kelly DJ, J Tissue Eng Regen M 2018, 12, E1826. [DOI] [PubMed] [Google Scholar]
- [197].Compaan AM, Christensen K, Huang Y, Acs Biomater Sci Eng 2017, 3, 1519. [DOI] [PubMed] [Google Scholar]
- [198].Muller WEG, Schroder HC, Feng QL, Schlossmacher U, Link T, Wang XH, J Tissue Eng Regen M 2015, 9, E39. [DOI] [PubMed] [Google Scholar]
- [199].Isaacson A, Swioklo S, Connon CJ, Exp Eye Res 2018, 173, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Yoon S, Park JA, Lee HR, Yoon WH, Hwang DS, Jung S, Adv Healthc Mater 2018, 7. [DOI] [PubMed] [Google Scholar]
- [201].Duin S, Schutz K, Ahlfeld T, Lehmann S, Lode A, Ludwig B, Gelinsky M, Adv Healthc Mater 2019, 8. [DOI] [PubMed] [Google Scholar]
- [202].Roth AD, Lama P, Dunn S, Hong S, Lee MY, Mat Sci Eng C-Mater 2018, 90, 634. [DOI] [PubMed] [Google Scholar]
- [203].Li HJ, Tan YJ, Li L, Carbohyd Polym 2018, 198, 261. [DOI] [PubMed] [Google Scholar]
- [204].Chimene D, Peak CW, Gentry JL, Carrow JK, Cross LM, Mondragon E, Cardoso GB, Kaunas R, Gaharwar AK, Acs Appl Mater Inter 2018, 10, 9957. [DOI] [PubMed] [Google Scholar]
- [205].Sorkio A, Koch L, Koivusalo L, Deiwick A, Miettinen S, Chichkov B, Skottman H, Biomaterials 2018, 171, 57. [DOI] [PubMed] [Google Scholar]
- [206].Hedegaard CL, Collin EC, Redondo-Gomez C, Nguyen LTH, Ng KW, Castrejon-Pita AA, Castrejon-Pita JR, Mata A, Adv Funct Mater 2018, 28. [Google Scholar]
- [207].Ahlfeld T, Cidonio G, Kilian D, Duin S, Akkineni AR, Dawson JI, Yang S, Lode A, Oreffo ROC, Gelinsky M, Biofabrication 2017, 9. [DOI] [PubMed] [Google Scholar]
- [208].Dubbin K, Hori Y, Lewis KK, Heilshorn SC, Adv Healthc Mater 2016, 5, 2488. [DOI] [PubMed] [Google Scholar]
- [209].Hart LR, Li SW, Sturgess C, Wildman R, Jones JR, Hayes W, Acs Appl Mater Inter 2016, 8, 3115. [DOI] [PubMed] [Google Scholar]
- [210].Invernizzi M, Turri S, Levi M, Suriano R, Eur Polym J 2018, 101, 169. [Google Scholar]
- [211].Shao Y, Li C, Zhou X, Chen P, Yang ZQ, Li ZB, Liu DS, Acta Chim Sinica 2015, 73, 815. [Google Scholar]
- [212].Kuo CY, Eranki A, Placone JK, Rhodes KR, Aranda-Espinoza H, Fernandes R, Fisher JP, Kim PCW, Acs Biomater Sci Eng 2016, 2, 1817. [DOI] [PubMed] [Google Scholar]
- [213].Levato R, Webb WR, Otto IA, Mensinga A, Zhang YD, van Rijen M, van Weeren R, Khan IM, Malda J, Acta Biomater 2017, 61, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Meng FB, Meyer CM, Joung D, Vallera DA, McAlpine MC, Panoskaltsis-Mortari A, Adv Mater 2019, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Kerscher P, Kaczmarek JA, Head SE, Ellis ME, Seeto WJ, Kim J, Bhattacharya S, Suppiramaniam V, Lipke EA, Acs Biomater Sci Eng 2017, 3, 1499. [DOI] [PubMed] [Google Scholar]
- [216].McBeth C, Lauer J, Ottersbach M, Campbell J, Sharon A, Sauer-Budge AF, Biofabrication 2017, 9; S. W. Sawyer, S. V. Shridhar, K. R. Zhang, L. D. Albrecht, A. B. Filip, J. A. Horton, P. Soman, Biofabrication 2018, 10. [DOI] [PubMed] [Google Scholar]
- [217].Yin J, Yan ML, Wang YC, Fu JZ, Suo HR, Acs Appl Mater Inter 2018, 10, 6849. [DOI] [PubMed] [Google Scholar]
- [218].Miri AK, Nieto D, Iglesias L, Hosseinabadi HG, Maharjan S, Ruiz-Esparza GU, Khoshakhlagh P, Manbachi A, Dokmeci MR, Chen SC, Shin SR, Zhang YS, Khademhosseini A, Adv Mater 2018, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Jia WT, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu WJ, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Khademhosseini A, Biomaterials 2016, 106, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Koffler J, Zhu W, Qu X, Platoshyn O, Dulin JN, Brock J, Graham L, Lu P, Sakamoto J, Marsala M, Chen SC, Tuszynski MH, Nat Med 2019, 25, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Costantini M, Idaszek J, Szoke K, Jaroszewicz J, Dentini M, Barbetta A, Brinchmann JE, Swieszkowski W, Biofabrication 2016, 8. [DOI] [PubMed] [Google Scholar]
- [222].Kuss M, Kim J, Qi DJ, Wu SH, Lei YG, Chung S, Duan B, Acta Biomater 2018, 71, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Daly AC, Pitacco P, Nulty J, Cunniffe GM, Kelly DJ, Biomaterials 2018, 162, 34. [DOI] [PubMed] [Google Scholar]
- [224].Mouser VHM, Melchels FPW, Visser J, Dhert WJA, Gawlitta D, Malda J, Biofabrication 2016, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Kosik-Koziol A, Costantini M, Mroz A, Idaszek J, Heljak M, Jaroszewicz J, Kijenska E, Szoke K, Frerker N, Barbetta A, Brinchmann JE, Swieszkowski W, Biofabrication 2019, 11. [DOI] [PubMed] [Google Scholar]
- [226].Liu X, Carter SSD, Renes MJ, Kim J, Rojas-Canales DM, Penko D, Angus C, Beirne S, Drogemuller CJ, Yue ZL, Coates PT, Wallace GG, Adv Healthc Mater 2019, 8. [DOI] [PubMed] [Google Scholar]
- [227].Ojansivu M, Rashad A, Ahlinder A, Massera J, Mishra A, Syverud K, Finne-Wistrand A, Miettinen S, Mustafa K, Biofabrication 2019, 11. [DOI] [PubMed] [Google Scholar]
- [228].Daly AC, Cunniffe GM, Sathy BN, Jeon O, Alsberg E, Kelly DJ, Adv Healthc Mater 2016, 5, 2353. [DOI] [PubMed] [Google Scholar]
- [229].Ma YF, Ji Y, Huang GY, Ling K, Zhang XH, Xu F, Biofabrication 2015, 7. [DOI] [PubMed] [Google Scholar]
- [230].Wang ZJ, Abdulla R, Parker B, Samanipour R, Ghosh S, Kim K, Biofabrication 2015, 7. [DOI] [PubMed] [Google Scholar]
- [231].Ma YF, Ji Y, Zhong TY, Wan WT, Yang QZ, Li A, Zhang XH, Lin M, Acs Biomater Sci Eng 2017, 3, 3534. [DOI] [PubMed] [Google Scholar]
- [232].Zhou X, Cui HT, Nowicki M, Miao SD, Lee SJ, Masood F, Harris BT, Zhang LG, Acs Appl Mater Inter 2018, 10, 8993. [DOI] [PubMed] [Google Scholar]
- [233].Thakur A, Jaiswal MK, Peak CW, Carrow JK, Gentry J, Dolatshahi-Pirouz A, Gaharwar AK, Nanoscale 2016, 8, 12362. [DOI] [PubMed] [Google Scholar]
- [234].Cui XF, Breitenkamp K, Lotz M, D’Lima D, Biotechnol Bioeng 2012, 109, 2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Cui XF, Breitenkamp K, Finn MG, Lotz M, D’Lima DD, Tissue Eng Pt A 2012, 18, 1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Pescosolido L, Schuurman W, Malda J, Matricardi P, Alhaique F, Coviello T, van Weeren PR, Dhert WJA, Hennink WE, Vermonden T, Biomacromolecules 2011, 12, 1831. [DOI] [PubMed] [Google Scholar]
- [237].Gao GF, Schilling AF, Yonezawa T, Wang J, Dai GH, Cui XF, Biotechnol J 2014, 9, 1304. [DOI] [PubMed] [Google Scholar]
- [238].Muller M, Becher J, Schnabelrauch M, Zenobi-Wong M, Biofabrication 2015, 7. [DOI] [PubMed] [Google Scholar]
- [239].Kirillova A, Maxson R, Stoychev G, Gomillion CT, Ionov L, Adv Mater 2017, 29. [DOI] [PubMed] [Google Scholar]
- [240].Kim SH, Yeon YK, Lee JM, Chao JR, Lee YJ, Seo YB, Sultan MT, Lee OJ, Lee JS, Yoon SI, Hong IS, Khang G, Lee SJ, Yoo JJ, Park CH, Nat Commun 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Wang Y, Wu SH, Kuss MA, Streube PN, Duan B, Acs Biomater Sci Eng 2017, 3, 826. [DOI] [PubMed] [Google Scholar]
- [242].Yang WG, Yu HB, Li GX, Wang YC, Liu LQ, Small 2017, 13; M. Costantini, S. Testa, P. Mozetic, A. Barbetta, C. Fuoco, E. Fornetti, F. Tamiro, S. Bernardini, J. Jaroszewicz, W. Swieszkowski, M. Trombetta, L. Castagnoli, D. Seliktar, P. Garstecki, G. Cesareni, S. Cannata, A. Rainer, C. Gargioli, Biomaterials 2017, 131, 98. [Google Scholar]
- [243].Jin YF, Liu CC, Chai WX, Compaan A, Huang Y, Acs Appl Mater Inter 2017, 9, 17457. [Google Scholar]
- [244].Hsieh FY, Lin HH, Hsu SH, Biomaterials 2015, 71, 48. [DOI] [PubMed] [Google Scholar]
- [245].Rhee S, Puetzer JL, Mason BN, Reinhart-King CA, Bonassar LJ, Acs Biomater Sci Eng 2016, 2, 1800. [DOI] [PubMed] [Google Scholar]
- [246].Jakab K, Norotte C, Damon B, Marga F, Neagu A, Besch-Williford CL, Kachurin A, Church KH, Park H, Mironov V, Markwald R, Vunjak-Novakovic G, Forgacs G, Tissue Eng Pt A 2008, 14, 413. [DOI] [PubMed] [Google Scholar]
- [247].Liu WJ, Heinrich MA, Zhou YX, Akpek A, Hu N, Liu X, Guan XF, Zhong Z, Jin XY, Khademhosseini A, Zhang YS, Adv Healthc Mater 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Schuurman W, Levett PA, Pot MW, van Weeren PR, Dhert WJA, Hutmacher DW, Melchels FPW, Klein TJ, Malda J, Macromol Biosci 2013, 13, 551. [DOI] [PubMed] [Google Scholar]
- [249].Zhao Y, Yao R, Ouyang LL, Ding HX, Zhang T, Zhang KT, Cheng SJ, Sun W, Biofabrication 2014, 6. [DOI] [PubMed] [Google Scholar]
- [250].Mozetic P, Giannitelli SM, Gori M, Trombetta M, Rainer A, J Biomed Mater Res A 2017, 105, 2582. [DOI] [PubMed] [Google Scholar]
- [251].Yan M, Lewis PL, Shah RN, Biofabrication 2018, 10. [DOI] [PubMed] [Google Scholar]
- [252].Wang LL, Highley CB, Yeh YC, Galarraga JH, Uman S, Burdick JA, J Biomed Mater Res A 2018, 106, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Shi L, Hu YQ, Ullah MW, Ullah I, Ou H, Zhang WC, Xiong LM, Zhang XL, Biofabrication 2019, 11. [DOI] [PubMed] [Google Scholar]
- [254].Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, Nat Biotechnol 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [255].Dai XL, Ma C, Lan Q, Xu T, Biofabrication 2016, 8. [DOI] [PubMed] [Google Scholar]
- [256].Benning L, Gutzweiler L, Trondle K, Riba J, Zengerle R, Koltay P, Zimmermann S, Stark GB, Finkenzeller G, J Biomed Mater Res A 2017, 105, 3231. [DOI] [PubMed] [Google Scholar]
- [257].Schoneberg J, De Lorenzi F, Theek B, Blaeser A, Rommel D, Kuehne AJC, Kiessling F, Fischer H, Sci Rep-Uk 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Jang J, Kim TG, Kim BS, Kim SW, Kwon SM, Cho DW, Acta Biomater 2016, 33, 88. [DOI] [PubMed] [Google Scholar]
- [259].Cui HT, Zhu W, Holmes B, Zhang LG, Adv Sci 2016, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Sakai S, Ueda K, Gantumur E, Taya M, Nakamura M, Macromol Rapid Comm 2018, 39. [DOI] [PubMed] [Google Scholar]


