Abstract
Purpose:
Computable phenotypes are constructed to utilize data within the electronic health record (EHR) to identify patients with specific characteristics; a necessary step for researching a complex disease state. We developed computable phenotypes for resistant hypertension (RHTN) and stable controlled hypertension (HTN) based on the National Patient-Centered Clinical Research Network (PCORnet) common data model (CDM). The computable phenotypes were validated through manual chart review.
Methods:
We adapted and refined existing computable phenotype algorithms for RHTN and stable controlled HTN to the PCORnet CDM in an adult HTN population from the OneFlorida Clinical Research Consortium (2015–2017). Two independent reviewers validated the computable phenotypes through manual chart review of 425 patient records. We assessed precision of our computable phenotypes through positive predictive value (PPV) and test validity through interrater reliability (IRR).
Results:
Among the 156,730 HTN patients in our final dataset, the final computable phenotype algorithms identified 24,926 patients with RHTN and 19,100 with stable controlled HTN. The PPV for RHTN in patients randomly selected for validation of the final algorithm was 99.1% (n=113, CI: 95.2%–99.9%). The PPV for stable controlled HTN in patients randomly selected for validation of the final algorithm was 96.5% (n=113, CI: 91.2%–99.0%). IRR analysis revealed a raw percent agreement of 91% (152/167) with Cohen’s kappa statistic = 0.87.
Conclusions:
We constructed and validated a RHTN computable phenotype algorithm and a stable controlled HTN computable phenotype algorithm. Both algorithms are based on the PCORnet CDM, allowing for future application to epidemiological and drug utilization based research.
Keywords: electronic health records, computable phenotypes, hypertension, resistant hypertension, pharmacoepidemiology
1. INTRODUCTION
Resistant hypertension (RHTN) describes a complex hypertension that is unresponsive to multiple antihypertensive medications, which is classically defined as requiring four or more antihypertensive medications from different antihypertensive classes to achieve blood pressure (BP) control.1,2 It is estimated that RHTN occurs in ~14–20% of those with treated hypertension (HTN).3,4 Because uncontrolled BP is associated with increased risk for adverse cardiovascular outcomes (e.g. stroke, myocardial infarction (MI), death), it is important to identify individuals at risk for, or with RHTN.5–11 Currently, there is no definitive way to identify which hypertensive patients will ultimately be classified as RHTN, or determine whether use of particular antihypertensive drugs will affect risk for RHTN. If those identified as at-risk for RHTN fail to achieve BP control with usual antihypertensive regimens, targeted pharmacotherapy with a mineralocorticoid receptor antagonist (e.g. spironolactone),12–15 adherence counseling,16–20 or alternative non-pharmacological therapies (e.g. renal denervation) could be prescribed sooner.21–23
RHTN is difficult to study since there is not a diagnostic code and usually multiple observations over a period of time are required to make a diagnosis. Complex phenotypes like this can benefit from computable phenotypes algorithms, which utilize structured and/or unstructured data from the electronic health record (EHR) to identify patients with a specific disease or trait.24–27 The basic definition of RHTN has been established in statements from the American Heart Association (AHA).2,28 Additionally, the electronic MEdical Records & GEnomics (eMERGE) Network has developed and validated computable phenotypes for RHTN and controlled HTN based on International Classification of Diseases (ICD)-9-Clinical Modification (CM) codes, medication information, and use for genome-wide association studies (GWAS).29,30 However, additional work is necessary to adapt and validate these phenotypes to ICD-10-CM codes, common data models such as the PCORnet (The National Patient-Centered Clinical Research Network) common data model (CDM), standardized prescription classification, and study designs other than GWAS that may offer more flexibility on inclusion and exclusion criteria. To fill this need, we developed and validated two computable phenotype algorithms using the PCORnet CDM: an RHTN phenotype and a stable controlled HTN phenotype. These validated algorithms can be applied to determine drug utilization in the RHTN and controlled HTN populations, and used to conduct other needed epidemiological and comparative effectiveness research.
2. METHODS
2.1. Study population
Data Source.
The OneFlorida Clinical Research Consortium is a statewide clinical research network that operates the OneFlorida Data Trust, a repository of longitudinal EHR data mapped to the PCORnet CDM.31 The PCORnet CDM was adapted from the Mini-Sentinel CDM, and outlines specifications for the representation of EHR data and claims data.32 This study only included data from one partner within the OneFlorida Data Trust, University of Florida (UF) Health. This study was approved by the Institutional Review Board (IRB) at UF.
Patient Population.
The HTN population was defined as all adults (age ≥ 18 years) with ≥1 HTN diagnosis from an outpatient encounter, defined as ICD-9-CM code 401.x or ICD-10-CM code I10.
Data fields.
The data on the patient population were extracted from the OneFlorida Data Trust on June 12th, 2018 in the PCORnet CDM, version 4.0, and included EHR data from January 1, 2015 to December 31, 2017. Data and fields were included from the following PCORnet CDM tables: Demographic, Encounter, Diagnosis, Procedures, Vital, Condition, and Prescribing.
2.2. Computable phenotype algorithms
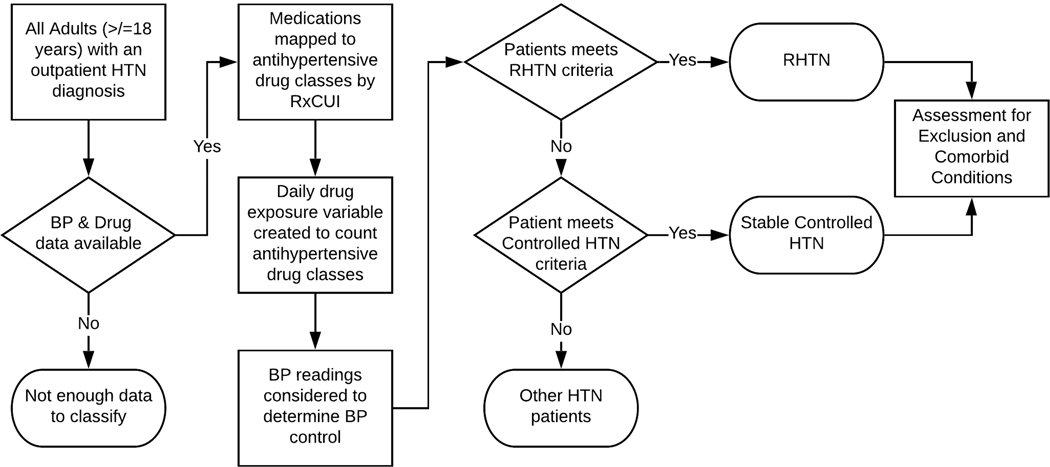
Computable phenotypes for RHTN and stable controlled HTN were developed and validated through an iterative process, employing rounds of manual chart review, with revisions to the computable phenotype algorithms in between rounds based on the findings. Prior studies have shown that this process improves algorithm performance and increases algorithm sharing.30,33,34 A summary of the data preparation and algorithm steps are shown in the Figure. Patients were included in the cohort if they had prescription information available from the Prescribing table, and blood pressure (BP) readings available from the Vital table. For patients with all necessary data, the following data preparation steps were conducted: 1) all medications were mapped to antihypertensive drug classes based on RxNorm Concept Unique Identifiers (RxCUI) utilizing our previously developed map and methodology,35 2) a drug exposure variable was created to count the number of antihypertensive drug classes prescribed at any point during the study period (by day), and 3) BP data was considered to determine systolic and diastolic BP control (defined as <140/<90 mm Hg) during the study time period when antihypertensive medications were prescribed. Our data preparation steps assume patients are exposed to all antihypertensive medications they are prescribed.
Figure.
Data preparation and algorithm steps. Workflow for the resistant hypertension computable phenotype and the controlled HTN computable phenotype.
RHTN.
A version 1 algorithm for RHTN was constructed by adapting the 2008 AHA RHTN definition (140/90 BP threshold), and the RHTN computable phenotype developed by the eMERGE Network to the PCORnet CDM. We further adapted the algorithm to include ICD-10-CM codes and a standardized prescription drug mapping (Algorithm 1).1,29 Patients were classified as RHTN if they met either of the following criteria during at least two drug exposure-by-day time points, at least 30 days apart during the study period (2015–2017): 1) four simultaneous antihypertensive prescriptions from different drug classes, or 2) three simultaneous antihypertensive prescriptions from different drug classes and BP ≥ 140/90 at least 30 days after the start of the third antihypertensive prescription. Finally, patients were assessed for the presence of potential exclusion and comorbid diagnoses, including secondary forms of HTN, during the study period (Supplemental Table 1). The version 2 algorithm included all elements of Algorithm 1 and added Current Procedural Terminology (CPT) codes to indicate routine outpatient care, available in Supplemental Table 2 (Algorithm 2). The final algorithm included all elements of Algorithm 2 and refined drug counting (Final Algorithm).
Stable Controlled HTN.
A version 1 of the computable phenotype was created based on the phenotype developed by the eMERGE Network.29 Our adaption included less restrictive BP control, mapping to the PCORnet CDM, ICD-10-CM codes, and standardized prescription drug mapping (Algorithm 1). Patients were classified as stable controlled HTN patients if the following criteria were met during at least two drug-exposure-by-day time points, at least 30 days apart during the study period (2015–2017): one or two simultaneous antihypertensive prescriptions from different classes AND BP < 140/90 at least 30 days after the start of the antihypertensive prescription. Then, patients were assessed for potential exclusion and comorbid diagnoses (Supplemental Table 1). The version 2 algorithm was modified to 80% BP control over the study period (2015–2017) plus CPT codes to indicate routine outpatient care, available in Supplemental Table 2 (Algorithm 2), and the version 3 algorithm was modified to include refined drug counting (Final Algorithm).
Validation through manual chart review.
Blinded chart review was conducted on patient records from UF Health. Two pharmacy students (KB and KC) were trained using the guide shown in the Supplement. The chart review was conducted independently, using an iterative process with fewer charts during the first rounds, and more during the later rounds, as recommended by the best practices established by eMERGE.30 In total 425 charts were reviewed, 75 charts during round one, 100 charts during round two, and 250 charts during round three. For each patient selected for chart review, information in the EHR during the study period (2015–2017) was abstracted and collected in a standardized and customized REDCap data collection instrument.36 Based on diagnoses, prescriptions, BP values, and free text clinic notes, the reviewer made a final determination of the status for each patient: RHTN, stable controlled HTN, or other HTN patient. The reviewers overlapped on 167 total charts: 75 (100%) in round one, 32 (32%) in round two, and 60 (24%) in round three. In cases of discordance, a chart was reviewed by a third reviewer (CWM), and discussed among all three reviewers for a final determination. The two reviewers could also flag difficult patients for review by the third reviewer. All reviewers were blinded to the computable phenotype assignment until the end of review round.
2.3. Statistical analysis
Following each round of chart review, the positive predictive value (PPV) of each algorithm was calculated. Additionally, 95% confidence intervals for the PPV was calculated using Wilson’s formula.37 When the algorithm classification differed from the classification by manual chart review, the EHR was further reviewed by CWM to identify the reason for discordance, and the algorithms were updated accordingly. Interrater reliability (IRR) was assessed through raw agreement between the two reviewers, as well as the Cohen’s Kappa statistic. All analyses and algorithm coding were conducted using SAS version 9.4. The code for the final algorithms are shown in the Supplement and are available at (github.com/caitrinmcd/RHTN_CP).
3. RESULTS
3.1. Sample characteristics
Data were extracted from the OneFlorida Data Trust on June 12, 2018, and included 202,174 patients from UF Health with an ICD-9-CM or ICD-10-CM diagnosis code for HTN. A total of 156,730 patients had the required BP and prescription data available (Table 1). The majority of the patients were ≥ 60 years old (56.3%), female (55.3%), and white (62.6%). Almost one-third of patients were Black or African American (30.2%).
Table 1.
Sample Demographics and Characteristics
| Characteristic | Overall n=156,730 | RHTN* n=24,926 | Stable Ctrl HTN** n=19,100 |
|---|---|---|---|
| Age in Years | |||
| 18–29 | 3,969 (2.5) | 109 (0.4) | 376 (2.0) |
| 30–39 | 10,292 (6.6) | 603 (2.4) | 1,003 (5.3) |
| 40–49 | 19,196 (12.3) | 2,105 (8.4) | 2,431 (12.7) |
| 50–59 | 35,030 (22.4) | 5,350 (21.5) | 4,887 (25.6) |
| 60–69 | 41,674 (26.6) | 7,572 (30.4) | 5,591 (29.3) |
| 70–79 | 29,751 (19.0) | 5,875 (23.6) | 3,396 (17.8) |
| 80–89 | 13,320 (8.5) | 2,723 (10.9) | 1,160 (6.1) |
| 90+ | 3,498 (2.2) | 589 (2.4) | 256 (1.3) |
| Female | 86,615 (55.3) | 13,906 (55.8) | 10,645 (55.7) |
| Race | |||
| Black | 47,334 (30.2) | 11,362 (45.6) | 4,414 (23.1) |
| Missing | 485 (0.3) | 39 (0.2) | 21 (0.1) |
| Other | 10,753 (6.9) | 1,214 (4.9) | 1,483 (7.8) |
| White | 98,158 (62.6) | 12,311 (49.4) | 13,182 (69.0) |
| Hispanic | 6,188 (4.0) | 760 (3.1) | 819 (4.3) |
| Body Mass Index, kg/m2 | |||
| <25.0 | 33,606 (21.7) | 3,992 (16.2) | 4,529 (23.8) |
| 25.0-<30.0 | 45,060 (29.1) | 6,260 (25.5) | 5,876 (30.9) |
| ≥30.0 | 76,244 (49.2) | 14,324 (58.3) | 8,606 (45.3) |
| Missing | 1,820 (1.2) | 350 (1.4) | 89 (0.5) |
| Smoking Status | |||
| Current Smoker | 27,381 (17.7) | 3,867 (15.6) | 3,063 (16.1) |
| Former Smoker | 50,975 (32.9) | 9,269 (37.4) | 6,324 (33.2) |
RHTN: Resistant Hypertension, Ctrl: Control, HTN: Hypertension
Presumptive RHTN patients identified using the RHTN Final Algorithm (RHTN + CPT codes + drug counting)
Stable Ctrl HTN patients identified using Stable Ctrl HTN Final Algorithm (80% BP Control + CPT codes + drug counting)
3.2. Positive predictive value of the computable phenotypes
EHR abstraction was completed for 425 patients. The chart review was conducted using an iterative process shown in Supplemental Table 3.
RHTN.
The RHTN algorithm 1 identified 27,978 presumptive RHTN patients (Table 2). Of the 25 patients reviewed, 21 were confirmed [PPV: 84.0% (CI 63.9–95.5)]. The main issues identified from Algorithm 1 were inclusion of patients who were not established patients (e.g. only one procedure based encounter), and inclusion of BP values from non-routine outpatient care (e.g. from outpatient procedures). These issues were addressed by adding CPT codes to indicate routine outpatient visits, only including patients with these CPT codes, and BP values from encounters associated with these CPT codes. The RHTN algorithm 2 identified 25,933 presumptive RHTN patients (Table 2). Thirty-three of the 37 patients reviewed were confirmed RHTN [PPV: 89.2% (CI 74.6–97.0)]. The main issue identified dealt with the counting of drug classes when patients were switched between single pill and combination pill preparations of the same antihypertensive drug class. The algorithm was updated to allow single pill preparations and combination pill preparations to combine into the same drug class. The final algorithm, identified 24,926 presumptive RHTN patients (Table 2). Out of the 113 patients that were reviewed, 112 were confirmed as RHTN [PPV: 99.1% (CI 95.2–99.9)].
Table 2.
Performance of the resistant hypertension (RHTN) computable phenotype algorithms
| CP | Presumptive RHTN, n | Subset of charts reviewed, n | Confirmed RHTN, n (%) | PPV (95% CI) |
|---|---|---|---|---|
| Algorithm 1* | 27,978 | 25 | 21 (84.0) | 84.0 (63.9–95.5) |
| Algorithm 2** | 25,933 | 37 | 33 (89.2) | 89.2 (74.6–97.0) |
| Final Algorithm*** | 24,926 | 113 | 112 (99.1) | 99.1 (95.2–99.9) |
RHTN: Resistant Hypertension, PPV: Positive predictive value, CI: confidence interval
Algorithm 1: RHTN algorithm based on the 2008 AHA statement on resistant hypertension and the eMERGE RHTN algorithm (RHTN)
Algorithm 2: Algorithm 1 + CPT codes algorithm added CPT codes to identify patients with routine outpatient encounters, and only use blood pressure readings from routine outpatient encounters (RHTN + CPT codes)
Final Algorithm: Algorithm 2 + drug counting algorithm added additional consideration for patients switching between single pill and combination pill preparations of the same antihypertensive drug class (RHTN + CPT codes + drug counting)
Stable Controlled HTN.
The stable controlled HTN algorithm 1 identified 53,634 presumptive stable controlled HTN patients (Table 3). Of the 25 patients reviewed, only 13 were confirmed as stable controlled HTN [PPV: 52.0% (CI 31.3–72.2)]. The main issues found in algorithm 1 dealt with the level of BP control required to be called a stable controlled HTN patient, the inclusion of patients who were not established patients, and inclusion of BP values from non-routine outpatient care. These were addressed by adding the 80% BP control level, and CPT codes to identify patients with routine outpatient encounters and BP values from those encounters. Algorithm 2 identified 17,341 presumptive stable controlled HTN patients (Table 3). Of the 37 who were reviewed, 35 were confirmed as stable controlled HTN [PPV: 94.6% (CI 81.8–99.3)]. Correct drug counting for single and combination drug products was also an issue. When the drug counting portion of the algorithm was refined, the final algorithm identified 19,100 presumptive stable controlled HTN patients (Table 3). One-hundred and thirteen patients were reviewed and 109 were confirmed as stable controlled HTN [PPV: 96.5% (CI 91.2–99.0)].
Table 3.
Performance of stable controlled HTN computable phenotype algorithms
| Algorithm | Presumptive Controlled HTN, n | Subset of charts reviewed, n | Confirmed Controlled HTN, n (%) | PPV (95% CI) |
|---|---|---|---|---|
| Algorithm 1* | 53,634 | 25 | 13 (52.0) | 52.0 (31.3–72.2) |
| Algorithm 2** | 17,341 | 37 | 35 (94.6) | 94.6 (81.8–99.3) |
| Final Algorithm*** | 19,100 | 113 | 109 (96.5) | 96.5 (91.2–99.0) |
HTN: Hypertension, PPV: Positive predictive value, CI: confidence interval
Algorithm 1: Any BP Control algorithm allowed any level of BP control (Any BP Control)
Algorithm 2: 80% BP Control + CPT codes algorithm required patients have 80% controlled BP during the study period and added CPT codes to identify patients with routine outpatient encounters, and only use blood pressure readings from routine outpatient encounters (80% BP Control + CPT codes)
Final Algorithm: Model 2 + drug counting algorithm added additional consideration for patients switching between single pill and combination pill preparations of the same antihypertensive drug class (80% BP Control + CPT codes + drug counting)
3.3. Interrater reliability
The agreement between the two individuals who performed the chart review is shown in Supplemental Table 4. Out of the 425 charts that were reviewed, 167 charts were reviewed by both reviewers. The reviewers’ raw percent agreement was 152/167 = 91.0%, and the Cohen’s kappa statistic = 0.87, indicating high interrater reliability.
3.4. Characteristics of the RHTN and Stable Controlled HTN populations
The characteristics of the RHTN population and the stable controlled HTN population using the final validated algorithms are shown in Table 1. The majority of the RHTN patients and stable controlled HTN patients were also ≥ 60 years old (67.2% and 54.5%, respectively) and female (55.8% and 55.7%, respectively, Table 1). For the stable controlled HTN population, the majority of the patients were white (69.0%, Table 1). However, for the RHTN population, 49.4% of the patients were white and 45.6% were Black or African American (Table 1). A summary of the final data preparation steps and final algorithm steps is shown in the Supplemental Figure.
4. DISCUSSION
We developed and validated a computable phenotype for RHTN and a computable phenotype for stable controlled HTN. Both computable phenotypes are based on the PCORnet CDM and include standardized prescription drug mapping, allowing for broader application and use. Additionally, we assessed the performance of our algorithms through PPV and IRR. Our final RHTN algorithm showed similar prevalence (15.9%) and PPV (99.1%) to prior literature (prevalence: 14–20%; PPV: 84.4–100%).3,4,30
Through our validation process, we identified major areas that could lead to the misclassification of patients. Broadly, the issues found were encounter related, BP related, or medication related. We were able to address nearly all these issues through modifications to the computable phenotype algorithms. First, we added CPT codes to identify routine outpatient care. We used these CPT codes to filter encounters, and only utilized BP values from encounters with these select CPT codes. Additionally, we used these CPT codes to filter out patients who were only seen within the healthcare system for an outpatient consultation, and not routine care. We also refined our medication counting algorithms. With RHTN, it is necessary to properly assign each antihypertensive medication to a medication class or classes in order to determine if a patient meets the definition for RHTN. This assignment is complicated due to the multiple classes of antihypertensive drugs with different mechanisms of action,2,38 and the switching of antihypertensive prescriptions that occurs over a patients treatment course. Particular care must be taken to ensure that drug classes combine and count correctly, regardless of preparation (single drug product vs. combination drug product). We relied on our prior work on antihypertensive medication classification to ensure proper grouping and counting of the prescription data.35
The final algorithms for RHTN and stable controlled HTN still classify a small number (5/250=2%) of patients as either RHTN or stable controlled HTN when they should be other HTN patients. From our work, four out of five of the patients who were misclassified during the final round appeared to be due to our method of handling medication start and end dates for historical medications and/or discontinued medications. We designed our algorithm to calculate a prescription end date if one was not provided in the EHR. The end date is calculated based on the prescription information (quantity, refill, start date, etc.). If this information is blank, the end date is calculated as one year from the start date, as that is the maximum amount of time prescriptions can legally be filled. Through our chart review, overall this method was accurate. However, there were some cases where a patient was seen at UF Health in a specialty outpatient clinic and their primary care provider was outside the UF Health system. This pattern of healthcare access resulted in most to all of the patient’s antihypertensive prescriptions being prescribed outside of the UF Health system, and only documented within UF Health as historical medications. Often the documentation of historical medications was correct; however, there were cases when patients would remain on their historical medications, verified through clinic notes, but they were only recorded once, over one year ago. Cases like this would cause underestimating the number of medications a patient was prescribed. We also observed cases of overestimation, when a historical medication was discontinued before the recorded end date in the patient’s EHR. Without reading the clinic notes, it is impossible to know if historical medications are accurate as recorded within the EHR. While this only affected a small number of patients using our final algorithms (4/250 =1.6%), we recommend others assess the impact of historical medications within their health systems and data structures.
The two pharmacy students who conducted the chart review had high interrater reliability; however, there were some common issues that lead to disagreement. These included clinic notes related to medication adherence and the aforementioned issues with historical medications. There is a growing body of data suggesting that many HTN patients classified as RHTN are non-adherent (non-adherence rates of 34–80%).39–42 The wide ranges in non-adherence may reflect some of the limitations of studies to date, but also highlight the need for standardized ways to measure adherence, and perhaps including adherence information in the EHR.43–46
Our study is not without limitations. One limitation is our method to determine controlled BP. BP is a variable phenotype influenced by many factors such as smoking, diet, physical activity, sex, stress, age, socioeconomic status, family history, and comorbidities (diabetes, chronic kidney disease, and obesity).38 Given the number of factors influencing BP, it is possible that a patient’s BP value is higher or lower than normal on any given day. With this in mind, we selected a value that permitted a patient to have an occasional BP value of >140/90 at some visits, and yet be considered well controlled overall across multiple visits. We selected an 80% threshold based on the Million Hearts Hypertension Control Challenge and literature in other areas of pharmacotherapy.47,48 However, this is an arbitrary number and could be shifted, which would impact our phenotype definition. We recognize that this threshold is strict; the final stable controlled HTN algorithm prevalence was 12.2%. Prior work across PCORnet, including OneFlorida, has shown the percent of patients with their last BP reading <140/90 was 58%.49 Future work could include investigating different BP control levels and their effects on the computable phenotype’s precision. Additionally, our current RHTN computable phenotype does not include a measure of adherence because data on adherence are unavailable as structured data in either EHRs generally or the PCORnet CDM. Future work includes estimating the level of adherence by using the proportion of days covered (PDC) measure in individuals with linked EHR-based data and Florida Medicaid claims data in the OneFlorida Data Trust.50,51 Other limitations with our algorithms included the coding systems and the temporal aspects. By using both ICD-9-CM and ICD-10-CM codes in the identification of our study population, we are unable to distinguish the performance between coding systems. Furthermore, our algorithms classify patients to RHTN or stable controlled HTN utilizing data over the entire study period (2015–2017), and our manual chart review also considered data over the entire study period (2015–2017). With this methodology we did not validate an index date, but rather if a patient met the algorithm criteria during the study period. Finally, while our algorithms are based on the PCORnet CDM, our manual chart review was only conducted at a single site. This may impact generalizability and future studies are also needed to validate the performance of the final algorithms at other PCORnet sties.
In conclusion, we constructed two computable phenotypes: one for RHTN and one for stable controlled HTN. Through manual chart review, we were able to refine these algorithms to high PPV in identifying RHTN cases and stable controlled HTN patients. In the future, we plan to utilize these algorithms to conduct additional epidemiological research.
Supplementary Material
Key Points:
Complex phenotypes like resistant hypertension (RHTN) can benefit from computable phenotypes.
The positive predictive value for RHTN in patients randomly selected for validation of the final algorithm was 99.1%, using manual chart review of the electronic health record as the reference.
The positive predictive value for stable controlled hypertension (HTN) in patients randomly selected for validation of the final algorithm was 96.5%, using manual chart review of the electronic health record as the reference.
The major areas that could lead to the misclassification of patients were encounter related, BP related, or medication related.
The final validated algorithms can be applied to electronic health record based data to determine drug utilization in the RHTN and controlled HTN populations, and used to conduct other needed epidemiological and comparative effectiveness research.
ACKNOWLEDGEMENTS
Support for this project comes from NIH grants KL2 TR001429 (CWM) and K01 HL141690 (CWM). Additionally, the research reported in this publication was supported in part by the OneFlorida Clinical Data Network, funded by the Patient-Centered Outcomes Research Institute #CDRN-1501-26692, in part by the OneFlorida Cancer Control Alliance, funded by the Florida Department of Health’s James and Esther King Biomedical Research Program #4KB16, in part by the University of Florida Clinical and Translational Science Institute, which is supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427, and in part by the Clinical and Translational Science Collaborative of Cleveland, UL1TR0002548 from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Patient-Centered Outcomes Research Institute (PCORI), its Board of Governors or Methodology, the OneFlorida Clinical Research Consortium, the University of Florida’s Clinical and Translational Science Institute, the Florida Department of Health, or the National Institutes of Health.
Funding and prior posting statement: This manuscript has not been posted or presented elsewhere, and was funded through NIH grants KL2 TR001429 (CWM) and K01 HL141690 (CWM), and supported in part by the OneFlorida Clinical Data Network, funded by the Patient-Centered Outcomes Research Institute #CDRN-1501-26692.
Footnotes
ETHICS STATEMENT
The study protocol was approved the University of Florida Institutional Review Board.
CONFLICT OF INTEREST STATEMENT
DCC is a research consultant for Merck Sharp & Dohme Corp., a subsidiary of Merck & Co. Inc. All other authors have no conflicts of interest to declare.
REFERENCES
- 1.Calhoun DA, Jones D, Textor S, et al. Resistant hypertension: diagnosis, evaluation, and treatment. A scientific statement from the American Heart Association Professional Education Committee of the Council for High Blood Pressure Research. Hypertension. June 2008;51(6):1403–1419. [DOI] [PubMed] [Google Scholar]
- 2.Carey RM, Calhoun DA, Bakris GL, et al. Resistant Hypertension: Detection, Evaluation, and Management: A Scientific Statement From the American Heart Association. Hypertension. November 2018;72(5):e53–e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achelrod D, Wenzel U, Frey S. Systematic review and meta-analysis of the prevalence of resistant hypertension in treated hypertensive populations. Am J Hypertens. March 2015;28(3):355–361. [DOI] [PubMed] [Google Scholar]
- 4.Carey RM, Sakhuja S, Calhoun DA, Whelton PK, Muntner P. Prevalence of Apparent Treatment-Resistant Hypertension in the United States. Hypertension. February 2019;73(2):424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith SM, Gong Y, Handberg E, et al. Predictors and outcomes of resistant hypertension among patients with coronary artery disease and hypertension. J Hypertens. March 2014;32(3):635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch AI, Irvin MR, Davis BR, Ford CE, Eckfeldt JH, Arnett DK. Genetic and Adverse Health Outcome Associations with Treatment Resistant Hypertension in GenHAT. Int J Hypertens. 2013;2013:578578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muntner P, Davis BR, Cushman WC, et al. Treatment-resistant hypertension and the incidence of cardiovascular disease and end-stage renal disease: results from the Antihypertensive and Lipid-Lowering Treatment to Prevent Heart Attack Trial (ALLHAT). Hypertension. November 2014;64(5):1012–1021. [DOI] [PubMed] [Google Scholar]
- 8.Irvin MR, Booth JN 3rd, Shimbo D, et al. Apparent treatment-resistant hypertension and risk for stroke, coronary heart disease, and all-cause mortality. Journal of the American Society of Hypertension : JASH. June 2014;8(6):405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith SM, Huo T, Delia Johnson B, et al. Cardiovascular and mortality risk of apparent resistant hypertension in women with suspected myocardial ischemia: a report from the NHLBI-sponsored WISE Study. Journal of the American Heart Association. February 2014;3(1):e000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Judd E, Calhoun DA. Apparent and true resistant hypertension: definition, prevalence and outcomes. J Hum Hypertens. August 2014;28(8):463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sim JJ, Bhandari SK, Shi J, et al. Comparative risk of renal, cardiovascular, and mortality outcomes in controlled, uncontrolled resistant, and nonresistant hypertension. Kidney Int. September 2015;88(3):622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerman LO, Textor SC. Hypertension in 2015: Resistant hypertension: impact and evolving treatment options. Nat Rev Nephrol. February 2016;12(2):70–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosa J, Widimsky P, Waldauf P, et al. Role of Adding Spironolactone and Renal Denervation in True Resistant Hypertension: One-Year Outcomes of Randomized PRAGUE-15 Study. Hypertension. February 2016;67(2):397–403. [DOI] [PubMed] [Google Scholar]
- 14.Rosa J, Widimsky P, Waldauf P, et al. Renal denervation in comparison with intensified pharmacotherapy in true resistant hypertension: 2-year outcomes of randomized PRAGUE-15 study. J Hypertens. May 2017;35(5):1093–1099. [DOI] [PubMed] [Google Scholar]
- 15.Oliveras A, Armario P, Clara A, et al. Spironolactone versus sympathetic renal denervation to treat true resistant hypertension: results from the DENERVHTA study - a randomized controlled trial. J Hypertens. September 2016;34(9):1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma C, Zhou Y, Zhou W, Huang C. Evaluation of the effect of motivational interviewing counselling on hypertension care. Patient Educ Couns. May 2014;95(2):231–237. [DOI] [PubMed] [Google Scholar]
- 17.Volpp KG. The Counseling African Americans to Control Hypertension study and ways to enhance the next wave of behavioral interventions. Circulation. Vol 129 United States2014:2002–2004. [DOI] [PubMed] [Google Scholar]
- 18.Ruppar TM, Dunbar-Jacob JM, Mehr DR, Lewis L, Conn VS. Medication adherence interventions among hypertensive black adults: a systematic review and meta-analysis. J Hypertens. June 2017;35(6):1145–1154. [DOI] [PubMed] [Google Scholar]
- 19.Hedegaard U, Kjeldsen LJ, Pottegard A, et al. Improving Medication Adherence in Patients with Hypertension: A Randomized Trial. Am J Med. December 2015;128(12):1351–1361. [DOI] [PubMed] [Google Scholar]
- 20.Grant AB, Seixas A, Frederickson K, et al. Effect of Expectation of Care on Adherence to Antihypertensive Medications Among Hypertensive Blacks: Analysis of the Counseling African Americans to Control Hypertension (CAATCH) Trial. J Clin Hypertens (Greenwich). July 2016;18(7):690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coppolino G, Pisano A, Rivoli L, Bolignano D. Renal denervation for resistant hypertension. Cochrane Database Syst Rev. February 21 2017;2:Cd011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krum H, Schlaich M, Whitbourn R, et al. Catheter-based renal sympathetic denervation for resistant hypertension: a multicentre safety and proof-of-principle cohort study. Lancet. April 11 2009;373(9671):1275–1281. [DOI] [PubMed] [Google Scholar]
- 23.Schlaich MP, Schmieder RE, Bakris G, et al. International expert consensus statement: Percutaneous transluminal renal denervation for the treatment of resistant hypertension. J Am Coll Cardiol. December 2013;62(22):2031–2045. [DOI] [PubMed] [Google Scholar]
- 24.Kohane IS. HEALTH CARE POLICY. Ten things we have to do to achieve precision medicine. Science. July 03 2015;349(6243):37–38. [DOI] [PubMed] [Google Scholar]
- 25.Weber GM, Mandl KD, Kohane IS. Finding the missing link for big biomedical data. Jama. June 25 2014;311(24):2479–2480. [DOI] [PubMed] [Google Scholar]
- 26.Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calhoun DA, Jones D, Textor S, et al. Resistant hypertension: diagnosis, evaluation, and treatment: a scientific statement from the American Heart Association Professional Education Committee of the Council for High Blood Pressure Research. Circulation. June 24 2008;117(25):e510–526. [DOI] [PubMed] [Google Scholar]
- 29.Dumitrescu L, Ritchie MD, Denny JC, et al. Genome-wide study of resistant hypertension identified from electronic health records. PLoS One. 2017;12(2):e0171745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. June 2013;20(e1):e147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shenkman E, Hurt M, Hogan W, et al. OneFlorida Clinical Research Consortium: Linking a Clinical and Translational Science Institute With a Community-Based Distributive Medical Education Model. Acad Med. October 17 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. Jul-Aug 2014;21(4):578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirby J, Speltz P, Rasmussen L, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. Journal of the American Medical Informatics Association. November 2016 2016;23(6):1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peissig PL, Rasmussen LV, Berg RL, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J Am Med Inform Assoc. Mar-Apr 2012;19(2):225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonough CW, Smith SM, Cooper-DeHoff RM, Hogan WR. Optimizing Antihypertensive Medication Classification in Electronic Health Record-Based Data: Classification System Development and Methodological Comparison. JMIR Med Inform. 2020;8(2):e14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. April 2009;42(2):377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niesner K, Murff HJ, Griffin MR, et al. Validation of VA administrative data algorithms for identifying cardiovascular disease hospitalization. Epidemiology. Vol 24 United States2013:334–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whelton PK, Carey RM, Aronow WS, et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension. November 13 2017. [DOI] [PubMed] [Google Scholar]
- 39.Azizi M, Pereira H, Hamdidouche I, et al. Adherence to Antihypertensive Treatment and the Blood Pressure-Lowering Effects of Renal Denervation in the Renal Denervation for Hypertension (DENERHTN) Trial. Circulation. September 20 2016;134(12):847–857. [DOI] [PubMed] [Google Scholar]
- 40.de Jager RL, de Beus E, Beeftink MM, et al. Impact of Medication Adherence on the Effect of Renal Denervation: The SYMPATHY Trial. Hypertension. April 2017;69(4):678–684. [DOI] [PubMed] [Google Scholar]
- 41.Bunker J, Chang CL, Chapman N, et al. True Resistant Hypertension Following Observed Drug Ingestion: A Systematic Evaluation. J Clin Hypertens (Greenwich). March 2017;19(3):250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmieder RE, Ott C, Schmid A, et al. Adherence to Antihypertensive Medication in Treatment-Resistant Hypertension Undergoing Renal Denervation. J Am Heart Assoc. February 12 2016;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berra E, Azizi M, Capron A, et al. Evaluation of Adherence Should Become an Integral Part of Assessment of Patients With Apparently Treatment-Resistant Hypertension. Hypertension. August 2016;68(2):297–306. [DOI] [PubMed] [Google Scholar]
- 44.Morisky DE, Ang A, Krousel-Wood M, Ward HJ. Predictive validity of a medication adherence measure in an outpatient setting. J Clin Hypertens (Greenwich). May 2008;10(5):348–354. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Voils CI, Maciejewski ML, Hoyle RH, et al. Initial validation of a self-report measure of the extent of and reasons for medication nonadherence. Med Care. December 2012;50(12):1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voils CI, King HA, Neelon B, et al. Characterizing weekly self-reported antihypertensive medication nonadherence across repeated occasions. Patient Prefer Adherence. 2014;8:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.CDC. Hypertension Control Challenge Rules & Eligibility. Million Hearts 2018; https://millionhearts.hhs.gov/partners-progress/champions/rules.html. Accessed November 15, 2018. [Google Scholar]
- 48.Khunti K, Seidu S, Kunutsor S, Davies M. Association Between Adherence to Pharmacotherapy and Outcomes in Type 2 Diabetes: A Meta-analysis. Diabetes Care. November 2017;40(11):1588–1596. [DOI] [PubMed] [Google Scholar]
- 49.Pletcher MJ, Fontil V, Carton T, et al. The PCORnet Blood Pressure Control Laboratory: A Platform for Surveillance and Efficient Trials. Circ Cardiovasc Qual Outcomes. March 2020;13(3):e006115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kho AN, Cashy JP, Jackson KL, et al. Design and implementation of a privacy preserving electronic health record linkage tool in Chicago. J Am Med Inform Assoc. September 2015;22(5):1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diagnosis and classification of diabetes mellitus. Diabetes Care. January 2014;37 Suppl 1:S81–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.