Abstract
The COVID-19 pandemic has claimed more than a million lives worldwide within a short time span. Due to the unavailability of specific antiviral drugs or vaccine, the infections are causing panic both in general public and among healthcare providers. Therefore, an urgent discovery and development of effective antiviral drug for the treatment of COVID-19 is highly desired. Targeting the main protease (Mpro) of the causative agent, SARS-CoV-2 has great potential for drug discovery and drug repurposing efforts. Published crystal structures of SARS-CoV-2 Mpro further facilitated in silico investigations for discovering new inhibitors against Mpro. The present study aimed to screen several libraries of synthetic flavonoids and benzisothiazolinones as potential SARS-CoV-2 Mpro inhibitors using in silico methods. The short-listed compounds after virtual screening were filtered through SwissADME modeling tool to remove molecules with unfavorable pharmacokinetics and medicinal properties. The drug-like molecules were further subjected to iterative docking for the identification of top binders of SARS-CoV-2 Mpro. Finally, molecular dynamic (MD) simulations and binding free energy calculations were performed for the evaluation of the dynamic behavior, stability of protein–ligand complex, and binding affinity, resulting in the identification of thioflavonol, TF-9 as a potential inhibitor of Mpro. The computational studies further revealed the binding of TF-9 close to catalytic dyad and interactions with conserved residues in the S1 subsite of the substrate binding site. Our in-silico study demonstrated that synthetic analogs of flavonoids, particularly thioflavonols, have a strong tendency to inhibit the main protease Mpro, and thereby inhibit the reproduction of SARS-CoV-2.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, COVID-19, flavonoids, flavonols, thioflavonols, oxoaurones, thioaurones, 3-benzyloxyflavones, 3-O-flavonol glycosides, benzisothiazolinones, molecular docking
1. Introduction
The novel coronavirus disease (COVID-19) is an infectious and contagious disease, caused by the severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2). Since its first appearance in Wuhan city, Hubei, China dates back in December 2019, the infection has spread exponentially in about 200 countries due to international travel. Because of the rapid transmission and absence of pre-existing immunity against the new virus in humans, the outbreak was declared a pandemic by WHO on 11th March 2020 (Mahase, 2020; Wang et al., 2020; Whitworth, 2020). As per the weakly situation report, released by the WHO on 11th October 2020, more than 37 million confirmed cases of COVID-19 infections have been reported worldwide with over a million global deaths (WHO, 2020). After the virus transmission in humans, the initial symptoms appears within an incubation period of 2–14 days, varying from mild flu-like symptoms to moderate acute severe respiratory syndrome characterized by a high grade fever, cough, extreme tiredness, body aches, and difficulty in breathing. In some patients, the infection can lead to a fatal and severe pneumonia-like illness requiring hospitalization (Song et al., 2020; Wu et al., 2020; Zhou et al., 2020; Zhu et al., 2020; Zou et al., 2020).
Currently, there exists no specific antiviral drug for the treatment or vaccine for the prevention of COVID-19. Under the severity of infection and increased threat of hospitalization due to COVID-19, several efforts have been launched for the development of potential therapeutics to combat SARS-CoV-2 (Ciliberto et al., 2020; Hage-Melim et al., 2020; Standing, 2020; Villoutreix et al., 2020). By using both in vitro experiments and in silico virtual screenings, many drug discovery and drug repurposing efforts have been initiated by exploiting the viral entry mechanism or essential viral enzymes as drug target (Hage-Melim et al., 2020; Muralidharan et al., 2020; Shah et al., 2020).
Targeting the virus-specific proteins has proven to be a very effective drug discovery strategy for the development of direct-acting antivirals (DAAs) (Ertekin et al., 2020; Holmes & Chung, 2020). Analogously, discovery and development of DAAs against SARS-CoV-2 could provide efficient drugs for the treatment of COVID-19 infection. SAR-CoV-2 genome is based on a positive-sense single-stranded RNA (ssRNA+) containing about 32,000 bases. The release of viral RNA into the cytoplasm of the host cells is followed by the translation of RNA into the large (∼790 kDa) viral polyprotein 1ab. For proper functioning and reproduction of the virus, the polyprotein needs to be processed by viral proteases. Together with another virus encoded Papain-like protease, the main protease (Mpro, also known as 3CLpro) of SARS-CoV-2 cleaves the viral polyprotein into several structural and non-structural components. The SARS-CoV-2 Mpro cleaves the polyprotein at ∼11 sites, mostly with the recognition sequence of Leu-Gln↓(Ser, Ala, Gly) (where ↓ indicates the cleavage site) (Zhang et al., 2020) and plays a central role in the maturation of viral proteins. Due to the essential role in the life-cycle of the virus, Mpro has become a key protein for the discovery and development of DAAs for COVID-19 (Dai et al., 2020; Jin et al., 2020; Mengist et al., 2020; Zhang et al., 2020).
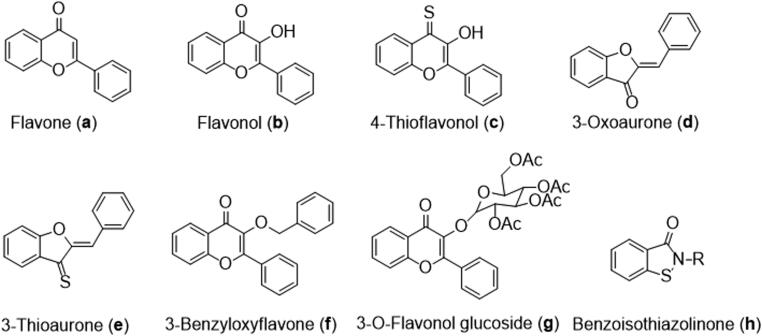
In this context, natural products and their synthetic analogs can offer a huge repertoire for the identification of leads for further development of anti-SARS-CoV-2 therapeutics. Flavonoids account for a large and significant group of plant-derived polyphenolic secondary metabolites with a benzo-γ-pyrone (C6-C3-C6) structure, and broad-spectrum pharmacological and biological activities (Ibrahim et al., 2020; Jimenez-Alberto et al., 2020; Joshi et al., 2020; Sies & Parnham, 2020; Tripathi et al., 2020). They are ubiquitously present in the plant kingdom and have many health benefits due to their antioxidant activity (Chae et al., 2013; Panche et al., 2016; Pietta, 2000) and potent in vitro and in vivo antiviral properties (Lalani & Poh, 2020; Zakaryan et al., 2017). Based on their chemical structure, flavonoids may be categorized into various subclasses, such as flavones, flavonols, flavanone, isoflavones, and aurones, which are found in a wide variety of plants as shown in Figure 1 (a–g).
Figure 1.
Chemical structures of representative flavone (a), 3-hydroxyflavone/flavonol (b), 4-thioflavonol (c), 3-oxoaurone (d), 3-thioaurone (e), 3-benzyloxyflavone (f), 3-O-flavonol glycoside (g), and 1,2-benzisothiazolinone (h).
Recently, Sies and Parnham (Sies & Parnham, 2020) have reviewed the potential of ebselen (an organoselenium compound) to inhibit Mpro of SARS-CoV-2, reporting it as a promising lead compound for the inhibition of the protease. The ebselen imparts its inhibitory action, in part, by making a covalent bond between selenium and Cys-145 of the catalytic dyad. Similarly, the covalent bond formation between sulfur of BITs and cysteine residue in the substrate binding site of enzymes is well-documented in literature (King et al., 2009). We envisioned that benzisothiazolinones (BITs) could also inhibit the Mpro of SARS-CoV-2 via similar covalent bond formation, and thus warrant in silico investigations.
In present research work, we have screened several synthetic flavonoids with diverse structures (Mughal et al., 2017, 2018, 2019) and BIT-analogs (Viani et al., 2017), and explored the in silico inhibition of Mpro of SAR-CoV-2 by 3-hydroxyflavones, 4-thioflavonols, 3-O-flavonol glucosides, 3-benzyloxyflavones, 3-oxoaurones, 3-thioaurones, and 1,2-benzisothiazol-3(2H)-ones. The virtual ADME/T properties were employed to assess drug-likeness and to filter molecules with unfavorable pharmacokinetics and medicinal properties, leading to identification of thioflavonols as potent inhibitors of the Mpro. The potential of synthetic flavonoids reported in this article has not been, to the best of our knowledge, previously reported.
2. Materials and methods
2.1. Synthesis of compound
The syntheses of flavonoid derivatives presented in this article have been published elsewhere (Mughal et al., 2017, 2018, 2019). The synthesis of 1,2-benzisothiazol-3(2H)-ones (BITs) was accomplished via a simple reported method (Viani et al., 2017).
2.2. Conformational analyses, geometry optimization, and energy minimization of structures
All calculations were run on a Window based PC system [Intel(R) Core™ i7-8550U CPU, 4 Core(s), 8 Logical processor(s) containing 8 GB memory]. Structures were drawn using ChemDraw and imported into Maestro (Version 17.2, Schrödinger) for 3 D conversion and optimization. For each molecule, conformational analysis was conducted by the Monte Carlo multiple minimum (MCMM) method embedded in the MacroModel tool of Maestro for the generation of low-energy conformations using the OPLS3 force field. The torsions were set at default values of 0˚and ±180˚ for each rotatable bond. The maximum number of steps were set at 1000, and the energy window for saving structures was set at 25.0 kJ/mol. All redundant and duplicate conformers were eliminated by setting the maximum atom deviation cut-off to 0.5 Å. After conformational screening, the lowest-energy conformer of each molecule was finally subjected to optimization and energy minimization.
2.3. High-throughput virtual screening
The energy-minimized and optimized conformer of each molecule was subjected to the LigPrep tool of Maestro, where the OPLS3 force field was employed for the preparation of ligands at pH 7.0 ± 1.0 using the Epik algorithm. Chirality of the 3 D structures were maintained. When we started this project, the only crystal structure for SARS-CoV-2 Mpro reported was PDB ID: 6LU7. For the preparation of the receptor, co-crystal monomeric structure (PDB ID: 6LU7) was imported from the Protein Databank and processed using the protein preparation wizard. The missing side chain (if any) were filled using Prime. All water molecules and heteroatoms (except covalently bound ligand) were removed. Hydrogen bonding assignment was optimized using PROPKA at pH 7.0 and finally the protein was minimized using the OPLS3 force field. For the preparation of docking grid, the covalently linked cognate N3 ligand was first extracted from the binding pocket of the protein and then remerged with the protein so that the ligand occupies the same position in the pocket but without the covalent bond with Cys-146. This process is important, because without it, the exclusion of the bound ligand from the substrate binding site would be difficult during the grid generation process. The centroid of the cognate ligand was selected as the grid center having x=-9.76 Å, y = 11.29 Å and z = 68.96 Å as xyz coordinates, respectively. The library of 101 compounds was virtually screened with flexible ligand sampling. Other than choosing the post-docking minimization, all other options were kept at the default values. Short-listing of the compounds was made with the help of a scatter plot between docking score and ligand efficiency Ln value. The molecules falling in the box (Figure S2) drawn by setting the cut-off values of the docking score of −6.0 kcal/mol and ligand efficiency Ln value of −1.35, were selected for the next phase.
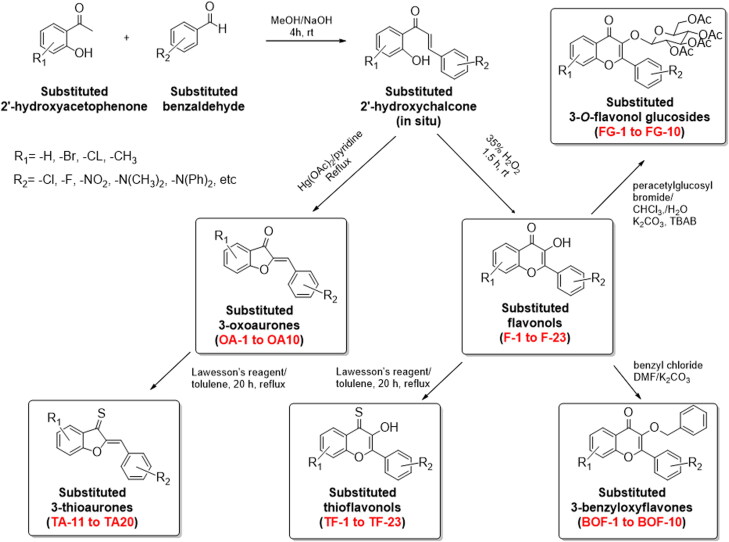
Figure 2.
Synthetic route to the substituted 3-oxoaurones (OA1-10), flavonoles (F1-23), 3-thioaurones (TA1-10), thioflavones (TF1-23), 3-benyloxyflavones (BOF1-10), and 3-O-flavonol glucosides (FG1-10).
2.4. Assessment of ADME properties of the screened molecules
The short-listed candidates from the virtual screening were analyzed by a Swiss-ADME modeling tool for the prediction of ADME data and toxicity of the selected compounds (Daina et al., 2017). The server uses its large database to estimate the pharmacokinetic behavior, drug-likeliness, physicochemical properties, lipophilicity, water solubility and medicinal properties with high precision. The molecules exhibiting the medicinal properties violations (Lipinski, etc.), PAINS behavior and/or potential of PgP substrate were filtered off at this stage.
2.5. Iterative docking analyses
The molecules showing favorable bioavailability and medicinal properties were subjected to iterative docking analyses using the AutoDock 4.2.6 program (Morris et al., 2009). PDBQT files of the shortlisted molecules and the receptor were prepared by using AutoDock Tool (ADT). The grid parameter file (gpf) was prepared by setting the same grid-center coordinates as used before for virtual screening. The box was set large enough to encompass the whole cognate N3 ligand within its boundary, while keeping the search spacing at 0.375 Å. For docking analyses, the Lamarckian genetic algorithm method was employed by using 100 runs (iterations) with a population size of 300 and 270,000 generations.
2.6. Analysis of the docking results and selection of the best binders
The binding energies of the docked structures were predicted by the AutoDock program. The docked structures of each ligand were clustered by using the integrated clustering tool of the AutoDock 4.2.6 software on the basis of root mean square deviation (RMSD) of 1.0 Å. A cluster containing 30% or more population and with a binding energy score of −8.0 kcal/mol was used as a cut-off for the ranking. For the illustration of hydrogen bonding and hydrophobic interactions of the docked structure with the receptor, we used the Chimera software. Finally, the top binders were further filtered on the basis of the favorable interactions (quality and quantity of hydrogen bonding and hydrophobic interactions) with the binding pocket residues as well as the shape complementarity.
2.7. Molecular dynamics (MD) simulation
All simulations were carried out with the GPU implementation of the PMEMD module in Amber18. The topological parameters of monomeric protein structure were processed by ff14SB force field while General Amber force filed (GAFF) was used for the parameterization of all the ligands. The AM1-BCC partial charges were assigned to all the ligands using Antechamber. The systems were solvated using TIP3P water model by placing into a truncated water box of 8.0 Å with periodic boundary conditions. Subsequently, the systems were neutralized by the addition of counter ions. The cut-off for non-bonded interactions were set to 12.0 Å and for the calculation of long-range electrostatic interactions, Particle Ewald Mesh algorithms were used while SHAKE algorithm was employed to constrain all the bonds involving hydrogen bonds.
Prepared systems were minimized to remove the bad contacts and to correct the geometry by using 2500 cycles of steepest descent method followed by 2500 cycles of conjugate gradient method. During minimization the force restrain was gradually decreased to minimize the solvent and solute atoms. The minimized systems were gradually heated from 0 to 300 K, in an NVT ensemble followed by NPT ensemble at pressure of 1 atm to get the stable systems. Berendsen thermostat and barostat was used to maintain the temperature and pressure during NVT and NPT ensemble, respectively. Finally, the production run of 100 ns for each equilibrated system was carried out with the time step of 2 fs. The stability for all the simulated systems were analyzed by RMSD, RMSF, and RoG.
2.8. Binding free energy calculation
Theoretical calculation of binding free energy is an efficient method to estimate the binding affinities between two molecules. In this present study, we calculate the binding affinities of all the four simulated systems by using MMGBSA and MMPBSA using following equation:
where ΔGbind is the binding free energy between simulated receptor and a small molecule. ΔEMM represented the change in the molecular mechanics energy of the gas phase, while ΔGsol and TΔS represent the change in the solvation free energy and entropy change upon binding of ligands, respectively. ΔEMM includes the sum of internal energy (angle, bond and dihedral energies), electrostatic and van der Waals energies. ΔGsol includes the sum of electrostatic solvation energy as polar contribution while non-electrostatic energy as non-polar contribution. GB or PB model is used to calculate polar component while SASA (solvent accessible surface area) is used to estimate non-polar contribution. For the calculation of binding free energy, snapshots collected at every 100 ps were used from the simulation trajectories.
3. Results and discussion
3.1. Chemistry
The syntheses of all compounds were accomplished as per the Scheme shown in Figure 2 and have been reported elsewhere.
3.2. Computational studies
The in silico binding analyses are becoming a hot area of scientific investigation, especially in the post COVID-19 era, for screening drugs (Ancy et al., 2020; Khan et al., 2020; Kumar et al., 2020; Marinho et al., 2020), synthetic compounds (Niu et al., 2008; Sepay et al., 2020; Ton et al., 2020; Wang et al., 2017) and natural products (Ghosh et al., 2020; Gupta et al., 2020; Gurung et al., 2020; Joshi et al., 2020; Narkhede et al., 2020) to measure the potential of these compounds for binding with the SARS-CoV-2 main protease. In the field of computer-aided drug designing (CADD), primarily used for the identification of a lead compound, the molecular docking analyses have been enormously employed for mapping and delineating the atomic level details of binding interaction between ligand and the receptor. Among different types of interactions, the hydrogen bonding, hydrophobic interaction, π-π interactions between ligand and residues in the binding site have been instrumental in explaining the binding efficacy of a receptor–ligand complex. The binding energy data (in kcal/mol) allows us to compare and rank the binding affinity of different ligands or compounds with their corresponding target receptor. A lower binding energy indicates a higher affinity of the ligand for the receptor. The ligand with the highest affinity (more negative docking score) can be chosen as the potential lead for further studies. In this study, we have applied typical drug discovery approach in silico by the initial short-listing of the potential inhibitors of Mpro from our in-house library of 101 compounds, passing them through computational ADME investigations, and finally selecting nine molecules with high docking score obtained by iterative docking analyses.
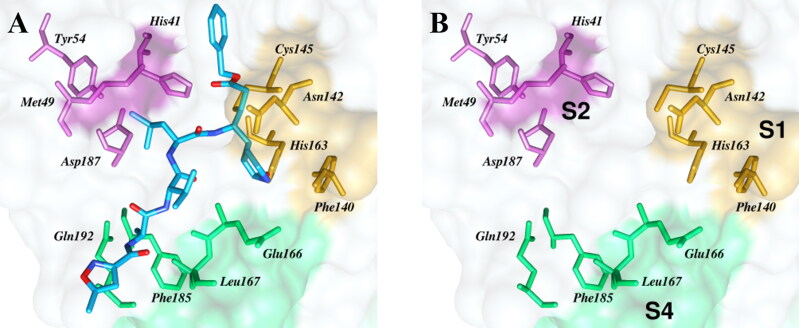
For the in silico analyses, we used a recently reported crystal structure of Mpro from SARS-CoV-2, containing a covalently linked ligand N3 (PDB ID: 6LU7) (Jin et al., 2020). The protease contains three domains: domain I from residue 8 to101, domain II from residue 102 to 184, and domain III from residue 201 to 303. The domain I and II comprise of two antiparallel β-barrels and the third domain comprises of a globular cluster of five α-helices. A shallow cleft between the domain I and II functions as a substrate binding site for Mpro, where a dyad of His-41 and Cys-145 catalyzes the proteolytic cleavage of the viral polyprotein into several structural and non-structural viral proteins. The substrate binding site is mainly composed of hydrophobic residues, and its chemical landscape can be further subdivided into several subsites (cavities) based on the orientation of the side chains (P1′ to P4) of the peptide backbone of the bound N3 ligand in the protease as shown in the Figure 3.
Figure 3.
Substrate binding Pocket of Mpro from SARS-CoV-2 and location of subsites. A) Display of the covalently linked N3 ligand (cyan) co-crystalized with the protease (shown as grey surface). The lactam moiety at P1 is located in a cavity of S1 (yellow), whereas the leucine side chain is deeply buried in the cavity of S2 (magenta). The P3 (valine) is solvent exposed and alanine at P4 is located in the S4 pocket (green). The residue lining the subsites are shown in matching color sticks. B) More open view of the subsites of the substrate binding sites.
3.3. Optimization and energy minimization of structures
Computational calculations are very sensitive to minor variations in the molecular structure and affect docking score and ultimately the reproducibility of the data. To minimize the introduction of random errors, we performed extensive and exhaustive structural optimizations. First, we investigated the conformational space of each molecule and used the conformer of the lowest potential energy, assuming that it is closer to the global minimum of the molecule. Apart from the symmetric structure of BIT5n, all molecules showed more than one allowed conformation, yielding the energy differences ranging from −0.001 to −20.995 kcal/mol between the highest and the lowest allowed conformers. In general, the larger the number of rotatable bonds a molecule has, the larger the number of allowed conformers exists and hence the more time-intensive and occasionally challenging it is to predict the global minimum. As expected, each member of the FG-series demonstrated more than 240 conformers each with an average energy difference of ∼ −20.8 kcal/mol. The results of the conformational search for each molecule of the tested libraries are given in Table S1–S6. Only the conformers with the minimum energy (global minimum) were used for the virtual screening and iterative docking studies.
3.4. In silico screening and initial selection of hits
The optimized structures were screened in silico for binding at the SARS-CoV-2 protease active site. The docking score and ligand efficiency of the tested molecules were calculated and drawn in XY scatter plot to facilitate the selection of potential hits. The ligand efficiency or binding efficiency is a normalized property of the ligand molecule and most commonly defined as the ratio of the binding free energy to the number of heavy atoms (non-hydrogen atoms). The XY plot between the docking score and ligand efficiency showed a widely scattered distribution of molecules around the correlation line with a poor R2 coefficient. Interestingly, we observed that the XY plot between the docking score and another closely related “ligand efficiency Ln” gives a better correlation (R2 = 0.9) in the scatter plot (Figure S2). The ligand efficiency Ln was calculated by using the following formula:
The compounds showing a virtual docking score of −6.0 kcal/mol or better and a ligand efficiency Ln of −1.35 or better were short-listed as shown in the red box in the Figure S2. Consequently, screening of these 101 tested compounds resulted in short-listing of 59 compounds: 20 out of 23 (87%) from the thioflavonol (TF) library, 9 out of 10 (90%) from the flavonol glucoside (FG) library, 7 out of 10 (70%) from the benzyloxyflavone (BOF) library, 6 out of 10 (60%) from the oxoaurone (OA) and 8 out of 10 (80%) compounds from the thioaurone (TA) library as hits. The flavonol (F) library rendered only 7 out of 23 (30%) compounds as hits. Benzisothiazolinone (BIT) library showed very poor docking score for binding to the substrate-binding site of the SAR-CoV-2 Mpro and yielded only 2 out of 15 (13%) hits.
3.5. ADME screening to select lead-like molecules
Since an inhibitor must have favorable bioavailability and drug-likeness for its clinical development, we sought it imperative to filter the short-listed molecules through the ADME screen so that molecules showing pharmacologically unfavorable absorption, distribution, metabolism, and excretion properties are removed in the earlier stage. To meet this end, the 59 short-listed compounds were subjected to ADME analyses using a web-based SwissADME modeling tool to predict the lipophilic character, water solubility, drug-likeness, etc. as described in the following sections.
The widely used standard descriptor of lipophilicity is the partition coefficient between n-octanol and water (log Po/w). The lipophilic profiles of our compounds were predicted using five predictive models including XLOGP3, WLOGP, MLOGP, SILICOS-IT and iLOGP. The purpose of using the multi model system is to minimize the bias in the estimation and evaluation of the consensus among the predicted values of log Po/w (Wang et al., 2015). As shown in the Table S7, the lipophilic character of all of the short-listed compounds varies from the consensus log Po/w value of +2.08 to +5.09, indicating that all of the short-listed compounds are highly lipophilic and meet the required criterion of drug molecule. In general, the presence of nitro group in a molecule significantly lowers the lipophilicity, whereas the presence of alkyl substituted amino group, and halogens significantly enhanced the lipophilic character. The change in lipophilic character due to the presence of methoxy group was not significant. Interestingly, the replacement of oxygen in the C = O group or in the ring B with sulfur also enhanced the lipophilicity of the molecule.
Water solubility is another important parameter to be considered to ensure the suitability of a drug molecule for oral availability or parenteral administration. We predicted the water solubility of the short-listed compounds using the SwissADME tool, which employs three different topological methods for estimating water solubility, including ESOL model, Ali model and SILICOS-IT. As shown in the Table S8, the log S values of the short-listed compounds were found in the range of − 2.9 (lower negative values indicate water solubility) to − 6.36 (higher negative values means poor water solubility) by ESOL model, − 3.04 to − 7.52 by Ali model, and − 3.25 to − 8.87 by SILICOS-IT method. Based on the calculation by all models, it turns out the majority of the short-listed compounds are moderately (log S between −4 to −6) water soluble, with some molecules showing good solubility (log S between −2 and −4) and some with poor water solubility (−6 to −9)
Prediction of absorption through gastrointestinal (GI) track, blood brain barrier (BBB) parameters and interaction with metabolic enzymes of the short-listed compounds (Table S9) further highlights the drug-like potential of these molecules. All short-listed flavonol glucosides (FG-series) and one thioflavonol, TF-14, showed very low GI absorption All other compounds showed high GI track absorption making them suitable for oral dosing. Almost half of the short-listed compounds passed the BBB permeability test. Except for FG-7 and TF-13, the short-listed compounds are not substrate of P-glycoprotein (Pgp). Many short-listed molecules show binding interactions with metabolic enzymes (CYP1A2, CYP2C19, CYP2C9, CYP2D6 and CYP3A4). Such interaction can cause toxicity and/or degradation of molecule. The log Kp parameter is helpful in the prediction of skin permeability. Higher negative values of log Kp make a molecule less permeable to penetrate the skin. The log Kp values of the short-listed compounds vary between −3.99 and −8.59 cm/s as shown in Table S9. The molecules showing unfavorable absorption, solubility and metabolism were filtered off from further evaluation.
It is quite interesting to note that the several molecules, despite having attractive docking score were filtered off in the light of the ADMT analyses. All compounds from the FG series were found to violate Lipinski rule, along with three compounds from the TA series (TA-2, TA-3 and TA-7), and were thus filtered off. The remaining compounds from the TA series showed at least one PAINS alert and were thus also dropped. Finally, the list was further refined to remove those molecules that showed more than 1 violation against lead-likeness. Thus, four molecules from the BOF series, one molecule from F series, two molecules from OA series, and four molecules from the TF series were dropped from further analyses. After careful structural inspection and removing the molecules showing unfavorable pharmacokinetics and medicinal properties, only 31 compounds were taken to the last step, where iterative docking analyses were conducted to select the molecule with a potential to inhibit the Mpro of SARS-CoV-2.
3.6. Iterative docking of the selected top molecules
The compounds passing through the filter of computational pharmacokinetics and bioavailability analyses were further subjected to iterative docking and clustering analyses (Table S11). The molecules showing a docking score better than −8.0 kcal/mol and clustering of more than 30% of the population resulted in the identification of nine compounds as potential inhibitors of Mpro of SARS-CoV-2 as shown in the Table 1. Again, most of these compounds were from TF-series (seven compounds) and one compound each from the BOF and F series.
Table 1.
Selected potential of Mpro of SARS-CoV-2.
| Entry | Compound | Docking score (kcal/mol) |
Cluster size (tolerance = 1Å) |
|---|---|---|---|
| 1 | TF-9 | −8.7 | 44 |
| 2 | TF-7 | −8.5 | 87 |
| 3 | TF-6 | −8.5 | 66 |
| 4 | BOF-9 | −8.3 | 34 |
| 5 | TF-19 | −8.2 | 70 |
| 6 | F-7 | −8.2 | 91 |
| 7 | TF-2 | −8.1 | 93 |
| 8 | TF-22 | −8.1 | 73 |
| 9 | TF-4 | −8.0 | 93 |
| 10 | N3 | −5.4 | 25 |
All the selected compounds were found to dock at the substrate-binding site, close to the catalytic dyad of His-41 and Cys-145 as shown in the Figure 4. The ring A of all molecules was found to be deep inside the hydrophobic cavity of S2 surrounding by the side chains of His-41, Met-54, Tyr-54 and Met-165 and supported further by the alkyl portion of Asp-187. Position of ring B of all molecules expect TF-9 were oriented at the S4 where the side chains of Met-165, Leu-167, Phe-185, Gln-182 form as small hydrophobic cavity. In case of TF-9, the ring B and in case of BOF-9, the benzyl group (ring C) was oriented at the S1 subsite, which is surrounded by the side chains of Phe-140, Asn-142, Glu-166, His-163 and His-172.
Figure 4.
3D Structures of the molecules docked at the substrate binding site and their orientation with respect to subsites S1 (yellow), S2 (magenta), and S4 (green).
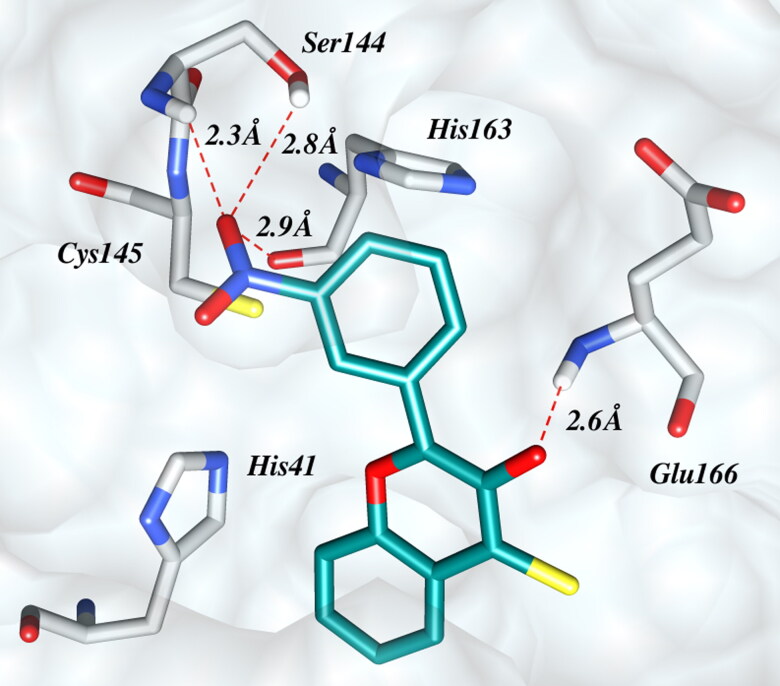
TF-9, containing a nitro group at the meta-position of the ring B is predicted to be the best inhibitor of SARS-CoV-2 Mpro with a favorable binding energy of −8.70 kcal/mol along with appropriate pharmacokinetics and drug-like properties. The benzene ring of the chromenthione was found resting in the S2 cavity and exhibited hydrophobic interactions with side chain of Met165 , Asp187 and Arg188. Most importantly the meta-nitro group of ring B is involved in multiple hydrogen bond interactions with Ser144 and Cys145 in the S1 subsite as shown in Figure 5. Additionally, the hydroxyl group is engaged in hydrogen bonding with the backbone –NH of Glu166 of S1 subsite. Both aromatic rings A and B demonstrate π − π interactions with His-41 and His-163, respectively. It is quite interesting to note that the residues Ans-142 and Gln-189 seem to hold both ring A and B of TF-9 in their respective cavities and functioning as lids of the S1 and S2 subsites.
Figure 5.
Binding of TF-9 at the active site of Mpro of SARS-CoV-2. The shallow surface of the active site of the protease as white surface. The catalytic dyad of His41 and Cys145 is shown as sticks. The ligand (TF-9, docking score -8.7 kcal/mol) is shown as dark cyan tube with conventional colors for oxygen (red), sulfur (yellow) and nitrogen (blue). Red dotted lines indicate hydrogen bonding interactions. For the clarity Asn142, Gln189, Asp187 and Asp188 were hidden.
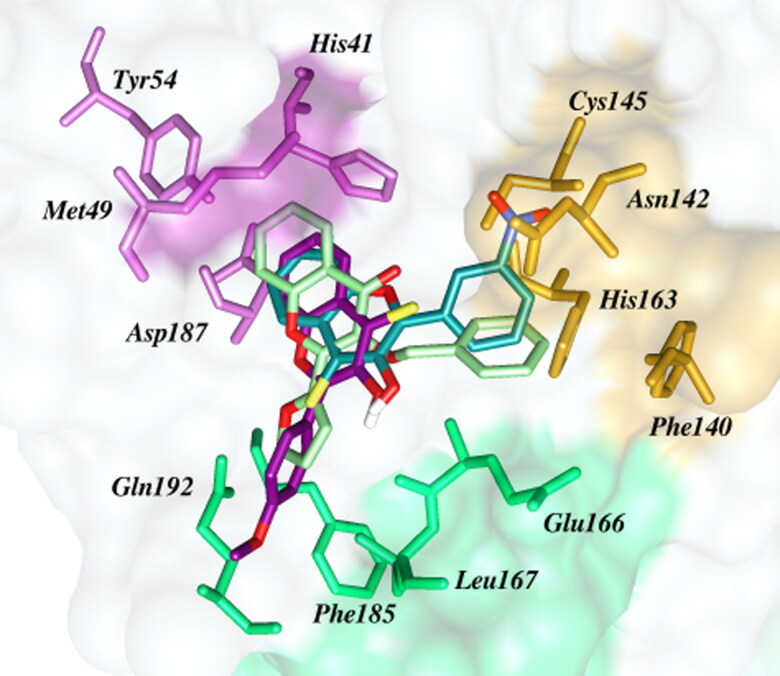
Binding features of other thioflavonols, TF-2, TF-4, TF-6, TF-7, TF-19 and TF- 22 were mutually identical but in stark contrast to the binding pattern of TF-9. Contrary to TF-9, the chromonthione moiety of these molecules, while still in the S2 subsite, but appeared to be flipped 180˚ around the C3 hydroxyl group to position the ring B of these molecules towards the S4 subsite. The docking scores of these flavonols were found in the range of −8.0 kcal/mol to −8.50 kcal/mol as shown in Figure 6 and S3–S8. The thio-ketone moiety was pointing towards the S1 subsite of the active site and the neighboring hydroxyl group was involved in hydrogen bonding interactions with backbone C = O and –NH of Glu-166. The substituent at the para position of ring B were involved in favorable hydrogen bonding interactions with the backbone –NH group of Gln-192 at a distance of 3.56 Å. The strength of hydrogen bonding with Gln-192 seems to depend on the charge density and polarity of the substituents. In case of electron rich –NO2, and –COO- substituents in TF-6 and TF-19 the hydrogen bonding interactions were shown at relatively smaller distances of 2.88 Å and 2.83 Å, respectively (Figures S3 and S4). The F-substituent of TF-22 also acted as hydrogen bond acceptor and demonstrated a long distance (3.11 Å) F˖˖˖H-N interaction with the backbone of Gln-192 (Figure S7). The ortho- and meta-CF3 substituent on the ring B of TF-2 and TF-4 protrude in the solvent exposed S3 subsite and thus yielded a higher binding energy score (Table 1). The residues showing important hydrophobic interactions are shown in the panel B of Figures 6 and S3–S8.
Figure 6.
Binding of TF-7 at the active site of Mpro of SARS-CoV-2. The ligand (TF-7, docking score -8.5 kcal/mol) is shown as dark magneta tube with conventional colors for oxygen (red), sulfur (yellow) and nitrogen (blue). The red dotted lines indicate hydrogen bonding interactions.
Figure 7.
Binding of BOF-9 at the active site of Mpro of SARS-CoV-2. The ligand (BOF-9, docking score -8.3 kcal/mol) is shown as lime green tube. The red dotted lines indicate hydrogen bonding interactions.
One molecule from the benzyloxyflavone (BOF) series, i.e. BOF-9 demonstrated very good docking score (-8.36 kcal/mol), as well as favorable pharmacokinetics and medicinal properties (Tables S7–S10). The binding pattern is reminiscent to TF-7; however due to the presence of an additional ring C at the C-3 position of the chromone moiety, the S1 subsite was also engaged in the binding. The benzyl ring (ring C) of BOF-9 occupies the S1 subsite, where the backbone –NH group of residue Glu-166 assist in a hydrogen bonding interaction with oxygen of the benzyloxy group as shown in Figure 7. All other hydrophobic interactions were more or less identical to the previously discussed molecules. Finally, the oxygen analog of TF-7, i.e. F-7, showed the binding energy score of −8.22 kcal/mol indicating the presence of C = S bond in place of C = O in flavonoids can increase the binding affinity by lowering the binding score up to ∼0.3 kcal/mol. All hydrogen bonding and hydrophobic interactions of F-7 (see Figure S5) were found identical to those observed for TF-7.
3.7. MD simulation
For the determination of structural stability, multiple molecular dynamic simulations were run with the most stable docked models. On the bases of docking scores, two compounds from TF-series (TF-9 and TF-7), one compound from BOF-series (BOF-9) and one compound from F-series (F-7) were subjected to MD simulation. The stability of the simulated systems was evaluated by calculating RMSD, root mean square fluctuation (RMSF) and radius of gyration (RoG).
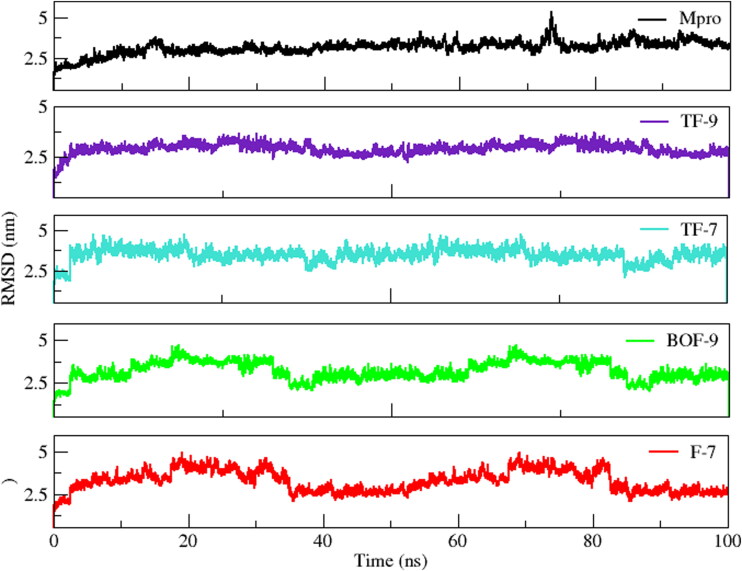
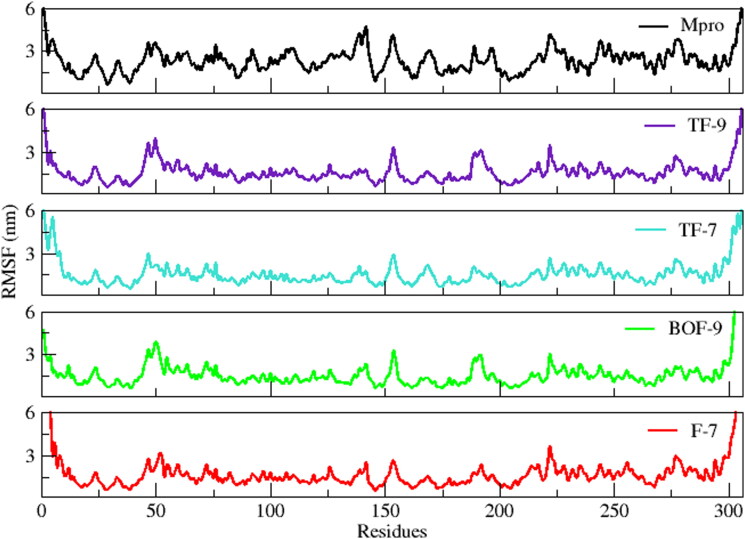
RMSD represents the average deviation from the initial structure over time and stability of the system. The evaluation of RMSD of all the four simulated complexes revealed that all the systems were significantly stable with inconsiderable fluctuations during the course of 100 ns simulation (Figure 8). The average RMSDs of Mpro, TF-9, TF-7, BOF-9, and F-7 were 3.15 ± 0.42 nm, 2.88 ± 0.33 nm, 3.40 ± 0.48 nm, 3.15 ± 0.57 nm and 3.24 ± 0.63 nm, respectively. The trajectory of simulations showed constant presence of inhibitors at the binding site. In case of TF-9, the system gradually reached to the equilibrium in 5 ns and remained stable throughout the 100 ns of simulation. Contrarily, in case of TF-7, BOF-9 and F-7 minor fluctuations were observed in comparison to Mpro and TF-9. However, all the three systems were stabilized after 90 ns and remained stable till the end of the simulation. The inhibitor F-7 fluctuated more as compare to other simulated systems while TF-9 was less deviated from the initial structure, attributing to the favorable and tight binding of this compound. The evaluation of fluctuation or flexibility of the amino acid residues upon binding of the ligands showed similar pattern for all the four simulated systems expect for Mpro, which showed variable fluctuation throughout the simulation. As evident from Figure 9, the regions with most fluctuations, i.e. the loop containing the residue 43-55, loop-antiparallel β-sheets-loop containing the residues 148–160, and the loop containing the residues 185–200, reside close to the active site of Mpro. These loop regions near active site fluctuate to accommodate ligands in the active site which indicate that the regions could be essential for the interaction of the inhibitors with Mpro. With the exception of these loop regions, the RMSF analysis indicated stable interactions for each simulated complex in comparison of Mpro. Thus, both RMSD and RMSF analyses revealed stable interaction of all the ligands upon binding at Mpro.
Figure 8.
Root mean square deviation (RMSD) of all the simulated systems with respect to time.
Figure 9.
Root mean square fluctuation (RMSF) of all the simulated systems with respect to time.
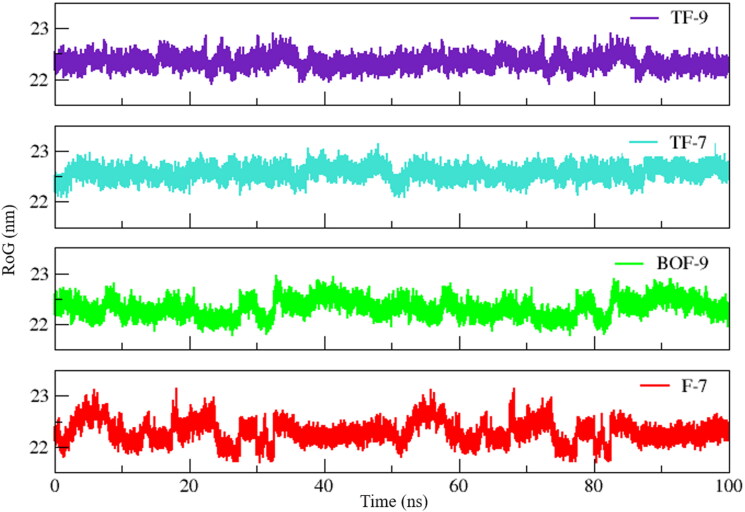
To determine the compactness of the simulated systems RoG was calculated. The results of RoG demonstrated that all the four simulated systems remained significantly compact during the course of 100 ns simulation (Figure 10). However F-7 showed some variable fluctuation but after 85 ns the system gain stability and remain stable till the end of the simulation. The average RoG for TF-9, TF-7, BOF-9 and F-7 was found to be 22.34 ± 0.12, 22.56 ± 0.12, 22.31 ± 0.17 and 22.28 ± 0.21 nm, respectively. Consequently, the analysis of time-dependent behavior of all the four systems indicate that all the ligands reside well in the active site and stabilize the protein.
Figure 10.
Radius of gyration (RoG) of all the simulated systems with respect to time.
3.8. Binding free energy
All the four simulated systems were further subjected to binding free energy calculations by using MM-GBSA and MM-PBSA methods. The binding affinities or ΔG Binding (GB) for TF-9, TF-7, BOF-9 and F-7 were found to be −29.07, −11.55, −6.30 and −25.75 kcal/mol, respectively, indicating that TF-9 was more tightly bound to Mpro than other inhibitors. The contribution of individual energy components upon binding of the ligands are enlisted in Table 2. Analysis of the individual components indicated that the EvdW was the most favorable contributing factor in the overall binding energy of all the tested models. This implies that non-polar and medium polar residues of loop 130-147, i.e. Leu141, Asn142, Gly143, Cys-145, and of β12, i.e. His163, Met-165, Glu166 provide major contribution in binding. In case of TF-9 EvdW interactions play major role in binding of the ligand. Nevertheless, the electrostatic interaction between the polar substituents of TF-9 and Glu166, Gln189 and Gln192 of the binding pocket showed significant contributions. The inhibitor F-7 showed the second strongest binding interaction with Mpro (ΔGB Bind = −25.3 kcal/mol), showing contributions of EvdW and Eele analogous to TF-9. Overall, the results indicated that electrostatic and van der Waals interactions were the predominant factors for the binding of the inhibitors to the SARS-CoV-2 Mpro.
Table 2.
Results of binding free energy calculations.
| System | EvdW | EELE | EGB | ESURF | ΔG Binging (GB) | ΔG Binding (PB) |
|---|---|---|---|---|---|---|
| TF-9 | −40.8 | −12.5 | 22.4 | −4.4 | −35.5 | −29.2 |
| TF-7 | −18.4 | −4.4 | 11.7 | −2.0 | −13.2 | −12.0 |
| BOF-9 | −26.8 | −4.1 | 15.5 | −3.2 | −18.7 | −15.6 |
| F-7 | −34.0 | −7.7 | 20.6 | −4.1 | −25.3 | −19.7 |
4. Conclusions
SARS-CoV-2 has appeared as a potential threat to human health globally. In this study, we have conducted in silico screening of 101 synthetic derivatives of natural products and identified TF-9 as a highly promising inhibitor of Mpro of SARS-CoV-2 with predicted binding energy of −8.70 kcal/mol along with 8 other inhibitors with the binding energy between −8.0 to −8.50 kcal/mol. The docking analyses indicated that the identified compounds bind at the substrate binding site, specifically in the S1, S2 and S4 subsites, exhibiting critical hydrophobic, electrostatic and hydrogen bonding interactions with conserved residues of the active site of SARS-CoV-2 Mpro. The identified compounds further demonstrated favorable pharmacokinetics properties and drug-like behavior. Finally, MD simulations revealed the stability of the docked structures at the binding sites. Moreover, binding free energy analysis indicated the significant contribution of van der Waals and electrostatic interactions in the ligand binding. . Our findings suggest that the thioflavonols have the potential to inhibit SARS-CoV-2 Mpro, thereby inhibiting the replication of the virus and can be a potential candidate against SARS-CoV-2.
Supplementary Material
Acknowledgements
The authors are grateful to Shelvia Malik from Schrodinger Inc. for providing Glide evaluation license and invaluable instructions about the software
Glossary
Abbreviations
- ADME
Absorption, Distribution, Metabolism, and Excretion
- Asn
Asparagine
- Asp
Aspartic acid
- BOF
Benzyloxyflavone
- CADD
Computer-aided drug designing
- Cys
Cysteine
- F
3-Hydroxyflavone/Flavonol
- FG
Flavonol glycoside
- Glu
Glutamic acid
- Gln
Glutamine
- GPF
Grid parameter file
- His
Histidine
- Leu
Leucine
- MCMM
Monte Carlo multiple minimum method
- Mpro
Main protease
- Met
Methionine
- OA
3-Oxoaurone
- PBD
Protein Data Bank
- Phe
Phenylalanine
- SAR-CoV-2
Severe acute respiratory syndrome coronavirus 2
- TA
3-Thioaurone
- TF
3-Thioflavonol
- Tyr
Tyrosine
- WHO
World health organization
Funding Statement
The study was supported by Faculty Initiative Funding (FIF-642) from Lahore University of Management Sciences
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
F.B. ran iterative docking experiments and ADME analyses. A.S and N.N. draw chemical structures and performed experimental work. A. J. formed some docking analyses. M. S. and E. U. M. conceived idea, supervised and wrote manuscript. M. S. designed computational study, analyzed data, and edited the manuscript. K. Z. and Z.U.-H. performed MD simulation study.
References
- Ancy, I., Sivanandam, M., & Kumaradhas, P. (2020). Possibility of HIV-1 protease inhibitors-clinical trial drugs as repurposed drugs for SARS-CoV-2 main protease: A molecular docking, molecular dynamics and binding free energy simulation study. Journal of Biomolecular Structure and Dynamics, 1–8. 10.1080/07391102.2020.1786459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, S. C., Lee, J. H., & Park, S. U. (2013). Recent studies on flavonoids and their antioxidant activities. EXCLI Journal, 12, 226–230. [PMC free article] [PubMed] [Google Scholar]
- Ciliberto, G., Mancini, R., & Paggi, M. G. (2020). Drug repurposing against COVID-19: Focus on anticancer agents. Journal of Experimental & Clinical Cancer Research: Cr, 39(1), 86. 10.1186/s13046-020-01590-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, W., Zhang, B., Jiang, X.-M., Su, H., Li, J., Zhao, Y., Xie, X., Jin, Z., Peng, J., Liu, F., Li, C., Li, Y., Bai, F., Wang, H., Cheng, X., Cen, X., Hu, S., Yang, X., Wang, J., … Liu, H. (2020). Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science (New York, N.Y.).), 368(6497), 1331–1335. 10.1126/science.abb4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717. 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertekin, S. S., Morgado-Carrasco, D., Forns, X., & Mascaro, J. M. Jr. (2020). Complete remission of hypertrophic discoid cutaneous lupus erythematosus after treatment of chronic hepatitis C with direct-acting antivirals. JAMA Dermatology, 156(4), 471–472. 10.1001/jamadermatol.2020.0013 [DOI] [PubMed] [Google Scholar]
- Ghosh, R., Chakraborty, A., Biswas, A., & Chowdhuri, S. (2020). Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors - An in silico docking and molecular dynamics simulation study. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1779818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Singh, A. K., Kushwaha, P. P., Prajapati, K. S., Shuaib, M., Senapati, S., & Kumar, S. (2020). Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1776157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung, A. B., Ali, M. A., Lee, J., Farah, M. A., & Al-Anazi, K. M. (2020). Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sciences, 255, 117831. 10.1016/j.lfs.2020.117831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage-Melim, L. I. d. S., Federico, L. B., de Oliveira, N. K. S., Francisco, V. C. C., Correia, L. C., de Lima, H. B., Gomes, S. Q., Barcelos, M. P., Francischini, I. A. G., & da Silva, C. H. T. d. P. (2020. a). Virtual screening, ADME/Tox predictions and the drug repurposing concept for future use of old drugs against the COVID-19. Life Sciences, 256, 117963. 10.1016/j.lfs.2020.117963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, J. A., & Chung, R. T. (2020). Shortening treatment with direct-acting antivirals in HCV-positive organ transplantation. The Lancet. Gastroenterology & Hepatology, 5(7), 626–627. 10.1016/S2468-1253(20)30154-0 [DOI] [PubMed] [Google Scholar]
- Ibrahim, M. A. A., Abdeljawaad, K. A. A., Abdelrahman, A. H. M., & Hegazy, M. F. (2020). Natural-like products as potential SARS-CoV-2 M(pro) inhibitors: In-silico drug discovery. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1790037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Alberto, A., Ribas-Aparicio, R. M., Aparicio-Ozores, G., & Castelan-Vega, J. A. (2020). Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Comput Biol Chem, 88, 107325. 10.1016/j.compbiolchem.2020.107325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Joshi, T., Joshi, T., Sharma, P., Mathpal, S., Pundir, H., Bhatt, V., & Chandra, S. (2020). In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. European Review for Medical and Pharmacological Sciences, 24(8), 4529–4536. 10.26355/eurrev_202004_21036 [DOI] [PubMed] [Google Scholar]
- Khan, S. A., Zia, K., Ashraf, S., Uddin, R., & Ul-Haq, Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- King, A. R., Lodola, A., Carmi, C., Fu, J., Mor, M., & Piomelli, D. (2009). A critical cysteine residue in monoacylglycerol lipase is targeted by a new class of isothiazolinone-based enzyme inhibitors. British Journal of Pharmacology, 157(6), 974–983. 10.1111/j.1476-5381.2009.00276.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, Y., Singh, H., & Patel, C. N. (2020). In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. Journal of Infection and Public Health, 13(9), 1210–1223. 10.1016/j.jiph.2020.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani, S., & Poh, C. L. (2020). Flavonoids as antiviral agents for enterovirus A71 (EV-A71). Viruses, 12(2), 184. 10.3390/v12020184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase, E. (2020). Covid-19: WHO declares pandemic because of "alarming levels" of spread, severity, and inaction. BMJ (Clinical Research ed.).), 368, m1036. 10.1136/bmj.m1036 [DOI] [PubMed] [Google Scholar]
- Marinho, E. M., Batista de Andrade Neto, J., Silva, J., Rocha da Silva, C., Cavalcanti, B. C., Marinho, E. S., & Nobre Junior, H. V. (2020). Virtual screening based on molecular docking of possible inhibitors of Covid-19 main protease. Microbial Pathogenesis, 148, 104365. 10.1016/j.micpath.2020.104365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengist, H. M., Fan, X., & Jin, T. (2020). Designing of improved drugs for COVID-19: Crystal structure of SARS-CoV-2 main protease Mpro). Signal Transduction and Target Therapy, 5(1), 67. 10.1038/s41392-020-0178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). Autodock4 and AutodockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughal, E. U., Javid, A., Sadiq, A., Murtaza, S., Zafar, M. N., Khan, B. A., … Khan, K. M. (2018). Synthesis, structure-activity relationship and molecular docking studies of 3-O-flavonol glycosides as cholinesterase inhibitors. Bioorg Med Chem, 26(12), 3696–3706. 10.1016/j.bmc.2018.05.050 [DOI] [PubMed] [Google Scholar]
- Mughal, E. U., Sadiq, A., Ashraf, J., Zafar, M. N., Sumrra, S. H., Tariq, R., Mumtaz, A., Javid, A., Khan, B. A., Ali, A., & Javed, C. O. (2019). Flavonols and 4-thioflavonols as potential acetylcholinesterase and butyrylcholinesterase inhibitors: Synthesis, structure-activity relationship and molecular docking studies. Bioorganic Chemistry, 91, 103124. 10.1016/j.bioorg.2019.103124 [DOI] [PubMed] [Google Scholar]
- Mughal, E. U., Sadiq, A., Murtaza, S., Rafique, H., Zafar, M. N., Riaz, T., Khan, B. A., Hameed, A., & Khan, K. M. (2017). Synthesis, structure-activity relationship and molecular docking of 3-oxoaurones and 3-thioaurones as acetylcholinesterase and butyrylcholinesterase inhibitors. Bioorg Med Chem, 25(1), 100–106. 10.1016/j.bmc.2016.10.016 [DOI] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics., 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Narkhede, R. R., Pise, A. V., Cheke, R. S., & Shinde, S. D. (2020). Recognition of natural products as potential inhibitors of COVID-19 main protease (Mpro): In-silico evidences. Natural Products and Bioprospecting, 10(5), 297–306. 10.1007/s13659-020-00253-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, C., Yin, J., Zhang, J., Vederas, J. C., & James, M. N. (2008). Molecular docking identifies the binding of 3-chloropyridine moieties specifically to the S1 pocket of SARS-CoV Mpro. Bioorganic & Medicinal Chemistry, 16(1), 293–302. 10.1016/j.bmc.2007.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panche, A. N., Diwan, A. D., & Chandra, S. R. (2016). Flavonoids: An overview. Journal of Nutritional Science, 5, e47. 10.1017/jns.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietta, P. G. (2000). Flavonoids as antioxidants. Journal of Natural Products, 63(7), 1035–1042. 10.1021/np9904509 [DOI] [PubMed] [Google Scholar]
- Sepay, N., Sepay, N., Al Hoque, A., Mondal, R., Halder, U. C., & Muddassir, M. (2020). In silico fight against novel coronavirus by finding chromone derivatives as inhibitor of coronavirus main proteases enzyme. Structural Chemistry, 31(5), 1831–1840. 10.1007/s11224-020-01537-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, B., Modi, P., & Sagar, S. R. (2020). In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sciences, 252, 117652. 10.1016/j.lfs.2020.117652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies, H., & Parnham, M. J. (2020). Potential therapeutic use of ebselen for COVID-19 and other respiratory viral infections. Free Radical Bioloogy and Medicine, 156, 107–112. 10.1016/j.freeradbiomed.2020.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, F., Shi, N., Shan, F., Zhang, Z., Shen, J., Lu, H., Ling, Y., Jiang, Y., & Shi, Y. (2020). Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology, 295(1), 210–217. 10.1148/radiol.2020200274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standing, J. F. (2020). Quantitative clinical pharmacology input to SARS-CoV-2 therapeutics should be based on robust data. Clinical Pharmacology and Therapeutics, 108(2), 187. 10.1002/cpt.1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, A. T., Gentile, F., Hsing, M., Ban, F., & Cherkasov, A. (2020). Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Molecular Informatics, 39(8), e2000028. 10.1002/minf.202000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, M. K., Singh, P., Sharma, S., Singh, T. P., Ethayathulla, A. S., & Kaur, P. (2020). Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1790425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viani, F., Rossi, B., Panzeri, W., Merlini, L., Martorana, A. M., Polissi, A., & Galante, Y. M. (2017). Synthesis and anti-bacterial activity of a library of 1,2-benzisothiazol-3(2H)-one (BIT) derivatives amenable of crosslinking to polysaccharides. Tetrahedron, 73(13), 1745–1761. 10.1016/j.tet.2017.02.025 [DOI] [Google Scholar]
- Villoutreix, B. O., Beaune, P. H., Tamouza, R., Krishnamoorthy, R., & Leboyer, M. (2020). Prevention of COVID-19 by drug repurposing: Rationale from drugs prescribed for mental disorders. Drug Discov Today, 25(8), 1287–1290. 10.1016/j.drudis.2020.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Horby, P. W., Hayden, F. G., & Gao, G. F. (2020). A novel coronavirus outbreak of global health concern. Lancet (London, England), 395(10223), 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Bao, B.-B., Song, G.-Q., Chen, C., Zhang, X.-M., Lu, W., Wang, Z., Cai, Y., Li, S., Fu, S., Song, F.-H., Yang, H., & Wang, J.-G. (2017). Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: Chemical synthesis, biological evaluation, molecular docking and 3D-QSAR study. European Journal of Medicinal Chemistry, 137, 450–461. 10.1016/j.ejmech.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Xing, J., Xu, Y., Zhou, N., Peng, J., Xiong, Z., Liu, X., Luo, X., Luo, C., Chen, K., Zheng, M., & Jiang, H. (2015). In silico ADME/T modelling for rational drug design. Quarterly Reviews of Biophysics, 48(4), 488–515. 10.1017/S0033583515000190 [DOI] [PubMed] [Google Scholar]
- Whitworth, J. (2020). COVID-19: A fast evolving pandemic. Transactions of the Royal Society of Tropical Medicine and Hygiene, 114(4), 241–248. 10.1093/trstmh/traa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020, October 11). Coronavirus disease (COVID-19 ). https://www.who.int/docs/default-source/coronaviruse/situation-reports/20201012-weekly-epi-update-9.pdf
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaryan, H., Arabyan, E., Oo, A., & Zandi, K. (2017). Flavonoids: Promising natural compounds against viral infections. Archives of Virology, 162(9), 2539–2551. 10.1007/s00705-017-3417-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.).), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W.; China Novel Coronavirus Investigating and Research Team. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L., Ruan, F., Huang, M., Liang, L., Huang, H., Hong, Z., Yu, J., Kang, M., Song, Y., Xia, J., Guo, Q., Song, T., He, J., Yen, H.-L., Peiris, M., & Wu, J. (2020). SARS-CoV-2 viral load in upper respiratory specimens of infected patients. The New England Journal of Medicine, 382(12), 1177–1179. 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.