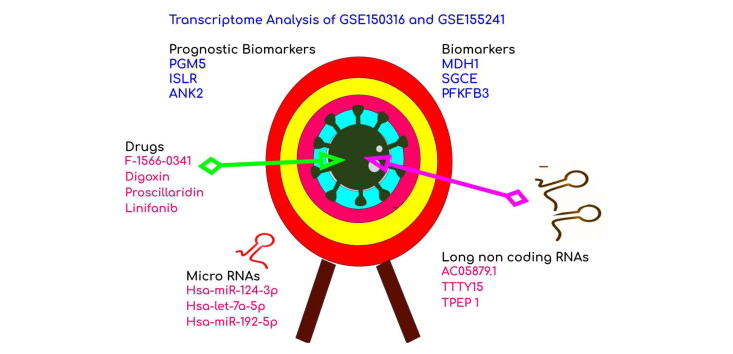
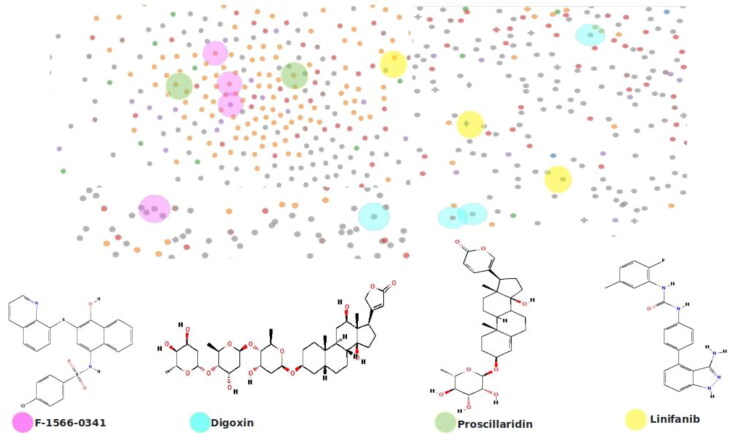
Abstract
The coronavirus disease, caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is a global health crisis that is being endured with an increased alarm of transmission each day. Though the pandemic has activated innumerable research attention to decipher an antidote, fundamental understanding of the molecular mechanisms is necessary to halt the disease progression. The study focused on comparison of the COVID-19 infected lung tissue gene expression datasets -GSE155241 and GSE150316 with the GEO2R-limma package. The significant up- and downregulated genes were annotated. Further evaluation of the enriched pathways, transcription factors, kinases, noncoding RNAs and drug perturbations revealed the significant molecular mechanisms of the host response. The results revealed a surge in mitochondrial respiration, cytokines, neurodegenerative mechanisms and deprived oxygen, iron, copper, and glucose transport. Hijack of ubiquitination by SARS-CoV-2, hox gene differentiation, histone modification, and miRNA biogenesis were the notable molecular mechanisms inferred. Long non-coding RNAs such as C058791.1, TTTY15 and TPTEP1 were predicted to be efficient in regulating the disease mechanisms. Drugs-F-1566-0341, Digoxin, Proscillaridin and Linifanib that reverse the gene expression signatures were predicted from drug perturbations analysis. The binding efficiency and interaction of proscillaridin and digoxin as obtained from the molecular docking studies confirmed their therapeutic potential. Two overlapping upregulated genes MDH1, SGCE and one downregulated gene PFKFB3 were appraised as potential biomarkers candidates. The upregulation of PGM5, ISLR and ANK2 as measured from their expressions in normal lungs affirmed their possible prognostic biomarker competence. The study explored significant insights for better diagnosis, and therapeutic options for COVID-19.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, bioconductor, differential gene expression, miRNAs, lncRNAs, drug perturbations
Graphical Abstract
1. Introduction
Coronavirus disease 2019 (COVID-19) is a prominent health distress that governs the contemporary era and is worsening every single human’s existence. The disease declared as a pandemic threat by the World Health Organization (WHO) has abruptly risen and has more than 27,738,179 confirmed active cases and 899,916 deaths spanning 216 countries and territories (Kuster et al., 2020). The vital information associated with the disease’s causative agent, symptoms and rate of mortality are numerous with little focus on the virus’s pathobiology and molecular mechanisms (Wu & McGoogan, 2020). Though the novel coronavirus (nCoV) shares 80% homology with the severe acute respiratory syndrome (SARS) coronavirus outbreak in 2002, there is no accurate prevention or treatment options implemented so far. It encompasses functional receptors that can differentially express pathogenesis in various parts of the respiratory, circulatory, central nervous systems and other vital organs of the body. A profoundly studied one such functional receptor is angiotensin-converting enzyme 2 (ACE2) targeted by the spike protein from SARS-CoV-2 (Chen et al., 2020; Li et al.. 2003; Walls et al., 2020). The structural and functional analysis of ACE2, showed high integrity and expression levels in the lung (epithelial cells), heart, ileum, kidney and bladder (Dong et al., 2018; Letko et al., 2020; Zou et al., 2020). Though there is profound research on various host receptors tantalizing the virus, significant focus on the mechanisms and other host responses are still lacking.
There is a vital need to analyse the disease in its cellular biology perspective which can point out the additional target sites preferred by the virus. This will definitely lead to a way that unravels the complications involved to produce an antidote. Gene expression profiles via high throughput RNA sequencing can trace back the rate of pathogenesis and develop targeted drugs against a specific ailment (Zhao et al., 2018). Gene chips along with statistical programs, offer a significant sustenance that can analyse gene expressions and ability to determine diagnostic and prognostic biomarkers to combat imminent diseases (Hou et al., 2019). Molecular mechanisms and pathology are interrelated and generally imperceptible. Elucidation of the gene expression profile discriminates between normal and disease conditions with an exploration of the molecular data (Zhou et al., 2016). RNA interference is one such molecular strategy adopted by both the host and the virus. Competing endogenous RNAs (ceRNA) are non-coding functional RNAs like microRNAs (miRNA) and long non-coding RNAs (lncRNA) which interact with the mRNA to either suppress or induce viral pathogenesis (Hsu et al., 2007; Wen et al., 2019). Recently, there are enormous research reports on evaluation of inhibitors based on machine learning, to name a few, drug repurposing with chloroquine (Liu et al., 2020), Withania somnifera (Kumar et al., 2020a), and quaternary ammonium compounds (Baker et al., 2020) to tackle the spread and pathogenesis. Conversely, the integration of molecular signatures, gene patterns, and enrichment of pathways, network biology and drug repositioning are reliable panacea. The current work is a result of such integrative approach that exposes the unique characteristics and underlying molecular mechanisms of COVID-19 which may lead to the development of more precise diagnostic biomarkers and effective therapeutic strategies.
The present study focused on statistically synchronized in silico methods to identify the differential gene expression studies of two datasets of Gene expression omnibus, GSE150316 and GSE155241. The datasets contained high throughput RNA sequencing data from the autopsy of various organs of patients succumbed to COVID-19. In order to eliminate poor reproducibility, intensive attention was given only on the lung tissues from both healthy and diseased samples which evaluated the differential expression of genes. Varied Integrated approaches applied in this study computed the enrichment of functions, pathways and non-coding RNAs which gained insights into molecular mechanisms underlying the pathogenesis. The assessment of protein–protein interactions performed augmented the hub genes and their respective association. Potential diagnostic and prognostic biomarkers were confirmed from the overlapping genes of the disastrous pandemic COVID-19 which can be developed into further treatment and therapeutic options.
2. Methodology
2.1. Data retrieval and normalization
The gene expression datasets of COVID-19 were retrieved from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with the accessions GSE150316 (Desai et al., 2020) and GSE155241 (Han et al., 2020). The datasets housed multiple samples and we chose only the lung tissues for our study. The platforms for GSE150316 was based on GPL15520 Illumina Miseq (Homo sapiens) and had 88 samples of various tissues. Among them 5 control and 16 lung tissue samples affected with COVID-19 were chosen. GSE155241 was based on the platform GPL24676 Illumina Novaseq6000 (H. sapiens) with 18 samples of lung autopsy tissues and organoids. Three samples each from healthy lung, COVID-19 affected lung autopsy tissues, mock control hpsc (human pluripotent stem cell) derived lung organoid and COVID-19 infected hpsc lung organoid samples were analysed in our study. Both the datasets were standardised, log transformed and scrutinised for the differentially expressed genes (DEGs) using the GEO2R module of Limma package of R. Statistics of Benjamini-Hochberg false discovery rate (Dubitzky et al., 2013) was applied with log2FC >0.5 (Perez et al., 2019) and p-value <0.05 thresholds (Stephens et al., 2019) to screen the significant genes. Heat maps of top 100 genes clustered with K-means and volcano plots representing the up- and downregulated DEGs were plotted using the bioinfokit module of python (Bedre, 2020). Venn diagrams were useful to determine the mutually regulated genes and the overlapping DEGs of both the datasets were identified using the Venny tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
2.2. Gene ontology and functional enrichment
Gene ontology (GO) enrichment, a remarkable strategy predicted the Biological Process, Molecular Function, and Cellular Component of the DEGs (Zhu et al., 2020). The DEGs of individual datasets were subjected to David functional annotation tool (https://davidd.ncifcrf.gov/) to annotate the GO functions and pathway terms from the KEGG (http://www.genome.jp/kegg/) (Kanehisa et al., 2016) and Reactome (https://reactome.org/) (Fabregat et al., 2018) databases. Relative gene annotation and pathway analysis was computed with ClueGo version 2.5.7 (Bindea et al., 2009)implemented in the cytoscape software version 3.5.8 based on Benjamini Hochberg method with kappa score of 3 and hypergeometric two-sided Statistical tests (Rivals et al., 2007). Enrichment of transcription factors and kinases of the significant DEGs were predicted from the Encode (https://genome.ucsc.edu/ENCODE) (Davis et al., 2018; Rivals et al., 2007) and KEA (http://www.maayanlab.net/KEA2/) (Lachmann & Ma’ayan, 2009) databases from Enrichr platform (Kuleshov et al., 2016).
2.3. Protein–protein interaction and hub gene analysis
Significant DEGs were evaluated for their encoding protein interactions to identify the hub genes using the STRING database (http://stringdb.org/) (Szklarczyk et al., 2017). The potential interactions were screened using the MCODE plugin installed in the cytoscape software. Cluster analysis of the protein–protein interaction network was performed with the networks scoring degree cutoff of 2, K-score of 2, and node score cutoff of 0.2 and identified the up- and downregulated hub genes (Bader & Hogue, 2003). The hub genes were further validated by predicting their expression patterns across the tissues of the respiratory and circulatory systems using the Expression Atlas (https://www.ebi.ac.uk/gxa/home) (Papatheodorou et al., 2019) and the human protein atlas platforms (http://www.proteinatlas.org). The transcript and encoding protein expression rates of the hub genes were predicted from the Genotype Expression dataset (GTEX) (GTEx Consortium, 2013) available in the human protein atlas database for their normal expression levels in lungs, bronchus and blood.
2.4. Network of mRNA-miRNA-lncRNA
MicroRNAs are small endogenous RNAs that are proven to suppress the target mRNAs by complementarity binding and hence they play a vulnerable role in viral pathogenesis (Li et al., 2006). Many evidence suggests that host miRNA plays a significant role in viral life cycle, replication and pathogenesis by a complicated regulatory mechanism (Hou et al., 2019). Hence, targeting the host miRNAs can certainly aid in reducing the disease progression. From the DEGs, targets of miRNAs were identified by referring to the Mirnet (http://www.mirnet.ca) (Chang et al., 2020). The DEGs with expression levels that are inversely proportional to the miRNAs were retained (Lei et al., 2019). Long non coding RNAs are reported to act on the miRNAs as sponges, to prevent miRNA–mRNA-binding and rescue mRNA levels to translation (Wang, 2018). The cross talk between lncRNAs and miRNAs were predicted from the DIANA lncBase (http://diana.imis.athena.innovation.gr) with scores of >0.99 (Yousefi et al., 2020). The network of competing endogenous RNAs (ceRNA) including the mRNA–miRNA–lncRNA network was constructed using cytoscape (Zhou et al., 2019).
2.5. Drug perturbation analysis on the transcriptome data
The significant overlapping and individual DEGs of the datasets were used to analyse the small compounds and drugs that can either mimic or reverse the gene expression pattern. L1000FWD (http://www.lincsproject.org/LINCS/dmoa) (Wang et al., 2018) platform computed the chemical perturbagens capable of reversing the DEGs (Ye et al., 2018). The potential drug candidates that reverse the DEG pattern on the lung cancer cell line A549 were identified based on p-value (Huang et al., 2018)
2.6. Molecular Docking studies of the predicted drugs
In order to validate the performance of the predicted drug perturbagens, molecular docking was performed with the four drug compounds against the extensively studied target of COVID-19, Main protease (MPro) enzyme. Three dimensional structure of the four drug compounds – Digoxin, Proscillaridin, Linifanib and F1566-1034 were retrieved from pubchem (http://www.pubchem.ncbi.nlm.nih.gov) and optimised with OPLS 2005 force field using the ligprep module of Schrodinger (version 2020-1) (Ogidigo et al., 2020). The 3D crystal structure of COVID-19 main protease (6LU7) was retrieved from PDB and prepared with protein preparation wizard of Schrodinger using the OPLS2005 force field (Shivanika et al., 2020). The active site of the protein was calculated using a sitemap module and the receptor grid was generated. Molecular docking of the optimised ligands with the target protein was performed in eXtra Precision docking (XP) mode of the Glide module. Docking of Drugs along with the complexed ligand–peptide-like inhibitor N3, enabled better comparison of the binding energy and interactions (Mittal et al., 2020).
3. Results
3.1. Data set and normalization
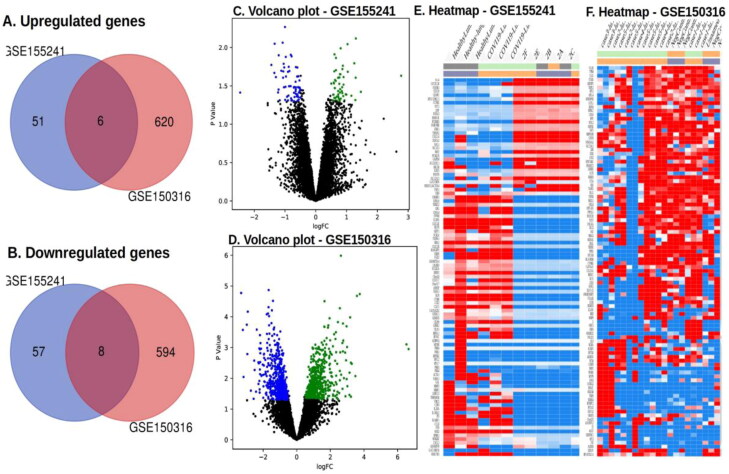
The raw data of GSE150316 and GSE155241 were downloaded from the GEOdatabase where the samples of lung were retained and the remaining tissues from both datasets were discarded for better comparison and reproducibility. The analysis was on 5 healthy controls, 16 samples of COVID-affected lung autopsy tissues from GSE150316. Tissues of healthy lung, COVID-19 affected lung autopsy tissues, mock control hpsc lung organoid and COVID-19-infected hpsc lung organoid samples (3 each) from GSE155241 were analysed. The selected sample sets were standardised and analysed with the GEO2R package to obtain DEGs with the threshold of p value <0.05 and log2FC >0.5. There were 51 up- and 57 downregulated DEGs obtained from GSE155241. A total of 620 upregulated and 594 downregulated DEGs acquired from GSE150316. Further a total of six upregulated and eight downregulated DEGs were identified that were mutual to both the datasets (Figure 1(A,B)). Volcano plots representing the up- and downregulated DEGs are shown in the Figure 1(C,D). For effective visualization, top 100 DEGs ranked based on their p values were clustered in a heatmap based on K-means clustering algorithm. Heatmaps of the top 100 DEGs of dataset GSE155241 and GSE150316 are shown in Figure 1(E,F), respectively. The overlapping upregulated genes were identified as AMZ2, MDH1, TMEM261, PHF14, PTGFRN and SGCE. The overlapping down regulated genes were UGCG, DUSP6, TIPARP, NUPL1, NOLC1, PFKFB3, ERRFI1 and SLC19A2.
Figure 1.
Differentially expressed genes of the datasets GSE155241 and GSE150316. (A) Venn diagrams representation of the commonly found upregulated DEGs of both datasets. (B) Venn diagrams representation of the commonly found downregulated DEGs of both datasets. (C)Volcano plots of the upregulated DEGs with log2FC >0.5 and p value (FDR) < 0.05. (D) Volcano plots of the downregulated DEGs with log2FC >0.5 and p value (FDR) < 0.05. Green and blue dots represent up- and downregulated genes, respectively. Black dots represent the remaining genes with no significant difference. (E) Heatmap representation of the top 100 DEGs of GSE155241 is shown. (F) Heatmap representation of the top 100 DEGs of GSE150316 is shown. DEGs: differentially expressed genes.
3.2. Gene ontology analysis
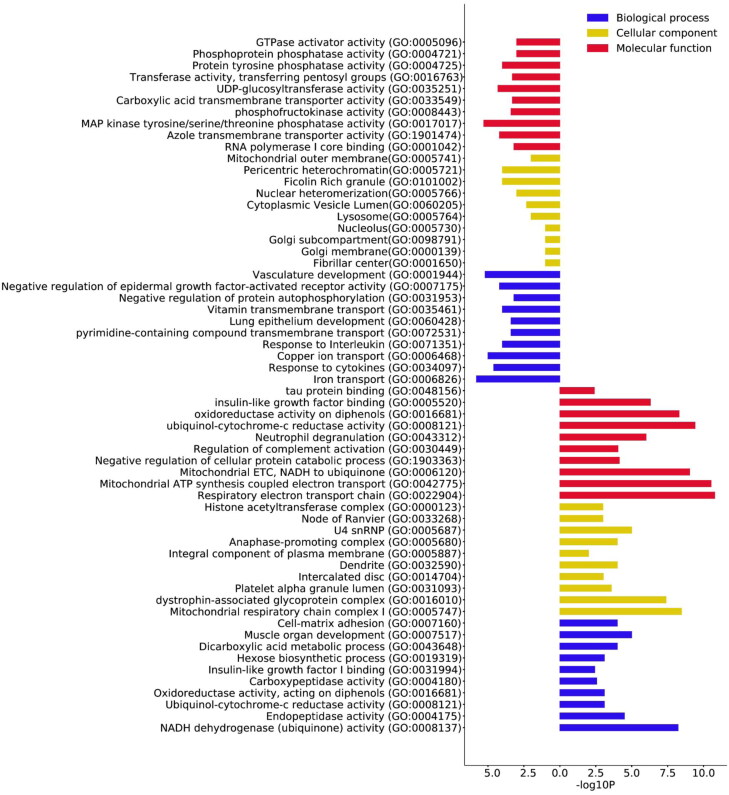
The significant up- and downregulated DEGs were annotated using the DAVID tool. The top 10 annotations were screened based on their pvalue including the sub-ontologies biological process (BP), cellular components (CC) and molecular functions (MF) (Saddala et al., 2020) and shown in Figure 2. For the upregulated BP, significant enrichment was found in NADH ubiquinone activity, endopeptidase activity, ubiquinol cytochrome-c reductase activity and cell matrix adhesion. Conversely, for CC, the enrichment was observed in mitochondrial respiratory complex I, dystrophin associated glycoprotein complex, platelet alpha granules lumen, dendrites and intercalated disc. Enrichment in the molecular functions like mitochondrial respiration, neutrophil degranulation, complement activation and tau protein binding were significantly enriched. The significant downregulated BP enrichment was seen in response to cytokines, interleukins, lung development, copper and iron transport. Lysosomes, golgi complex and nucleolus were down regulated CC and MF like GTPase activators, protein tyrosine phosphatase, MAP kinase activity and transporters like azole and carboxylic acid trans membrane transporters were significantly downregulated.
Figure 2.
Gene Ontology (GO) term enrichment analysis of upregulated DEGs. The top 10 annotations ranked based on p values are shown for three sub-ontologies, namely biological process, molecular function, and cellular component shown in the bars to the right. The top 10 downregulated annotations ranked based on p values are shown for three sub-ontologies, namely biological process, molecular function, and cellular component shown in the bars to the left.
3.3. Functional enrichment and interrelational analysis
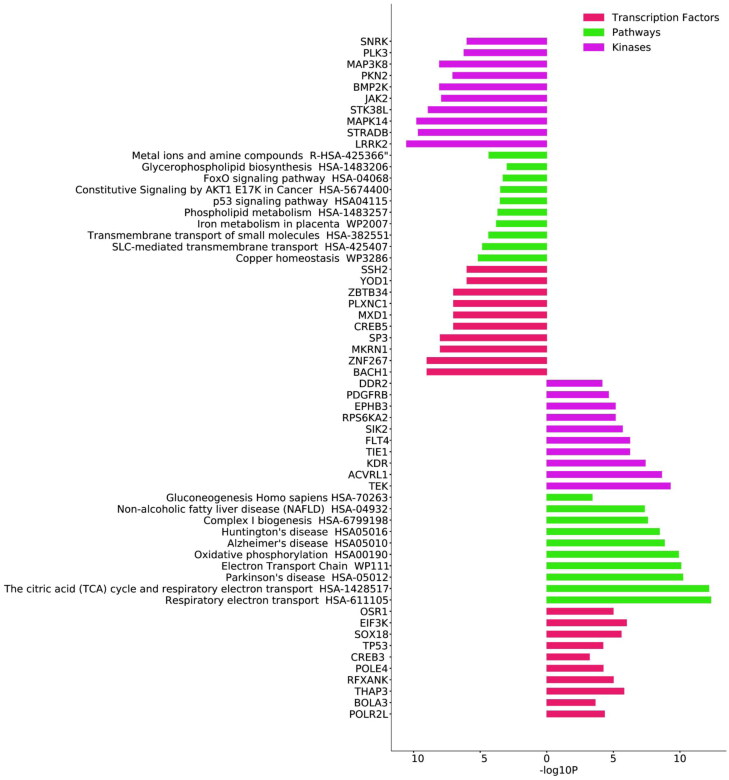
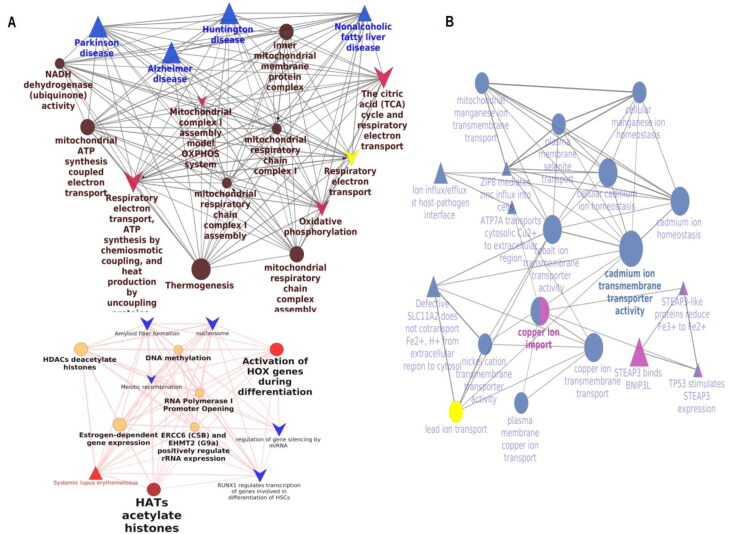
As shown in Figure 3, transcription factors like RNA polymerase 1, CREB, DNA polymerase E4, BOLA3 responsible for mitochondrial chain complex assembly were few upregulated transcription factors. Most of the overexpressed transcription factors and kinases play a role in angiogenesis and vasculature development. Kruppel like factors, BACH1, MKRN1, SP3 and various zinc finger domain transcription factors were downregulated. Plexin – a transcription factor that was reported to elicit interleukin production upon viral entry is remarkably downregulated. LRRK2, STK38L that were responsible for neuronal plasticity and differentiation of mature neurons were considerably under expressed. Pathway enrichment was performed from both KEGG and REACTOME databases. The top 10 pathways mutually identified in both databases and ranked by their p-value (Saddala et al., 2020) are shown in Figure 3. Electron transport chains, citric acid cycle, nicotinamide scavenging, Alzheimer's disease and Huntington’s disease pathways were upregulated and identified from both the databases. Copper homeostasis, FOXO signalling pathways, metal ion transporters, and various signalling pathways involved in cancers were all under expressed. Tyrosine kinases, active receptor like kinases, salt inducible kinases, and myosin light chain kinases were few of the kinases that were upregulated whereas ubiquitin kinase, protein kinase 3 and bone morphogenetic protein kinases were few of the downregulated kinases identified from the DGEs. Further to ensure consistency with the above results of annotations and functional enrichments, an interrelation analysis with ClueGO module was performed. Notable upregulation of mitochondrial respiration, electron transport chain, hijacking of ubiquitination by SARS-CoV-2, miRNA biogenesis, viral mRNA synthesis, histone acetyl transferases and activation of hox genes were identified. Pathological conditions like systemic lupus erythematosus, Parkinson’s, Alzheimers’, non-alcoholic fatty acid liver diseases were over expressed (Figure 4(A)). The relative analysis confirmed the downregulation of cadmium, copper, iron, zinc and manganese transport as seen in Figure 4(B).
Figure 3.
Signalling pathway enrichment analysis of overlapped DEGs. The pathway enrichment analysis with KEGG and REACTOME are shown for upregulated DEGs on the bars to the right. The pathway enrichment analysis with KEGG and REACTOME are shown for downregulated DEGs on the bars to the left.
Figure 4.
Interrelational pathway analysis and GO enrichment of the DEGs. (A) interrelational analysis of the upregulated DEGs. (B) interrelational analysis of the downregulated DEGs. The pathways are shown in arrow representation and the GO in ellipses.
3.4. Protein–protein interactions and hub gene validation
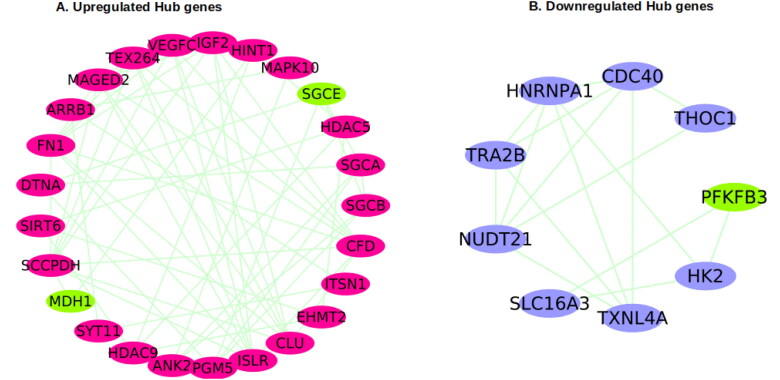
Protein- Protein interactions (PPI) encoded by the significant DEGs of both the datasets were identified from STRING database and analysed in cytoscape to predict the hub genes. From the overall up- and downregulated PPI obtained, MCODE module of cytoscape predicted 10 and 7 predominant clusters respectively. The top two upregulated clusters with 25 nodes with a score of 29 (Figure 5(A)) and a significant down regulated cluster with 9 nodes and a score of 17.34 (Figure 5(B)) were chosen to identify the hub genes. The selected clusters had two of the overlapping upregulated genes -SGCE and MDH1 and one overlapping down regulated gene PFKFB3, respectively.
Figure 5.
Protein–protein interaction network of the DEGs and identification of hub genes. (A) hub genes from the PPI network of the upregulated DEGs (B) hub genes from the PPI network of the downregulated DEGs. Overlapping genes are shown in green. PPI: protein–protein interaction. DEGs: differentially expressed genes.
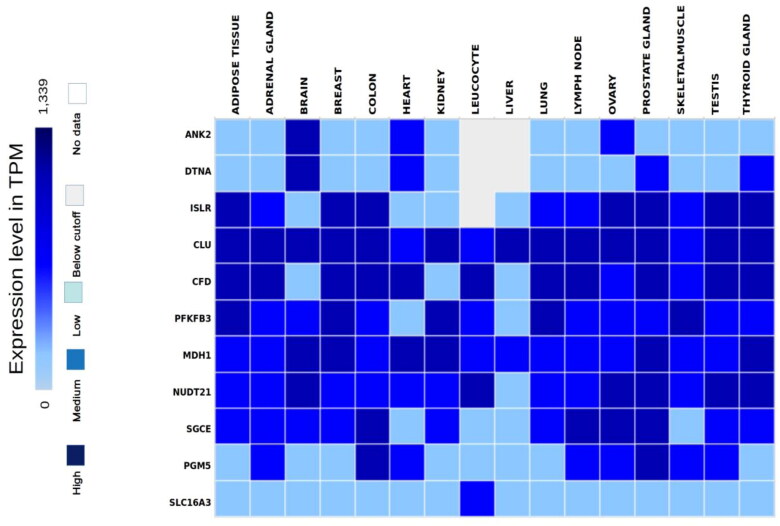
In order to validate the hub genes, expression levels of the hub genes from COVID-19 affected lungs, were compared with their expressions in healthy lungs from the expression atlas platform and human protein atlas platforms. The normal expressions were measured at 1339 TPM and were presented in Table 1 and Figure 6. As shown in Table 1, very low expressions of MDH1 and SGCE were identified with values of 29.2 and 44.7 TPM, respectively. Their encoding proteins were also found to be low in bronchus and lungs. On the contrary the two genes were significantly up regulated and occurred in both the datasets of COVID-affected lungs. Hence, the two genes were proclaimed to be potential biomarker candidates of COVID-19. Availability of them in blood MDH1 in T-cells and SGCE in B-cells make them more significant and measurable biomarkers. Expression of significantly downregulated hub gene PFKFB3, was very high in healthy lungs with 96.4 TPM and in neutrophils with 124.8 TPM. The contrast expression rates of PFKFB3 in healthy vs COVID-19 infected lungs were attributed to be a significant gene biomarker. Higher expression of SLC16A3 and medium expression of NUDT21 were observed in other tissues like brain, heart and adipose tissues and hence were not considered significant for COVID-19. Expression rates of CLU, CFD were not consistent with previous results and again did not hold significance. Genes PGM5, ISLR and ANK2 were not detected in bronchus or lungs in normal conditions whereas they were significantly upregulated in the lungs of COVID-19 affected samples. The expression patterns of the upregulated genes PGM5, ISLR and ANK2 were consistent (Table 1; Figure 6) and were observed due to the infection of COVID-19 which signified their role as prognostic biomarkers of COVID-19.
Table 1.
Expression levels of the hub genes in TPM obtained from human protein atlas database.
| Gene | RNA expression in TPM | Protein expression in TPM |
Availability in blood | Expression in TPM | |
|---|---|---|---|---|---|
| Lung | Bronchus | Lung | |||
| MDH1 | 29.8 | Low | Low | T cells | 75.8 |
| SGCE | 44.7 | Low | Low | B cells | 24.5 |
| PFKBF3 | 96.4 | Medium | High | Neutrophils | 124.2 |
| SLC16A3 | 69.4 | High | High | Neutrophils | 338.4 |
| NUDT21 | 55.6 | Medium | Medium | T cells | 124.1 |
| CFD | 39.8 | Not detected | Low | Basophils | 1440.3 |
| CLU | 30.5 | Not detected | Not detected | Peripheral blood mononuclear cells | 567.2 |
| PGM5 | 20.1 | Not detected | Not detected | Neutrophils | 7.2 |
| ISLR | 15.2 | Not detected | Low | – | – |
| ANK2 | 9.8 | Not detected | Low | – | – |
Figure 6.
The comparison of the expression levels of hub genes among the tissues identified from the expression atlas platform.
3.5. Enrichment of microRNAs and long non-coding RNAs
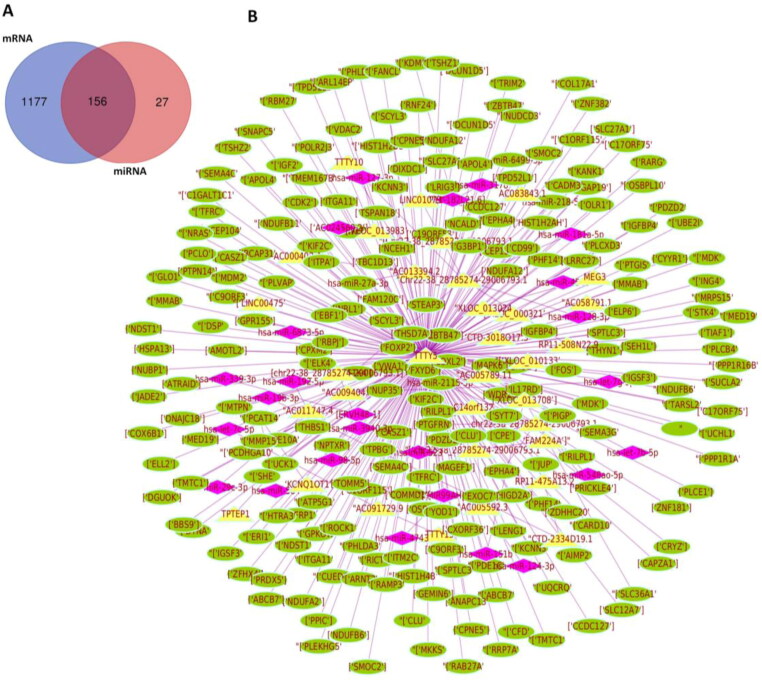
MicroRNAs are small endogenous RNAs that can post transcriptionally interact with the mRNAs to suppress them (Wu et al., 2019). Long non coding RNAs act as competing endogenous RNA (ceRNA), to inhibit the miRNA expression and regulate the gene expression (Wang, 2018). For the establishment of the miRNA-mRNA-lncRNA network, miRNAs were predicted from the DEGs and filtered based on the p value <0.05. There were a total of 1333 mRNAs and 183 miRNAs identified from Mirnet with 156 mutual targets from the DEGs (Figure 7(A)). Top 15 miRNAs, their target genes and lncRNAs are shown in Supplementary Table 2.
Figure 7.
Network construction of mRNA-miRNA-lncRNA. (A) Venn diagram representing the targets of mRNA and miRNA. (B) Network of competing endogenous RNA. mRNA are shown in green, miRNA in pink and lncRNA in yellow.
Three miRNAs – hsa-miR-124-3p, hsa-miR-192-5p and hsa-let-7b-5p were significant with higher degrees of 270, 236 and 187 and a maximum of 69, 55 and 48 target genes, respectively. A total of 129 co-expressed lncRNAs exempting the pseudogenes and unspliced introns with scores between 0.99 and 1.00 were predicted from DIANA lncbase. Three lncRNAs AC058791.1, TTTY15, and TPTEP1, had maximum degrees of target genes. A network of differentially expressed interacting miRNA–mRNA–lncRNA was constructed using cytoscape and is represented in the Figure 7(B).
3.6. Drug perturbation results of the transcriptome data
We further evaluated the drug perturbation signatures of the significant DEGs. Gene signatures are a set of genes that render a common expression pattern in independent conditions (Cantini et al., 2018). Firework display module of LINCS server was explored to predict the chemical perturbagens. 50 gene signatures with drug perturbagens that can reverse the observed gene expression patterns were envisaged. The perturbagens based on p-value were sorted and filtered out. Drugs acting on A549 lung adenocarcinoma cell lines with the capability to reverse the gene expressions patterns were identified for COVID-19 infected lungs. The genes GADD45B, SAT1, NUPL1 were common to all the four gene signature patterns. Growth arrest and DNA damage inducible beta (GADD45B) responsible for DNA damage response was reported in lung and hepatocellular cancer (Hou et al., 2019). Spermidine N1 acetyltransferase 1 (SAT1) involved in the metabolism of polyamines and transport was reported to be a potential target in Schizophrenia, anxiety and keratosis follicularis spinulosa decalvans, a rare genetic disorder (Bermudo-Soriano et al., 2009). Nucleoporin like protein 1 (NUPL1) was reported as an effective target of HIV, Influenza and COVID-19 (Gordon et al., 2020). Supplementary Table 2 represents the mutual gene lists of the four gene signatures. Table 2 represents the drug perturbations of the DEGs. Four drugs F-1566-0341, Digoxin, Proscillaridin and Linifanib were capable of reversing the set of gene expressions. F-1566-0341 was a reported STAT inhibitor with antiviral, antihistamine properties and prescribed to treat neurological disorders (Redell & Tweardy, 2005). Digoxin, an ATPase inhibitor (Ravi Kumar & Kurup, 2000) commonly used to treat heart failure, effectively reversed the gene signatures of COVID-19. Proscillaridin was reported as modulators of hypoxia via apoptosis (Costa et al., 2019) and linifanib for gastric cancer, and neurological disorder (Chen et al., 2016). They had remarkable activity in reversing the gene expression patterns of COVID-19. Conversely, Proscillaridin and linifanib were also reported as ocular and anti-glaucoma agents to treat ocular cancer. These four drugs as shown in Figure 8, had considerable perturbation effects on the gene signatures observed from the COVID-19 affected lung samples and obviously act as repurposed drugs to against COVID-19.
Table 2.
Drug perturbation analysis of the overlapped DEGs.
| Gene signature | Drug name | Reported for | Mechanism of action | Phase of clinical trial | p value |
|---|---|---|---|---|---|
| CPC013_A549_6H:BRD-K69852452 (452 genes) | F-1566-0341 | Antiviral Antihistamine Drugs to treat neurological disorders | STAT inhibitor | Unknown | 1.03 × 10−13 |
| CPC017_A549_24H:BRD-K23478508 (456 genes) | Digoxin | Congestive heart failure atrial fibrillation | ATPase inhibitor | Phased | 1.39 × 10−13 |
| CPC015_A549_24H:BRD-A34806832 (446 genes) | Proscillaridin | Ocular cancer modulators of hypoxia inducible factors | Unknown | Unknown | 9.34 × 10−7 |
| CPC014_A549_24H:BRD-K99749624 (445 genes) | Linifanib | Antiarthritis Disorders of nervous system Antiglaucoma agent Ophthalmic agent |
PDGFR tyrosine kinase receptor inhibitor, VEGFR inhibitor | Phase III | 2.27 × 10−6 |
Figure 8.
Drug perturbations on A-549 cell lines that reverse the gene signatures observed in COVID-19. Perturbations of F-1566-0341 shown in pink ellipses. Perturbation of Digoxin in blue; Perturbation of Proscillaridin in green and Perturbation of Linifanib in yellow.
3.7. Molecular docking analysis of the drug perturbagens
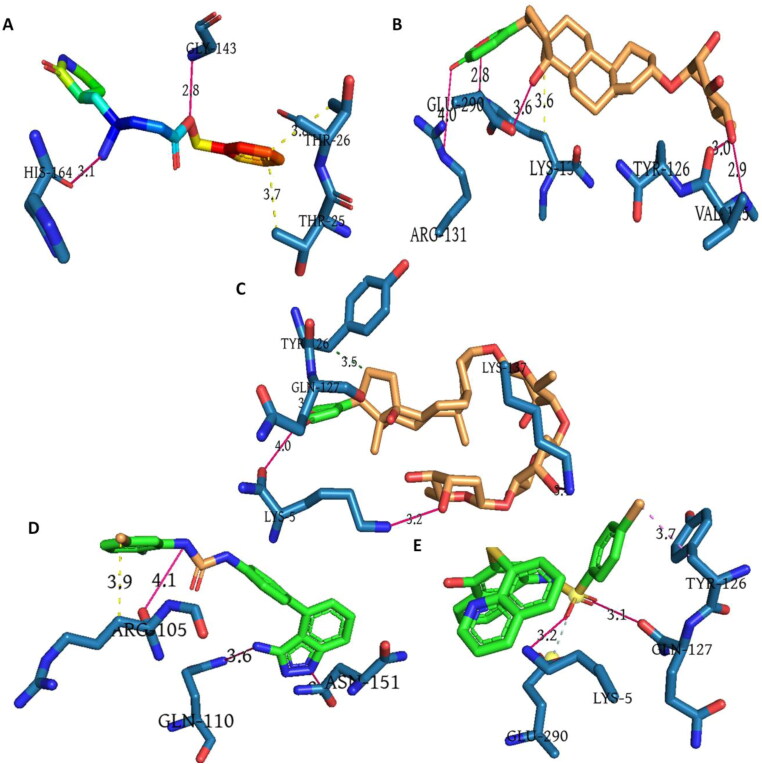
Molecular docking was performed for the aforementioned drug perturbagens to validate their potential. The four drugs were compared with the original ligand N3 complexed with the chosen target Mpro and the docking energy along with interacting residues are shown in Table 3. Ligand (Figure 9(A)) exhibited binding energy value of −82.29 Kcal/mol with two hydrogen bond interactions of 2.80 A0 and 3.07 A0 with Gly 143 and His 164 residues, respectively. The ligand also exhibited hydrophobic interactions with Thr 25 and Thr 26 at distances of 3.73 A0 and 3.86 A0, respectively. The drug Proscillaridin exhibited binding energy of −80.06 Kcal/mol with six interactions to the target. As seen in Figure 9(B), proscillaridin, interacted with N atom of Val 125 at 2.93 A0and O atom of Val 125 at 3.03 A0. It also showed an interaction with N atom and O atom of Lys 137 at a distance of 2.83 A0 each. Interactions of Glu 290 with 3.59 A0 and Arg 131 at 4.04 A0 were also found but are exceeding the good Hydrogen bond interactions. Proscillaridin was also found to possess hydrophobic interactions with Lys 137 at 3.63 A0. Digoxin followed next with an energy of −79.16 Kcal/mol and hydrophobic interaction of 3.49 A0 with Tyr 126. As seen in Figure 9(C), digoxin also possessed interactions with N and O atoms of Lys 5 at a distance of 3.16 A0 each . Interaction of digoxin with residues Glu 127 and Lys 137 were also found at distances of 3.27 A0 and 3.39 A0 respectively. As seen in Figure 9(D), linifanib showed a slightly higher energy of −74.89 Kcal/mol with interacting residues Arg 105, Gln 110 and Asn 151 at distances of 4.08 A0, 3.63 A0 and 3.14 A0 respectively. The drug F-1566-0341 (Figure 9(E)) exhibited an energy of −76.89 Kcal/mol and interactions of 3.16 A0 and 3.70 A0 with 5 Lys and 3.15 A0 with 127 Gln. From the results of docking, it was inferred that the drugs proscillaridin and digoxin exhibited a comparable lower binding energy value with the control ligand N3 and had better interactions with the target. Though Linifanib and F-1566-0341 had slightly higher binding energy values, the interactions were better when compared to the control ligand N3. These drugs possessed better interactions with the ideal target Mpro thereby can be accounted for repurposing.
Table 3.
Molecular docking results of drugs with the Mpro enzyme of COVID-19.
| S. No. | Ligand | Glide score | Glide energy (Kcal/mol) | H bond residues | H bond interaction (A0) | Hydrophobic residues | Hydrophobic interaction (A0) |
|---|---|---|---|---|---|---|---|
| 1 | Ligand N3 | –7.784 | –82.29 | Gly 143(NH…O) | 2.80 | Thr 25 | 3.73 |
| (OH…O) His 164 | 3.07 | Thr 26 | 3.86 | ||||
| 2 | Digoxin | –6.34 | –79.16 | Lys 5 (NH…O) | 3.16 | Tyr 126 | 3.49 |
| (OH…N) Lys 5 | 3.16 | ||||||
| Gln 127 (NH…O) | 3.27 | ||||||
| Lys 137 (NH…O) | 3.39 | ||||||
| 3 | Proscillaridin | –7.12 | –80.06 | 125 Val (NH…O) | 2.93 | Lys 137 | 3.63 |
| 125 Val (OH…O) | 3.03 | ||||||
| 131 Arg (OH…N) | 4.04 | ||||||
| Lys 137 (NH…O) | 2.83 | ||||||
| Lys 137 (OH…O) | 2.83 | ||||||
| Glu 290 (OH…O) | 3.59 | ||||||
| 4 | Linifanib | –6.78 | –74.89 | (OH…O) Arg 105 | 4.08 | Arg 105 | 3.85 |
| 110 Gln (NH…O) | 3.63 | ||||||
| 151 Asn (NH…O) | 3.14 | ||||||
| 5 | F-1566-0341 | –7.79 | –76.89 | 5Lys (NH…O) | 3.16 | 126 Tyr | 3.65 |
| 5 Lys (OH…N) | 3.70 | ||||||
| (OH…O) 127 Gln | 3.15 |
Figure 9.
Interactions of drug perturbagens with the COVID-19 Mpro. (A) Original inhibitor N3 in rainbow color, (B) Proscillaridin in brown, (C) Digoxin in brown, (D) Linifanib in green and (E) F-1566-0341 in green. Interacting residues of target is shown in blue.
4. Discussion
COVID-19 is a rapidly disseminated pandemic that has spread to 216 countries globally within a very short span since its origin in December 2019. Treatment has become more challenging due to rapid transmission and severe progression of the disease (Kumar et al., 2020b). Even with the fast paced continuous scientific research, on development of vaccines, repurposed drugs and diagnosis techniques, there is a lack of significant treatment options. The current study was conducted with a specific focus on human responses to enumerate multifactorial approaches that are involved in pathogenesis and control of COVID-19 (Kumar et al., 2020c). With an aim to identify candidate gene biomarkers, two transcriptome data sets GSE155241 and GSE150316 were analysed and compared. Subsequently integrated Bioinformatics tools were used to predict the molecular mechanisms, ceRNA networks and chemical perturbagens to aid in the better understanding of COVID-19 pathogenesis. Analysis of GSE155241 attained 51 and 57 up- and downregulated genes whereas, the dataset of GSE150316 revealed 620 and 524 up- and downregulated genes, respectively. Significant upregulated overlapping genes of the two datasets were identified as AMZ2 (Archaelysin Family Metallopeptidase 2), MDH1 (Malate Dehydrogenase 1), TMEM261 (Transmembrane protein 261), PHF14 (PHD finger protein 14), PTGFRN (Prostaglandin F2 receptor inhibitor), and SGCE (Sarcoglycan epsilon). Eight overlapping genes -UGCG (UDP-Glucose Ceramide Glucosyltransferase), DUSP6 (Dual Specificity Phosphatase 6), TIPARP (TCDD Inducible Poly(ADP-Ribose) Polymerase), NUPL1 (Nucleoporin 58), NOLC1 (Nucleolar And Coiled-Body Phosphoprotein 1), PFKFB3 (6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase 3), ERRFI1 (ERBB Receptor Feedback Inhibitor 1), SLC19A2 (Solute Carrier Family 19 Member 2) were significantly downregulated.
The contributors of the dataset GSE150316, reported temporal spatial heterogeneity analysis of host response to SARS-CoV-2 pulmonary infection. As per their results, there was a surge in immune responses especially interferon pathways, JAK STAT pathway, collagen synthesis and antiviral genes. The study also confirmed the heterogeneity of SARS-CoV-2 infection associated with viral load and interferon pathways in multiple organs (Desai et al., 2020). The second dataset GSE155241 was established from RNA sequences from lung biopsy tissues and organoid models acquired from human pluripotent stem cells. The study reported that Proximal airway cells are critical for viral infectivity and distal alveolar cells for emulating host response along with twenty predominant genes involved in severity of the disease (Han et al., 2020). Though both the studies suggest ample clinical evidence of SARS-CoV-2 infection, current comparative transcriptome analysis that deduce differential gene expression would enumerate and augment both diagnostic and therapeutic efficacy. This study strongly supports the clinical findings of the contributors Desai et al. and Han et al. and extends additional evidence of molecular mechanisms such as increase in mitochondrial respiration, endopeptidase activity, ubiquinone activity, complement activation, neutrophil degranulation and tau protein binding. There was a predominant impairment related to the transport mechanism of copper, iron, zinc, cobalt, cadmium, and manganese. It is evident that there is a serious downregulation of GTPase activators, protein tyrosine phosphatase, response to cytokines, interleukins, MAP kinase activity along with transporters like azole and carboxylic acid trans membrane transporters. Cellular components like dystrophin associated glycoprotein complex, dendrites, platelet alpha granular lumen were relatively found to be more when compared with lysosomes, golgi and nucleolus. Immense amount of functional non coding RNAs (LINC00493) and small RNAs (SNORD variants) were expressed across the tissue samples whose functions are yet to be understood. The overexpression of LINC00493 is a reported significant biomarker in ailments like muscular dystrophy and urothelial cancer. It also has been categorised to originate from the family of small integral membrane protein 26 transcript (Cogill & Wang, 2014). Pathway annotations from KEGG, and REACTOME suggested that most of the genes involved in mitochondrial respiratory chain complex biogenesis, assembly, degradation, respiratory electron transport systems and Insulin like growth factors were predominantly upregulated. Phospholipid metabolism, immune reactions to pathogenesis, like response to cytokine stimulation, interleukins, neutrophils, and metal ion (copper and iron) transport components were all drastically downregulated. Captivating enrichment of neurodegenerative pathways like Alzheimer’s, Parkinson’s, Huntington’s and alcoholic fatty acid diseases were noted as an assert in the study. Pathways responsible for signal transduction, iron and copper metabolisms, transport of sugars, bile and organic acids were principally downregulated. The gene annotations were evident that the pathology of COVID-19 is due to high respiration rate along with the lack of glucose, ions and organic acid transportation (MacKenzie et al., 2008). COVID-19 is a multi-ailment disability that threatens the foremost physiological systems like the CNS where the chances of severe neurodegeneration is elevated over the course of infection. In consistency with the above mentioned enrichment evaluations, three distinct signalling pathways such as over-expression of miRNA biogenesis, hijack of ubiquitination by SARS-CoV-2, and angiogenesis-vasculature development were annotated. Perceived from interrelation studies, HIV arrest and its associated discovery pathways are considered to be momentous realization.
Subsequently, we identified predominant upregulated hub genes PGM5, MDH1, SGCE, ANK2, DTNA, CFD, CLU, and ISLR and downregulated PFKFB3, SLC16A3, NUDT21, from the protein-protein interaction analysis. Further expression analysis of GTEx data confirmed the higher expression levels of PFKFB3 in normal healthy lungs which was contrary to the lower expression rate in COVID-19 lungs. Lower expression of Phosphofructo kinase bisphosphatase 3 (PFKFB3) function was attributed to autophagy and death of lung tissues (Sears & Jacko, 2009). Similarly, normal healthy lungs and bronchus had very low levels of MDH1 and SGCE, whereas they were significantly upregulated overlapping genes of COVID-19 lungs. Hence these three genes are considered significant biomarkers of COVID-19. Higher levels of Malate Dehydrogenase 1 (MDH1) was associated with schizophrenia (Ritsner, 2011), prions disease (Zerr et al., 2019) and early onset of encephalopathy (Broeks et al., 2019). Elevation of SGCE Sarcoglycan-epsilon gene was attributed to muscular dystrophy (Emery, 2001). As per the GteX data, higher expression levels of PGM5, ISLR and ANK2 were generally not detected in healthy lungs, in contrast to their higher expression levels in COVID-19 affected lungs. In normal conditions, phosphoglucomutase 5 (PGM5) responsible for cell differentiation was observed high in muscles, and heart but not in lungs (Sun et al., 2019). Immunoglobulin super family leucine rich repeats (ISLR) were generally present in brain and related tissues but not in lungs. They were elevated in response to calcium levels (Nagasawa et al., 1999) . Ankyrin 2 (ANK2), responsible for stabilization of ion transporters and attributed to cardiac arrhythmia was prevalent only in normal brain and heart tissues but not detected in lungs (Mohler et al., 2007). These three genes had strikingly significant attributes of being prognostic biomarkers of COVID-19 due to their unnatural expressions in COVID-19 infected lungs.
In addition to the application of bioinformatics approaches the competing endogenous RNA (ceRNA) network of COVID-19 pathogenesis was evaluated. Three potential miRNAs-hsa-miR-124-3p, hsa-miR-192-5p and hsa-let-7b-5p, showed significant interactions with 56 strong mRNA candidates. The miRNAs, hsa-miR-124-3p, and hsa-miR-192-5p were reported in the negative regulation of Alzheimer’s disease, interleukin 21 and cell matrix adhesion, respectively (Enomoto et al., 2017). Hsa-let-7b-5p was supposed to negatively regulate autophagy and cysteine type endopeptidase activity and increase angiogenesis (Ham et al., 2015). A similar scenario is instigated in this study where miRNA is considered as a motive to intensify the severity of the pathogenicity. Conversely, the work elucidated an interesting fact that three lncRNAs that interacted in higher degrees with the mRNA-miRNA network to release the mRNA for translation. The top three lncRNAs with maximum target genes were identified as AC058791.1 with unknown activity, TTTY15 reported to regulate proteolysis, ubiquitin dependent catabolism (Lai et al., 2019) and TPTEP1 inhibitor of STAT3 (Ding et al., 2019). Both miRNA and lncRNA functions were consistent with the DEG enrichment and necessitated the regulations of COVID-19 pathology. The studies were extended to explore the action of chemical perturbagens on the observed gene patterns of the COVID-19 datasets. A reversal mechanism of the gene expression patterns was elicited by four chemical perturbagens, F-1566-0341, Digoxin, Proscillaridin and Linifanib. Originally, F-1566-0341 were reported as antiviral drugs and digoxin to treat heart failure. Proscillaridin and linifanib were reported as hypoxia modulators and inhibitors of neurological disorders, respectively. The distinguished gene expression reversal mechanism exhibited by the four drugs establish a strong reposition effect on the COVID-19 gene expressions.
In conclusion it is reported that there can be severe hypoxia due to failure of iron transport, and imbalance of copper homeostasis (Prohaska, 2008) which leads to severe complications in respiration, liver, blood clotting, and neuroendocrine peptides (Cagliani et al., 2020). Obviously the pathways identified were involved with the neurodegenerative disorders and increased oxidative phosphorylation. With increased respiratory transport chains and low oxygen uptake, the damage caused to the cells are severe and irreparable which could be the evident reason of death of COVID affected patients. Increase in kinases like TIE1, KDRL and TEK are either directly or indirectly linked to production of angiopoietin 1 thereby rise in angiogenesis is evident (Ahmad et al., 2001). Hijacking of ubiquitination by the SARS-CoV-2 was a striking upregulated phenomenon commonly adopted by viruses to disseminate pathogenesis and digoxin has been reported to prevent hijack by HIV and influenza (Amarelle et al., 2017). Linifanib, a receptor tyrosine kinase inhibitor, though terminated after phase II trials, had exhibited effectiveness in inhibiting neurological disorders like Parkinson’s and against chronic hepatitis viral infection (Monroig-Bosque et al., 2018). Digoxin and proscillaridin, are approved FDA drugs and are protective in heart ailments with the capacity of inhibiting hypoxia, were also proved against the HIV and HBV pathogenesis (Wong et al., 2013). The drug perturbations were validated with a molecular docking approach against the crystal structure of the main protease enzyme of the target. Proscillaridin and digoxin exhibited lower binding energy and better interactions with the active site of the target. Though linifanib and F-1566-0341, showed significant interactions compared to the original control ligand N3, their energy levels were high. Hence from both the drug perturbations on the gene expressions and molecular docking studies, it is concluded that digoxin and proscillaridin can effectively be repurposed to treat advanced COVID-19 infections. Further clinical research is warranted to establish the prognostic biomarkers identified in the current study. The study is significant and sheds light on the pivotal molecular mechanisms and host responses to the pandemic COVID-19.
Conclusion
The novel coronavirus has caused devastating effects on human health across the world. Though several clinical trials are in progress with a focus on finding drugs and vaccines to treat SARS-CoV-2, very little focus has been perceived in unravelling the molecular mechanisms and host responses. In order to fetch appropriate solutions, it is significant to develop intervening strategies based on the host responses and viral pathology. To overcome the shortcomings, an integrated computational analysis of the transcriptome data of COVID-19 infected lung tissues was performed. Interrelational approaches were employed to annotate differential gene expressions, small RNA enrichment, overexpressed pathways and kinases involved in the host responses of COVID-19 pathogenesis. The study identified multi-faceted complications integrated with COVID-19 that evoked devastating distress. Thereby this work has laid a pedal stone to identify potential candidate gene biomarkers, competing endogenous RNA network and strategise the mechanism of drug repurposing to combat COVID-19. Hence it is substantial that this offers comprehensive solutions to tackle the pandemic by revolutionising innovative diagnostic and therapeutic interventions.
Supplementary Material
Acknowledgements
The authors are obligated to the management of Stella Maris College Chennai, Tamil Nadu, India. The facility provided by seed research grant is immensely accredited. The Schrodinger software used in the study was procured from the grant of Department of Science and Technology (DST), New Delhi, India, through FIST Programme (Level -0, No: SR/FIST/College-252-C.Dy.No.3555/IFD/2016-2017), to Stella Maris College, Chennai, India. DST-FIST grant is greatly acknowledged. The support extended by the CAS in Crystallography and Biophysics, University of Madras, Chennai, India and Bishop Heber College, Tiruchirappalli is extensively gratified.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ahmad, S. A., Liu, W., Jung, Y. D., Fan, F., Wilson, M., Reinmuth, N., Shaheen, R. M., Bucana, C. D., & Ellis, L. M. (2001). The effects of angiopoietin-1 and -2 on tumor growth and angiogenesis in human colon cancer. Cancer Research, 61(4), 1255–1259.s [PubMed] [Google Scholar]
- Amarelle, L., Lecuona, E., & Sznajder, J. I. (2017). Anti-influenza treatment: drugs currently used and under development. Archivos de Bronconeumología (English Edition), 53(1), 19–26. 10.1016/j.arbr.2016.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader, G. D., & Hogue, C. W. V. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 4(2), 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, N., Williams, A. J., Tropsha, A., & Ekins, S. (2020). Repurposing quaternary ammonium compounds as potential treatments for COVID-19. Pharmaceutical Research, 37(6), 104. 10.1007/s11095-020-02842-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedre, R. (2020). Reneshbedre/Bioinfokit: Bioinformatics Data Analysis and Visualization Toolkit, 10.5281/zenodo.3965241 [DOI] [Google Scholar]
- Bermudo-Soriano, C. R., Vaquero-Lorenzo, C., Diaz-Hernandez, M., Perez-Rodriguez, M. M., Fernandez-Piqueras, J., Saiz-Ruiz, J., & Baca-Garcia, E. (2009). SAT-1 -1415T/C polymorphism and susceptibility to schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry, 33(2), 345–348. 10.1016/j.pnpbp.2008.12.015 [DOI] [PubMed] [Google Scholar]
- Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., Fridman, W.-H., Pagès, F., Trajanoski, Z., & Galon, J. (2009). ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics (Oxford, England)), 25(8), 1091–1093. 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks, M. H., Shamseldin, H. E., Alhashem, A., Hashem, M., Abdulwahab, F., Alshedi, T., Alobaid, I., Zwartkruis, F., Westland, D., Fuchs, S., Verhoeven-Duif, N. M., Jans, J. J. M., & Alkuraya, F. S. (2019). MDH1 deficiency is a metabolic disorder of the malate-aspartate shuttle associated with early onset severe encephalopathy. Human Genetics, 138(11-12), 1247–1257. 10.1007/s00439-019-02063-z [DOI] [PubMed] [Google Scholar]
- Cagliani, R., Forni, D., Clerici, M., & Sironi, M. (2020). Computational inference of selection underlying the evolution of the novel coronavirus, severe acute respiratory syndrome coronavirus 2. Journal of Virology, 94(12). 10.1128/JVI.00411-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantini, L., Calzone, L., Martignetti, L., Rydenfelt, M., Blüthgen, N., Barillot, E., & Zinovyev, A. (2018). Classification of gene signatures for their information value and functional redundancy. NPJ Systems Biology and Applications, 4(2), 2. 10.1038/s41540-017-0038-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, L., Zhou, G., Soufan, O., & Xia, J. (2020). miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res, 48(W1), W244–W251. 10.1093/nar/gkaa467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Guo, J., Chen, Z., Wang, J., Liu, M., & Pang, X. (2016). Linifanib (ABT-869) potentiates the efficacy of chemotherapeutic agents through the suppression of receptor tyrosine kinase-mediated AKT/mTOR signaling pathways in gastric cancer. Scientific Reports, 6, 29382. 10.1038/srep29382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Guo, Y., Pan, Y., & Zhao, Z. J. (2020). Structure analysis of the receptor binding of 2019-nCoV. Biochemical and Biophysical Research Communications, 525(1), 135–140. 10.1016/j.bbrc.2020.02.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogill, S. B., & Wang, L. (2014). Co-expression network analysis of human lncRNAs and cancer genes. Cancer Informatics, 13s5, CIN.S14070. CIN.S14070). 10.4137/CIN.S14070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, E. M. D., Da Costa, E. M., Armaos, G., McInnes, G., Beaudry, A., Moquin-Beaudry, G., Bertrand-Lehouillier, V., Caron, M., Richer, C., St-Onge, P., Johnson, J. R., Krogan, N., Sai, Y., Downey, M., Rafei, M., Boileau, M., Eppert, K., Flores-Díaz, E., Haman, A., … -M. Raynal, N. J. (2019). Heart failure drug proscillaridin A targets MYC overexpressing leukemia through global loss of lysine acetylation. Journal of Experimental & Clinical Cancer Research 38 (1). 10.1186/s13046-019-1242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, C. A., Hitz, B. C., Sloan, C. A., Chan, E. T., Davidson, J. M., Gabdank, I., Hilton, J. A., Jain, K., Baymuradov, U. K., Narayanan, A. K., Onate, K. C., Graham, K., Miyasato, S. R., Dreszer, T. R., Strattan, J. S., Jolanki, O., Tanaka, F. Y., & Cherry, J. M. (2018). The Encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res, 46(D1), D794–D801. 10.1093/nar/gkx1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, N., Neyaz, A., Szabolcs, A., Shih, A. R., Chen, J. H., Thapar, V., Nieman, L. T., Solovyov, A., Mehta, A., Lieb, D. J., Kulkarni, A. S., Jaicks, C., Pinto, C. J., Juric, D., Chebib, I., Colvin, R. B., Kim, A. Y., Monroe, R., Warren, S. E., … Deshpande, V. (2020). Temporal and spatial heterogeneity of host response to SARS-CoV-2 Pulmonary Infection. medRxiv: The Preprint Server for Health Sciences. 10.1101/2020.07.30.20165241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, H., Liu, J., Zou, R., Cheng, P., & Su, Y. (2019). Long non-coding RNA TPTEP1 inhibits hepatocellular carcinoma progression by suppressing STAT3 phosphorylation. Journal of Experimental & Clinical Cancer Research: CR, 38(1), 189. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dong, L., Li, H., Zhang, S., & Su, L. (2018). Identification of genes related to consecutive trauma-induced sepsis via gene expression profiling analysis. Medicine, 97(15), e0362. 10.1097/md.0000000000010362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubitzky, W., Wolkenhauer, O., Yokota, H., & Cho, K.-H. (2013). Encyclopedia of systems biology. Springer. [Google Scholar]
- Emery, A. E. H. (2001). The muscular dystrophies. Oxford Medical Publications. [Google Scholar]
- Enomoto, Y., Takagi, R., Naito, Y., Kiniwa, T., Tanaka, Y., Hamada-Tsutsumi, S., Kawano, M., Matsushita, S., Ochiya, T., & Miyajima, A. (2017). Identification of the novel 3' UTR sequences of human IL-21 mRNA as potential targets of miRNAs. Scientific Reports, 7(1), 7780. 10.1038/s41598-017-07853-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat, A., Sidiropoulos, K., Viteri, G., Marin-Garcia, P., Ping, P., Stein, L., D'Eustachio, P., & Hermjakob, H. (2018). Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics (Oxford, England)), 34(7), 1208–1214. 10.1093/bioinformatics/btx752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., O'Meara, M. J., Rezelj, V. V., Guo, J. Z., Swaney, D. L., Tummino, T. A., Hüttenhain, R., Kaake, R. M., Richards, A. L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., … Krogan, N. J. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816), 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. (2013). The Genotype-Tissue Expression (GTEx) project. Nature Genetics, 45(6), 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham, O., Lee, S.-Y., Lee, C. Y., Park, J.-H., Lee, J., Seo, H.-H., Cha, M.-J., Choi, E., Kim, S., & Hwang, K.-C. (2015). Let-7b suppresses apoptosis and autophagy of human mesenchymal stem cells transplanted into ischemia/reperfusion injured heart 7by targeting caspase-3. Stem Cell Research & Therapy, 6, 147. 10.1186/s13287-015-0134-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y., Yang, L., Duan, X., Duan, F., Nilsson-Payant, B. E., Yaron, T. M., Wang, P., Tang, X., Zhang, T., Zhao, Z., Bram, Y., Redmond, D., Houghton, S., Nguyen, D., Xu, D., Wang, X., Uhl, S., Huang, Y., Johnson, J. L., … Chen, S. (2020). Identification of candidate COVID-19 therapeutics using hpsc-derived lung organoids. The Preprint Server for Biology. 10.1101/2020.05.05.079095 [DOI] [Google Scholar]
- Hou, T., Ma, J., Hu, C., Zou, F., Jiang, S., Wang, Y., Han, C., & Zhang, Y. (2019). Decitabine reverses gefitinib resistance in PC9 lung adenocarcinoma cells by demethylation of RASSF1A and GADD45β promoter. International Journal of Clinical and Experimental Pathology, 12(11), 4002–4010. [PMC free article] [PubMed] [Google Scholar]
- Hsu, P. W.-C., Lin, L.-Z., Hsu, S.-D., Hsu, J. B.-K., & Huang, H.-D. (2007). ViTa: Prediction of host microRNAs targets on viruses. Nucleic Acids Res, 35, D381–D385. 10.1093/nar/gkl1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Tong, T.-T., Yau, L.-F., Chen, C.-Y., Mi, J.-N., Wang, J.-R., & Jiang, Z.-H. (2018). LC-MS based sphingolipidomic study on A549 human lung adenocarcinoma cell line and its taxol-resistant strain. BMC Cancer, 18(1). 10.1186/s12885-018-4714-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., & Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research, 44(D1), D457–D462. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., Koplev, S., Jenkins, S. L., Jagodnik, K. M., Lachmann, A., McDermott, M. G., Monteiro, C. D., Gundersen, G. W., & Ma'ayan, A. (2016). Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research, 44(W1), W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D., Kumari, K., Jayaraj, A., Kumar, V., Kumar, R. V., Dass, S. K., Chandra, R., & Singh, P. (2020. a). Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1752310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D., Kumari, K., Vishvakarma, V. K., Jayaraj, A., Kumar, D., Ramappa, V. K., Patel, R., Kumar, V., Dass, S. K., Chandra, R., & Singh, P. (2020. b). Promising inhibitors of main protease of novel corona virus to prevent the spread of COVID-19 using docking and molecular dynamics simulation. Journal of Biomolecular Structure & Dynamics, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V., Dhanjal, J. K., Bhargava, P., Kaul, A., Wang, J., Zhang, H., Kaul, S. C., Wadhwa, R., & Sundar, D. (2020. c). Withanone and Withaferin-A are predicted to interact with transmembrane protease serine 2 (TMPRSS2) and block entry of SARS-CoV-2 into cells. Journal of Biomolecular Structure & Dynamics, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster, G. M., Pfister, O., Burkard, T., Zhou, Q., Twerenbold, R., Haaf, P., Widmer, A. F., & Osswald, S. (2020). SARS-CoV2: Should inhibitors of the renin-angiotensin system be withdrawn in patients with COVID-19? European Heart Journal, 41 (19), 1801–1803. 10.1093/eurheartj/ehaa235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann, A., & Ma'ayan, A. (2009). KEA: Kinase enrichment analysis. Bioinformatics (Oxford, England)), 25(5), 684–686. 10.1093/bioinformatics/btp026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, I.-L., Chang, Y.-S., Chan, W.-L., Lee, Y.-T., Yen, J.-C., Yang, C.-A., Hung, S.-Y., & Chang, J.-G. (2019). Male-specific long noncoding RNA TTTY15 inhibits non-small cell lung cancer proliferation and metastasis via TBX4. International Journal of Molecular Sciences, 20(14), 3473. 10.3390/ijms20143473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, F., Zhang, H., & Xie, X. (2019). Comprehensive analysis of an lncRNA-miRNA-mRNA competing endogenous RNA network in pulpitis. PeerJ, 7, e7135. 10.7717/peerj.7135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko, M., Marzi, A., & Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology, 5(4), 562–569. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.-C., Pan, C.-Y., & Lin, W.-C. (2006). Bioinformatic discovery of microRNA precursors from human ESTs and introns. BMC Genomics, 7, 164. 10.1186/1471-2164-7-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Cao, R., Xu, M., Wang, X., Zhang, H., Hu, H., Li, Y., Hu, Z., Zhong, W., & Wang, M. (2020). Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discovery, 6, 16. 10.1038/s41421-020-0156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., Choe, H., & Farzan, M. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie, E. L., Iwasaki, K., & Tsuji, Y. (2008). Intracellular iron transport and storage: From molecular mechanisms to health implications. Antioxid Redox Signal, 10(6), 997–1030. 10.1089/ars.2007.1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal, L., Kumari, A., Srivastava, M., Singh, M., & Asthana, S. (2020). Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. Journal of Biomolecular Structure & Dynamics, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler, P. J., Healy, J. A., Xue, H., Puca, A. A., Kline, C. F., Allingham, R. R., Kranias, E. G., Rockman, H. A., & Bennett, V. (2007). Ankyrin-B syndrome: Enhanced cardiac function balanced by risk of cardiac death and premature senescence. PloS One, 2(10), e1051. 10.1371/journal.pone.0001051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroig-Bosque, P. d C., Shah, M. Y., Fu, X., Fuentes-Mattei, E., Ling, H., Ivan, C., Nouraee, N., Huang, B., Chen, L., Pileczki, V., Redis, R. S., Jung, E.-J., Zhang, X., Lehrer, M., Nagvekar, R., Mafra, A. C. P., Monroig-Bosque, M. d M., Irimie, A., Rivera, C., … Calin, G. A. (2018). OncomiR-10b hijacks the small molecule inhibitor linifanib in human cancers. Scientific Reports, 8(1). 10.1038/s41598-018-30989-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa, A., Kudoh, J., Noda, S., Mashima, Y., Wright, A., Oguchi, Y., & Shimizu, N. (1999). Human and mouse ISLR (immunoglobulin superfamily containing leucine-rich repeat) genes: Genomic structure and tissue expression. Genomics, 61(1), 37–43. 10.1006/geno.1999.5934 [DOI] [PubMed] [Google Scholar]
- Ogidigo, J. O., Iwuchukwu, E. A., Ibeji, C. U., Okpalefe, O., & Soliman, M. E. S. (2020). Natural phyto, compounds as possible noncovalent inhibitors against SARS-CoV2 protease: Computational approach. Journal of Biomolecular Structure and Dynamics, 1–18. 10.1080/07391102.2020.1837681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatheodorou, I., Moreno, P., Manning, J., Fuentes, A. M.-P., George, N., Fexova, S., Fonseca, N. A., Füllgrabe, A., Green, M., Huang, N., Huerta, L., Iqbal, H., Jianu, M., Mohammed, S., Zhao, L., Jarnuczak, A. F., Jupp, S., Marioni, J., Meyer, K., … Brazma, A. (2019). Expression Atlas update: From tissues to single cells. Nucleic Acids Research. 10.1093/nar/gkz947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, S., Kaspi, A., Domovitz, T., Davidovich, A., Lavi-Itzkovitz, A., Meirson, T., Holmes, J. A., Dai, C.-Y., Huang, C.-F., Chung, R. T., Nimer, A., El-Osta, A., Yaari, G., Stemmer, S. M., Yu, M.-L., Haviv, I., & Gal-Tanamy, M. (2019). Hepatitis C virus leaves an epigenetic signature post cure of infection by direct-acting antivirals. In PLoS Genetics, 15(6), e1008181. 10.1371/journal.pgen.1008181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohaska, J. R. (2008). Role of copper transporters in copper homeostasis. American Journal of Clinical Nutrition, 88(3), 826S–829S. 10.1093/ajcn/88.3.826S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi Kumar, A., & Kurup, P. A. (2000). Digoxin and membrane sodium potassium ATPase inhibition in cardiovascular disease. Indian Heart Journal, 52(3), 315–318. [PubMed] [Google Scholar]
- Redell, M., & Tweardy, D. (2005). Targeting transcription factors for cancer therapy. Current Pharmaceutical Design, 11(22), 2873–2887. 10.2174/1381612054546699 [DOI] [PubMed] [Google Scholar]
- Ritsner, M. S. (2011). Handbook of schizophrenia spectrum disorders, volume ii: phenotypic and endophenotypic presentations. Springer Science & Business Media. [Google Scholar]
- Rivals, I., Personnaz, L., Taing, L., & Potier, M.-C. (2007). Enrichment or depletion of a GO category within a class of genes: Which test? Bioinformatics (Oxford, England)), 23(4), 401–407. 10.1093/bioinformatics/btl633 [DOI] [PubMed] [Google Scholar]
- Saddala, M. S., Lennikov, A., Bouras, A., & Huang, H. (2020). RNA-Seq reveals differential expression profiles and functional annotation of genes involved in retinal degeneration in Pde6c mutant Danio rerio. BMC Genomics, 21(1), 132. 10.1186/s12864-020-6550-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddala, M. S., Lennikov, A., & Huang, H. (2020). RNA-seq data from C-X-C chemokine receptor type 5 (CXCR5) gene knockout aged mice with retinal degeneration phenotype. Data in Brief, 31, 105915. 10.1016/j.dib.2020.105915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, A., & Jacko, J. A. (2009). Human-computer interaction: Development process. CRC Press. [Google Scholar]
- Shivanika, C., Kumar, S. D., Ragunathan, V., Tiwari, P., Sumitha, A., & Brindha Devi , P. (2020). Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. Journal of Biomolecular Structure and Dynamics, 1–27. 10.1080/07391102.2020.1815584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, K. E., Zhou, W., Ji, Z., Chen, Z., He, S., Ji, H., Guan, Y., & Taverna, S. D. (2019). Sex differences in gene regulation in the dorsal root ganglion after nerve injury. BMC Genomics, 20(1). 10.1186/s12864-019-5512-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Long, H., Sun, L., Sun, X., Pang, L., Chen, J., Yi, Q., Liang, T., & Shen, Y. (2019). PGM5 is a promising biomarker and may predict the prognosis of colorectal cancer patients. Cancer Cell International, 19(1), 253. 10.1186/s12935-019-0967-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., Santos, A., Doncheva, N. T., Roth, A., Bork, P., Jensen, L. J., & von Mering, C. (2017). The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res, 45(D1), D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishvakarma, V. K., Bahadur, I., Chandra, R., Kumari, K., & Singh, P. (2020). Thiazolidinones: Potential human novel coronavirus (SARS-CoV-2) protease inhibitors against COVID-19. ChemRxiv. Preprint. 10.26434/chemrxiv.12891656.v1 [DOI] [Google Scholar]
- Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. (2018). The opening of pandora's box: An emerging role of long noncoding RNA in viral infections. Frontiers in Immunology, 9, 3138. 10.3389/fimmu.2018.03138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Lachmann, A., Keenan, A. B., & Ma'ayan, A. (2018). L1000FWD: Fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics (Oxford, England)), 34(12), 2150–2152. 10.1093/bioinformatics/bty060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, J., Hall, B., & Shi, X. (2019). A network view of microRNA and gene interactions in different pathological stages of colon cancer. BMC Medical Genomics, 12(S7). 10.1186/s12920-019-0597-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, R. W., Balachandran, A., Ostrowski, M. A., & Cochrane, A. (2013). Digoxin suppresses HIV-1 replication by altering viral RNA processing. PLoS Pathogens, 9(3), e1003241. 10.1371/journal.ppat.1003241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M., Li, W., Huang, F., Sun, J., Li, K. P., Shi, J., Yang, J., Li, J., Li, Y., Hu, N., & Hu, Y. (2019). Comprehensive analysis of the expression profiles of long non-coding RNAs with associated ceRNA network involved in the colon cancer staging and progression. Scientific Reports, 9(1), 16910. 10.1038/s41598-019-52883-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., & McGoogan, J. M. (2020). Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA, 323(13), 1239–1242. 10.1001/jama.2020.2648 [DOI] [PubMed] [Google Scholar]
- Ye, C., Ho, D. J., Neri, M., Yang, C., Kulkarni, T., Randhawa, R., Henault, M., Mostacci, N., Farmer, P., Renner, S., Ihry, R., Mansur, L., Keller, C. G., McAllister, G., Hild, M., Jenkins, J., & Kaykas, A. (2018). DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nature Communications, 9(1), 4307. 10.1038/s41467-018-06500-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi, H., Maheronnaghsh, M., Molaei, F., Mashouri, L., Reza Aref, A., Momeny, M., & Alahari, S. K. (2020). Long noncoding RNAs and exosomal lncRNAs: Classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene, 39(5), 953–974. 10.1038/s41388-019-1040-y [DOI] [PubMed] [Google Scholar]
- Zerr, I., Villar-Piqué, A., Schmitz, V. E., Poleggi, A., Pocchiari, M., Sánchez-Valle, R., Calero, M., Calero, O., Baldeiras, I., Santana, I., Kovacs, G. G., Llorens, F., & Schmitz, M. (2019). Evaluation of human cerebrospinal fluid malate dehydrogenase 1 as a marker in genetic prion disease patients. Biomolecules, 9(12), 800. 10.3390/biom9120800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, P., Hu, W., Wang, H., Yu, S., Li, C., Bai, J., Gui, S., & Zhang, Y. (2018). Corrigendum to “identification of differentially expressed genes in pituitary adenomas by integrating analysis of microarray data". International Journal of Endocrinology, 2018, 6069189. 10.1155/2018/6069189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Qiu, Z., Gao, S., Chen, Q., Li, S., Tan, W., Liu, X., & Wang, Z. (2016). Integrated analysis of expression profile based on differentially expressed genes in middle cerebral artery occlusion animal models. International Journal of Molecular Sciences, 17(5), 776. 10.3390/ijms17050776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q., Yu, Q., Gong, Y., Liu, Z., Xu, H., Wang, Y., & Shi, Y. (2019). Construction of a lncRNA-miRNA-mRNA network to determine the regulatory roles of lncRNAs in psoriasis. Experimental and Therapeutic Medicine, 18(5), 4011–4021. 10.3892/etm.2019.8035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Rhee, J.-W., Cheng, P., Waliany, S., Chang, A., Witteles, R. M., Maecker, H., Davis, M. M., Nguyen, P. K., & Wu, S. M. (2020). Correction to: Cardiovascular complications in patients with COVID-19: Consequences of viral toxicities and host immune response. Current Cardiology Reports, 22(5), 36. 10.1007/s11886-020-01302-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, X., Chen, K., Zou, J., Han, P., Hao, J., & Han, Z. (2020). Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Frontiers of Medicine, 14(2), 185–192. 10.1007/s11684-020-0754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.