Abstract
Spike protein and main proteases of SARS-CoV-2 have been identified as potential therapeutic targets and their inhibition may lead to the reticence of viral entry and replication in the host body. Despite several efforts; till now no specific drugs are available to treat SARS-CoV-2. Considering all these challenges, the main objective of the present study was to establish therapeutic potential of cordycepin against COVID-19 as a conventional therapeutic strategy. In the present study; molecular interaction study was performed to assess potential binding affinity of cordycepin with SARS-CoV-2 target proteins using computational approach. Additionally, network pharmacology was used to understand cordycepin-protein interactions and their associated pathways in human body. Cordycepin is under clinical trial (NCT00709215) and possesses structural similarity with adenosine except that, it lacks a 3′ hydroxyl group in its ribose moiety and hence it served as a poly(A) polymerase inhibitor and terminate premature protein synthesis. Additionally, it is known that functional RNAs of SARS-CoV-2 genome are highly 3’-plyadenylated and leading to synthesis of all viral proteins and if cordycepin can destabilize SARS-CoV-2 RNAs by inhibiting polyadenylation process then it may step forward in terms of inhibition of viral replication and multiplication in the host. Moreover, cordycepin showed strong binding affinity with SARS-CoV-2 spike protein (-145.3) and main proteases (-180.5) that further corroborate therapeutic potential against COVID-19. Since cordycepin has both pre-clinical and clinical information about antiviral activities, therefore; it is suggested to the world community to undertake repurposing cordycepin to test efficacy and safety for the treatment of COVID-19.
Keywords: Coronavirus, main proteases, 2019nCov, SARS-CoV-2, spike protein
Introduction
COVID-19, a new pandemic disease caused by an enveloped positive-sense single stranded RNA virus that belongs to coronaviridae family was first identified in November-December 2019 in Wuhan, China (Phelan et al., 2020). The International Committee on Taxonomy of Viruses named this novel coronavirus as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the symptoms are similar to pneumonia was designated as COVID-19 by the World Health Organization (WHO) on February 11, 2020 (Dai et al., 2020; World Health Organization (WHO), 2020). The disease spread rapidly and covered more than 212 countries and territories around the world and was declared a global health emergency by WHO. Globally, there have been 47,930,397 confirmed cases of COVID-19, including 1,221,781 deaths, reported to WHO (5 November 2020). The novel coronavirus, SARS-CoV-2 is 96.2% identical to a bat coronavirus and shares significant identity (79.5%) with 2002 SARS-CoV in terms of genetic makeup and use almost similar mechanisms to enter into the host cell although with higher affinity. Both the viruses make entry into the host body by binding of the viral spike glycoprotein S1 to the host receptor, angiotensin converting enzyme 2 (ACE2). An alluring drug target among coronaviruses is the ∼306 amino acid long main protease (Mpro, 3CLpro), to forestall the spread of disease by restraining the cleavage of the viral polyprotein. Mpro is essential for processing the polyproteins that led to the proteolytic activation of the viral functional proteins (Zhang et al., 2020). Targeting entry of the virus has a better advantage than inhibiting the later stages of the viral life cycle.
Till now, no clinically proven specific vaccines or antiviral drugs are available for the prevention and treatment of COVID-19 pandemic. Due to the gravity of the situation and rapid worldwide spread of SARS-CoV-2; urgent and complementary efforts from researchers are necessary to find therapeutic agents and new preventive methods. Drugs like hydroxychloroquine, arbidol, remdesivir, and favipiravir are currently undergoing clinical trials to test their effectiveness and safety in the treatment of COVID-19 and some promising results have been achieved thus far (Aanouz et al., 2020; Dong et al., 2020; Boopathi et al., 2020). Recently, Indian Council of Medical Research, under the Ministry of Health and Family Welfare (MHFW), has suggested the use of hydroxychloroquine (400 mg twice on day 1, then 400 mg once a week thereafter) as chemoprophylaxis for asymptomatic health-care workers who are directly involved in treating COVID-19 patients with suspected or confirmed COVID-19. Seeing the potent antiviral activities of cordycepin against several human viruses including influenza virus, human immunodeficiency virus, murine leukemia virus, plant viruses and epstein-barr virus (Chanda et al., 2015; Ohta et al., 2007; Ryu et al., 2014) it was considered to evaluate inhibitory potential against SARS-CoV-2 using in silico approach.
As it is evident that spike protein receptor-binding domain (RBD) and main protease play essential roles during viral infection, replication and multiplication in the host body (Wang et al., 2020; Wrapp et al., 2020; Zhang et al., 2020). Therefore, in the present study, these protein targets have been utilized during molecular interactions simulation with cordycepin using a computational approach. The present paper highlights the therapeutic potential of cordycepin and hence it is suggested to the world community that drug repurposing would be a more effective method to develop drugs against SARS-CoV-2.
Materials and methods
Collection of drug and target proteins
SARS-CoV-2 associated target proteins, namely spike protein (ID: 6VW1) (Shang et al., 2020) and main protease (6LU7) (Jin et al., 2020) were used in the present study for molecular interaction simulation. The 3 D crystal structures were downloaded from the RCSB Protein Data Bank (PDB) (Source: http://www.rcsb.org/ pdb/home/home.do). The target protein has been reported for their significant roles during infection, replication, survival and multiplication of SARS-CoV-2 in the host body (Liu et al., 2020; Xi et al., 2020). Protein preparation was carried out by the inbuilt program of MD algorithms. Cordycepin was downloaded from NCBI and the molecular arrangement and geometry were fully optimized using the semiempirical quantum chemistry method (PM3).
Molecular interaction simulation
The molecular interaction simulations between potent antiviral compounds, cordycepin and aforesaid SARS-CoV-2 target proteins were studied by using MD 2010.4.0 software for Windows (Bitencourt-Ferreira & De Azevedo, 2019; Kusumaningrum et al., 2014). The molecular simulation parameters were set considering a grid radius 15 Å with the number of runs: 10 runs; maximum interactions size: 1500; maximum population: 50; maximum steps used: 300; neighbor distance factor: 1.00; the maximum number of poses accepted: 5 to cover the ligand-biding site of the target proteins structure (Thomsen & Christensen, 2006). Moreover, post docking protein-ligand complex along with chemical interaction was further analyzed and visualized by Chimera software (https://www.cgl.ucsf.edu/chimera/) (Goddard et al., 2007) and Discovery Studio (BIOVIA, D.S., 2017) (https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/).
Pharmacology network of cordycepin
Interactions between proteins and bioactive compounds or drugs are an integral part of biological processes in living organisms. In the present study interaction network of potent compound was determined by STITCH (Search Tool for Interacting Chemicals) algorithm. The interactions between drugs and receptors include direct (physical) and indirect (functional) associations and generated by computational prediction from knowledge transfer between organisms, and from interactions aggregated from other databases (primary). Interactions in STITCH are derived from different sources such as genomic context predictions, genomic context predictions, (conserved) co-expression, automated textmining and previous knowledge in databases (Szklarczyk et al., 2016).
Results and discussion
Several pharmacologically active compounds have been reported from fungi, Cordyceps militaris; among those cordycepin has gained additional attention due to its broad-spectrum cellular and biological action (Tuli et al., 2014). It is known to regulate various biochemicals and molecular processes including purine biosynthesis, DNA/RNA and mTOR (mammalian target of rapamycin) signaling pathways (Holbein et al., 2009). Cordycepin and its related analogues have remarkable clinical health effects (Liu et al., 2015) including action on hepatic, renal, cardiovascular, respiratory, nervous, sexual, immunological systems, besides having anti-cancer, antiviral, antioxidant, anti-inflammatory and anti-microbial activities (Tuli et al., 2014). Potent antiviral activities of cordycepin and its analogs have been reported against various disease-causing human viruses (Chanda et al., 2015; Ohta et al., 2007; Ryu et al., 2014).
The large transmembrane spike glycoprotein (type I) of SARS-CoV-2 accounts for its notable feature and is heavily-glycosylated (Wrapp et al., 2020). However, it has been suggested that this virus and several others acquire a glycan coat that is adequate and similar to endogenous host protein glycosylation to serve as a glycan shield, which facilitates immune evasion by masking non-self-viral peptides with self-glycans (Watanabe, Allen et al., 2020; Watanabe, Berndsen et al., 2020). Virus proteins are usually glycosylated via one of three distinct pathways. These processes not only vary in the cellular enzymes that are involved, but also produce various forms of glycan structure (Sugrue, 2007). The resulting glycans are either N-linked, O-linked, or glycosylphosphatidyl inositol (GPI)-anchored. N-linked glycosylation occurs at protein sites where Asn-X-Ser/Thr amino acid sequence is present. O-linked glycans are typically bound to the chain by serine or threonine residue. In some cases, glycoproteins are bound to a lipid membrane through lipid linkage and this type of modification is often referred to as a glycosylphosphatidyl inositol (GPI) anchor (Sugrue, 2007).
The SARS-CoV-2 spike glycoprotein consists of two subunits, a receptor binding subunit (S1) and a membrane fusion subunit (S2) (Lu et al., 2020; Zhou et al., 2020L). The Spike glycoprotein assembles into stable homotrimers that together possess 66 canonical sequons for N-linked glycosylation (Asn-X-Ser/Thr, where X is any amino acid except Proline) as well as a number of potential O-linked glycosylation sites (Watanabe, Allen et al., 2020; Watanabe, Berndsen et al., 2020). Interestingly, coronaviruses virions bud into the lumen of the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), raising unanswered questions regarding the precise mechanisms by which viral surface glycoproteins are processed as they traverse the secretory pathway (Stertz et al., 2007; Zhao et al., 2020).
Like many viruses, SARS-CoV-2 utilizes a spike glycoprotein trimer for recognition and binding to the host cell surface receptor angiotensin converting enzyme 2 (ACE2) glycoprotein and facilitate host cell entry (Watanabe et al., 2019). Given the importance of viral spike proteins for targeting and entry into host cells along with their location on the viral surface, spike proteins are often used as immunogens for vaccines to generate neutralizing antibodies and frequently targeted for inhibition by small molecules that might block host receptor binding and/or membrane fusion (Li, 2016; Watanabe et al., 2019). Thus, understanding the glycosylation pattern of the viral spike trimer is essential for the development of efficacious vaccines, neutralising antibodies, and therapeutic inhibitors of such infection.
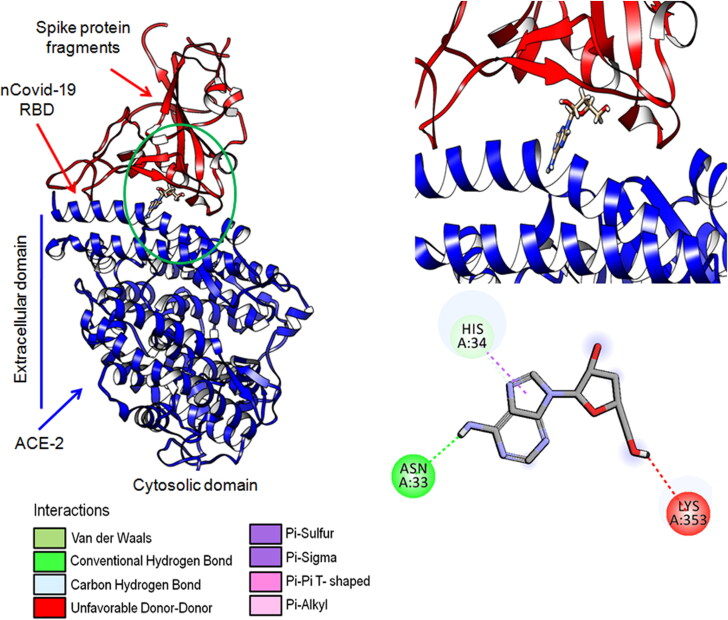
The molecular interaction study revealed that cordycepin has a strong binding affinity (-145.3) with SARS-CoV-2 RBD domain of spike protein (Figure 1). Cordycepin has shown chemical interactions (H-bond) in RBD domain-human ACE2 interface with Asn33, His34 and Lys353 (Figure 1). It is worth mentioning that all these amino acids are localized in the interface region of spike glycoprotein and host receptors that ultimately facilitates receptor- mediated endocytosis (Hoffmann et al., 2020) during primary infection. In a recent study, it has been reported that pan-coronavirus fusion inhibitor (EK1C4), targeting spike protein successfully restricted (IC50 range: 1.3 − 15.8 nM) viral entry into the host body in many coronaviruses such as SARS-CoV, MERS-CoV, SARS-CoV-2, HCoV-OC43 and SARSr-CoVs (Xia et al., 2020).
Figure 1.
Docking structure and chemical interactions of cordycepin have been shown along with ligand atoms and interacting amino acids in the binding sites of the SARS-CoV-2 spike protein RBD.
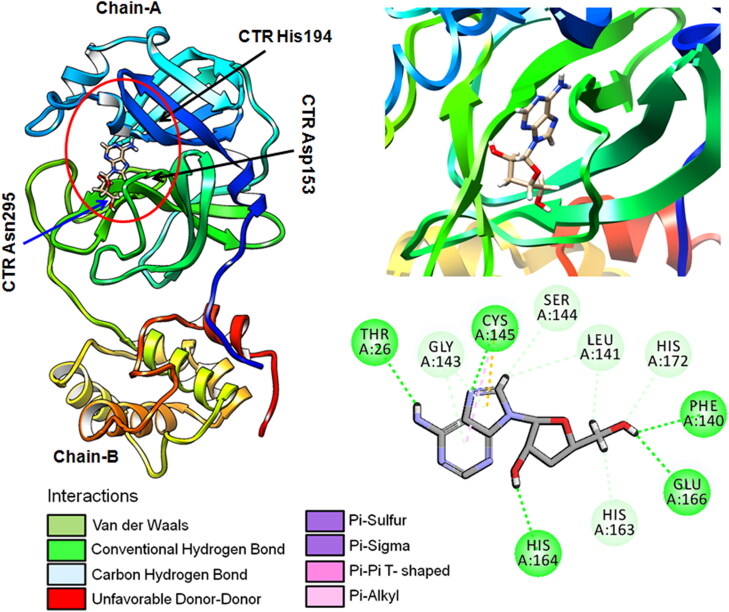
An interesting and additional drug target besides spike protein among coronaviruses is the ∼306 amino acid long main protease (Mpro, 3CLpro), to forestall the spread of disease by restraining the cleavage of the viral polyprotein. Mpro is essential for processing the polyproteins that led to the proteolytic activation of the viral functional proteins (Zhang et al., 2020) in SARS-CoV-2. An in silico analysis revealed that the cordycepin bind strongly (interaction score: −180.5) with Mpro (Figure 2). The Mpro active site amino acids such as Thr26, Gly143, Cys145, Ser144, Leu141, His172, Phe140, Glu166, His163 and His164 play a major role during chemical interactions with cordycepin (Figure 2).
Figure 2.
Docking structure and chemical interactions of cordycepin have been shown along with ligand atoms and interacting amino acids in the binding sites of Mpro.
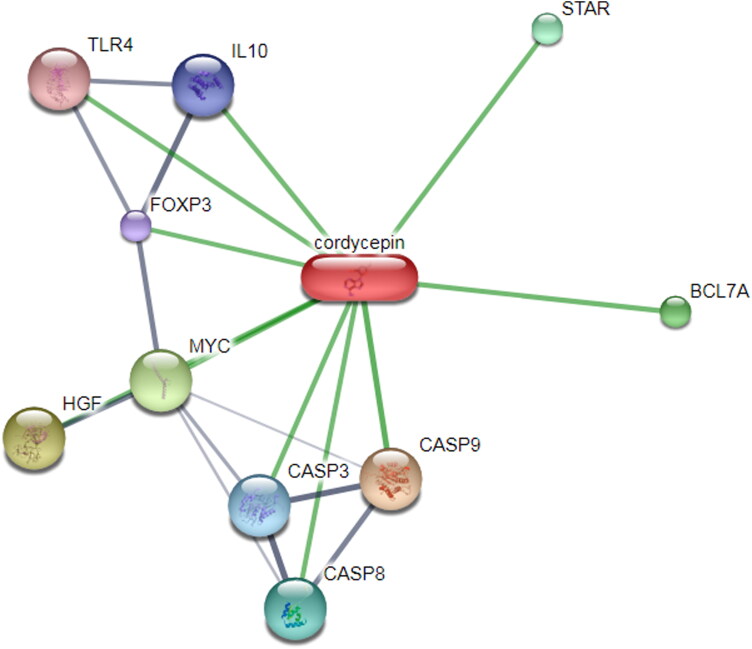
The biological response of small molecules in a living organisms is largely regulated by their interaction partners (Sharan et al., 2007). The role of the interaction network becomes more important in computer aided drug design (CADD), since diseases are often reflected by alterations of protein complex of the certain pathways (Gollapalli et al., 2020; Oti et al., 2006). The cordycepin-protein network data revealed that cordycepin can modulate multiple pathways associated with apoptosis, cancer, hepatitis B, tuberculosis, influenza A, herpes simplex infection and many more. The statistical significant cordycepin-protein network is found to be associated with herpes simplex infection (0.000562): CASP3 and CASP8; hepatitis B (2.38e-10): CASP3, CASP8, CASP9, MYC and TLR4 and tuberculosis (9.08e-10): CASP3, CASP8, CASP9, IL10 and TLR4 (Figure 3).
Figure 3.
The interaction network of cordycepin was determined by STITCH algorithm, showing the involvement of cordycepin in multiple pathways. This is the confidence view; stronger associations are represented by thicker lines. Protein-protein interactions are shown in grey, chemical-protein interactions in green and interactions between chemicals in red.
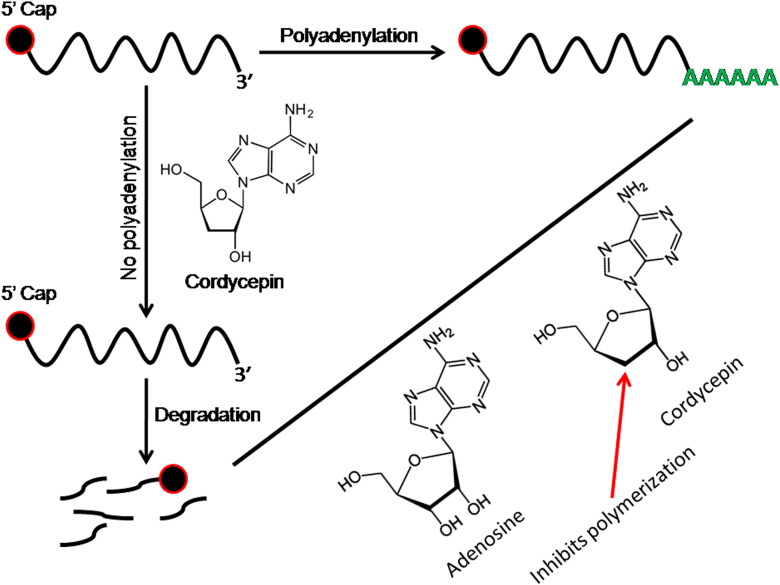
Polyadenylation is a vital process common to all viruses that increase the half life of the polyadenylated transcripts by protecting the RNA from degradation by the exonucleases (Ustyantsev et al., 2020). Polyadenylation at the 3′ end has also been reported in COVID-19 that plays major roles in pathogenesis and viral multiplication. During genome replication in SARS-CoV-2, full-length (-) RNA copies of the genome are synthesized leading to all structural proteins by discontinuous transcription (Hoffmann et al., 2020; Luk et al., 2019). Thereafter, viral nucleocapsids along with genomic RNA and R protein combine together in the cytoplasm and then channelized into the lumen of the endoplasmic reticulum. Virions are then released from the infected lungs tissues through exocytosis and can bind angiotensin-converting enzyme 2 (ACE2) receptor of other target tissues such as kidney cells, liver cells, intestines, and T lymphocytes, as well as the lower respiratory tract, where they form the main symptoms and signs including collapse of the immune system (Lambeir et al., 2003; Wrapp et al., 2020; Yan et al., 2020). In this context, cordycepin (3′-deoxyadenosine), a bioactive compound of Cordyceps militaris believed to be effective in controlling SARS-CoV-2 replication. Cordycepin possesses structural similarity with adenosine except that; it lacks a 3′ hydroxyl group in its ribose moiety. As cordycepin has a structural similarity with adenosine, some enzymes fails to discriminate between the two. It can, therefore, participate in certain biochemical reactions including poly(A) polymerase inhibition, shortening of poly(A) tails, destabilization of mRNAs, purine biosynthesis inhibition (Figure 4) and also resulting in premature termination of protein synthesis (Holbein et al., 2009; Overgaard-Hansen, 1964). This research is the first study of its kind that highlights the therapeutic potential of cordycepin against COVID-19. However, several research groups have also reported that cordycepin has antiviral activity against several viruses including influenza virus, human immunodeficiency virus, murine leukemia virus, plant viruses and epstein-barr virus (Ohta et al., 2007; Ryu et al., 2014). Cordyceps has a long history of use as a lung and kidney tonic, and for the treatment of chronic bronchitis, asthma, tuberculosis and other diseases of the respiratory system (Tuli et al., 2014). The folk healers of Sikkim use Cordyceps to cure 21 ailments including cancer, asthma, TB, diabetics, cough and cold, erectile dysfunction, female BHP, hepatitis, etc. (Panda & Swain, 2011). Cordycepin derived from Emericella nidulans, an endophytic fungus has been reported to have HCV NS3/4A protease inhibitory properties (Hawas et al., 2012; Suwannarach et al., 2020). High cordycepin concentrations selectively inhibit influenza viral genome replication (Pridgen, 1976). In another study it has been reported that cordycepin analogs inhibit purified HIV-1 reverse transcriptase (Ryu et al., 2014). In vitro RNA synthesis of tobacco mosaic virus and cowpea chlorotic mottle virus replicase are inhibited by high concentrations of cordycepin. Moreover, EBV-induced transformation of human lymphocytes is inhibited by cordycepin in the absence of interferons (Ryu et al., 2014). The molecular mechanisms by which cordycepin exerts its antiviral activities are yet to be determined. However, its effects on renal and hepatic function, immunomodulatory-related antitumor activities, poly(A) polymerase inhibition, shortening of poly(A) tails, destabilization of mRNAs and purine biosynthesis inhibition are most promising and deserve further attention.
Figure 4.
Cordycepin mediated biological response for SARS-CoV-2 RNA degradation.
Conclusion
Thus, based on the strong molecular interactions of cordycepin with SARS-CoV-2 spike protein and main proteases in addition to reported polyadenylation inhibition; suggesting a higher potential of cordycepin to inhibit virus entry and replication into the host body. It is, therefore, suggested that world community should undertake repurposing clinical studies to test efficacy and safety in the treatment of COVID-19 which is an urgent need of the hour. Moreover, the remarkable clinical health benefits of cordycepin including protective action on hepatic, renal, cardiovascular, respiratory, nervous, immunological systems, besides having anti-cancer, anti-oxidant, anti-inflammatory and anti-microbial activities is also supporting the present study because these are the tissues that are mostly affected by COVID-19 during the later phase of infection.
Author’s contribution
Study design, data collection, data analysis and manuscript writing was done by A. K. Verma.
Acknowledgements
Department of Zoology, Cotton University is highly acknowledged for all the supports related to the study. The author gratefully acknowledges Mr. Rohit Aggarwal (owner of Cosmic Cordycep Farms, Faridabad, Haryana, India) for sharing some basic information related to Cordyceps. No special fund allocated to the work.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-Ldrissi, M., & Bouachrine, M. (2020). Moroccan Medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIOVIA, D.S. (2017). Discovery studio visualizer, Release 2017, San Diego: Dassault Systèmes-2016. To be found under http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php
- Bitencourt-Ferreira, G., & De Azevedo, W. F. (2019). Molegro virtual docker for docking. In W. de Azevedo, Jr. (Ed.), Docking screens for drug discovery. Methods in molecular biology (Vol. 2053, pp. 149–167). New York, NY: Humana. [DOI] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda, S., Banerjee, A., Nandi, S., Chakrabarti, S., & Sarkar, M. (2015). Cordycepin an adenosine analogue executes anti rotaviral effect by stimulating induction of type I interferon. Journal of Virology and Antiviral Research, 4(2), 1–12. [Google Scholar]
- Dai, W., Zhang, B., Jiang, X.-M., Su, H., Li, J., Zhao, Y., Xie, X., Jin, Z., Peng, J., Liu, F., Li, C., Li, Y., Bai, F., Wang, H., Cheng, X., Cen, X., Hu, S., Yang, X., Wang, J., … Liu, H. (2020). Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science, 368(6497), 1331–1335. 10.1126/science.abb4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, L., Hu, S., & Gao, J. (2020). Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discoveries & Therapeutics, 14(1), 58–60. 10.5582/ddt.2020.01012 [DOI] [PubMed] [Google Scholar]
- Goddard, T. D., Huang, C. C., & Ferrin, T. E. (2007). Visualizing density maps with UCSF Chimera. Journal of Structural Biology, 157(1), 281–287. 10.1016/j.jsb.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Gollapalli, P., Sharath, B. S., Rimac, H., Patil, P., Nalilu, S. K., Kandagalla, S., & Shetty, P. K. (2020). Pathway enrichment analysis of virus-host interactome and prioritization of novel compounds targeting the spike glycoprotein receptor binding domain–human angiotensin-converting enzyme 2 interface to combat SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1841681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawas, U. W., El Kassem, L. T. A., Ahmed, E. F., & Emam, M. (2012). In-vitro bioassays on the metabolites of the fungus Emericella nidulans isolated from the Egyptian red sea algae. Egyptian Pharmaceutical Journal, 11, 124–128. [Google Scholar]
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N.-H., Nitsche, A., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbein, S., Wengi, A., Decourty, L., Freimoser, F. M., Jacquier, A., & Dichtl, B. (2009). Cordycepin interferes with 3' end formation in yeast independently of its potential to terminate RNA chain elongation. RNA, 15(5), 837–849. 10.1261/rna.1458909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Kusumaningrum, S., Budianto, E., Kosela, S., Sumaryono, W., & Juniarti, F. (2014). The molecular docking of 1, 4-naphthoquinone derivatives as inhibitors of Polo-like kinase 1 using Molegro Virtual Docker. Journal of Applied Sciences, 4, 47–53. [Google Scholar]
- Lambeir, A.-M., Durinx, C., Scharpé, S., & De Meester, I. (2003). Dipeptidyl-peptidase IV from bench to bedside: An update on structural properties, functions, and clinical aspects of the enzyme DPP IV. Critical Reviews in Clinical Laboratory Sciences, 40(3), 209–294. 10.1080/713609354 [DOI] [PubMed] [Google Scholar]
- Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, 3(1), 237–261. 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Morse, J. S., Lalonde, T., & Xu, S. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem, 21(5), 730–738. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Wang, J., Wang, W., Zhang, H., Zhang, X., & Han, C. (2015). The chemical constituents and pharmacological actions of Cordyceps sinensis. Evidence-Based Complementary and Alternative Medicine, 2015, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk, H. K., Li, X., Fung, J., Lau, S. K., & Woo, P. C. (2019). Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infection, Genetics and Evolution, 71, 21–30. 10.1016/j.meegid.2019.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, Y., Lee, J. B., Hayashi, K., Fujita, A., Park, D. K., & Hayashi, T. (2007). In vivo anti-influenza virus activity of an immunomodulatory acidic polysaccharide isolated from Cordyceps militaris grown on germinated soybeans. Journal of Agricultural and Food Chemistry, 55(25), 10194–10199. 10.1021/jf0721287 [DOI] [PubMed] [Google Scholar]
- Oti, M., Snel, B., Huynen, M. A., & Brunner, H. G. (2006). Predicting disease genes using protein-protein interactions. Journal of Medical Genetics, 43(8), 691–698. 10.1136/jmg.2006.041376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard-Hansen, K. (1964). The inhibition of 5-phosphoribosyl-1-pyrophosphate formation by cordycepin triphosphate in extracts of Ehrlich ascites tumor cells. Biochimica et Biophysica Acta (BBA)-Specialized Section on Nucleic Acids and Related Subjects, 80(3), 504–507. [DOI] [PubMed] [Google Scholar]
- Panda, A. K., & Swain, K. C. (2011). Traditional uses and medicinal potential of Cordyceps sinensis of Sikkim. Journal of Ayurveda and Integrative Medicine, 2(1), 9–13. 10.4103/0975-9476.78183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan, A. L., Katz, R., & Gostin, L. O. (2020). The novel coronavirus originating in Wuhan, China: Challenges for global health governance. JAMA, 323(8), 709–710. 10.1001/jama.2020.1097 [DOI] [PubMed] [Google Scholar]
- Pridgen, C. L. (1976). Influenza virus RNA's in the cytoplasm of chicken embryo cells treated with 3'-deoxyadenosine. Journal of Virology, 18(1), 356–360. 10.1128/JVI.18.1.356-360.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, E., Son, M., Lee, M., Lee, K., Cho, J. Y., Cho, S., Lee, S. K., Lee, Y. M., Cho, H., Sung, G.-H., & Kang, H. (2014). Cordycepin is a novel chemical suppressor of Epstein-Barr virus replication. Oncoscience, 1(12), 866–881. 10.18632/oncoscience.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., Geng, Q., Auerbach, A., & Li, F. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature, 581(7807), 221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan, R., Ulitsky, I., & Shamir, R. (2007). Network‐based prediction of protein function. Molecular Systems Biology, 3(1), 88. 10.1038/msb4100129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz, S., Reichelt, M., Spiegel, M., Kuri, T., Martínez-Sobrido, L., García-Sastre, A., Weber, F., & Kochs, G. (2007). The intracellular sites of early replication and budding of SARS-coronavirus. Virology, 361(2), 304–315. 10.1016/j.virol.2006.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue, R. J. (2007). Viruses and glycosylation: An overview. Methods in Molecular Biology, 379, 1–13. 10.1007/978-1-59745-393-6_1 [DOI] [PubMed] [Google Scholar]
- Suwannarach, N., Kumla, J., Sujarit, K., Pattananandecha, T., Saenjum, C., & Lumyong, S. (2020). Natural bioactive compounds from fungi as potential candidates for protease inhibitors and immunomodulators to apply for coronaviruses. Molecules, 25(8), 1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D., Santos, A., von Mering, C., Jensen, L. J., Bork, P., & Kuhn, M. (2016). STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Research, 44(D1), D380–D384. 10.1093/nar/gkv1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen, R., & Christensen, M. H. (2006). MolDock: A new technique for high-accuracy molecular docking. Journal of Medicinal Chemistry, 49(11), 3315–3321. 10.1021/jm051197e [DOI] [PubMed] [Google Scholar]
- Tuli, H. S., Sandhu, S. S., & Sharma, A. (2014). Pharmacological and therapeutic potential of Cordyceps with special reference to Cordycepin. 3 Biotech, 4(1), 1–12. 10.1007/s13205-013-0121-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustyantsev, I., Tatosyan, K., Stasenko, D., Kochanova, N., Borodulina, O., & Kramerov, D. (2020). Polyadenylation of sine transcripts generated by RNA polymerase III dramatically prolongs their lifetime in cells. Molecular Biology, 54(1), 67–74. 10.1134/S0026893319040150 [DOI] [PubMed] [Google Scholar]
- Wang, Q., Qiu, Y., Li, J. Y., Zhou, Z. J., Liao, C. H., & Ge, X. Y. (2020). A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virologica Sinica, 35(3), 337–339. 10.1007/s12250-020-00212-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., Allen, J. D., Wrapp, D., McLellan, J. S., & Crispin, M. (2020). Site-specific glycan analysis of the SARS-CoV-2 spike. Science, 369(6501), 330–333. 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., Berndsen, Z. T., Raghwani, J., Seabright, G. E., Allen, J. D., Pybus, O. G., McLellan, J. S., Wilson, I. A., Bowden, T. A., Ward, A. B., & Crispin, M. (2020). Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nature Communications, 11(1), 2688 10.1038/s41467-020-16567-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., Bowden, T. A., Wilson, I. A., & Crispin, M. (2019). Exploitation of glycosylation in enveloped virus pathobiology. Biochimica et Biophysica Acta. General Subjects, 1863(10), 1480–1497. 10.1016/j.bbagen.2019.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (2020). Coronavirus disease (COVID-19) pandemic 2020. Retrieved April 16, 2020, from https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, J., Xu, K., Jiang, P., Lian, J., Hao, S., Jia, H., Yao, H., Zhang, Y., Zheng, R., & Chen, D. (2020). Virus strain of a mild COVID-19 patient in Hangzhou representing a new trend in SARS-CoV-2 evolution related to Furin cleavage site. Emerging Microbes and Infections, 9, 1474–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., Qi, F., Bao, L., Du, L., Liu, S., Qin, C., Sun, F., Shi, Z., Zhu, Y., Jiang, S., & Lu, L. (2020). Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Research, 30(4), 343–355. 10.1038/s41422-020-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, P., Praissman, J. L., Grant, O. C., Cai, Y., Xiao, T., Rosenbalm, K. E., Aoki, K., Kellman, B. P., Bridger, R., Barouch, D. H., Brindley, M. A., Lewis, N. E., Tiemeyer, M., Chen, B., Woods, R. J., & Wells, L. (2020). Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host & Microbe, 28(4), 586–601. 10.1016/j.chom.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]