Abstract
The global emergence of novel coronavirus disease and its rapid global expansion over a short span of time require effective countermeasures to combat it. Development of a specific vaccine can induce an optimal antibody response, thus providing immunity against it. Our study proposes a detailed and comprehensive immunoinformatic approach that can be applied to the currently available coronavirus protein data in the online server for vaccine candidate development. We have identified the receptor binding domain (RBD) of structural spike protein (S1) as a potential target for immunity against COVID- 19 infection. Epitope prediction illustrated cytotoxic T-cell epitopes, helper T-cell epitopes, and B-cell epitopes associated with the target protein. These were joined through specific linkers along with adjuvant beta-defensin located at the N-terminal to create a multi epitope subunit vaccine (MESV). The specificity in the binding of the devised vaccine candidate to the TLR-3 immune cell receptor was evaluated via molecular docking interaction studies. Good docking score combined with robust interactions in the binding cavity certified the stringency of the engineered vaccine. Molecular dynamics simulation data showed minimal variation of the root-mean square deviations (RMSDs) and root-mean-square fluctuations (RMSFs) which confirmed the interaction stability. These results obtained from various in-silico experiments indicate the potency of this vaccine candidate as a probable therapeutic agent against COVID-19. Vaccination strategies targeting conserved epitope-based immune response would be beneficial in providing cross protection across beta-coronaviruses, and such vaccines would be resistant to the ever-evolving viruses.
Communicated by Ramaswamy H. Sarma
Keywords: Spike glycoprotein, receptor binding domain, subunit vaccine, COVID-19, immunoinformatics
1. Introduction
A novel strain of the coronavirus is suspected to have emerged in Wuhan, China in late 2019 and has resulted in an emergency situation throughout world. The first cases of the novel coronavirus were reported in the Hubei province and later named the disease as COVID-19 outbreak in China (Mackenzie & Smith, 2020). It was declared a pandemic disease on March 11, 2019 by the World Health Organization (WHO) with more than 21.2 million people being infected and resulting in more than 761,000 deaths worldwide to date (World Health Organization, 2020) due to a severe respiratory infection which induces systemic cytokine storm in the host (Song et al., 2020). The virus acts mainly by attacking the respiratory system in three key phases: viral replication, hyper-reactive immune response, and subsequent obstruction of pulmonary airflow. According to the reports, perforated holes are created in the lungs giving them a honey-comb like appearance upon viral infection.
The severe acute respiratory syndrome-corona virus-2 (SARS-CoV-2) is a key member from family Coronaviridae, order Nidovirales. It consists of a large genome comprising a positive-sense single-stranded RNA (Fauquet, 2008). The size of the virus is 65–125 nm with a crown like structure protruding on the surface. The coronaviridae family constitutes four subgroups: alpha, beta, gamma, and delta (Chan et al., 2015). The novel strain of the coronavirus belongs to beta group which has low pathogenicity (Enjuanes et al., 2006; Perlman & Netland, 2009). The virus has specific attributes that allow it to infect interspecies, and it infects a diverse range of hosts-birds to mammals and humans-and causes severe respiratory and enteric diseases (Lee, 2015). The symptoms are similar to those of the common flu, such as mild to moderate fever, cough, sore throat, and headache in addition to difficulty in breathing (Mäkelä et al., 1998; Owusu et al., 2014).
The virus is comprised of spike glycoprotein, membrane glycoprotein, nucleocapsid, and enveloped non-glycosylated protein (Gorbalenya et al., 2006). These are essential to synthesize a complete virus and its associated non-structural protein components, which are critical to the process of viral replication and transcription. The spike protein primarily determines the virus infection. Virus interaction is an essential step for initiating infection, and proteolytic cleavage plays a critical role in the peptide release in the host (Belouzard et al., 2012; Ye et al., 2004). The spike protein of the corona virus is a class I transmembrane that is trimeric, has a molecular weight of 180 kDa, and has a monomer that requires host protease cleavages for its activation, such as CoV which requires two specific cleavages for fusion of spike protein. This last factor is crucial for the interaction, fusion, and internalization of virus to host. The interaction of the spike is associated with cell and tissue tropism as well as pathogenesis. Moreover, tropism is associated with specific cleavage of spike protein, and mutants suggest the alternation in viral pathogenesis. The spike protein consists of two functional domains S1 and S2 (F. Li, 2016). The RBD of the S1 domain contains the receptor binding motif and plays a key role in host interaction (Tai et al., 2020). It acts together with the angiotensin converting enzyme-2 (ACE-2) receptor, a known efficient surface receptor of the host. It is the key target of antibodies (W. Li et al., 2003, 2005).
Currently, there is no specific remedial measure in the form of drug/medicine or vaccine available for the treatment or termination of the spread of SARS-CoV-2 infection. With the daily increasing cases of infection and the high death-toll, there is an imperative and immediate need to design and develop an efficient vaccine against the deadly virus to order control this global pandemic.
Antigenic epitopes of RBD of spike glycoprotein of novel coronavirus strain have been determined through utilizing immunoinformatic techniques which might be put to use for designing and developing a MESV peptide to suppress the viral infection through eliciting TLR-3 innate immune response. Epitopes, in general, are the short stretches of protein amino acid sequences which have a capability of eliciting a direct, specific and strong immune response, in comparison to the expected response generated by the complete protein itself (Palatnik-de-Sousa et al., 2018).
In the current study, a strong and prospective vaccine candidate was developed using an immunoinformatic approach, wherein we have identified antigenic epitopes against our target viral protein and combined them. The specificity in binding for the designed vaccine to the TLR-3 receptor was evaluated via molecular docking interaction. Good docking scores combined with robust interactions in the binding cavity certified the stringency of the engineered vaccine. Certain other parameters such as physicochemical properties, antigenicity, allergenicity, and toxicity were also examined as they are the requisites for being a candidate vaccine. Molecular dynamics (MD) simulation data have been generated to investigate the interaction stability. Moreover, strong immunological response was obtained via the T-cell and B-cell mediated humoral response through immune simulation. The results obtained from various in-silico experiments indicate the potency of our vaccine candidate as a probable therapeutic in case of COVID-19.
2. Methodology
2.1. Extraction of protein sequence and evaluation of the antigenic propensity
The spike protein sequence of the SARS coronavirus for subunit vaccine designing was retrieved from NCBI (https://www.ncbi.nlm.nih.gov) in FASTA format. This downloaded protein was cropped from residue 331–524 which corresponds to its receptor binding domain (RBD). After retrieval, the viral protein sequence was put through pBLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) to verify its similarity with human proteome. Next, the antigenic propensity of the protein sequence was done by using an online available antigenic peptides prediction server of the Immunomedicine group (http://imed.med.ucm.es/Tools/antigenic.pl).
2.2. Prediction and screening of cytotoxic T-lymphocyte (CTL), helper T-lymphocyte (HTL) epitopes with subsequent MHC alleles and B-cell epitopes
The prediction of cytotoxic T-lymphocyte (CTL) epitopes for the preferred SARS-CoV-2 RBD of the protein was done via NetCTL1.2 tool (http://www.cbs.dtu.dk/services/NetCTL/). The helper T-lymphocyte (HTL) epitopes were anticipated with Immune Epitope Databases’ (IEDB) MHC-II epitope prediction module (http://tools.iedb.org/mhcii/). The epitopes with low percentile rank were chosen for further consideration. In addition to that, these epitopes were scrutinized for their Th1 type immune response induction propensity following the production of IFN-ϒ. This step was achieved via IFN epitope server (http://crdd.osdd.net/raghava/ifnepitope/). Next, we deployed the ABCPred server (http://crdd.osdd.net/raghava/abcpred/) for the prediction of B-cell lymphocyte (BCL) epitopes. Finally, all identified epitopes of the CTL, HTL and BCL were examined for their non-toxic/toxic characteristics by deploying the ToxinPred server (http://crdd.osdd.net/raghava/toxinpred/multi_submit.php).
2.3. Designing of the multi-epitope subunit vaccine
The top selected epitopes for each cell type were connected sub sequentially to develop the final vaccine construct. Epitope separation was done by utilizing proper linkers/spacer amino acid sequences for the production of an efficient individual function. AAY, GPGPG and KK were the respective linkers employed for joining CTL, HTL and the B-cell epitopes. An immunostimulatory adjuvant (human β-defensin 3) was then added to the sequence towards the N-terminal region. This was followed by another linker sequence (EAAAK) which was added to join the adjuvant with the antigenic epitopes. A single potent vaccine construct was obtained upon subsequent fusion of the adjuvant, epitopes and their specific linkers in a structured way.
2.4. Immunological assessment of the designed vaccine model
The ANTIGENpro tool (http://scratch.proteomics.ics.uci.edu) has been deployed for prediction of antigenicity of the final vaccine candidate. This primordial server can predict the protein antigenicity on the basis of experimentally proven microarray analysis reactivity data. Antigenicity prediction of protein is alignment free and is done based on sequence. The server is pathogen independent and uses five algorithms of machine learning to make predictions. The non-antigenic/antigenic character of the protein is decided through SVM (Support Vector Machine) classifier (Magnan et al., 2010).
Next, we made use of two online servers, that is, Algpred (http://crdd.osdd.net/raghava/algpred/) along with AllerTop (https://www.ddg-pharmfac.net/AllerTOP/) to predict the allergenic nature of our newly developed vaccine candidate. The prediction of allergenicity via AlgPred server is based on matching the candidate vaccine sequence with that of the conventional epitope sequences (Saha & Raghava, 2006) while in case of the AllerTop server, machine learning techniques including k nearest neighbors (kNN), Logistic regression (LR), Decision tree (DT), Random forest (RF), Multilayer perceptron (MLP), and naive Bayes (NB) are utilized for prediction. Of all these machine learning techniques, only the kNN method showed five-fold cross validation with 85.3% maximum precision score (Dimitrov et al., 2013).
Additionally, the designed vaccine candidate was assessed for prediction of its toxicity which was done using ToxinPred server (http://crdd.osdd.net/raghava/toxinpred/). The toxicity prediction through this tool is done based on toxicity of peptides together with the whole protein sequences. It is fed with the input sequence in FASTA format which then works on scoring of position specific basis of the Quantitative Matrix that in return classifies the sequence toxicity (S. Gupta et al., 2015).
2.5. Evaluation of physicochemical properties
A range of different physicochemical features of this newly designed vaccine candidate were characterized through ProtParam tool (https://web.expasy.org/protparam/) which is present at the ExPASy (Expert Protein Analysis System) bioinformatics resource portal. Various protein parameters together with molecular weight of the peptide, theoretical pI, estimated half-life, extinction coefficient, aliphatic index, instability index and GRAVY index are computed through this server.
2.6. Prediction and validation of vaccine structure and molecular docking with human TLR-3 receptor
We made use of PSIPred 4.0 Protein Sequence Analysis Workbench (http://bioinf.cs.ucl.ac.uk/psipred/) to predict secondary structure features of subunit vaccine candidate. Furthermore, the modeling of tertiary structure was done with Swiss-Model web-server (https://swissmodel.expasy.org/) which has an automated system for generating a 3 D model of a given protein from the amino acid sequence using homology modelling techniques. The only requirement of this server is submission of amino acid sequence of protein under consideration. Selection of a template, its alignment and model building are done automatically by the server.
Afterwards, the designed model was evaluated for its quality by using computational approaches like PROCHECK and ProSA (Protein Structural Analysis). The PROCHECK program verifies the structure, figures and the stereochemical behavior of developed model by taking Ramachandran plots as reliable source. The ProSA technique operates by checking energy criteria of the model with respect to a large set of known protein structures of comparable sizes.
Molecular docking of vaccine construct with the human TLR-3 receptor was accomplished through HDOCK web server which was based on intrinsic scoring for determining the binding affinity of our ligand with receptor. The human TLR-3 receptor (PDB ID-3ULV) was downloaded from protein databank.
2.7. Molecular dynamics simulation studies
All-atomistic Molecular dynamics (MD) simulation was accomplished for the vaccine and TLR3 receptor complex using GROMACS 5.0 package which gave details on the dynamics and molecular interactions of the docking complex. Initial optimization of the vaccine-receptor complex was achieved with Schrödinger Maestro (Maestro, Schrödinger, New York) which was consequently adopted as the starting structure to conduct MD simulation studies. The system was immersed in a cube using simple point charge (SPC) water model and overall charges were neutralized by the addition of counter ions to the system. The energy minimization of this system was carried out by utilizing steepest descent algorithm for 50000 steps until it attained the maximum force < 10.0 kJ/mol/nm. The electrostatic interactions of a long-range were treated using Particle Mesh Ewald (PME) method. This energy minimized system was further subjected to NVT and NPT equilibration for 100 ps at a constant temperature set to 300 K and 1 bar constant pressure. During equilibration, v-rescale, a modified Brendsen thermostat was used for temperature coupling along with Parrinello-Rahman barostat for pressure coupling. The resulting system was subjected to 100 ns MD production run using leap-frog integrator with 2 fs integration time step. The linear constraint solver (LINCS) algorithm was used for restraining the bond lengths. The trajectories were saved every 10 ps and further structural analyses were done using various inbuilt GROMACS scripts. Moreover, the binding energies were evaluated using the molecular mechanics Poisson Boltzmann surface area (MM-PBSA) tool in Gromacs to get detailed insights into molecular interactions of the vaccine candidate and TLR3 complex. It is a widely used method for calculating the binding free energies of the complex from the non-covalent interactions (Kumari et al., 2014). The g_mmpbsa tool in GROMACS was used after molecular dynamics simulations, the output files obtained were used to post-process binding free energies by the single-trajectory MM-PBSA method. Specifically, for a non-covalent binding interaction in the aqueous phase the binding free energy, ΔGBinding, is: –
where ΔGBinding, ΔGReceptor and ΔGLigand are solvation free energies of complex, receptor and ligand, respectively.
2.8. Immune Simulation of vaccine construct
A significant way to comprehending the immune system is immune simulation which works by calculating the immune response profile and immunogenicity of probable vaccine construct. An agent-based immune simulator, C-ImmSim (http://kraken.iac.rm.cnr.it/C-IMMSIM/), has been used in our study to carry-out the immune simulation of our designed vaccine construct. The prediction of immune interactions and immune epitopes using C-ImmSim simulator derived from the machine learning techniques and position-specific scoring matrix (PSSM), respectively (Rapin et al., 2010). Three injections at a gap of four weeks each were given for target product profiling of the newly designed coronavirus vaccine construct. This time period is in accordance with the recommended time interval for most of the currently used vaccines doses that is four weeks. The HLA alleles of host were conserved and time step for injection were computed at 1, 84, and 168 (herein, every time step corresponds to 8 h from real-life, while the first step is injection at time = 0). The volume of simulation was set at 50, while the number of simulation steps was 1000. Remaining simulation specifications were set as default.
3. Results and discussion
3.1. Antigenic protein screening
The amino acid sequence of target surface spike glycoprotein of SARS-coronavirus was downloaded from National Centre for Biotechonology Information (NCBI). The protein is well-known to perform key function in interaction, fusion and internalization of virus to host, hence conferring the viral pathogenicity. This sequence was cropped and residues numbered 331 to 524 were retained which corresponds to the RBD of the protein of our interest (Tai et al., 2020). The viral sequence was checked for homology with the human proteome through the pBLAST server. The results showed no similarity with Homo sapiens. The similarity between pathogen and host should not be more than 40% to avoid chances of cross-reactivity. Thus, the results indicated that the antigenic viral proteins used in designing vaccine candidate will not cross-react upon administration in the host.
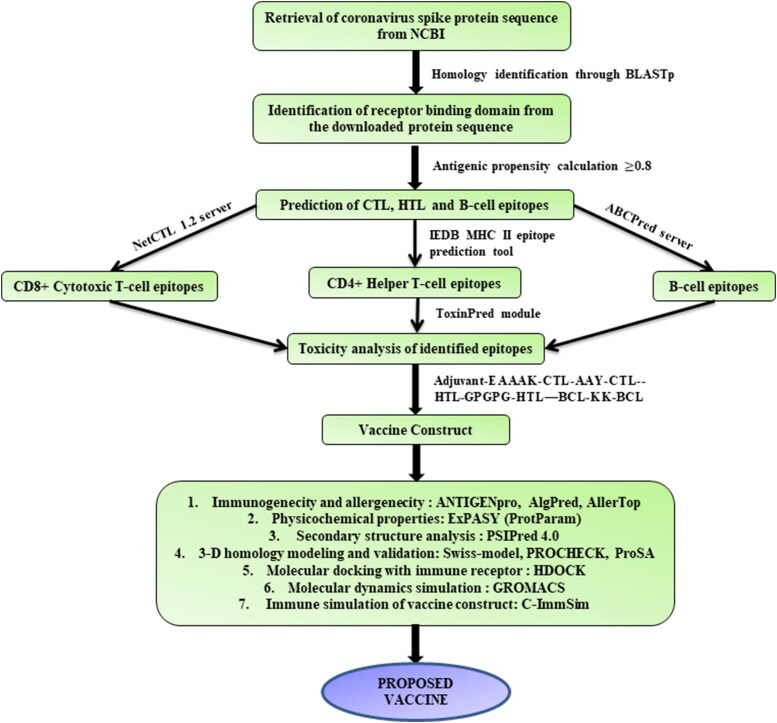
Next, the protein antigenic propensity average was calculated using the antigenic peptide prediction tool from the Immunomedicine group. It was found to be 1.03, indicating the highly antigenic nature of the peptide which can be further utilized for the prediction of epitopes for developing a multi-epitope vaccine. Figure 1 gives a schematically represents the methodology used for constructing the multi-epitope subunit vaccine.
Figure 1.
Schematic work flow used for designing the multi-epitope subunit vaccine candidate against the coronavirus (SARS-CoV-2).
3.2. Epitope prediction for cytotoxic T-lymphocyte (CTL), helper T-lymphocyte (HTL), and B-cell lymphocytes
The Cytotoxic T-lymphocytes (CTLs) have a critical role in inducing the MHC-I based cellular immune responses. These cells neutralize the viral particle infected or damaged host cells by producing the proteins such as perforins, granzymes, etc (Neurath, 2008). The CTL epitopes of the screened protein were predicted deploying the NetCTL1.2 server (http://www.cbs.dtu.dk/services/NetCTL/). We considered A2, A3, and B7 supertypes for the CTL epitopes determination as these supertypes majorly cover 88.3% of the total ethnic population (Sidney et al., 2008). The top ten epitopes obtained to be used for further vaccine designing are shown in Supplementary Table 1.
The Helper T-lymphocytes (HTLs) have a key role in adaptive immunity. These activate the B-cells along with the cytotoxic T cells for production antibody and eventually killing infected/damaged cells (Alberts et al., 2002). The HTL epitopes for the selected protein were calculated using the prediction tool for MHC-II epitope. The identified epitopes had lower percentile ranks. The HTL epitopes were additionally checked by their potency to induce IFN-ϒ production ability using IFN epitope server. The uppermost immunogenic epitopes of 15-mer were shortlisted based on basis of the percentile rank for each. Along with that, IFN epitope server output gave a positive score to all these epitopes (Supplementary Table 2).
Humoral immunity is mainly driven by B-cells which are responsible for the production of antibodies for recognizing and eliminating the antigens (Janeway et al., 2001). This necessitated the need for predicting the B-cell epitopes before designing our vaccine candidate. The ABCPred server (http://crdd.osdd.net/raghava/abcpred/) was deployed for prediction of these B-cell epitopes (Supplementary Table 3). Finally, the non-toxic/toxic characteristics of all chosen CTL, HTL, and B-cell epitopes were verified via ToxinPred module (http://crdd.osdd.net/raghava/toxinpred/multi_submit.php). The results for same have been demonstrated in Supplementary Table 4.
3.3. Construction of a multi-epitope subunit vaccine
Once the epitopes of different immune cells were predicted, we combined them using specific linker sequences in order to create a single vaccine construct. Each epitope of multi-epitope subunit vaccine should be should be having the capacity of inducing a potential immune response. The linkers play critical role in locating the epitope and are also key for conferring immunogenicity to the whole vaccine construct. These linker sequences are accountable for the separation of different domains combined in a unique single protein sequence without disrupting the functionality of that protein. Along with this, linkers are instrumental in maintaining stability and flexibility in the protein complex (Chen et al., 2013).
In the current work, we employed diverse varieties of linker sequences to connect the epitopes. The CTL epitopes were linked using “AAY” (Ojha et al., 2019) derived from the proteasomal cleavage site to eliminated the genesis of newer epitope, HTL epitopes were connected by “GPGPG” linkers (Livingston et al., 2002) used for improving the flexibility of protein and epitopes. The “KK” linkers (Yano et al., 2005), for the enhancement of the proteasome processing, were used for linking B-cell epitopes. In addition to these, another linker known to form helices, EAAAK, was used for designing the next-generation subunit vaccine.
A human beta-defensin-3 (45 amino acids) adjuvant was attached at the N-terminal location of the designed vaccine to confer immunogenicity to the vaccine construct. The human beta defensin 3 is a TLR-3 agonist capable of eliciting enhancement in the strength of T-cell and B-cell immune responses (Gupta et al., 2020; Mohan et al., 2013). A total of 10 CTL, 10 HTL, and 10 B-cell epitopes derivative of the receptor binding protein were combined for designing a subunit vaccine (487 aa residues) against SARS-CoV-2. Presently, we drafted the candidate vaccine molecule in the following sequence: “adjuvant-CTL-HTL-B-cell epitopes”. Therein, the initial 45 amino acids represent an adjuvant, followed by a 6 residue linker (EAAAK). The region from 51 to 167 amino acids was represented by CTL epitopes while 168–367 and 368–487 amino acid residues were covered by HTL and B-cell epitopes, respectively (Figure 2).
Figure 2.
Engineered vaccine candidate against non-glycosylated receptor binding domain of COVID-19 (SARS-CoV-2). The designed vaccine candidate is in the sequential manner like- Adjuvant-PADRE sequence-CTL epitopes-HTL epitopes-B-cell epitopes. In the vaccine sequence, the initial 45 amino acids are occupied by an adjuvant (Black color), followed by next 6 amino acids of EAAAK linker (Red color), then from number 51–167 amino acids represent the CTL epitopes (Blue color) whereas 168–367 and 368–487 are covered by HTL epitopes (Green color) and B-cell epitopes (Brown color), respectively. All the linkers have been represented in red color.
3.4. Immunogenicity and allergenicity prediction
An imperative consideration to consider in a vaccine candidate is its immunogenicity, that is, it should have the ability to induce an immune response (cell-mediated/humoral) against the targeted pathogenic organism. Computational findings from our work indicated that the antigenic characteristics of our candidate vaccine construct had a probability score of 0.854 as predicted by the ANTIGENPro server. The preferred average score of antigenic probability of the candidate to be used as vaccine is 0.8 (Pandey et al., 2018). Thus our obtained score for antigenic probability points towards the potentially antigenic nature of the newly designed subunit vaccine candidate. The antigenic probability for the sequence orientation used in the present study was higher than the cases where the epitopes were differently placed.
The AlgPred server was deployed for allergenicity assessment of the vaccine. The basis for data analysis through this server includes following six approaches:(i) IgE mapping, (ii) MEME/Mast motif, (iii, iv) SVM modules for both amino acid and dipeptide composition, (v) BLAST search on ARPs, and (vi) Fusion technique/hybrid approach involving all the parameters. Of the six approaches, the MEME/Mast motif, BLAST search on ARPs, and IgE mapping approaches indicate that the vaccine construct is non-allergenic in nature, whereas the SVM modules working with both amino acid and dipeptide compositions and hybrid approach recognized allergenicity of the vaccine construct. Therefore, we investigated further and performed the allergenicity analysis using the AllerTop server. The results obtained from this tool validated the non-allergen nature of the vaccine. Thus, analyzing the results from both the servers, we concluded that the vaccine candidate was most likely a non-allergen and it would not produce any allergic reaction during the vaccination protocol.
3.5. Assessment of the physicochemical properties
The newly designed vaccine candidate construct is comprised of 487 amino acids along with an approximate molecular weight of 53.64 kDa. Other physiochemical features of the vaccine candidate were computed to determine the vaccine characteristics. The instability index which should be less than 40 was 17.31, demonstrating the stability of our vaccine construct. The significantly basic nature of the vaccine was determined from the theoretical pI which was 9.84. Next, the protein half-life was computed on the basis of N-terminal residue. The data obtained showed that the vaccines half-life of >30 h in mammalian reticulocytes, suggestive of in vivo stability of the vaccine candidate.
The vaccine was found to be moderately thermostable in nature with the computed aliphatic index score of 57.97. The higher aliphatic index was suggestive of its thermostability. The estimated GRAVY (grand average of hydropathicity) score was −0.415. The negative GRAVY score indicated towards hydrophilic behavior having good interaction with the water molecules. Further evaluated parameters have been tabulated in Table 1.
Table 1.
Characteristics of the secondary structure of COVID-19 vaccine candidate calculated through ProtParam tool from IEDB.
| Characteristics of Vaccine | Assessment |
|---|---|
| Number of amino acids | 487 |
| Molecular weight (kDa) | 53.64 |
| Theoretical pI | 9.84 |
| Total number of negatively charged residues (Asp + Glu) |
25 |
| Total number of positively charged residues (Arg + Lys) |
73 |
| Formula | C2452H3685N667O668S14 |
| Extinction coefficient (M-1cm-1) | 94965 |
| Estimated half life | 30 h (mammalian reticulocytes) |
| Instability index | 17.31 |
| Aliphatic index | 57.97 |
| Grand average of hydropathicity (GRAVY) | −0.415 |
3.6. Vaccine structure prediction and validation followed by molecular docking interaction studies
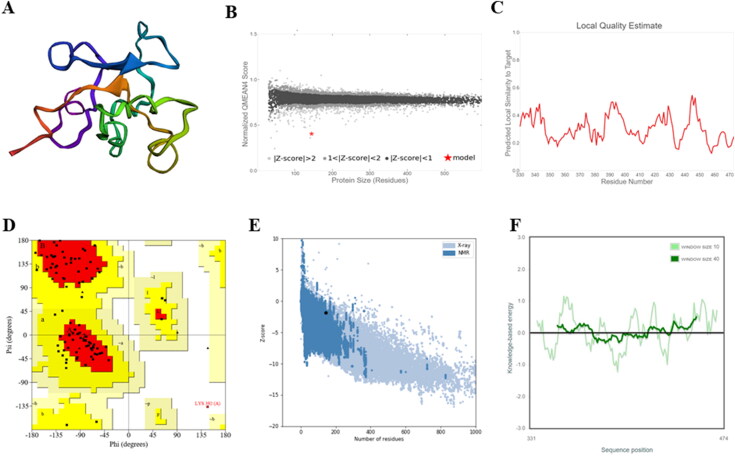
The amino acid sequence of our vaccine candidate was used for evaluating the secondary structural features of the model using PSIPred 4 server. Figure 3 illustrates the predicted results for secondary structure. Since the immunoinformatic approach is heavily dependent on the tertiary structural features of the protein, the 3 D-structure prediction is a crucial step in designing a vaccine. The homology modeling of protein structure is a commonly used technique to generate 3 D models for protein in the absence of experimental structures. Thus, the tertiary structure of the vaccine was estimated via employing an online tool SWISS-MODEL which is a fully automated server with a user-friendly web interface that generates reliable models (Figure 4(A)).
Figure 3.
The secondary structure calculations of multi-epitope vaccine construct using PSIPRED 4.0. The multi-epitope vaccine is comprised of 35.3% α-helix (H, cylinder), 13.5% β-strand (E, arrow), and 51% coil (C, line) as the elements of secondary structure. Percentage of confidence is represented through the bar chart.
Figure 4.
Prediction of tertiary structure model and its validation (A) 3 D model obtained for novel multi-epitope subunit vaccine protein. (B and C) Represents the Z-score and model quality estimation plots calculated using PROCHECK server (D) Ramachandran plot showing the presence of amino acid residues in favored (76.7%), allowed (22.1%), and outlier/disallowed regions (1.2%). (E) ProSA-web z-score (-1.84) plot for the predicted 3 D structure and (F) Energy plot for all residues in the predicted structure.
Various tools such as the PROCHECK and ProSA were used for validating the 3 D model by checking its structural integrity. Figure 4(B,D) gives information on the quality of our vaccine model generated by homology modelling. Ramachandran plot (Figure 4(D)) statistics were generated using the PROCHECK server. The output data generated for the vaccine candidate was indicative of a reasonable quality model of the construct with 76.7% (66 aa) of all the residues in the most favored regions and 22.1% (19 aa) were present in additionally permitted regions. Only 1.2% (1 aa) were occurring in the disallowed plot region.
Additionally, the ProSA server was employed to compute the Z-score and generate an energy plot for the 3 D model. The Z-score (dark spot) value was −1.84, which is well within the range defined for known proteins determined through NMR (dark blue) and X-ray (light blue) (Figure 4(E)), thus predicting a good quality model. The energy plot for the 3 D model has been shown in Figure 4(F) which illustrates the model quality via plotting energies as a function on the x-axis against the residue sequence position on the y-axis. Low/negative energy values of the residues indicated that the model was in the acceptable range.
3.7. Vaccine receptor docking
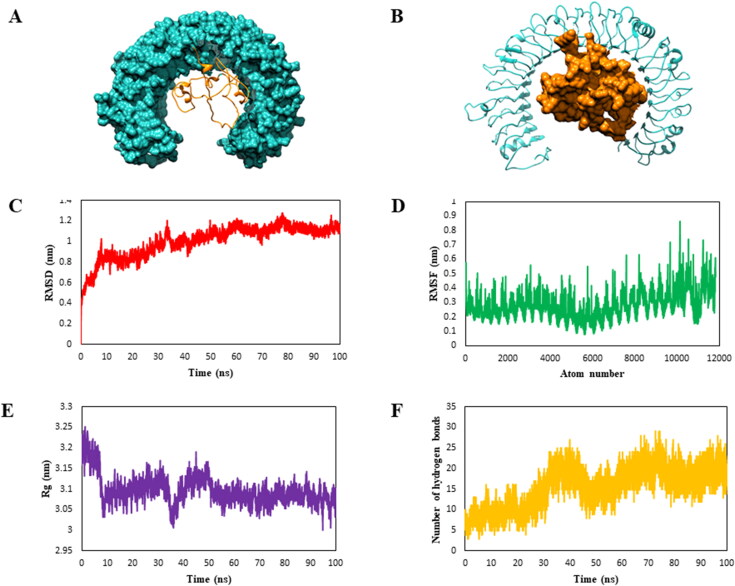
Docking is a molecular modeling method used for predicting the favored ligand molecule orientation to the target protein for building a stable adduct. Molecular docking studies were conducted to approximately calculate binding energy of our vaccine construct to the TLR-3 receptor (Figure 5(A,B)). The binding score of the complex was −217.8 kJ/mol. Furthermore, the vaccine-receptor complex was subjected to molecular dynamics simulation studies.
Figure 5.
Molecular docking and molecular dynamics simulations studies. (A and B) The molecular interaction studies of vaccine candidate with immune receptor TLR3 using HDOCK server was performed and different representation of the vaccine (orange) and TLR3 receptor (orange) are shown here. Molecular dynamics simulation of vaccine and immune receptor TLR3 complex was done for 100 ns using GROMACS. (A) RMSD (root mean square deviation) plots reflect the stability between the vaccine candidate and TLR-3 receptor whereas (B) RMSF (root mean square fluctuation) reflecting the flexibility and fluctuation of the residual atoms in the side chain of docked complex. (C) Radius of gyration is plotted as a function of time to know the structural compactness. (D) Hydrogen bonds of the complex are graphically represented to study the changes in stability of vaccine-receptor complex.
3.8. MD Simulations of vaccine construct and TLR-3 receptor complex
An all-atomistic MD simulation study was performed for 100 ns to evaluate the binding mode, molecular dynamics, and structural stability of our vaccine-TLR-3 docked complex (Figure 5). The interactive parameters between vaccine and TLR-3 were determined at atomic-level, and trajectory analysis was performed to examine the variations in root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and hydrogen bonding over a period of simulation.
The convergence of our complex system was studied using RMSD analysis of all the Cα atoms present in the simulated complex. The RMSD plot for vaccine-receptor complex showed high energy fluctuations up to 40 ns (Figure 5(C)), wherein the resulting RMSD was found to be in the range of 0.2–1.2 nm for the backbone. After 40 ns, the energy was observed to be consistent until the end of the simulation period (100 ns), thus resulting in RMSD values from 1.0–1.2 nm. The magnitude of fluctuations in the simulation trajectories was the basis for our studies. A stable RMSD plot provides for a firm base to perform advanced investigations. The RMSF values for the residual fluctuations were investigated in order to gain insights concerning the alterations in the dynamic behavior of atoms of the docked complex (Figure 5(D)).
We further analyzed the values for Rg to further study the comprehensive aspect of protein. The Rg is usually represented as the mass weight root mean square distance for set of atoms distanced based on their common center of mass. Figure 5(E) shows the Rg plot for the Cα atom of protein versus time at a given temperature (300 K). The hydrogen bonds were chiefly accountable for the maintenance of a stable configuration of the vaccine-receptor protein complex. Therefore, NH bond analysis of the vaccine-TLR3 complex was performed to determine the connection between NH bond formation and complex flexibility over a simulation time period of 100 ns. The number of intermolecular hydrogen bonds increased with time (Figure 5(F)). The results of H-bond plot were suggestive of reduced flexibility, increased strength of binding, and improved vaccine-receptor stability.
3.9. MM-PBSA binding free energy study
The binding energies were computed using the MM-PBSA tool to get detailed insights into molecular interactions of the vaccine candidate and TLR3 complex. The calculations were done for the equilibrium phase between 90–100 ns from the 100 ns output file of MD simulations. The interactions were found to be the most favorable in terms of the sum of various binding energies. The electrostatic energy distribution in molecular mechanics was derived through the high Vander Waal energy (VDW) _-738.16 ± 38.22 kJ/mol, electrostatic energy −8413.94 ± 346.10 kJ/mol and total binding energy of -6592.13 ± 261.07 kJ/mol, which are considered as most significant contributors in the binding of ligand to the target protein. The polar solvation energy (PBSOL) was calculated to be 2656.29 ± 143.86 kJ/mol and SASA energy was −96.33 ± 4.69 kJ/mol which added a vital contribution to the stable binding of the ligand to target protein.
3.10. In-silico analysis of the final vaccine construct using immune simulation
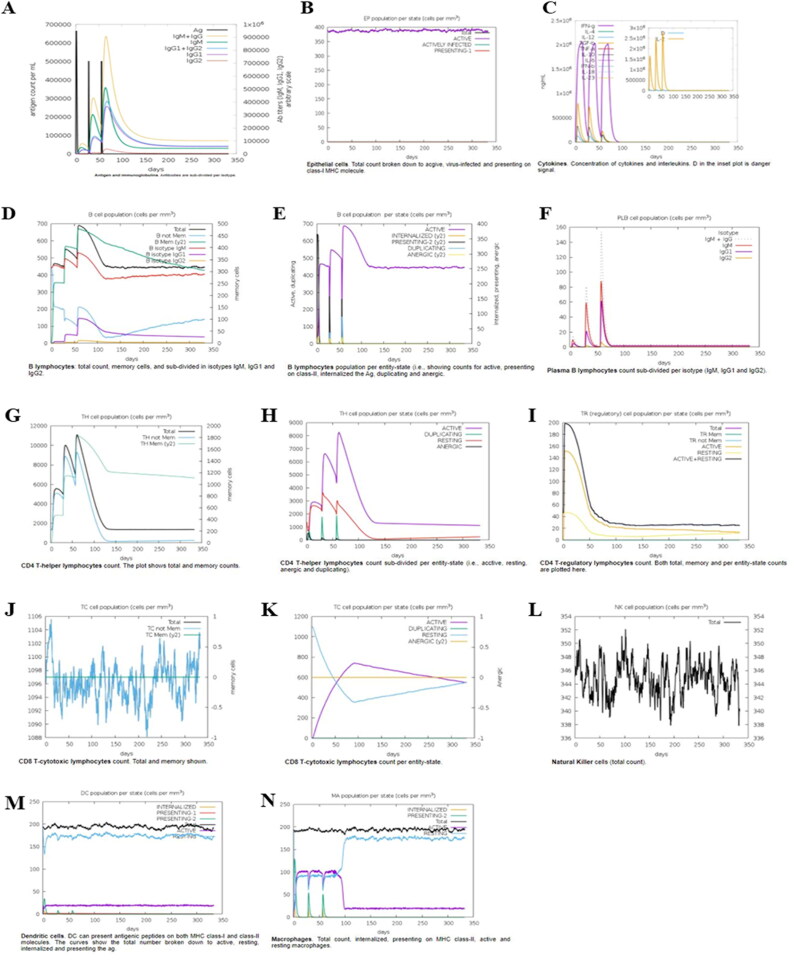
The designed vaccine construct was finally examined for its ability to generate immune response in the body. This study was performed with an online in-silico server for immune simulation (C-ImmSim). The results obtained were in accordance with a typical immune response generated in the body (Figure 6). Increased levels of IgM with every injection indicated the activation of primary immune response. All the associated immune responses (secondary and tertiary) were identified by looking at the elevated levels of IgM, IgG + IgM, and IgG1 + IgG2 antibodies which were followed by subsequent reduction in the concentration of antigen (Figure 6(A)). Additionally, long lasting B-cell isotypes were observed, thus indicating a potential to possibly switch the isotypes and memory formation (Figure 6(D,E)). Likewise, there were increased levels of the T-helper (TH) and cytotoxic (TC) cell populations with corresponding development of memory (Figure 6(G,J), respectively). The macrophage activity was found increasing (Figure 6(N)) whereas the activity levels of dendritic cells were consistent throughout the period of antigenic exposure (Figure 6(M)). The increased levels of interferon (IFN-γ) and interleukin (IL-2) production were indicative of efficient immunoglobulin production, thus implicating towards the generation of a humoral response. Furthermore, various other attributes were analyzed including plasma B lymphocytes (PLB) cell population, CD4+ T regulatory cell population (TR) per state, natural killer (NK) cell population, along with epithelial cells population (EP) per state (Figure 6), wherein a good immune response was observed in the cases of all these cell populations. Overall, the results demonstrated the development of immune memory along with subsequent antigen clearing potential at consecutive exposures.
Figure 6.
In-silico immune response simulation by the candidate vaccine. The simulation is performed for three injections at time steps of 1, 84, and 100. Every step corresponds to 8 h of real-life. (A) Antigen and immunoglobulins (antibodies are sub-divided per isotype); (B) Epithelial-cell population per state; (C) Concentration of cytokines and interleukins; T helper (TH) cell population. (D) B-lymphocytes count; (E) B-lymphocytes count per entity-state; (F) Plasma B-cell population; (G) CD4 T-helper lymphocytes count; (H) CD4 T-helper lymphocytes count per entity-state; (I) CD4 T-regulatory lymphocytes count; (J) CD8 T-cytotoxic lymphocytes count; (K) CD8 T-cytotoxic lymphocytes count per entity-state; (L) Natural killer cells total count; (M) Dendritic cell (DC) can present antigenic peptides on both MHC class-I and class-II molecules; (N) Macrophage cell population.
4. Conclusion
The inhibition of viral infections in humans can be best achieved through vaccination. The recent COVID-19 outbreak which has claimed lives worldwide has spurred the need for urgently developing an effective vaccine for treating this deadly infectious disease. Considering the relative ease of application in our investigative experimentations, we made use of the available immunoinformatic techniques for prospective vaccine candidate designing against the SARS-CoV2 antigen. A multi-epitope subunit peptide was constructed with addition of a suitable adjuvant (beta-defensin-3) at the N-terminal to ensure proper delivery along with proficient epitope immuno-processing. This was followed by connecting the CTL epitopes, HTL epitopes, and B-cell epitopes via suitable linker sequences. The effectiveness of the vaccine construct was assessed by analyzing the physicochemical properties through available online servers. Molecular docking interaction studies were performed to evaluate the specificity of binding for the vaccine candidate to the human TLR3-immune cell receptors. A low energy score indicated an efficient interaction that was both strong and specific between the vaccine candidate and interactive cavity of the human TLR-3 receptor. The MD simulations result showed stable and prolonged binding with non-significant deviation in RMS value. Moreover, a strong immunological response was obtained with the T-cell and B-cell mediated humoral response.
These encouraging findings through computational analysis are indicative that this newly designed multiple-epitope based subunit vaccine candidate has a high probability of being safe and effective in protecting humans from the SARS-CoV-2 infections. However, this proposed vaccine will need experimental validation in suitable animal models to determine its real potency in inducing immunity against the coronavirus. Prophylactic measures for self-defense against the virus include preventive use of animals or avian in human diet. Also, the maintenance of balance in the ecosystem should be reinvestigated.
In summary, the work done highlights a novel vaccine construct which can elicit a strong immune response against the current COVID-19 outbreak to stop the pandemic. This can serve as a prototype for developing vaccines against the other emerging infectious diseases.
Acknowledgements
PRS is thankful to Indian Council of Medical Research (ICMR), India for the senior research fellowship. JG is contended to DBT for fellowship. PS is grateful to TERI School of Advanced Studies for research and management. AG is thankful to the University Grant Commission, India for the faculty recharge position.
Disclosure statement
The authors declare that they have no conflicts of interest.
Author contributions
PRS, JG, PS, VKP and AG conceptualized the study and designed the methods and experimental setup. PRS conducted computational experiments, analyzed the results and drafted the manuscript. JG helped in manuscript compilation. All authors reviewed and approved the manuscript.
References
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., & Walter P. (2002). Lymphocytes and the cellular basis of adaptive immunity. In Molecular biology of the cell (4th ed.). Garland Science. https://www.ncbi.nlm.nih.gov/books/NBK26921/ [Google Scholar]
- Belouzard, S., Millet, J. K., Licitra, B. N., & Whittaker, G. R. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses, 4(6), 1011–1033. 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F. W., Lau, S. K. P., To, K. K. W., Cheng, V. C. C., Woo, P. C. Y., & Yuen, K.-Y. (2015). Middle East Respiratory Syndrome Coronavirus: Another Zoonotic Betacoronavirus Causing SARS-Like Disease. Clinical Microbiology Reviews, 28(2), 465–522. 10.1128/CMR.00102-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Zaro, J. L., & Shen, W.-C. (2013). Fusion protein linkers: Property, design and functionality. Advanced Drug Delivery Reviews, 65(10), 1357–1369. 10.1016/j.addr.2012.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov, I., Flower, D. R., & Doytchinova, I. (2013). AllerTOP - a server for in silico prediction of allergens. BMC Bioinformatics, 14(Suppl 6), S4. S4. 10.1186/1471-2105-14-S6-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes, L., Almazán, F., Sola, I., & Zuñiga, S. (2006). Biochemical aspects of coronavirus replication and virus-host interaction. Annual Review of Microbiology, 60, 211–230. 10.1146/annurev.micro.60.080805.142157 [DOI] [PubMed] [Google Scholar]
- Fauquet, C. M. (2008). Taxonomy, Classification and Nomenclature of Viruses. Encyclopedia of Virology (3rd ed), 9–23. 10.1016/B978-012374410-4.00509-4 [DOI] [Google Scholar]
- Gorbalenya, A. E., Enjuanes, L., Ziebuhr, J., & Snijder, E. J. (2006). Nidovirales: Evolving the largest RNA virus genome. Virus Research, 117(1), 17–37. 10.1016/j.virusres.2006.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N., Regar, H., Verma, V. K., Prusty, D., Mishra, A., & Prajapati, V. K. (2020). Receptor-ligand based molecular interaction to discover adjuvant for immune cell TLRs to develop next-generation vaccine. International Journal of Biological Macromolecules, 152, 535–545. https://doi.org/https//.org/10.1016/j.ijbiomac.2020.02.297 10.1016/j.ijbiomac.2020.02.297 [DOI] [PubMed] [Google Scholar]
- Gupta, S., Kapoor, P., Chaudhary, K., Gautam, A., Kumar, R., & Raghava, G. P. S. (2015). Peptide Toxicity Prediction BT - Computational Peptidology (Zhou P. & Huang J., Eds., pp. 143–157). Springer. 10.1007/978-1-4939-2285-7_7 [DOI] [PubMed] [Google Scholar]
- Janeway, C. A., Jr, Travers, P., Walport, M., Shlomchik, M. (2001). The humoral immune response. In Immunobiology: The immune system in health and disease (5th ed.). Garland Science. https://www.ncbi.nlm.nih.gov/books/NBK10752/ [Google Scholar]
- Kumari, R., Kumar, R., & Lynn, A, Open Source Drug Discovery Consortium (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Lee, C. (2015). Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus. Virology Journal, 12, 193. 10.1186/s12985-015-0421-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, 3(1), 237–261. 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., Choe, H., & Farzan, M. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450–454. 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Zhang, C., Sui, J., Kuhn, J. H., Moore, M. J., Luo, S., Wong, S.-K., Huang, I.-C., Xu, K., Vasilieva, N., Murakami, A., He, Y., Marasco, W. A., Guan, Y., Choe, H., & Farzan, M. (2005). Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. The EMBO Journal, 24(8), 1634–1643. 10.1038/sj.emboj.7600640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston, B., Crimi, C., Newman, M., Higashimoto, Y., Appella, E., Sidney, J., & Sette, A. (2002). A Rational Strategy to Design Multiepitope Immunogens Based on Multiple Th Lymphocyte Epitopes. Journal of Immunology (Baltimore, Md.: 1950), 168(11), 5499–5506. 10.4049/jimmunol.168.11.5499 [DOI] [PubMed] [Google Scholar]
- Mackenzie, J. S., & Smith, D. W. (2020). COVID-19: A novel zoonotic disease caused by a coronavirus from China: What we know and what we don’t. Microbiology Australia, 41(1), 45. 10.1071/MA20013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan, C. N., Zeller, M., Kayala, M. A., Vigil, A., Randall, A., Felgner, P. L., & Baldi, P. (2010). High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics (Oxford, England), 26(23), 2936–2943. 10.1093/bioinformatics/btq551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkelä, M. J., Puhakka, T., Ruuskanen, O., Leinonen, M., Saikku, P., Kimpimäki, M., Blomqvist, S., Hyypiä, T., & Arstila, P. (1998). Viruses and bacteria in the etiology of the common cold. Journal of Clinical Microbiology, 36(2), 539–542. 10.1128/JCM.36.2.539-542.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan, T., Sharma, C., Bhat, A. A., & Rao, D. N. (2013). Modulation of HIV peptide antigen specific cellular immune response by synthetic α- and β-defensin peptides. Vaccine, 31(13), 1707–1716. https://doi.org/https//.org/10.1016/j.vaccine.2013.01.041 10.1016/j.vaccine.2013.01.041 [DOI] [PubMed] [Google Scholar]
- Neurath, A. R. (2008). Immune Response to Viruses: Antibody-Mediated Immunity. Encyclopedia of Virology (3rd ed), 56–70. 10.1016/B978-012374410-4.00591-4 [DOI] [Google Scholar]
- Ojha, R., Pareek, A., Pandey, R. K., Prusty, D., & Prajapati, V. K. (2019). Strategic Development of a Next-Generation Multi-Epitope Vaccine To Prevent Nipah Virus Zoonotic Infection. ACS Omega, 4(8), 13069–13079. 10.1021/acsomega.9b00944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu, M., Annan, A., Corman, V. M., Larbi, R., Anti, P., Drexler, J. F., Agbenyega, O., Adu-Sarkodie, Y., & Drosten, C. (2014). Human coronaviruses associated with upper respiratory tract infections in three rural areas of Ghana. PloS One, 9(7), e99782 10.1371/journal.pone.0099782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik-de-Sousa, C. B., Soares, I. d S., & Rosa, D. S. (2018). Editorial: Epitope Discovery and Synthetic Vaccine Design. Frontiers in Immunology, 9, 826. https://www.frontiersin.org/article/10.3389/fimmu.2018.00826 10.3389/fimmu.2018.00826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R. K., Bhatt, T. K., & Prajapati, V. K. (2018). Novel Immunoinformatics Approaches to Design Multi-epitope Subunit Vaccine for Malaria by Investigating Anopheles Salivary Protein. Scientific Reports, 8(1), 1125. 10.1038/s41598-018-19456-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman, S., & Netland, J. (2009). Coronaviruses post-SARS: Update on replication and pathogenesis. Nature Reviews Microbiology, 7(6), 439–450. 10.1038/nrmicro2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapin, N., Lund, O., Bernaschi, M., & Castiglione, F. (2010). Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLOS One, 5(4), e9862. 10.1371/journal.pone.0009862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S., & Raghava, G. P. S. (2006). AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Research, 34(Web Server issue), W202–W209. 10.1093/nar/gkl343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney, J., Peters, B., Frahm, N., Brander, C., & Sette, A. (2008). HLA class I supertypes: A revised and updated classification. BMC Immunology, 9(1), 1. 10.1186/1471-2172-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, P., Li, W., Xie, J., Hou, Y., & You, C. (2020). Cytokine storm induced by SARS-CoV-2. Clinica Chimica Acta; International Journal of Clinical Chemistry, 509, 280–287. 10.1016/j.cca.2020.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, W., He, L., Zhang, X., Pu, J., Voronin, D., Jiang, S., Zhou, Y., & Du, L. (2020). Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cellular & Molecular Immunology, 17(6), 613–620. 10.1038/s41423-020-0400-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . (2020). Coronavirus disease (COVID-19). Weekly Epidemiological Update, August 16. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200817-weekly-epi-update-1.pdf?sfvrsn=b6d49a76_4
- Yano, A., Onozuka, A., Asahi-Ozaki, Y., Imai, S., Hanada, N., Miwa, Y., & Nisizawa, T. (2005). An ingenious design for peptide vaccines. Vaccine, 23(17–18), 2322–2326. 10.1016/j.vaccine.2005.01.031 [DOI] [PubMed] [Google Scholar]
- Ye, R., Montalto-Morrison, C., & Masters, P. S. (2004). Genetic analysis of determinants for spike glycoprotein assembly into murine coronavirus virions: Distinct roles for charge-rich and cysteine-rich regions of the endodomain. Journal of Virology, 78(18), 9904–9917. 10.1128/JVI.78.18.9904-9917.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]