Abstract
The Coronavirus Disease 2019, caused by the severe acute respiratory syndrome coronavirus 2 is an exceptionally contagious disease that leads to global epidemics with elevated mortality and morbidity. There are currently no efficacious drugs targeting coronavirus disease 2019, therefore, it is an urgent requirement for the development of drugs to control this emerging disease. Owing to the importance of nucleocapsid protein, the present study focuses on targeting the N-terminal domain of nucleocapsid protein from severe acute respiratory syndrome coronavirus 2 to identify the potential compounds by computational approaches such as pharmacophore modeling, virtual screening, docking and molecular dynamics. We found three molecules (ZINC000257324845, ZINC000005169973 and ZINC000009913056), which adopted a similar conformation as guanosine monophosphate (GMP) within the N-terminal domain active site and exhibiting high binding affinity (>−8.0 kcalmol−1). All the identified compounds were stabilized by hydrogen bonding with Arg107, Tyr111 and Arg149 of N-terminal domain. Additionally, the aromatic ring of lead molecules formed π interactions with Tyr109 of N-terminal domain. Molecular dynamics and Molecular mechanic/Poisson–Boltzmann surface area results revealed that N-terminal domain – ligand(s) complexes are less dynamic and more stable than N-terminal domain – GMP complex. As the identified compounds share the same corresponding pharmacophore properties, therefore, the present results may serve as a potential lead for the development of inhibitors against severe acute respiratory syndrome coronavirus 2.
Communicated by Ramaswamy H. Sarma
Keywords: Coronavirus disease 2019, N-terminal domain, guanosine monophosphate, pharmacophore modeling, molecular dynamics simulation
1. Introduction
Coronaviruses (CoVs) belong to the family of Coronaviridae and the order of Nidovirales, which include nonsegmented, enveloped, single-stranded, positive-sense RNA viruses and cause various infectious diseases in vertebrates and humans (Cavanagh, 2007; Fehr & Perlman, 2015; Gorbalenya et al., 2006). According to the latest International Committee of Taxonomy of Viruses (ICTV), CoVs are classified into four genera, namely Alphacoronaviruses, Betacoronaviruses, Gammacoronaviruses and Deltacoronaviruses (King et al., 2018). Alphacoronaviruses and Betacoronaviruses infect the humans and mammals mainly with the gastrointestinal, respiratory and central nervous system, while the gammacoronaviruses and deltacoronaviruses mainly infect birds (Cavanagh, 2007; Fehr & Perlman, 2015; Perlman & Netland, 2009; Woo et al., 2012). Some members of coronaviruses, such as human alphacoronaviruses, 229E (HCoV-229E) and NL63 (HCoV-NL63) and human betacoronaviruses, OC43 (HCoV-OC43) and HKU1 (HCoV-HKU1) are commonly found worldwide and cause mild upper respiratory diseases in most individuals. The other two betacoronaviruses, including severe acute respiratory syndrome coronavirus (SARS-CoV) and middle-east respiratory syndrome coronavirus (MERS-CoV) have emerged in 2002–2003 and 2012, respectively, and both are highly pathogenic to humans (Lancet, 2013; Lee et al., 2003; Marra et al., 2003).
Recently, a novel coronavirus named as 2019 novel coronavirus (2019-nCoV/SARS-CoV-2) has been identified as a causing pathogen for the coronavirus disease 2019 (abbreviated as COVID-19) (Chen et al., 2020; Zhou et al., 2020; Zhu et al., 2020). 2019-nCoV belongs to the betacoronavirus and closely related more to SARS-CoV than MERS-CoV (Wu et al., 2020; Zhou et al., 2020). HCoVs genome consists of open reading frames for five major proteins, which plays a significant role in the virus structure assembly and viral replication, including replicase complex (ORF1ab), spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins (Forni et al., 2017). Among these, N-protein is a major structural protein as it produces the ribonucleoprotein complex upon the binding of the viral RNA genome (Hsieh et al., 2005). It plays an essential role in the regulation of viral RNA synthesis and has functional importance in fundamental aspects of the CoV life cycle, such as encapsidation and replication of virus genomes (Schelle et al., 2005). Moreover, it has the ability to regulate the cellular processes during viral pathogenesis, including actin reorganization, host cell cycle progression and apoptosis (Du et al., 2008; Surjit et al., 2006). Besides these roles, there is less variability in the viral N-gene sequence, and thus, it is a genetically stable protein, an essential prerequisite of an effective drug target candidate (Chang et al., 2014).
Coronavirus N-proteins are organized into three domains, namely an N-terminal RNA-binding domain (NTD), a poorly structured central Ser/Arg (SR)-rich linker and a C-terminal dimerization domain (CTD) (Chang et al., 2013; Chen et al., 2013; Lo et al., 2013). Previous mutagenic studies showed that CoVs NTD were linked at 3′end of the viral genome via electrostatic interactions (Chen et al., 2013; Grossoehme et al., 2009; Keane et al., 2012; Saikatendu et al., 2007). Kang et al. (2020) showed that native NTD has more capability to bind the RNA in the binding pocket as compared to the mutant NTD (Kang et al., 2020). Furthermore, complex crystal structures of HCoV-OC43 N-NTD also provide structural clues about the binding of ribonucleoside 5′-monophosphates, including guanosine monophosphate (GMP), uridine monophosphate (UMP), cytidine monophosphate (CMP) and adenosine monophosphate (AMP) to NTD of N-protein (Lin et al., 2014). Specific molecules (PJ34, H3) were designed using this knowledge against NTD of HCoV-OC43 N-protein, which were verified by the in vitro experiments (Chang et al., 2016; Lin et al., 2014). Due to its importance to the virus cycle and pathogenicity to the host, N-protein of SARS-CoV-2 has become an enticing and essential target for drug development.
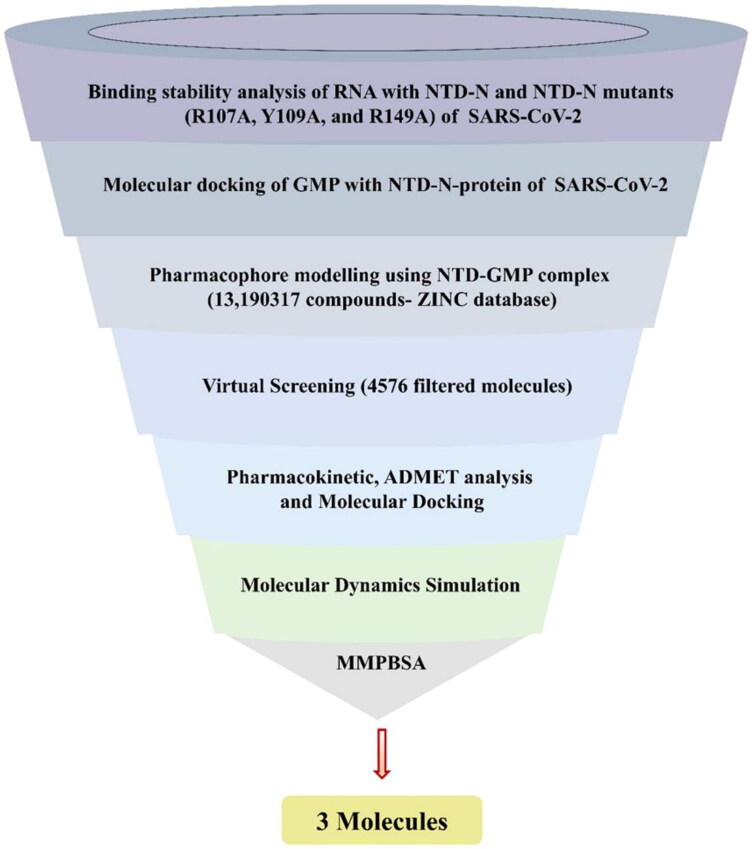
Here, we report the potent molecules against NTD-N-protein of SARS-CoV-2. We have used the GMP as a pharmacophore template in virtual screening for the identification of novel potent compounds. To screen a set of molecules, pharmacophore features of GMP were considered and filtered through the ADMET profile and drug-like properties. Of these, three pharmacokinetically hits (ZINC000257324845, ZINC000005169973 and ZINC000009913056) have the binding affinity higher than GMP with NTD. Molecular dynamics and Molecular mechanic/Poisson–Boltzmann Surface Area (MMPBSA) revealed the stability of NTD–GMP and NTD–ligand(s) complexes. The sequential work done to screen potent molecules against NTD-N-protein of SARS-CoV-2 is represented in Figure 1. Our findings will assist in the development of new drugs, which can interfere with the binding of viral RNA to N-protein.
Figure 1.
Schematic representation implemented to identify the lead molecules against N-terminal domain (NTD) from SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2).
2. Material methods
2.1. Retrieving crystallographic structures
The crystal structure of N-terminal domain (NTD) of nucleocapsid protein (N-protein) of SARS-CoV-2 (PDB ID 6VYO) was downloaded from the RCSB-PDB database to find its potential inhibitors. Due to the existence of this structure in native form, the protein sequence similarity search was performed by NCBI-BLAST server (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against the PDB (https://www.rcsb.org/) in order to screen the complex homologous structure (Altschul et al., 1990). The structure of N-protein-NTD of HCoV-OC43 complexed with its substrate GMP (PDB ID 4LM9) was considered as a point of reference to analyse the active site (Lin et al., 2014).
2.2. Stability of RNA with NTD
Molecular docking of ssRNA with the NTD and the NTD mutant variants was performed using HADDOCK (High Ambiguity Driven protein-protein DOCKing), as done by Chen et al. (2013), De Vries et al. (2010), Kang et al. (2020), Kurkcuoglu et al. (2018), Lin et al. (2014), Saikatendu et al. (2007), Singh et al. (2019). The 3D structure of RNA (5′-UUCGU-3′) was predicted from RNA composer and SimRNAweb (Biesiada et al., 2016; Magnus et al., 2016). NTD mutants (R107A, Y109A and R149A) were generated using COOT (Emsley & Cowtan, 2004). The active site residues (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149) were considered as active residues in HADDOCK to monitor the molecular interactions between RNA and NTD and generated clusters were analyzed and inspected in PyMOL (De Vries et al., 2010; DeLano, 2002). Further, molecular dynamics simulation was performed using amber99sb force-field in GROningen Machine for Chemical Simulations (GROMACS) 2019.2 to determine the stability of protein–RNA complexes (Hornak et al., 2006; van der Spoel et al., 2005). The protonation states of the NTD residues were generated by H++ server (http://biophysics.cs.vt.edu/) at pH 7.5, and the resulted protein was considered for molecular simulation, as done by Anandakrishnan et al. (2012) and Kumari et al. (2020). The systems were placed in a triclinic box with a minimum distance of 1.5 nm between protein and edge of the box. The systems were solvated with TIP3P water model, and genion tool was used to add the counterions to neutralize the systems. The steepest descent algorithm was utilized for 50,000 steps to minimize the protein up to an energy cut of 10.0 kJmol−1. Two-phase of equilibration: constant number of particles, volume and temperature (NVT) and a constant number of particles, pressure and temperature (NPT) were performed for 5,000,000 steps. The systems were kept at NVT using the Parrinello–Rahman barostat pressure coupling method at 300 K for 1 ns (Parrinello & Rahman, 1981). NPT was done for 1 ns using Berendsen thermostat (Berendsen et al., 1984). The covalent bonds were constrained by Linear Constraints Solver (LINCS) (Hess et al., 1997). Long-range interactions were determined using Particle Mesh Ewald (PME) (Abraham & Gready, 2011). Radius cut-off 12 Å was used to compute the short-range (Lennard–Jones and Coulomb) interactions. The final molecular dynamics run was kept for 100 ns, and trajectories were retrieved to generate the Root mean square deviations (RMSD), radius of gyration (Rg), solvent accessible surface area (SASA) and hydrogen bond numbers of NTD–RNA and NTD mutants–RNA complexes, as done by Malik et al. (2019).
2.3. MMPBSA binding free energy calculation
The binding energy of NTD–RNA, R107A–RNA, Y109A–RNA and R149A–RNA complexes were calculated by Molecular Mechanics/Poisson–Boltzmann Surface Area (MMPBSA) suite in GROMACS, as done by Kumari et al. (2014) and Pandit et al. (2018). Molecular simulation trajectories generated at every 10 ps of protein–RNA complexes were used for MMPBSA. In GROMACS, the g_mmpbsa tool was used to compute the binding free energy of protein–RNA complexes as:
Where, ΔGcomplex, ΔGprotein and ΔGligand is the total free energy of protein–RNA complex, protein and ligand in a solvent, respectively.
Here, we have used 2000 frames extracted from the last 20 ns (80–100 ns) of molecular dynamics to compute the solvation free energy (polar and nonpolar) and molecular mechanics potential energy (electrostatic and van der Waals interactions) for estimation of total binding affinity of NTD–RNA and NTD mutants–RNA complexes. Per-residue decomposition analysis is a quantitative analysis to assess the contribution of important residues in protein–ligand complexes. In this study, binding energy decomposition analysis was done from the last 20 ns of molecular dynamics to determine the binding potential of crucial important residues of the active site (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149) with RNA in NTD–RNA and NTD mutants–RNA complexes.
2.4. Pharmacophore modeling
For the screening of potent compounds against NTD, GMP was taken as a template molecule for the pharmacophore model. The 3D structure of SARS-CoV-2 (PDB ID 6VYO) was used as a starting model for further in silico studies. The water and the other molecules were omitted, and the protein structure was prepared by adding the essential hydrogen atoms and Kollman charges (8.0) using the AutoDock Tools (Morris et al., 2009). GMP was extracted from the PDB ID 4LM9 and processed by adding polar hydrogens and gasteiger charges (0.5003). The protein and ligand were saved in .pdbqt and the molecular docking was performed using AutoDock Vina and AutoDock Tools (Morris et al., 2009; Trott & Olson, 2010). The 50 conformations of GMP were predicted using the Lamarckian genetic algorithm in AutoDock Tools, as done by Singh et al. (2017). The best conformation of GMP with NTD was selected on the basis of binding energy and corresponding interactions with NTD. The multiple sequence alignment (MSA) of NTD-N-protein of SARS-CoV-2 with other NTD from other viruses was generated using Clustal Omega and illustrated using ESPript (Gouet et al., 1999; Sievers et al., 2011).
Further, the structure-based pharmacophore modeling was conducted by the Pharmit server, which directly works on the ligand–protein complex (Sunseri & Koes, 2016). The 3D structure of NTD–GMP complex was loaded into the Pharmit web server as a query to identify the compounds with pharmacophore features from the ZINC library (Sterling & Irwin, 2015). Shape filters were applied for the protein and ligand to filter out the compounds in the database before conducting the pharmacophore search. Further, the total 4576 compounds with RMSD less than 1.5 Å from the pharmacophore features of query molecule were saved and considered for virtual screening.
2.5. Virtual screening
Structure-based virtual screening (SBVS) was performed by utilizing the AutoDock Vina in PyRx 0.8 platform (Dallakyan & Olson, 2015; Trott & Olson, 2010). The total 4576 hit compounds with the pharmacophore properties of GMP, were considered as ligands for the virtual screening against NTD. All the ligands were energy minimized by applying the Universal Force Field (UFF) and converted into .pdbqt by OpenBabel (O'Boyle et al., 2011). The grid map was centred with a dimension of 58 × 60 × 57 Å that covers the active site residues of the protein. Each ligand was docked in the binding site of NTD and scored with binding affinity. Top-ranked molecules were analyzed using PyMOL, and best conformations with high affinity were saved (DeLano, 2002).
2.6. Pharmacokinetic and ADMET analysis
The pharmacophore screened compounds were refined by means of Lipinski’s rule of five for the drug-like criteria, including molecular weight, log P, number of rotatable bonds, hydrogen bond acceptors and donors (Lipinski, 2004). Further, to evaluate the possible impact of the selected compounds on humans, the ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) studies were performed by the pkCSM server, as done by Dhankhar et al. (2020), Dhankhar et al. (2020), Pires et al. (2015). The filtered compounds, with Lipinski’s rule of five and ADMET properties, were further chosen for the molecular docking to determine the appropriate orientation and binding affinities with NTD.
2.7. Molecular docking
Molecular docking simulation studies for three lead compounds (ZINC000257324845, ZINC000005169973 and ZINC000009913056) were performed using the AutoDock Tools. Hydrogen atoms and Kollman charges were added to the protein receptor. Polar hydrogen atoms and Gasteiger charges for ZINC000257324845 (0.0007), ZINC000005169973 (0.0002) and ZINC000009913056 (–0.0002) were added and saved in .pdbqt format. The molecular grid was created around the active site residues (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149) of NTD-N-protein of SARS-CoV-2. The grid box dimensions were set to be 60 × 60 × 60 Å and centre coordinates were −7.93 × 2.48 × 3.81 with 0.375 Å grid space value. All the generated 50 conformations for each lead compound during molecular docking were analyzed, and figures were prepared in PyMOL 2.0.5 and Maestro (DeLano, 2002; Release, 2016).
2.8. Molecular dynamics and MMPBSA
Molecular dynamics simulation of NTD with GMP, ZINC000257324845, ZINC000005169973 and ZINC000009913056 were performed to assess the structural and dynamics variation at an atomistic level. Protein topologies and coordinates files were generated using GROMACS 2019.2 along with GROMOS96 43a1 force field, as done by Kesari et al. (2020), Singh et al. (2018), van Gunsteren et al. (1996), van der Spoel et al. (2005). Total four systems (NTD–GMP, NTD–ZINC000257324845, NTD–ZINC000005169973 and NTD–ZINC000009913056) were generated and kept for molecular simulation of 100 ns. The topology of ligands (GMP, ZINC000257324845, ZINC000005169973 and ZINC000009913056) were predicted by PRODRG webserver, as done by Kumar et al. (2020), Schüttelkopf and Van Aalten (2004). The partial atomic charges for ligands were computed by density functional theory (DFT) analysis using Lee–Yang–Parr correlation functional (B3LYP) method with a 6-311 G (d,p) basis set in Gaussian 16, as done by Frisch et al. (2016), Jain et al. (2004), Lee et al. (1988), Saini et al. (2019), Schlegel (1982). The protein–ligand complexes were solvated using a simple point charge (SPC) water model in a triclinic box of volume (234.61 nm3) with a minimum distance of 1.0 nm between atoms of protein and edge of the box. Three chlorides (Cl–) ions were added using genion tool to neutralize the system. The final production run was done for 100 ns, and coordinates were updated every 10 ps, as done by Kesari et al. (2020). The trajectories were analyzed by visual molecular dynamics (VMD) and XMGRACE (Humphrey et al., 1996). Further details of the molecular simulation are aforementioned section (stability of RNA with NTD). In the present study, the binding energy calculations of protein–ligand complexes were done using MMPBSA, as aforementioned in subsection 2.3 (MMPBSA binding free energy calculation) (Kumari et al., 2014). We have not calculated the entropy as it is computational expensive and we were interested in relative binding of GMP and the identified compounds with NTD. Further, the contributions of active site residues (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149), per-residue decomposition analysis were determined from the last 20 ns of molecular simulation of NTD–GMP and NTD–inhibitor(s) complexes.
3. Results
3.1. Retrieving crystallographic structures
As mentioned earlier, NTD-N-protein CoV is a potential target for the treatment of CoV diseases. The crystal structure of NTD-N-protein of SARS-CoV-2 (PDB ID 6VYO) was obtained from the PDB and used as the target sequence. NTD structure consists of antiparallel β sheets and three regions named as: basic finger, basic palm and acidic wrist. Right-handed (loops-β-sheet core-loops) sandwiched fold is conserved among the NTD of CoVs (Kang et al., 2020). NTD structure from HCoV-OC43 (PDB-ID 4LM9) complexed with RNA substrate (GMP) reported that residues Ser64, Phe66, Gly68, His104, Arg122, Tyr124, Tyr126 and Arg164 were noteworthy interacting residues for the formation of NTD–GMP complex (Lin et al., 2014). The structural superposition of NTD-N-protein of SARS-CoV-2 (PDB ID 6VYO) with HCoV-OC43 (PDB ID 4LM9) complexed with RNA substrate (GMP) showed that residues (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149) in the NTD-N-SARS-CoV-2 are the corresponding to GMP interacting residues in NTD-HCoV-OC43. Figure S1 Supporting Information illustrates that these residues are conserved among all the NTD of CoV.
3.2. Stability of RNA with NTD
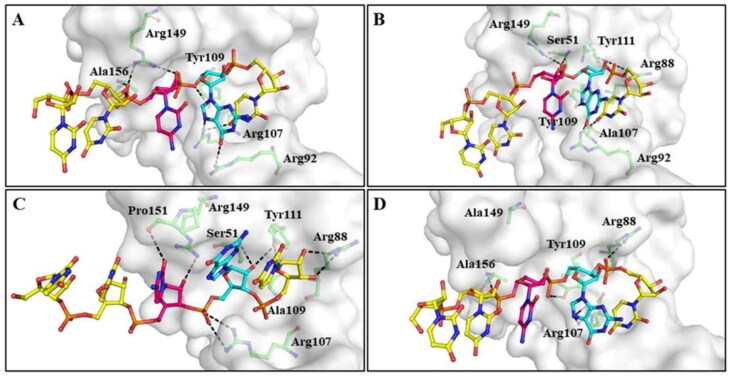
The predicted 3D structure of RNA was docked with the RNA binding region (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149) of NTD and their mutants. From the obtained clusters, the best cluster was selected based on their HADDOCK score, RMSD from the overall lowest energy structure, cluster size and corresponding interactions between protein and RNA. Table 1 represents the hydrogen-bonding interactions and binding energy of the selected clusters. The docking results revealed that RNA was interacted with the wild-type NTD of N-protein more strongly as compared to the mutants NTD. Wild-type NTD–RNA complex was stabilized by the formation of hydrogen bonds with Arg92, Arg107, Tyr109, Arg149 and Ala156 of protein with RNA (Figure 2(A)). On the mutation of R107A, RNA was interacted with Ser51, Arg88, Arg92, Tyr109, Tyr111 and Arg149 residues through hydrogen bonds (Figure 2(B)). While in the case of Y109A, RNA was hydrogen-bonded with amino acid residues (Ser51, Arg88, Arg107, Tyr111, Arg149 and Pro151) of protein (Figure 2(C)). On the mutation of R149A, hydrogen bonds were formed by the RNA with Arg88, Arg107, Tyr109 and Ala156 residues of protein (Figure 2(D)). The molecular docking results revealed that residues (Ser51, Arg88, Arg92, Arg107, Tyr109, Tyr111, Arg149 and Ala156) are essential for the binding of RNA to the NTD of SARS-CoV-2.
Table 1.
Molecular docking of NTD with RNA.
| S.No. | Complex | Native–RNA | R107A–RNA | Y109A–RNA | R149A–RNA |
|---|---|---|---|---|---|
| 1 | H.S. | – 60.5 +/– 2.9 | – 45.9 +/– 3.1 | – 46.4 +/– 7.2 | – 46.7 +/– 1.2 |
| 2 | RMSD | 1.2 +/– 0.2 | 3.3 +/– 0.7 | 3.1 +/– 0.2 | 3.0 +/– 0.4 |
| 3 | V.E | – 31.9 +/– 5.5 | – 26.8 +/– 3.8 | – 27.3 +/– 5.5 | – 26.9 +/– 2.5 |
| 4 | E.E | – 186.7 +/– 20.0 | – 133.3 +/– 20.2 | – 150.7 +/– 21.8 | – 136.9 +/– 13.8 |
| 5 | D.E | 8.8 +/– 3.0 | 6.7 +/– 1.5 | 11.1 +/– 2.3 | 5.7 +/– 1.8 |
| 6 | Hydrogen bond interacting residue | Arg92, Arg107, Tyr109, Arg149 and Ala156 | Ser51, Arg88, Arg92, Tyr109, Tyr111 and Arg149 | Ser51, Arg88, Arg107, Tyr111, Arg149 and Pro151 | Arg88, Arg107, Tyr109 and Ala156 |
Molecular docking results show the Haddock score (H.S.), RMSD from overall lowest energy structure (RMSD), van der Waals energy (V.E.), Electrostatic Energy (E.E.) and Desolvation Energy (D.E.). All the energy mentioned in the table is in kcalmol−1.
Figure 2.
Molecular interaction between RNA and active site residues of SARS-CoV-2 NTD and NTD mutants. (A) Wild-type NTD, (B) NTD–R107A (C) NTD–Y109A and (D) NTD–R149A. A single-stranded RNA (5′-UUCGU-3′) was predicted from RNA composer and SimRNAweb. NTD (white surface) was represented with active site residues in green ball and sticks. Riboucleotides of RNA were displayed in ball and sticks (Uracil in yellow, cytosine in magenta and guanine in cyan color). Hydrogen bonds were shown with black dashed lines.
Further, the stability of NTD–RNA and NTD mutants–RNA complexes were assessed using molecular dynamics in triplicate by amber99sb force field in GROMACS. RMSD for Cα backbone atoms for NTD, NTD mutants, NTD–RNA and NTD mutants–RNA complexes were determined, as shown in Supporting Information Figure S2. NTD and NTD mutants achieved convergence at 22 ns around 0.19 nm and remained stable up to the end of molecular simulation, as shown in Supporting Information Figure S2(A). While NTD mutants–RNA complexes showed higher RMSD as compared to NTD and NTD–RNA complexes (Supporting Information Figure S2(B)). Average RMSD of NTD, R107A, Y109A, R149A, NTD–RNA, R107A–RNA, Y109A–RNA and R149A–RNA complexes are 0.18 ± 0.03, 0.20 ± 0.03, 0.19 ± 0.03, 0.19 ± 0.03, 0.16 ± 0.03, 0.28 ± 0.05, 0.22 ± 0.04 and 0.21 ± 0.03 nm, respectively, as shown in Supporting Information Table S1. In Supporting Information Figure S3(A), it can be clearly observed that NTD and NTD mutants were compact and stable. Supporting Information Figure S3(B) shows the higher Rg values of NTD mutants–RNA complexes than NTD and NTD–RNA complex. NTD mutants–RNA complexes exhibit average Rg values in the range of 1.59 ± 0.02 and 1.60 ± 0.02 nm, as shown in Supporting Information Table S1. The Rg analysis suggested that binding of RNA to NTD mutants form a less stable NTD mutants–RNA complex than NTD–RNA complex.
Less SASA value of NTD–RNA complex indicated that binding of RNA to NTD form a stable protein–RNA complex, as shown in Supporting Information Figure S4. SASA plot reveals that the mutants–RNA complexes are less stable as compared to NTD and NTD–RNA complex. Further, intraprotein and intermolecular hydrogen bond plots of NTD, NTD mutants, NTD–RNA and NTD mutants–RNA complexes are shown in Supporting Information Figure S5. NTD–RNA complex exhibit a higher number of intraprotein hydrogen bonds as compared to NTD, NTD mutants and NTD mutants–RNA complex, as shown in Supporting Information Figure S5(A,B). The average number of intraprotein hydrogen bonds for NTD, R107A, Y109A, R149A, NTD–RNA, R107A–RNA, Y109A–RNA and R149A–RNA complexes are 77.32 ± 4.74, 76.93 ± 4.71, 76.78 ± 4.81, 77.21 ± 4.59, 79.71 ± 4.83, 72.39 ± 4.64, 71.85 ± 4.17 and 71.84 ± 4.75, respectively, as shown in Supporting Information Table S1. Supporting Information Figure S5(C) shows that the NTD–RNA complex is stabilized by a higher number of intermolecular hydrogen bonds than NTD mutants–RNA complexes. In Supporting Information Figure S5(D), the presence of more high-affinity intermolecular hydrogen bonds in NTD–RNA complex than NTD mutants–RNA complexes affirms that the protein–RNA complex is more stable than protein mutants–RNA complexes. Overall, molecular docking and simulation results confirm that active site residues (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149) of NTD play a major role in the formation of stable NTD–RNA complex.
3.3. MMPBSA binding free energy calculation
Molecular mechanic/Poisson–Boltzmann Surface Area (MMPBSA) was used to calculate the binding free energy of NTD–RNA and NTD mutants–RNA complexes. The binding affinity of NTD–RNA, R107A–RNA, Y109A–RNA and R149A–RNA complexes are −973.97 +/– 8.92, −677.67 +/– 9.13, −614.28 +/– 7.16 and −690.63 +/– 7.73 kJmol−1, respectively, as shown in Table 2. In order to quantify the interactions of important amino acid residues of NTD with RNA towards binding affinity, per residue decomposition analysis, was done using MMPBSA. We have computed the binding affinity contributions of active site residues (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149) of NTD in NTD–RNA, R107A–RNA, Y109A–RNA and R149A–RNA complexes. In Supporting Information Table S2, it can be clearly seen that binding energy contribution of active site residues of protein for RNA is decreased in NTD mutants–RNA complexes as compared to NTD–RNA complex. In NTD–RNA complex, Arg107 and Arg149 of NTD contributed 109.92 and 107.52 kJmol−1 for binding of RNA, which is decreased by 97% in R109A–RNA and R149A–RNA complex, respectively. Hence, MMPBSA analysis confirmed that active site residues (Arg107, Tyr109 and Arg149) of NTD played a major role in the binding of RNA to form a lower energy stable NTD–RNA complex.
Table 2.
MMPBBSA binding free energy (kJmol−1) of NTD–RNA, R107A–RNA, Y109A–RNA and R149A–RNA calculated by MMPBSA from the last 20 ns (80–100 ns) of molecular simulation.
| Energy (kJmol-1) | NTD–RNA | R107A–RNA | Y109A–RNA | R149A–RNA |
|---|---|---|---|---|
| van der Waals energy | −158.52 +/– 1.93 | −121.12 +/– 3.38 | −102.38 +/– 5.62 | −106.71 +/– 3.85 |
| Electrostatic energy | –1570.49 +/– 7.86 | –1512.59 +/– 7.82 | –1421.19 +/– 9.27 | –1352.85 +/– 8.63 |
| Polar solvation energy | 780.86 +/– 8.78 | 972.46 +/– 9.92 | 929.56 +/– 8.86 | 792.17 +/– 7.35 |
| SASA energy | −25.82 +/– 0.61 | −16.42 +/– 0.46 | −22.27 +/– 0.32 | −23.24 +/– 0.28 |
| Binding energy | −973.97 +/– 8.92 | −677.67 +/– 9.13 | −616.28 +/– 7.16 | −690.63 +/– 7.73 |
3.4. Pharmacophore modeling
The GMP, a ribonucleotide, was used as a template to screen the potent compounds against NTD of SARS-CoV-2. The molecular docking study of GMP with NTD-N-SARS-CoV-2 demonstrated that GMP was interacted with the expected residues of NTD-N-SARS-CoV-2 with −5.0 kcalmol−1 binding energy, as shown in Table 3. The best-docked conformation of GMP in the respective pocket of NTD-N-SARS-CoV-2 was shown in Figure 3(A) and Supporting Information Figure S6. Further, the structure-based pharmacophore modeling was performed using the validated complex of NTD-N-SARS-CoV-2 with GMP, where the NTD-N-SARS-CoV-2 and GMP were used as receptor and ligand, respectively. Total five pharmacophore properties, including three hydrogen bond acceptors (orange), 1 aromatic group (purple) and 1 hydrophobic feature (green), with the radius of 1.0 Å were considered to screen the compounds, as shown in Supporting Information Figure S7. ZINC database was screened using the constructed pharmacophore model and 8192 compounds were identified, which shares the pharmacophore features. Further, these identified compounds were refined with the Lipinski’s rule of five, RMSD and energy minimization. With the RMSD of less than 1.5 Å and binding affinity of more than −5.0 kcalmol−1, a total of 4576 hits were eventually saved (Supporting Information file 2).
Table 3.
The binding affinities (kcalmol−1) and molecular interactions of the selected compounds with active site residues of NTD.
| S. No. | Compound | Binding energy (kcalmol–1) |
|
|---|---|---|---|
| AutoDock VINA | AutoDock Tool | ||
| 1 | GMP | −5.5 | −2.6 |
| 2 | ZINC000257324845 | −8.3 | −7.4 |
| 3 | ZINC000005169973 | −8.1 | −6.4 |
| 4 | ZINC000009913056 | −8.0 | −5.6 |
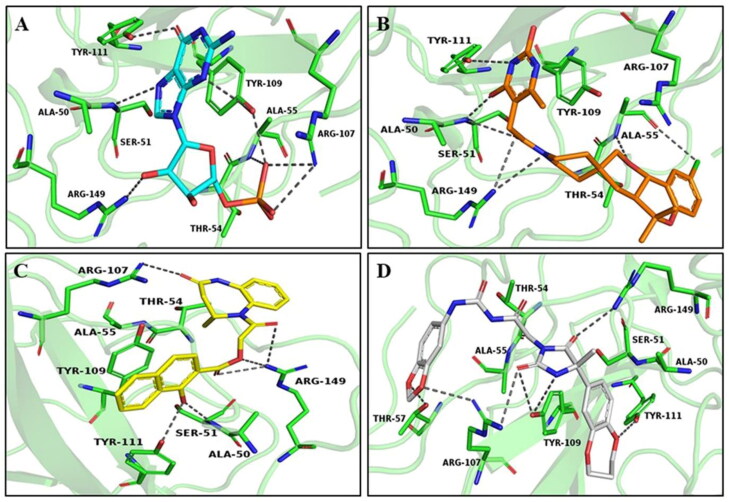
Figure 3.
Binding interaction of best-docked pose of compounds with NTD protein. (A) GMP (cyan), (B) ZINC000257324845 (orange), (C) ZINC000005169973 (yellow) and (D) ZINC000009913056 (grey). NTD is displayed as the cartoon, and key residues are shown in green. Hydrogen bonds are represented with gray dashed lines.
3.5. Virtual screening
The virtual screening was performed to identify novel compounds against the NTD-N-protein, using the AutoDock Vina in PyRx 0.8. The downloaded selected compounds from the ZINC database having a high negative score and less RMSD values were converted from .sdf format to .pdbqt format using Open Babel. Further, these compounds were docked into the GMP binding site and ranked on the basis of binding energy with the NTD-N-protein. Ligands showing binding affinity in the range of −7.5 to −8.3 kcalmol−1 were considered for further study.
3.6. Pharmacokinetic and ADMET analysis
In silico ADMET prediction was performed for the top-ranked results using the pkCSM server to predict the overall risks of absorption, distribution, metabolism, excretion and toxicity. Three hits (ZINC000257324845, ZINC000005169973 and ZINC000009913056) were successfully fulfilled the drug-like properties as per the Lipinski’s rule of five, including molecular weight, Log P, hydrogen bond donor and acceptor (Supporting Information Table S3). A promising ADMET profile is required for the compounds in drug discovery (Hop et al., 2008). For this purpose, pharmacokinetic parameters such as aqueous solubility (log S), skin permeability coefficient (log Kp), log BB, CNS permeability, number of metabolic reactions, and so forth were calculated and enlisted in Supporting Information Table S4. The results indicated that all three compounds were soluble in water, and the aqueous solubility (S) of a compound substantially affects its absorption and distribution properties. All compounds showed skin permeability and also absorbed by the human intestine. In addition, they were able to penetrate the BBB and CNS after oral administration. CYP enzymes, various CYP450 substrates and inhibitors played a fundamental role in the metabolism of the drug. Further, metabolism analysis revealed that none of the compounds were substrates of CYP2D6 substrate. Toxicity assessment has shown that all compounds are nontoxic in nature to humans.
3.7. Molecular docking
The molecular docking study of all three pharmacokinetically screened compounds with NTD revealed that all compounds showed higher binding energy than GMP with NTD and presented in Table 3. ZINC000257324845 was found stable at the active site of NTD by forming H-bonds (Ser51, Ala55, Tyr111 and Arg149), π-π (Tyr109) and π-alkyl (Tyr109 and Ala156), as shown in Figure 3(B) and Supporting Information S6(B). ZINC000005169973 was able to make H-bond (Ser51, Arg107, Tyr111 and Arg149), π- donor (Tyr109, Arg107 and Arg149), π- sigma (Ala156) and π-alkyl (Ala50 and Ala90) (Figure 3(C) and Supporting Information Figure S6(C)). ZINC000009913056 was able to form H-bond (Thr57, Arg107, Tyr109, Tyr111 and Arg149), π- stacked (Tyr109) and π-alkyl (Ala50 and Ala90) (Figure 3(D) and Supporting Information Figure S6(D)).
3.8. Molecular dynamics and MMPBSA
Molecular dynamics simulation was carried out in triplicates to assess the flexibility and stability of NTD with identified inhibitor(s). Therefore, in this study, molecular dynamics were employed to evaluate the stable and static interactions of protein–ligand complexes by examining the various molecular simulation results like RMSD, Root mean square fluctuation (RMSF), Rg, SASA, hydrogen bond formation and PCA.
3.8.1. Root mean square deviation
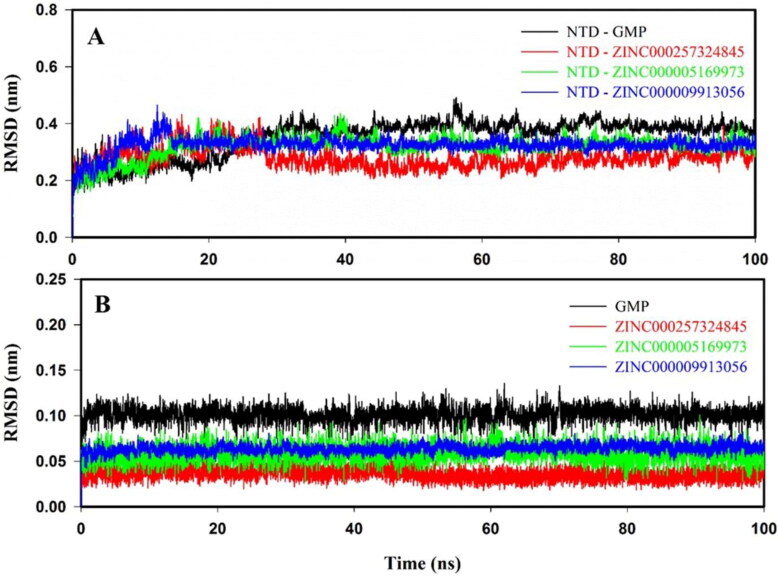
The atomistic dynamics movements and conformational variations of Cα backbone atoms of NTD–GMP and NTD–inhibitor(s) complexes were calculated by RMSD. In Figure 4(A), it is clearly seen that all the protein–ligand complexes showed an initial sharp in RMSD values during 5–10 ns and attained equilibrium at 30 ns, and systems remained stable throughout the molecular simulation of 100 ns. NTD–inhibitor(s) complexes exhibited an RMSD in the range of 0.22 to 0.34 nm, which is lesser than NTD–GMP (0.37 nm) complex. The average RMSD values of NTD–GMP and NTD–inhibitor(s) complexes are shown in Supporting Information Table S5. Ligand RMSD of GMP and inhibitors are represented in Figure 4(B). The average ligand RMSD for GMP, ZINC000257324845, ZINC000005169973 and ZINC000009913056 are 0.10 ± 0.02, 0.04 ± 0.02, 0.06 ± 0.01 and 0.07 ± 0.01 nm, as shown in Supporting Information Table S5. Overall, RMSD results illustrated that the binding of identified inhibitor(s) at the active site of NTD is stable and formed more stable NTD–inhibitor(s) complexes as compared to NTD–GMP complex.
Figure 4.
Root mean square deviation (RMSD) of protein–ligand (s) complexes and ligands only. RMSD plots: (A) NTD–GMP (black), NTD–ZINC000257324845 (red), NTD–ZINC000005169973 (green) and NTD–ZINC000009913056 (blue); (B) Ligands: GMP (black), ZINC000257324845 (red), ZINC000005169973 (green) and ZINC000009913056 (blue) for the molecular dynamics of 100 ns at 300 K.
3.8.2. Root mean square fluctuation
The RMSF profile of NTD–inhibitor(s) complexes is almost comparable to NTD–GMP, as shown in Supporting Information Figure S8. The average RMSF values for NTD–GMP, NTD–ZINC000257324845, NTD–ZINC000005169973 and NTD–ZINC000009913056 are 0.17 ± 0.07, 0.15 ± 0.06, 0.15 ± 0.07 and 0.16 ± 0.07 nm, respectively, as shown in Supporting Information Table S5. RMSF analysis suggested that inhibitor(s) were well fitted at the active site of NTD and form a stable NTD–inhibitor(s) complex.
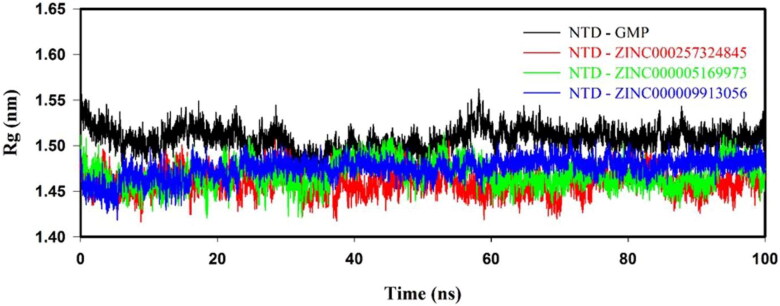
3.8.3. Radius of gyration
NTD–inhibitor(s) complex showed a lesser Rg value than NTD–GMP complex, as shown in Figure 5. NTD–inhibitor(s) complexes showed average Rg values in the range of 1.46 ± 0.01 to 1.47 ± 0.01 nm, which was lesser than NTD–GMP (1.50 ± 0.01 nm) complex, as shown in Supporting Information Table S5. Rg results indicated that the binding of inhibitor(s) to NTD form a higher stable NTD–inhibitor(s) complex as compared to NTD–GMP complex.
Figure 5.
Radius of gyration (Rg) profile of NTD with GMP, ZINC000257324845, ZINC000005169973 and ZINC000009913056 from the molecular simulation of 100 ns at 300 K.
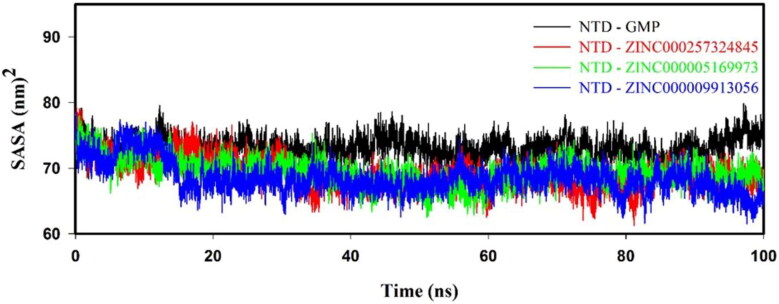
3.8.4. Solvent accessible surface area
SASA plot reveals that values of NTD–inhibitor(s) complexes are smaller than NTD–GMP complex, as shown in Figure 6. Average SASA of NTD–GMP, NTD–ZINC000257324845, NTD–ZINC000005169973 and NTD–ZINC000009913056 complexes were 73.24 ± 1.82, 69.41 ± 2.26, 69.68 ± 2.31 and 68.77 ± 2.12 nm2, respectively, as shown in Supporting Information Table S5. SASA analysis implied that NTD–inhibitor(s) complexes were more stable and compact than NTD–GMP complex.
Figure 6.
Solvent accessible surface area (SASA) graph for protein–ligand (NTD–GMP, NTD–ZINC000257324845, NTD–ZINC000005169973 and NTD–ZINC000009913056) complexes.
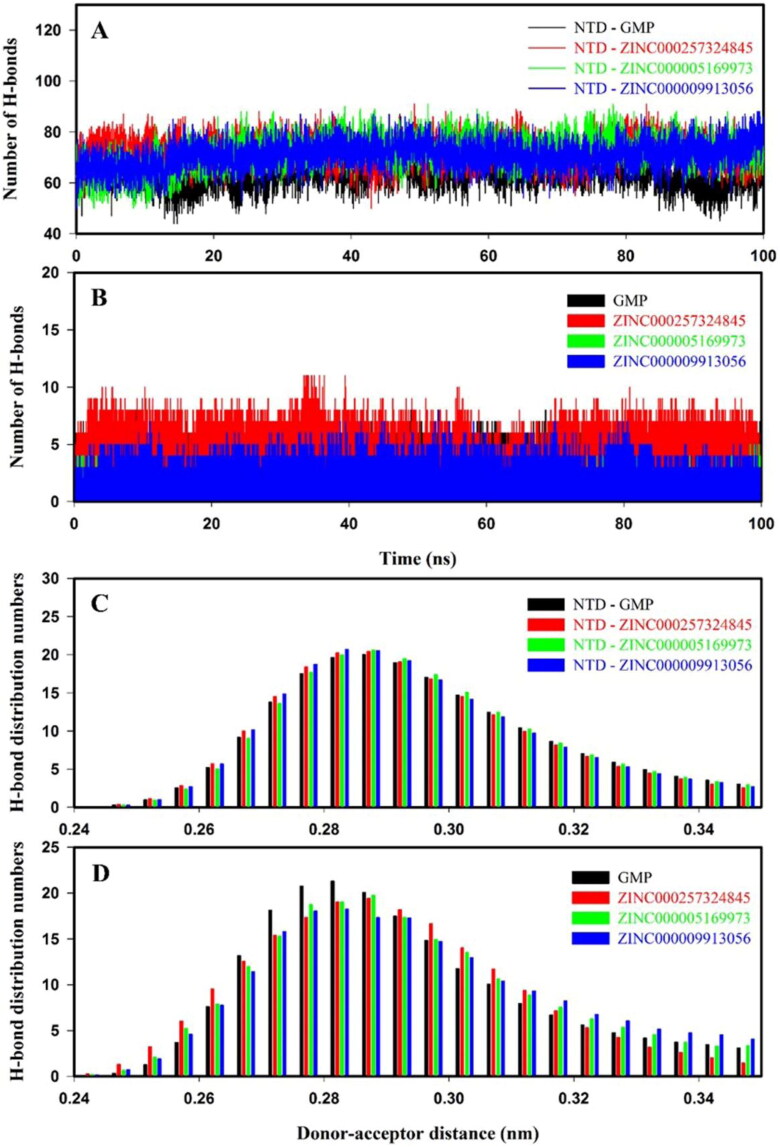
3.8.5. Hydrogen bond analysis
Intraprotein and intermolecular hydrogen bond plots of NTD–GMP and NTD–inhibitor(s) complexes are shown in Figure 7. NTD–inhibitor(s) complexes have higher intraprotein hydrogen bonds as compared to NTD–GMP as shown in Figure 7(A). The average number of intraprotein hydrogen bonds between protein–ligand(s) complexes were in a range of 63.59 ± 5.41 to 72.11 ± 5.71, as shown in Supporting Information Table S5. As shown in Figure 7(B), intermolecular hydrogen bonds between NTD and inhibitor(s) were sustained during the course of molecular simulation. The average number of intermolecular hydrogen bonds between NTD–GMP and NTD–inhibitor(s) complexes are shown in Supporting Information Table S5. The hydrogen bonding affinity of NTD–inhibitor(s) complexes were similar to NTD–GMP complex, as shown in Figure 7(C,D). Overall, hydrogen bond results affirmed that binding of inhibitor(s) to NTD form stable NTD–inhibitor(s) complexes.
Figure 7.
Hydrogen bond numbers and distribution patterns of NTD–GMP (black), NTD–ZINC000257324845 (red), NTD–ZINC000005169973 (green) and NTD–ZINC000009913056 (blue) for molecular dynamics of 100 ns at 300 K. The number of hydrogen bonds analysis: (A) Intraprotein hydrogen bonds and (B) intermolecular hydrogen bonds. Hydrogen bonds distribution pattern: (C) Intraprotein hydrogen bonds and (D) intermolecular hydrogen bonds.
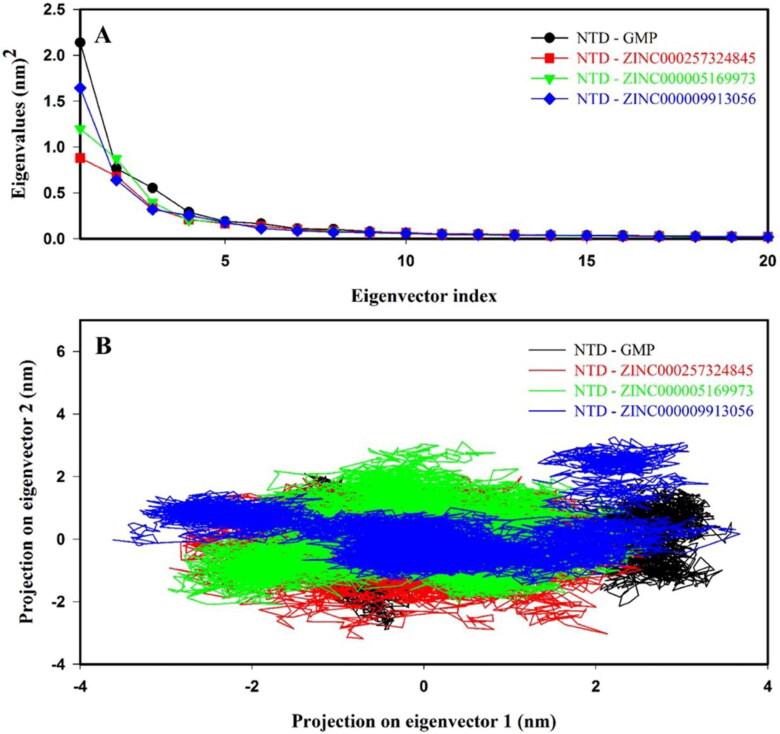
3.8.6. Principal component analysis
Principal component analysis (PCA) reveals the overall expansion of protein during the molecular simulation. The dynamical differences of 372 eigenvectors were generated and framed in a covariance matrix. Figure 8(A) shows the first 20 eigenvectors vs. eigenvalues for NTD–GMP and NTD–inhibitor(s) complexes. It can be clearly observed that first ten eigenvectors contributed 81.48%, 75.72%, 79.62% and 80.17% for NTD–GMP, NTD–ZINC000257324845, NTD–ZINC000005169973 and NTD–ZINC000009913056 complexes, respectively. The NTD–inhibitor(s) complexes showed very few motions as compared to NTD–GMP complex while NTD–ZINC000257324845 complex exhibit least motion among NTD–inhibitor(s) complexes.
Figure 8.
Principal component analysis (PCA) of protein–ligand complexes for molecular dynamics of 100 ns. (A) Plot of eigenvector index versus eigenvalues for the first 20 eigenvectors. (B) Projection of motion of NTD–GMP and NTD–inhibitor(s) complexes along PC1 and PC2. The color code for NTD–GMP, NTD–ZINC000257324845, NTD–ZINC000005169973 and NTD–ZINC000009913056 is black, red, green and blue color, respectively.
First two eigenvectors contributed 53.61%, 42.75%, 48.86% and 51.75% for NTD–GMP, NTD–ZINC000257324845, NTD–ZINC000005169973 and NTD–ZINC000009913056 complexes, respectively. Directional movements contributed by the first eigenvector (PC1) and second eigenvector (PC2) were represented in a two-dimensional projection, as shown in Figure 8(B). The two-dimensional projection PCA results showed that binding of inhibitor(s) to NTD results in the formation of stable NTD–inhibitor(s) complexes. Further, the superposition of NTD–GMP and NTD–inhibitor(s) complexes before and after MD confirmed that key interactions between NTD and ligands remained stable during the MD simulation and binding of inhibitor(s) to NTD form the stable complexes (Supporting Information Figure S9).
3.8.7. MMPBSA binding free energy calculation
The molecular dynamics trajectory from last 20 ns (2000 frames) were used to generate the binding affinity of NTD–GMP and NTD–inhibitors(s) complexes. The binding affinity of NTD–GMP, NTD–ZINC000257324845, NTD–ZINC000005169973 and NTD–ZINC000009913056 complexes were −98.67 +/– 1.84, −172.07 +/– 1.65, −159.94 +/– 1.42 and −142.61 +/– 1.96 kJmol−1, respectively, as shown in Table 4.
Table 4.
Binding free energy calculations of NTD–GMP and NTD–inhibitor(s) complexes computed from the last 20 ns (80–100 ns) from molecular dynamics. van der Waal, electrostatic, polar solvation, SASA and binding energy in kJmol−1 of NTD–GMP, NTD–ZINC000257324845, NTD–ZINC000005169973 and NTD–ZINC000009913056 complexes predicted by MMPBSA.
| Energy (kJmol–1) | GMP | ZINC000257324845 | ZINC000005169973 | ZINC000009913056 |
|---|---|---|---|---|
| van der Waals energy | –162.58 +/– 2.32 | –195.39 +/– 1.64 | –173.86 +/– 1.72 | –158.72 +/– 1.92 |
| Electrostatic energy | −67.21 +/– 2.30 | −62.36 +/– 1.27 | −57.29 +/– 1.29 | −51.12 +/– 1.36 |
| Polar solvation energy | 143.53 +/– 3.13 | 104.54 +/– 1.72 | 88.73 +/– 1.13 | 82.49 +/– 1.52 |
| SASA energy | −12.41 +/– 0.19 | −18.86 +/– 0.42 | −17.52 +/– 0.34 | −15.26 +/– 0.25 |
| Binding energy | −98.67 +/– 1.84 | –172.07 +/– 1.65 | –159.94 +/– 1.42 | –142.61 +/– 1.96 |
Per residue interactions, binding energy was calculated to determine the participation of important residues in a protein–ligand complex. Here, the per residues interaction profile of Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149 were computed using MMPBSA, as shown in Supporting Information Table S6. The binding affinity of active site residues of NTD for inhibitor(s) was higher as compared to GMP. Therefore, the MMPBSA results confirmed that the identified potent molecules bind at the active site of NTD with a higher affinity as compared to GMP and results in the formation of a higher stable NTD–inhibitor(s) complex than NTD–GMP complex.
4. Discussion
Currently, the global pandemic disease called coronavirus disease 2019 (COVID-19) is posing serious survival threats to the world population (Wu et al., 2020). COVID-19 is a zoonotic disease and caused by a third highly pathogenic human betacoronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wu et al., 2020; Zhou et al., 2020). There is no effective treatment or medicine for this disease. Therefore, it is imperative to deploy new antiviral drugs for SARS-CoV-2.
Previous studies have shown that N-protein can be used as a target for the development of an antiviral drug against viral infections (Chenavas et al., 2013; Lin et al., 2014; Lo et al., 2013; Monod et al., 2015). To inhibit the function of N-protein, two strategies were reported: one target the RNA binding site of NTD while another to inhibit the oligomerization of C-terminal domain (Cianci et al., 2013).
Here, NTD-N-protein from SARS-CoV-2 was used as a target to inhibit the viral cycle of SARS-CoV-2 by finding potential lead molecules. Molecular docking of RNA with NTD and NTD mutants (R107A, Y109A and R149A) reveal that RNA bind with a good binding affinity at the active site of NTD. Molecular dynamics analysis like RMSD, Rg, SASA and hydrogen bond results of NTD, NTD mutants, NTD–RNA and NTD mutants–RNA complexes affirm that NTD mutants are stable. However, NTD mutants–RNA complexes are less stable than NTD–RNA complex due to devoid of side chains of corresponding important residues like Arg107, Tyr109 or Arg149 in mutants. MMPBSA binding energy confirms that RNA is stabilized by interactions contributed by active site residues of NTD in NTD–RNA complex. A recent study on SARS-CoV-2 revealed that mutation of Tyr109 affects the RNA-binding affinity (Kang et al., 2020). Lin et al. (2014) also reported that R122A, Y124A and R164A in HCoV-OC43, which corresponds to Arg107, Tyr109 or Arg149 in SARS-CoV-2 play significant importance in the RNA binding (Lin et al., 2014). Our results showed the binding mode of RNA via important residues of NTD to make a stable protein–RNA complex.
Further molecular docking and dynamics results of nucleotides (AMP, CMP, GMP and UMP) with NTD revealed that GMP showed a higher binding affinity as compared to other (AMP, CMP and UMP) nucleotides. GMP at the active site of NTD-N-protein from SARS-CoV-2 confirms that identified residues are involved in holding the GMP at this site via hydrogen and hydrophobic interactions. In addition, molecular dynamics simulation confirms the stability of complexed NTD-N-protein with GMP. Following the validation of GMP interactions with NTD-N-protein, GMP was used by the ligand structure-based strategy of pharmacophore modeling to build a pharmacophore model.
The final hits from pharmacophore screening were utilized for virtual screening using the crystal structure of NTD-N-protein. Virtual screening, molecular docking and dynamics have been widely used techniques to identify the potent lead molecules and assess the stability of protein–ligand complexes (Dalal et al., 2019; Dhankhar, Dalal, Mahto, et al., 2020; Gau et al., 2018; Meng et al., 2015). Out of the top 50 molecules, three molecules (ZINC000257324845, ZINC000005169973 and ZINC000009913056) were successfully passed the ADMET filters and also having drug-like properties, which were analysed by Lipinski’s rule of five. The docking analysis for all three pharmacokinetically screened compounds indicated that all molecules were bound to the active site with high binding energy in the range of −5.6 to −7.4 kcalmol−1, which was higher than the binding energy (–2.6 kcalmol−1) of GMP. All three compounds and GMP form a stable protein–ligand complex by establishing a network of molecular interactions, including hydrogen bonds and the hydrophobic interactions with the key-residues (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149) of NTD as shown in Figure 3 and Supporting Information Figure S6. Several small molecules such as ZINC00003118440, ZINC0000146942, 6-chloro-7-(2-morpholin-4-yl-ethylamino) quinoxaline-5,8-dione, Hydroxychloroquine, chloroquine, 5817, 6797, 6799, 37542 (ribavirin), 72187 (zidovudine),135413535 (valganciclovir), PJ34 (N-(6-oxo-5,6-dihydrophenanthridin-2-yl)(N,N-dimethylamino)acetamide hydrochloride), H3 (6-chloro-7-(2-morpholin-4-yl-ethylamino) quinoxaline-5,8-dione), P1 (Benzyl-2-(hydroxymethyl)-1-indolinecarboxylate) and P3 (5-benzyloxygramine) have been reported against NTD (Amin & Abbas, 2020; Chang et al., 2016; Lin et al., 2014, 2020; Sarma et al., 2020; Yadav et al., 2020). The details of the molecular docking of these molecules with NTD, is shown in Table S7. All three (ZINC000257324845, ZINC000005169973 and ZINC000009913056) identified compounds showed a higher binding affinity than earlier reported molecules, as shown in Table 3 and Supporting Information Table S7. Henceforth, we speculate that these compounds may bind specifically to the NTD protein to inhibit its viral cycle activity.
RMSD and RMSF results indicated that binding of identified molecules (ZINC000257324845, ZINC000005169973 and ZINC000009913056) to NTD tends to form the higher stable NTD–inhibitor(s) complexes than NTD–GMP complex. The smaller Rg and SASA values of NTD–inhibitor(s) complex as compared to NTD–GMP complex revealed that identified compounds result in the formation of a higher stable protein–ligand complex than NTD–GMP complex. Further hydrogen bonding and PCA results affirmed that binding of identified molecules with NTD forms the stable and compact protein–ligand complex. MMPBSA results confirmed that binding of identified molecules to NTD is stabilized by interactions contributed by active site residues (Ser51, Phe53, Ala55, Ala90, Arg107, Tyr109, Tyr111 and Arg149) for the formation of lower energy NTD–inhibitor(s) complex as compared to NTD–GMP complex. Overall molecular dynamics and MMPBSA results concluded that all the identified compounds are potent molecules and might be used to inhibit the binding of viral RNA genome to the RNA binding region of NTD, and it may ultimately stop the essential functions of NTD, including ribonucleoprotein formation, viral cycle, which are required for the survival of virus into the host.
5. Conclusion
This study utilized molecular docking and dynamics analysis to analyze the interactions involved in the binding of RNA with NTD of N-protein of SARS-CoV-2. Various computational approaches, including pharmacophore-based virtual screening, molecular docking, molecular dynamics and MMPBSA, have been used for the identification of the potent molecules that can interact with RNA binding region named NTD of N-protein of SARS-CoV-2 to inhibit its function. Based on the NTD-N-GMP complexed structure, the pharmacophore model of GMP was generated by considering the features of GMP, including one aromatic ring of GMP, three hydrogen bond acceptors, one hydrophobic and one aromatic group. After the virtual screening against ZINC database, three molecules (ZINC000257324845, ZINC000005169973 and ZINC000009913056) were screened, which follow the Lipinski’s rule of five and fulfill the ADMET properties. Further, molecular docking results revealed that these molecules interact with the NTD-N-protein via hydrogen bonding and hydrophobic interactions. These molecules have higher binding affinities than earlier reported compounds for NTD. The molecular dynamics and MMPBSA study verified that the selected compounds efficiently bind to NTD and form stable NTD–ligand complexes. These lead molecules can be further investigated their importance through in vitro studies and can be used for the development of antiviral compounds against SARS-CoV-2 in the future.
Supplementary Material
Acknowledgements
We are thankful to the Department of chemistry, IIT Roorkee, to provide the Gaussian software. Authors thank Macromolecular Crystallographic Unit (MCU), a Central Facility at Institute Instrumentation Centre (IIC), IIT Roorkee, for computational work.
Glossary
Abbreviations
- 2019-nCoV
2019 novel coronavirus
- °
degree
- Å
angstrom
- ADMET
absorption, distribution, metabolism, excretion and toxicity
- AMP
adenosine triphosphate
- B3LYP
Lee–Yang–Parr correlation functional
- BBB
blood–brain barrier
- BLAST
basic local alignment search tool
- CMP
cytidine monophosphate
- CNS
central nervous system
- COVID-19
coronavirus disease 2019
- CoVs
coronaviruses
- CTD
C-terminal domain
- CYP
Cytochrome P450
- DFT
density functional theory
- E
envelope
- etc
et cetera
- GMP
guanosine monophosphate
- GROMACS
groningen machine for chemical simulations
- HADDOCK
high ambiguity driven protein–protein docking
- H-bond
hydrogen-bond
- HCoV
human coronavirus
- HIA
human intestinal absorption
- ICTV
International Committee of Taxonomy of Viruses
- kcalmol–1
kilocalorie per mole
- kJmol–1
kilojoule per mole
- LINCS
linear constraints solver
- M
membrane
- MERS-CoV
middle-east respiratory syndrome coronavirus
- MMPBSA
molecular mechanic/Poisson–Boltzmann surface area
- MSA
multiple sequence alignment
- N
nucleocapsid
- NCBI
National Center for Biotechnology Information
- nm
nanometer
- NPT
number of particles, pressure and temperature
- ns
nanosecond
- NTD
N-terminal domain
- NVT
number of particles, volume and temperature
- ORF
open reading frame
- PC
principal component
- PCA
principal component analysis
- PDB
protein data bank
- pdbqt
protein data bank, partial charge (q), & atom type (t))
- PME
particle mesh Ewald
- PPB
plasma protein binding
- ps
picosecond
- RCSB
research collaboratory for structural bioinformatics
- Rg
radius of gyration
- RMSD
root mean square deviations
- RMSF
root mean square fluctuation
- RNA
ribonucleic acid
- S
spike
- SARS-CoV
severe acute respiratory syndrome coronavirus
- SARS-CoV-2
severe acute respiratory syndrome coronavirus-2
- SASA
solvent accessible surface area
- SBVS
structure-based virtual screening
- sdf
structure data file
- SPC
simple point charge
- SR
Ser/arg
- UFF
universal force field
- UMP
uridine monophosphate
- VMD
visual molecular dynamics
Funding Statement
PK & ST would like to acknowledge Science and Engineering Research Board, Department of Science & Technology, Government of India [DST-SERB IPA/2020/000054]. PD, VD and VS thanks Ministry of Human Resource Development, Department of Biotechnology [DBT/2015/IIT-R/349] and University Grant Commission for financial support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abraham, M. J., & Gready, J. E. (2011). Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. Journal of Computational Chemistry, 32(9), 2031–2040. 10.1002/jcc.21773 [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amin, M., & Abbas, G. (2020). Docking study of chloroquine and hydroxychloroquine interaction with RNA binding domain of nucleocapsid phospho-protein-an in silico insight into the comparative efficacy of repurposing antiviral drugs. Journal of Biomolecular Structure & Dynamics, 1–13. 10.1080/07391102.2020.1775703 [DOI] [PubMed] [Google Scholar]
- Anandakrishnan, R., Aguilar, B., & Onufriev, A. V. (2012). H++ 3.0: Automating p K prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Research, 40(W1), W537–W541. 10.1093/nar/gks375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J., Postma, J. v., van Gunsteren, W. F., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Biesiada, M., Purzycka, K. J., Szachniuk, M., Blazewicz, J., & Adamiak, R. W. (2016). Automated RNA 3D structure prediction with RNAComposer RNA structure determination (pp. 199–215). Springer. [DOI] [PubMed] [Google Scholar]
- Cavanagh, D. (2007). Coronavirus avian infectious bronchitis virus. Veterinary Research, 38(2), 281–297. 10.1051/vetres:2006055 [DOI] [PubMed] [Google Scholar]
- Chang, C.-K., Chen, C.-M. M., Chiang, M.-H., Hsu, Y.-L., & Huang, T.-H. (2013). Transient oligomerization of the SARS-CoV N protein–implication for virus ribonucleoprotein packaging. PLoS One, 8(5), e65045. 10.1371/journal.pone.0065045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.-K., Hou, M.-H., Chang, C.-F., Hsiao, C.-D., & Huang, T.-H. (2014). The SARS coronavirus nucleocapsid protein-forms and functions. Antiviral Research, 103, 39–50. 10.1016/j.antiviral.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.-K., Jeyachandran, S., Hu, N.-J., Liu, C.-L., Lin, S.-Y., Wang, Y.-S., Chang, Y.-M., & Hou, M.-H. (2016). Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Molecular Biosystems, 12(1), 59–66. 10.1039/c5mb00582e [DOI] [PubMed] [Google Scholar]
- Chen, I.-J., Yuann, J.-M. P., Chang, Y.-M., Lin, S.-Y., Zhao, J., Perlman, S., Shen, Y.-Y., Huang, T.-H., & Hou, M.-H. (2013). Crystal structure-based exploration of the important role of Arg106 in the RNA-binding domain of human coronavirus OC43 nucleocapsid protein. Biochimica et Biophysica Acta, 1834(6), 1054–1062. 10.1016/j.bbapap.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., Qiu, Y., Wang, J., Liu, Y., Wei, Y., Xia, J., Yu, T., Zhang, X., & Zhang, L. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenavas, S., Crépin, T., Delmas, B., Ruigrok, R. W., & Slama-Schwok, A. (2013). Influenza virus nucleoprotein: Structure, RNA binding, oligomerization and antiviral drug target. Future Microbiology, 8(12), 1537–1545. 10.2217/fmb.13.128 [DOI] [PubMed] [Google Scholar]
- Cianci, C., Gerritz, S. W., Deminie, C., & Krystal, M. (2013). Influenza nucleoprotein: Promising target for antiviral chemotherapy. Antiviral Chemistry and Chemotherapy, 23(3), 77–91. 10.3851/IMP2235 [DOI] [PubMed] [Google Scholar]
- Dalal, V., Kumar, P., Rakhaminov, G., Qamar, A., Fan, X., Hunter, H., Tomar, S., Golemi-Kotra, D., & Kumar, P. (2019). Repurposing an ancient protein core structure: Structural studies on FmtA, a novel esterase of Staphylococcus aureus. Journal of Molecular Biology, 431(17), 3107–3123. 10.1016/j.jmb.2019.06.019 [DOI] [PubMed] [Google Scholar]
- Dallakyan, S., & Olson, A. J. (2015). Small-molecule library screening by docking with PyRx Chemical biology (pp. 243–250). Springer. [DOI] [PubMed] [Google Scholar]
- De Vries, S. J., Van Dijk, M., & Bonvin, A. M. (2010). The HADDOCK web server for data-driven biomolecular docking. Nature Protocols, 5(5), 883–897. 10.1038/nprot.2010.32 [DOI] [PubMed] [Google Scholar]
- DeLano, W. L. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography, 40(1), 82–92. https://www.ccp4.ac.uk/newsletters/newsletter40/11_pymol.pdf [Google Scholar]
- Dhankhar, P., Dalal, V., Kotra, D., & Kumar, P. (2020). In-silico approach to identify novel potent inhibitors against GraR of S. aureus. Frontiers in Bioscience (Landmark Edition), 25, 1337–1360. 10.2741/4859 [DOI] [PubMed] [Google Scholar]
- Dhankhar, P., Dalal, V., Mahto, J. K., Gurjar, B. R., Tomar, S., Sharma, A. K., & Kumar, P. (2020). Characterization of dye-decolorizing peroxidase from Bacillus subtilis. Archives of Biochemistry and Biophysics, 693, 108590. 10.1016/j.abb.2020.108590 [DOI] [PubMed] [Google Scholar]
- Dhankhar, P., Singh, V., & Tomar, S. (2020). Computational guided identification of novel potent inhibitors of NTD-N-protein of SARS-CoV-2. 10.26434/chemrxiv.12280532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L., Zhao, G., Lin, Y., Chan, C., He, Y., Jiang, S., Wu, C., Jin, D.-Y., Yuen, K.-Y., Zhou, Y., & Zheng, B.-J. (2008). Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine, 26(13), 1644–1651. 10.1016/j.vaccine.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P., & Cowtan, K. (2004). Coot: Model-building tools for molecular graphics. Acta Crystallographica Section D Biological Crystallography, 60(12), 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis coronaviruses (pp. 1–23). Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni, D., Cagliani, R., Clerici, M., & Sironi, M. (2017). Molecular evolution of human coronavirus genomes. Trends in Microbiology, 25(1), 35–48. 10.1016/j.tim.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Petersson, G. A., Nakatsuji, H., Li, X., Caricato, M., Marenich, A. V., Bloino, J., Janesko, B. G., Gomperts, R., Mennucci, B., Hratchian, H. P., Ortiz, J. V., … Fox, D. J. (2016). Gaussian 16. Gaussian, Inc. [Google Scholar]
- Gau, D., Lewis, T., McDermott, L., Wipf, P., Koes, D., & Roy, P. (2018). Structure-based virtual screening identifies a small-molecule inhibitor of the profilin 1-actin interaction. The Journal of Biological Chemistry, 293(7), 2606–2616. 10.1074/jbc.M117.809137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Enjuanes, L., Ziebuhr, J., & Snijder, E. J. (2006). Nidovirales: Evolving the largest RNA virus genome. Virus Research, 117(1), 17–37. 10.1016/j.virusres.2006.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet, P., Courcelle, E., Stuart, D. I., & Métoz, F. (1999). ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics, 15(4), 305–308. 10.1093/bioinformatics/15.4.305 [DOI] [PubMed] [Google Scholar]
- Grossoehme, N. E., Li, L., Keane, S. C., Liu, P., Dann, C. E., III, Leibowitz, J. L., & Giedroc, D. P. (2009). Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. Journal of Molecular Biology, 394(3), 544–557. 10.1016/j.jmb.2009.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, B., Bekker, H., Berendsen, H. J., & Fraaije, J. G. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. [DOI] [Google Scholar]
- Hop, C. E. C. A., Cole, M. J., Davidson, R. E., Duignan, D. B., Federico, J., Janiszewski, J. S., Jenkins, K., Krueger, S., Lebowitz, R., Liston, T. E., Mitchell, W., Snyder, M., Steyn, S. J., Soglia, J. R., Taylor, C., Troutman, M. D., Umland, J., West, M., Whalen, K. M., … Zhao, S. X. (2008). High throughput ADME screening: Practical considerations, impact on the portfolio and enabler of in silico ADME models. Current Drug Metabolism, 9(9), 847–853. 10.2174/138920008786485092 [DOI] [PubMed] [Google Scholar]
- Hornak, V., Abel, R., Okur, A., Strockbine, B., Roitberg, A., & Simmerling, C. (2006). Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Structure, Function, and Bioinformatics, 65(3), 712–725. 10.1002/prot.21123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, P.-K., Chang, S. C., Huang, C.-C., Lee, T.-T., Hsiao, C.-W., Kou, Y.-H., Chen, I.-Y., Chang, C.-K., Huang, T.-H., & Chang, M.-F. (2005). Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. Journal of Virology, 79(22), 13848–13855. 10.1128/JVI.79.22.13848-13855.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jain, R., Ahuja, B., & Sharma, B. (2004). Density-functional thermochemistry. III. The role of exact exchange. Indian Journal of Pure & Applied Physics, 42, 43–48. 10.1063/1.464913 [DOI] [Google Scholar]
- Kang, S., Yang, M., Hong, Z., Zhang, L., Huang, Z., Chen, X., He, S., Zhou, Z., Zhou, Z., Chen, Q., Yan, Y., Zhang, C., Shan, H., & Chen, S. (2020). Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceutica Sinica B, 10(7), 1228–1238. 10.1016/j.apsb.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane, S. C., Liu, P., Leibowitz, J. L., & Giedroc, D. P. (2012). Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein. The Journal of Biological Chemistry, 287(10), 7063–7073. 10.1074/jbc.M111.287763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesari, P., Pratap, S., Dhankhar, P., Dalal, V., Mishra, M., Singh, P. K., Chauhan, H., & Kumar, P. (2020). Structural characterization and in-silico analysis of Momordica charantia 7S globulin for stability and ACE inhibition. Scientific Reports, 10(1), 1–13. 10.1038/s41598-020-58138-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, A. M. Q., Lefkowitz, E. J., Mushegian, A. R., Adams, M. J., Dutilh, B. E., Gorbalenya, A. E., Harrach, B., Harrison, R. L., Junglen, S., Knowles, N. J., Kropinski, A. M., Krupovic, M., Kuhn, J. H., Nibert, M. L., Rubino, L., Sabanadzovic, S., Sanfaçon, H., Siddell, S. G., Simmonds, P., … Davison, A. J. (2018). Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Archives of Virology, 163(9), 2601–2631. 10.1007/s00705-018-3847-1 [DOI] [PubMed] [Google Scholar]
- Kumar, P., Dalal, V., Kokane, A., Singh, S., Lonare, S., Kaur, H., Ghosh, D. K., Kumar, P., & Sharma, A. K. (2020). Mutation studies and structure-based identification of potential inhibitor molecules against periplasmic amino acid binding protein of Candidatus Liberibacter asiaticus (CLasTcyA). International Journal of Biological Macromolecules, 147, 1228–1238. 10.1016/j.ijbiomac.2019.09.250 [DOI] [PubMed] [Google Scholar]
- Kumari, N., Dalal, V., Kumar, P., & Rath, S. N. (2020). Antagonistic interaction between TTA-A2 and paclitaxel for anti-cancer effects by complex formation with T-type calcium channel. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1839558 [DOI] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., Open Source Drug Discovery Consortium , & Lynn, A. (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Kurkcuoglu, Z., Koukos, P. I., Citro, N., Trellet, M. E., Rodrigues, J. P. G. L. M., Moreira, I. S., Roel-Touris, J., Melquiond, A. S. J., Geng, C., Schaarschmidt, J., Xue, L. C., Vangone, A., & Bonvin, A. M. J. J. (2018). Performance of HADDOCK and a simple contact-based protein-ligand binding affinity predictor in the D3R Grand Challenge 2. Journal of Computer-Aided Molecular Design, 32(1), 175–185. 10.1007/s10822-017-0049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancet, T. (2013). MERS-CoV: A global challenge. Lancet (London, England), 381(9882), 1960. 10.1016/s0140-6736(13)61184-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C., Yang, W., & Parr, R. G. (1988). Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Physical Review B, 37(2), 785–789. 10.1103/PhysRevB.37.785 [DOI] [PubMed] [Google Scholar]
- Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G. M., Ahuja, A., Yung, M. Y., Leung, C. B., To, K. F., Lui, S. F., Szeto, C. C., Chung, S., & Sung, J. J. Y. (2003). A major outbreak of severe acute respiratory syndrome in Hong Kong. The New England Journal of Medicine, 348(20), 1986–1994. 10.1056/NEJMoa030685 [DOI] [PubMed] [Google Scholar]
- Lin, S.-M., Lin, S.-C., Hsu, J.-N., Chang, C.-K., Chien, C.-M., Wang, Y.-S., Wu, H.-Y., Jeng, U.-S., Kehn-Hall, K., & Hou, M.-H. (2020). Structure-based stabilization of non-native Protein-protein interactions of coronavirus nucleocapsid proteins in antiviral drug design. Journal of Medicinal Chemistry, 63(6), 3131–3141. 10.1021/acs.jmedchem.9b01913 [DOI] [PubMed] [Google Scholar]
- Lin, S.-Y., Liu, C.-L., Chang, Y.-M., Zhao, J., Perlman, S., & Hou, M.-H. (2014). Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. Journal of Medicinal Chemistry, 57(6), 2247–2257. 10.1021/jm500089r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, C. A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today. Technologies, 1(4), 337–341. 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Lo, Y.-S., Lin, S.-Y., Wang, S.-M., Wang, C.-T., Chiu, Y.-L., Huang, T.-H., & Hou, M.-H. (2013). Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Letters, 587(2), 120–127. 10.1016/j.febslet.2012.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus, M., Boniecki, M. J., Dawson, W., & Bujnicki, J. M. (2016). SimRNAweb: A web server for RNA 3D structure modeling with optional restraints. Nucleic Acids Research, 44(W1), W315–W319. 10.1093/nar/gkw279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, A., Dalal, V., Ankri, S., & Tomar, S. (2019). Structural insights into Entamoeba histolytica arginase and structure-based identification of novel non-amino acid based inhibitors as potential antiamoebic molecules. The FEBS Journal, 286(20), 4135–4155. 10.1111/febs.14960 [DOI] [PubMed] [Google Scholar]
- Marra, M. A., Jones, S. J., Astell, C. R., Holt, R. A., Brooks-Wilson, A., Butterfield, Y. S., Khattra, J., Asano, J. K., Barber, S. A., Chan, S. Y., Cloutier, A., Coughlin, S. M., Freeman, D., Girn, N., Griffith, O. L., Leach, S. R., Mayo, M., McDonald, H., Montgomery, S. B., … Roper, R. L. (2003). The genome sequence of the SARS-associated coronavirus. Science, 300(5624), 1399–1404. 10.1126/science.1085953 [DOI] [PubMed] [Google Scholar]
- Meng, F., Cheng, S., Ding, H., Liu, S., Liu, Y., Zhu, K., Chen, S., Lu, J., Xie, Y., Li, L., Liu, R., Shi, Z., Zhou, Y., Liu, Y.-C., Zheng, M., Jiang, H., Lu, W., Liu, H., & Luo, C. (2015). Discovery and optimization of novel, selective histone methyltransferase SET7 inhibitors by pharmacophore- and docking-based virtual screening. Journal of Medicinal Chemistry, 58(20), 8166–8181. 10.1021/acs.jmedchem.5b01154 [DOI] [PubMed] [Google Scholar]
- Monod, A., Swale, C., Tarus, B., Tissot, A., Delmas, B., Ruigrok, R. W., Crépin, T., & Slama-Schwok, A. (2015). Learning from structure-based drug design and new antivirals targeting the ribonucleoprotein complex for the treatment of influenza. Expert Opinion on Drug Discovery, 10(4), 345–371. 10.1517/17460441.2015.1019859 [DOI] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3(1), 33. 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit, S., Dalal, V., & Mishra, G. (2018). Identification of novel phosphatidic acid binding domain on sphingosine kinase 1 of Arabidopsis thaliana. Plant Physiology and Biochemistry, 128, 178–184. 10.1016/j.plaphy.2018.04.039 [DOI] [PubMed] [Google Scholar]
- Parrinello, M., & Rahman, A. (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics, 52(12), 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Perlman, S., & Netland, J. (2009). Coronaviruses post-SARS: Update on replication and pathogenesis. Nature Reviews. Microbiology, 7(6), 439–450. 10.1038/nrmicro2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, D. E., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry, 58(9), 4066–4072. 10.1021/acs.jmedchem.5b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Release, S. (2016). 2: Maestro. Schrödinger, LLC. [Google Scholar]
- Saikatendu, K. S., Joseph, J. S., Subramanian, V., Neuman, B. W., Buchmeier, M. J., Stevens, R. C., & Kuhn, P. (2007). Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. Journal of Virology, 81(8), 3913–3921. 10.1128/JVI.02236-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini, G., Dalal, V., Savita, B. K., Sharma, N., Kumar, P., & Sharma, A. K. (2019). Molecular docking and dynamic approach to virtual screen inhibitors against Esbp of Candidatus Liberibacter asiaticus. Journal of Molecular Graphics & Modelling, 92, 329–340. 10.1016/j.jmgm.2019.08.012 [DOI] [PubMed] [Google Scholar]
- Sarma, P., Shekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., Singh, S., Kumar, H., Prakash, A., Dhibar, D. P., & Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelle, B., Karl, N., Ludewig, B., Siddell, S. G., & Thiel, V. (2005). Selective replication of coronavirus genomes that express nucleocapsid protein. Journal of Virology, 79(11), 6620–6630. 10.1128/JVI.79.11.6620-6630.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel, H. B. (1982). Optimization of equilibrium geometries and transition structures. Journal of Computational Chemistry, 3(2), 214–218. 10.1002/jcc.540030212 [DOI] [Google Scholar]
- Schüttelkopf, A. W., & Van Aalten, D. M. (2004). PRODRG: A tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallographica Section D Biological Crystallography, 60(8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, J. D., & Higgins, D. G. (2011). Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7(1), 539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N., Dalal, V., & Kumar, P. (2018). Structure based mimicking of Phthalic acid esters (PAEs) and inhibition of hACMSD, an important enzyme of the tryptophan kynurenine metabolism pathway. International Journal of Biological Macromolecules, 108, 214–224. 10.1016/j.ijbiomac.2017.12.005 [DOI] [PubMed] [Google Scholar]
- Singh, N., Dalal, V., Kumar, V., Sharma, M., & Kumar, P. (2019). Characterization of phthalate reductase from Ralstonia eutropha CH34 and in silico study of phthalate dioxygenase and phthalate reductase interaction. Journal of Molecular Graphics & Modelling, 90, 161–170. 10.1016/j.jmgm.2019.05.002 [DOI] [PubMed] [Google Scholar]
- Singh, N., Dalal, V., Mahto, J. K., & Kumar, P. (2017). Biodegradation of phthalic acid esters (PAEs) and in silico structural characterization of mono-2-ethylhexyl phthalate (MEHP) hydrolase on the basis of close structural homolog. Journal of Hazardous Materials, 338, 11–22. 10.1016/j.jhazmat.2017.04.055 [DOI] [PubMed] [Google Scholar]
- Sterling, T., & Irwin, J. J. (2015). ZINC 15-ligand discovery for everyone. Journal of Chemical Information and Modeling, 55(11), 2324–2337. 10.1021/acs.jcim.5b00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunseri, J., & Koes, D. R. (2016). Pharmit: Interactive exploration of chemical space. Nucleic Acids Research, 44(W1), W442–W448. 10.1093/nar/gkw287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit, M., Liu, B., Chow, V. T., & Lal, S. K. (2006). The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. The Journal of Biological Chemistry, 281(16), 10669–10681. 10.1074/jbc.M509233200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, F., Billeter, S., Eising, A., Hünenberger, P., Kruger, P., Mark, E., … Tironi, I. (1996). Biomolecular simulation: The GROMOS96 manual and user guide.
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren, W. F., S. R. Billeter, A. A. Eising, P. H. Hünenberger, P. K. H. C. Krüger, A. E. Mark, W. R. P. Scott, and I. G. Tironi . (1996). Biomolecular simulation: The GROMOS96 manual and user guide. Verlag der Fachvereine Hochschulverlag AG an der ETH Zurich. [Google Scholar]
- van der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Woo, P. C. Y., Lau, S. K. P., Lam, C. S. F., Lau, C. C. Y., Tsang, A. K. L., Lau, J. H. N., Bai, R., Teng, J. L. L., Tsang, C. C. C., Wang, M., Zheng, B.-J., Chan, K.-H., & Yuen, K.-Y. (2012). Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. Journal of Virology, 86(7), 3995–4008. 10.1128/JVI.06540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, R., Imran, M., Dhamija, P., Suchal, K., & Handu, S. (2020). Virtual screening and dynamics of potential inhibitors targeting RNA binding domain of nucleocapsid phosphoprotein from SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1778536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.