Abstract
Background
Susceptibility to metabolic diseases may be influenced by mitochondrial genetic variability among people living with human immunodeficiency virus (HIV; PLWH), but remains unexplored in populations with African ancestry. We investigated the association between mitochondrial DNA (mtDNA) haplogroups and the homeostatic model assessments of β-cell function (HOMA-B) and insulin resistance (HOMA-IR), as well as incident diabetes mellitus (DM), among Black women living with or at risk for HIV.
Methods
Women without DM who had fasting glucose (FG) and insulin (FI) data for ≥2 visits were included. Haplogroups were inferred from genotyping data using HaploGrep. HOMA-B and HOMA-IR were calculated using FG and FI data. Incident DM was defined by a combination of FG ≥ 126 mg/dL, the use of DM medication, a DM diagnosis, or hemoglobin A1c ≥ 6.5%. We compared HOMA-B, HOMA-IR, and incident DM by haplogroups and assessed the associations between HOMA-B and HOMA-IR and DM by haplogroup.
Results
Of 1288 women (933 living with HIV and 355 living without HIV), PLWH had higher initial HOMA-B and HOMA-IR than people living without HIV. PLWH with haplogroup L2 had a slower decline in HOMA-B per year (Pinteraction = .02) and a lower risk of incident DM (hazard ratio [HR], 0.51; 95% confidence interval [CI], .32–.82) than PLWH with other haplogroups after adjustments for age, body mass index, combination antiretroviral therapy use, CD4 cell counts, and HIV RNA. The impact of HOMA-IR on incident DM was less significant in those with haplogroup L2, compared to non-L2 (HR, 1.28 [95% CI, .70–2.38] vs 4.13 [95% CI, 3.28–5.22], respectively; Pinteraction < .01), among PLWH.
Conclusions
Mitochondrial genetic variation is associated with β-cell functions and incident DM in non-Hispanic, Black women with HIV and alters the relationship between insulin resistance and DM.
Keywords: mitochondrial genetics, diabetes mellitus, HIV, aging
Among women living with human immunodeficiency virus, African mitochondrial DNA haplogroup L2 was associated with a lower risk of diabetes mellitus; slower age-related declines in β-cell functions; and an attenuated impact of insulin resistance on incident diabetes mellitus.
People living with human immunodeficiency virus (HIV; PLWH) may have an increased risk of diabetes mellitus (DM) [1–4] and multiple metabolic changes [5], compared to their counterparts living without HIV. DM and HIV infection [6] are independent risk factors for cardiovascular disease, which is a leading cause of death among virally suppressed combination antiretroviral therapy (cART)-treated PLWH [7]. Therefore, understanding the pathophysiology of DM is critical for the development of HIV long-term management strategies and interventions as PLWH age.
Mitochondria produce cellular energy and play a central role in aerobic metabolism [8]. Mitochondrial dysfunction is associated with an increased risk of DM through direct impacts on both insulin action at the level of the target tissue (insulin resistance) and insulin release (ß-cell function) [9]. Mitochondrial genetic variation, which influences mitochondrial function and protein production [8], is likely to provide important insights into the pathophysiology of DM and treatment. Data from the Cohorts for Heart and Ageing Research in Genome Epidemiology mtDNA+ Consortium, a large-scale 45-cohort collaboration, confirmed that mitochondrial DNA (mtDNA) variation was associated with glucose and insulin metabolism, the risk for DM, and cardiovascular disease [10]. Ancestry-based, associated mtDNA variations, such as the mtDNA haplogroup, affect mitochondrial functions [11], and have been associated with several HIV and cART-related outcomes [12–15]. Studies in the general population suggest that mtDNA haplogroups are associated with incident DM [16–18]; however, few studies have addressed these associations among PLWH. In addition, previous studies have only examined the association within European and Asian populations [16–18], and the results have been inconsistent [19–21]. No study has investigated this association in an African-ancestry population. Moreover, there is no study evaluating the trajectory of insulin resistance and ß-cell functions over time by different mtDNA haplogroups.
We sought to study the association between mtDNA haplogroups and incident DM in a multicenter, cohort study of women living with or at risk of HIV infection. We further evaluate the associations between mtDNA haplogroups and dynamic changes in insulin resistance and ß-cell function, as well as their interrelation with incident DM.
METHODS
Study Population
We used data collected in the Women’s Interagency HIV Study (WIHS), a multicenter, prospective, cohort study that recruited women living with or at risk for HIV infection beginning in 1994. Institutional review boards at each study location approved the study protocol, and each participant provided written informed consent. The WIHS study design, enrollment, and data collection protocols have been previously reported [22, 23]. In short, participants attend routine, semiannual visits and complete a comprehensive physical examination, provide biological specimens, and complete interviewer-administered standardized questionnaires. The majority of the WIHS participants are of African descent [1]. Inclusion criteria for this study were WIHS women who self-reported as non-Hispanic and Black, who had an mtDNA haplogroup determination, and whose records contained simultaneous fasting glucose (FG) and fasting insulin (FI) data for ≥2 visits between 1999 (when WIHS started to collect FG and FI) and 2015. The first visit with FG and FI data available was referred to as the index visit. Women with prevalent DM at the index visit and women who were pregnant were excluded from the analysis.
Ascertainment of Incident Diabetes Mellitus and Homeostatic Model Assessment Measurements
Data collection for hemoglobin A1c (HbA1c) and glucose, as well as the ascertainment of incident DM in WIHS, have been previously described [1]. Incident DM cases were defined by 1 of the following: (1) the first report of anti-DM medication after the index visit; (2) HbA1c ≥ 6.5%, with confirmation by FG ≥ 126 mg/dL or anti-DM medication; (3) an FG ≥ 126 mg/dL with subsequent confirmation by an FG ≥ 126 mg/dL, anti-DM medication use, or HbA1c ≥6.5%; or (4) a self-report of DM with confirmation of anti-DM medication or 2 visits with either an FG ≥ 126 mg/dL or a combination of HbA1c ≥ 6.5% and an FG ≥ 126 mg/dL. The homeostatic model assessment of insulin resistance (HOMA-IR) and the homeostatic model assessment of ß-cell function (HOMA-B) were calculated using FG and FI data [24].
Mitochondrial DNA Haplogroup Determination
Mitochondrial and nuclear single nucleotide polymorphisms (SNPs) were genotyped using the Illumina HumanOmni2.5-quad beadchip (Illumina, San Diego, CA) for all WIHS women enrolled in the 1994–1995 and 2001–2002 recruitment waves who provided informed consent for genetic testing (n = 3353). Genetic data were assessed in WIHS and the quality control of the data has been described previously [25]. Excluded from the analytic data set were SNPs with a genotype call rate <95% and SNPs that failed our in-house quality control criteria. Specifically, SNPs with at least 2 discordant genotypes among greater than 20 duplicate samples were excluded. The HumanOmni2.5-quad beadchip included 256 mitochondrial SNPs. The mtDNA haplogroups were inferred using HaploGrep [26], which determines the most probable mtDNA haplogroups based on the phylogenetic tree of global mtDNA variation [27]. The majority of the participants were identified as 1 of 3 African haplogroups: L0L1 (combining haplogroup L1 with the neighboring, rarer haplogroup of L0), L2, and L3. The remaining participants were considered as being in an “other” haplogroup.
Covariates
Sociodemographic (age, self-reported race) data were collected at the baseline visit. Body height and weight measurements were used to calculate the body mass index (BMI; kg/m2). The HIV status in people living without HIV was collected annually. In women with HIV, CD4+ cell counts and HIV RNA were collected semi-annually. The presence of detectable plasma HIV RNA was defined as HIV RNA > 400 copies/mL. An interviewer-administered questionnaire collected participants’ self-reports of cART use, and the definitions were adapted from the Department of Health and Human Services/Kaiser Panel guidelines, updated in 2016. Fasting lipid profiles (total cholesterol, low-density lipoprotein, high-density lipoprotein, and triglycerides) were obtained annually.
Statistical Analyses
All analyses were conducted using SAS version 9.4 (SAS Institute, Cary, NC). Index visit characteristics were compared by HIV serostatus, and differences were evaluated using the Chi-square test for categorical variables and the Wilcoxon-Mann-Whitney test for continuous variables. Incidence rates of diabetes and incidence rate ratios by haplogroups and HIV status were assessed using Poisson regression models controlled for covariates. Person-time at risk was defined as the time from the index visit to the earliest date of 31 December 2015, incident DM, death, or loss to follow-up. We determined associations between mtDNA haplogroups with incident DM using Cox proportional hazard regression models with time-dependent covariates. We explored the dynamic associations of HOMA-B and HOMA-IR with the probability of DM by HIV serostatus using 3-dimensional graphs. In order to study the association between HOMA-B and HOMA-IR, DM, mtDNA haplogroups, and HIV infection, we conducted the following analysis. First, associations of haplogroups with rates of HOMA-B and HOMA-IR changes over time (including interactions between haplogroup and age) were assessed using random-effects linear regression models with random slopes after controlling for time-varying covariates: BMI and, for women with HIV, cART treatment, CD4+ cell counts, and the presence of detectable HIV RNA. Second, mtDNA haplogroup associations with incident DM were analyzed using Cox proportional hazard regression models, adjusting for the same covariates as above. Third, the relationships between HOMA-B/HOMA-IR and incident DM was assessed using Cox proportional hazard regression models and were stratified by different haplogroups and HIV status, adjusting for the same covariates as above. The covariates in adjusted models were selected based on research questions and significance in the univariate models.
RESULTS
Index Visit Characteristics
A total of 1288 self-reported non-Hispanic, Black women were included in this study and contributed 11 272 person-years of follow-up (median duration of follow-up, 10.4 years). Among them, 933 living with HIV had a median age of 40 and a median BMI of 27.8 kg/m2. Table 1 presents demographic information and laboratory results at the index visit by participants’ HIV status. Overall, women with HIV were more likely to be older, had lower high-density lipoprotein cholesterol, higher triglycerides, higher insulin, higher HOMA-IR, and higher HOMA-B than women without HIV at the index visit. The L3 haplogroup was the most common haplogroup (37.1%), followed by the L2 haplogroup (28.5%), the L0L1 haplogroup (23.3%), and all other haplogroups (11.1%; Table 1). We did not observe any differences in demographics, BMI, lipid profile, FG, or insulin resistance by haplogroup at the index visit between women with and without HIV (Supplementary Tables 1 and 2). Participants with haplogroup L2 had higher median HOMA-B at the index visit compared to other haplogroups in both women with and without HIV; however, these differences did not reach statistical significance (P-values = .63 and .55, respectively).
Table 1.
Demographic Information, Laboratory Data at Index Visit, and Mitochondrial DNA Haplogroup Frequencies
| Variables | Overall | HIV Seronegative, n = 355 | HIV Seropositive, n = 933 | P Valuea |
|---|---|---|---|---|
| Age | 39.6 (33.3–46.2) | 38.3 (29.0–45.5) | 40.0 (34.2–46.4) | <.01 |
| BMI, kg/m2 | 27.8 (23.6–33.1) | 28.2 (23.5–33.6) | 27.6 (23.7–33.0) | .65 |
| CD4 + count, cells/mm3 | 515 (317–765) | 912 (752–1191) | 444 (280–635) | <.01 |
| Detectable plasma HIV RNA, >400 cps/mL | … | … | 505 (54) | |
| On ART at index visit | … | … | 500 (54) | |
| Plasma HIV RNA, copies/mL | … | … | 770 (80–9900) | |
| Total cholesterol, mg/dL | 175 (15–202) | 177 (152–202) | 174 (150–203) | .43 |
| LDL cholesterol, mg/dL | 99 (79–126) | 99 (82–125) | 99.0 (79–126) | .99 |
| HDL cholesterol, mg/dL | 50 (40–62) | 56 (46–66) | 48 (37–60) | <.01 |
| Triglycerides, mg/dL | 94 (68–139) | 77 (56–111) | 101 (73–150) | <.01 |
| Glucose, mg/dL | 84 (78–91) | 83 (76–91) | 84 (78–91) | .09 |
| Insulin, mlU/L | 10 (6–16) | 8 (5–15) | 10 (6–17) | <.01 |
| HOMA-IR | 2.0 (1.2–3.5) | 1.7 (1.0–3.1) | 2.1 (1.3–3.6) | <.01 |
| HOMA-B | 171.8 (110.2–276.1) | 152.9 (100.2–253.1) | 180.0 (114.5–288.0) | <.01 |
| African mtDNA haplogroups | ||||
| L0L1 | 300 (23.3) | 81 (22.8) | 219 (23.5) | .30 |
| L2 | 367 (28.5) | 98 (27.6) | 269 (28.8) | |
| L3 | 478 (37.1) | 127 (35.8) | 351 (37.6) | |
| Other | 143 (11.1) | 49 (13.8) | 94 (10.1) |
Data are among non-Hispanic, Black WIHS participants, by HIV serostatus. Values shown are median (IQR) or n (%), unless otherwise indicated.
Abbreviations: ART, antiretroviral therapy; BMI, body mass index; HDL, high-density lipoprotein; HIV, human immunodeficiency virus; HOMA-B, homeostatic model assessment of β-cell function; HOMA-IR, homeostatic model assessment of insulin resistance; IQR, interquartile range; LDL, low-density lipoprotein; mtDNA, mitochondrial DNA; WIHS, Women’s Interagency HIV Study.
aThe P values were evaluated by the χ2 test for categorical variables and the Wilcoxon-Mann-Whitney test for continuous variables.
Associations Between Mitochondrial DNA Haplogroup and Changes in Homeostatic Model Assessments Over Time
Table 2 shows the comparison of the linear trajectory of HOMA-B changes over time by mtDNA haplogroups in random-effect linear regression models controlled for covariates. Models adjusted for genetic ancestry components, in addition to other covariates, had similar estimations, compared to models without adjustment (Supplementary Table 5). Among all groups, HOMA-B declined significantly as participants aged (P < .01). Among women with HIV, HOMA-B declined significantly slower over time among women with L2 than those with non-L2 haplogroups (Pinteraction = .02). Among women without HIV, we observed individuals with L0L1 had lower index visit HOMA-B (P = .01), but a slower decline in HOMA-B than non-L0L1 haplogroups over time (Pinteraction < .01). Also, women without HIV with L3 haplogroup had significantly higher index visit HOMA-B (P = .04), but a faster decline in HOMA-B over time (Pinteraction = .02) than women in non-L3 haplogroups. HOMA-IR increased significantly with age among all participants (Supplementary Table 3). However, we did not observe significant differences in the trajectory of HOMA-IR changes over time by haplogroup.
Table 2.
Longitudinal Change of Homeostatic Model Assessment of β-cell Function by Mitochondrial DNA Haplogroups
| HIV Seronegative | HIV Seropositive | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crude Estimation | Full Estimationa | Crude Estimation | Full Estimationb | ||||||||||
| Models | Variables | Estimates | SE | P Value | Estimates | SE | P Value | Estimates | SE | P Value | Estimates | SE | P Value |
| L0L1 vs non-L0L1 | Age | −.0306 | .0025 | <.01 | −.0245 | .0020 | <.01 | −.0196 | .0016 | <.01 | −.0153 | .0014 | <.01 |
| L0L1 | −.4572 | .2324 | .05 | −.4693 | .1829 | .01 | .2987 | .1660 | .07 | .0877 | .1319 | .51 | |
| Age*L0L1 | .0141 | .0051 | <.01 | .0129 | .0040 | <.01 | −.0061 | .0035 | .08 | −.0019 | .0028 | .50 | |
| L2 vs non-L2 | Age | −.0268 | .0026 | <.01 | −.0205 | .0020 | <.01 | −.0231 | .0017 | <.01 | −.0173 | .0014 | <.01 |
| L2 | .0439 | .2172 | .84 | .0934 | .1700 | .58 | −.2918 | .1476 | .05 | −.2113 | .1166 | .07 | |
| Age*L2 | −.0012 | .0048 | .79 | −.0025 | .0037 | .50 | .0078 | .0032 | .01 | .0061 | .0025 | .02 | |
| L3 vs non-L3 | Age | −.0247 | .0027 | <.01 | −.0185 | .0021 | <.01 | −.0206 | .0019 | <.01 | −.0150 | .0015 | <.01 |
| L3 | .2699 | .2002 | .18 | .3190 | .1572 | .04 | −.0071 | .1387 | .96 | .0247 | .1100 | .82 | |
| Age*L3 | −.0076 | .0045 | .09 | −.0086 | .0036 | .02 | −.0006 | .0030 | .85 | −.0016 | .0024 | .51 |
Abbreviations: BMI, body mass index; HIV, human immunodeficiency virus; HOMA-B, homeostatic model assessment of β-cell function; HOMA-IR, homeostatic model assessment of insulin resistance; SE, standard error.
aModels adjusted for BMI and time-varying HOMA-IR
bModels adjusted for BMI, time-varying HOMA-IR, CD4 counts, detectable HIV RNA, and combination antiretroviral therapy use.
Incidence Rates of Diabetes Mellitus by Mitochondrial DNA Haplogroup
The incidence rates of DM among women with and without HIV were similar (1.59 per 100 person-years and 1.53 per 100 person-years, respectively; adjusted incidence rate ratio, 1.08; 95% confidence interval [CI], .77–1.5). But the incidence rates of DM by mtDNA haplogroups varied between women with and without HIV (Table 3). Specifically, individuals with haplogroup L2 exhibited the lowest incidence rate (1.0 per 100 person-years) of DM among women with HIV, but the highest incidence rate (1.86 per 100 person-years) among women without HIV.
Table 3.
Diabetes Incidence Rate by African Haplogroups
| HIV Negative, n = 355 | HIV Positive, n = 933 | |||||
|---|---|---|---|---|---|---|
| Incidence DM | Person Years | Incidence Rate Per 100 Person-years (95% CI)a | Incidence DM | Person Years | Incidence Rate Per 100 Person-years (95% CI)b | |
| L0L1 | 11 | 722 | 1.52 (.84–2.75) | 27 | 1792 | 1.51 (1.03–2.20) |
| L2 | 17 | 915 | 1.86 (1.16–2.99) | 24 | 2396 | 1.00 (.67–1.49) |
| L3 | 19 | 1166 | 1.63 (1.04–2.55) | 63 | 3063 | 2.06 (1.61–2.63) |
| Others | 3 | 475 | .63 (.20–1.96) | 13 | 744 | 1.75 (1.01–3.01) |
| All non-Hispanic Black | 50 | 3277 | 1.53 (1.16–2.01) | 127 | 7995 | 1.59 (1.33–1.89) |
Data are among non-Hispanic, Black WIHS participants, by HIV serostatus.
Abbreviations: BMI, body mass index; CI, confidence interval; DM, diabetes mellitus; HIV, human immunodeficiency virus; WIHS, Women’s Interagency HIV Study.
aEstimation controlled for age at visit and BMI.
bEstimation controlled for age at visit, BMI, CD4 cell count, detectable HIV RNA, and combination antiretroviral therapy use.
Association Between Mitochondrial DNA Haplogroup and Incident Diabetes Mellitus
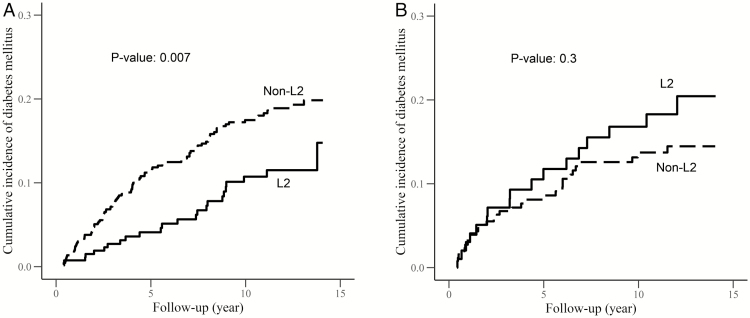
In adjusted Cox proportional hazards models, we observed that women living with HIV and haplogroup L2 had a 49% lower risk (hazard ratio [HR], .51; 95% CI, .32–.82) of developing DM during follow-up, after controlling for age, BMI, cART exposure, CD4+ cell counts, and the presence of detectable HIV RNA, as compared to women in non-L2 haplogroups (Figure 1A; Supplementary Table 4). Meanwhile, among women living with HIV, haplogroup L3 was associated with a 56% higher risk (HR, 1.56; 95% CI, 1.09–2.24) of developing DM during follow-up, after controlling for the same covariates (Supplementary Table 4; Supplementary Figure 3), as compared to women in non-L3 haplogroups. However, there were no observed differences in the risk of developing DM by mtDNA haplogroups among women living without HIV (Figure 1B; Supplementary Table 4). Overall, no difference in incident DM by haplogroup L0L1 was observed (Supplementary Table 4; Supplementary Figure 2).
Figure 1.
Cumulative incidence of diabetes mellitus and mitochondrial DNA haplogroups in Women’s Interagency HIV Study (WIHS) by human immunodeficiency virus (HIV) serostatus. A, Cumulative incidence of diabetes mellitus and mitochondrial DNA halogroups among WIHS participants living with HIV. B, Cumulative incidence of diabetes mellitus and mitochondrial DNA halogroups among WIHS participants living without HIV.
Associations of Homeostatic Model Assessments with Incident Diabetes Mellitus
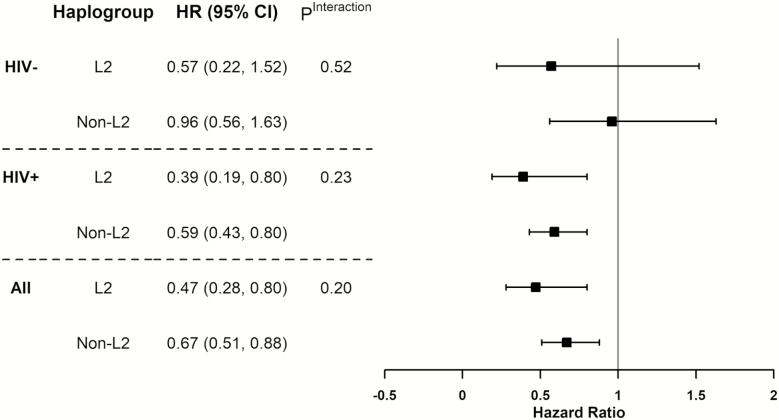
HOMA-B and HOMA-IR at index visits were associated with incident DM, with both lower HOMA-B and higher HOMA-IR associated with higher probabilities of DM during follow-up (Supplementary Figure 1). The associations were similar among women living with and without HIV. In Cox proportional hazard regression models, higher HOMA-IR was associated with a higher incidence of DM. Within the strata of women living with HIV, higher HOMA-IR was still associated with a 4-fold greater risk (HR, 4.13; 95% CI, 3.28–5.22) of incident DM among those with non-L2 haplogroups. In contrast, the association between HOMA-IR and the risk of DM was not significant (HR, 1.28; 95% CI, .69–2.38) among women with HIV and with the L2 haplogroup. The interaction between the L2 haplogroup and HOMA-IR was statistically significant (Pinteraction < .01; Figure 2). This difference was not observed among women living without HIV.
Figure 2.
The association between homeostatic model assessment of insulin resistance (HOMA-IR) and incidence diabetes mellitus in Women’s Interagency HIV Study (WIHS) by mitochondrial DNA haplogroup L2 and HIV status. Abbreviations: -, seronegative; +, seropositive; CI, confidence interval; HIV, human immunodeficiency virus; HR, hazard ratio.
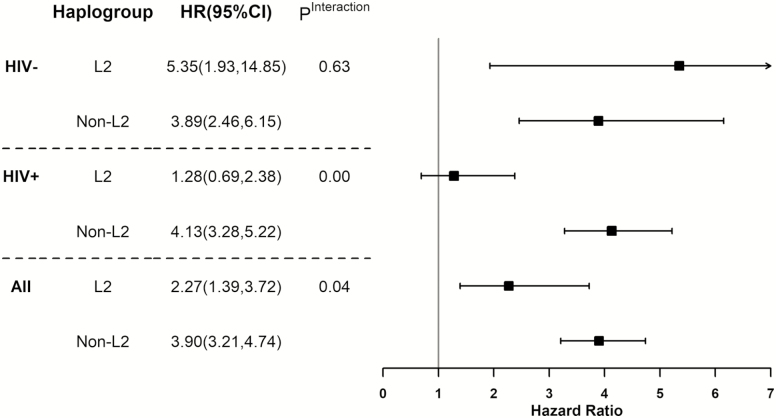
On the other hand, the magnitude of the protective effect of a higher ß-cell function (HOMA-B) on the risk of DM was stronger among women with haplogroup L2 than those with other haplogroups (HR for L2, 0.47 [95% CI, .28–.80] vs non-L2, .67 [95% CI, .51–.88]), although the interaction between haplogroup L2 and HOMA-B was not statistically significant (Figure 3). Overall, the difference was greater among women living with HIV.
Figure 3.
The association between homeostatic model assessment of β-cell function (HOMA-B) and incidence diabetes mellitus in Women’s Interagency HIV Study (WIHS) by mitochondrial DNA haplogroup L2 and HIV status. Abbreviations: -, seronegative; +, seropositive; CI, confidence interval; HIV, human immunodeficiency virus; HR, hazard ratio.
DISCUSSION
In the current study, we used data from a large, prospective cohort to demonstrate the contribution of mtDNA haplogroups to the trajectory of insulin resistance, ß-cell functions, and incident DM in non-Hispanic, Black women living with HIV. We observed that women living with HIV and haplogroup L2 had a slower decline in ß-cell functions, as well as a lower incidence rate and a lower hazard of DM, as compared to those with other haplogroups. Moreover, we found the impact of insulin resistance on incident DM was attenuated among women living with HIV and haplogroup L2. Among women living without HIV, the same haplogroup was not associated with a lower incidence rate, lower hazard, or slower decline of ß-cell functions, suggesting the possibility of a gene X environmental interaction between HIV or HIV treatment and the haplogroup. PLWH with the L3 haplogroup were associated with a higher risk of DM, as compared to those with non-L3 haplogroups. However, the results were not consistent in the trajectories of HOMA-IR and HOMA-B by the L3 haplogroup. Further investigation is warranted to identify the roles of haplogroup L3 and DM among PLWH. Our findings provide evidence to support the hypothesis that mtDNA haplogroups are associated with metabolic disorders among PLWH, and we further revealed variations in the long-term trajectories of ß-cell function and IR by different haplogroups.
Previous studies conducted in European-ancestry populations reported inconsistent results on the relationships between mtDNA haplogroups and metabolic changes in PLWH. Multiple studies have shown differences in metabolic changes among PLWH after initiating cART [13, 19, 20]. European haplogroup UK has been associated with insulin resistance (HOMA-IR ≥ 3.8), while haplogroups HV and H were associated with reduced odds of insulin resistance among people coinfected with HIV and hepatitis C virus [19]. In another study, haplogroup U was associated with greater increases in HOMA-IR after 24 weeks of ART [20]. In contrast, another study found no differences in metabolic changes between individuals with different European haplogroups [21]. However, the latter study was conducted in a population of patients living with HIV who had already developed ART-related lipodystrophy—a condition associated with metabolic disorders [28]—and, therefore, provided no statistical variability in metabolic profiles between those who had slow or later onset of lipodystrophy.
Through long-term use, ART can lead to metabolic complications, including dyslipidemia, insulin resistance, abnormal fat accumulation, and lipoatrophy [4, 29, 30]. Therapies such as first-generation nucleoside reverse transcriptase inhibitors have known mitochondrial toxicities [31, 32], which can be among the mechanisms that explain metabolic complications among PLWH with longer treatment durations. Genetic susceptibilities of certain mtDNA haplogroups could trigger mitochondrial dysfunctions and pose additional risks for the development of DM after treatment initiation. In our study, the protective effect of haplogroup L2 on DM was consistent: not only was haplogroup L2 associated with a slower decline in ß-cell functions, but even among individuals with higher insulin resistance at baseline, the risk of those with haplogroup L2 developing DM was attenuated. Further investigation into specific single-nucleotide polymorphisms and the genetic profile of haplogroup L2 might provide insights into understanding the pathogenesis of abnormal glucose metabolism among PLWH.
The vast majority of the mtDNA haplogroup studies have been conducted in European-ancestry populations, and only a few studies have examined the mtDNA variation among those with African ancestry [15, 33]. In a study conducted in the general population, researchers noted that African-ancestry mtDNA variations were associated with resting metabolic rates (RMRs) and total energy expenditure. African haplogroups L0, L2, and L3 were associated with lower RMRs, compared to European haplogroups H, JT, and UK. Among them, individuals with the L2 haplogroup have the lowest RMR, compared to other haplogroups [33]. Since the mtDNA haplogroup is associated with RMR, which is in turn associated with the development of DM [34], we speculate there might be a specific genetic predisposition among individuals with L2, making them resilient to metabolic changes and preventing them from progressing to DM. More studies conducted in African-ancestry haplogroups are warranted to confirm this hypothesis.
As with any observational study, our study has certain limitations. First, we did not investigate sub-haplogroups within each haplogroup; therefore, we cannot make inferences regarding potential, heterogeneous effects within each haplogroup. In addition, the haplogroup assignment using HaploGrep can only infer the most likely haplogroup based on the phylogenetic tree, and potential misclassifications of haplogroups is possible. Third, the generalizability of our study can be limited by the fact that all of the WIHS participants are from urban, US-based study sites and most had a BMI > 25. The results should be interpreted with caution. Last, cryptic relatedness was not assessed in the analysis, although all first-degree relatives were removed from the data set.
Our study also has several strengths. To our knowledge, this is the first longitudinal study investigating the contribution of African mtDNA haplogroups to incident DM, insulin resistance, and ß-cell functions. Black men and women are disproportionately affected by DM [35], but this population has not been well studied in the previous literature, and our study fills a gap in the literature. In addition, we used data from a large, prospective cohort, which gave us the opportunity to study incident DM among a disease-free population at the index visit, as well as the trajectory of ß-cell functions and insulin resistance over time using repeated measures. Furthermore, the participants within the cohort living with and without HIV are demographically and behaviorally similar, which decreases confounding, making the cohort ideal for studying genetic contributions to certain phenotypes. Last, the definition of incident DM in the WIHS cohort is consistent with the current American Diabetes Association guidelines [36], and the cohort used a combination of lab results and self-reported information to capture incident cases across all sites, giving a high accuracy of case definition.
In summary, we observed that African-ancestry mtDNA haplogroup L2 was associated with a lower risk of DM and slower age-related declines in ß-cell functions among women living with HIV. Mechanistic studies are needed to confirm our observations and to assess the functional differences between haplogroup L2 and other haplogroups. Investigating the underlying mechanisms of these associations could potentially bring new insights to the development of personalized DM treatment and prevention strategies in this and other high-risk populations.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Study group. Data in this manuscript were collected by the following Women’s Interagency Human Immunodeficiency Virus Study (WIHS) teams and principal investigators: University of Alabama-Birmingham-Mississippi WIHS, Mirjam-Colette Kempf and Deborah Konkle-Parker; Atlanta WIHS, Ighovwerha Ofotokun, Anandi Sheth, and Gina Wingood; Bronx WIHS, Kathryn Anastos and Anjali Sharma; Brooklyn WIHS, Deborah Gustafson and Tracey Wilson; Chicago WIHS, Mardge Cohen and Audrey French; Metropolitan Washington WIHS, Seble Kassaye and Daniel Merenstein; Miami WIHS, Maria Alcaide, Margaret Fischl, and Deborah Jones; University of North Carolina WIHS, Adaora Adimora; Connie Wofsy Women’s Human Immunodeficiency Virus Study, Northern California, Bradley Aouizerat and Phyllis Tien; WIHS Data Management and Analysis Center, Stephen Gange and Elizabeth Golub; Southern California WIHS, Joel Milam.
Author contributions. T. H., T. T. B., and J. S. developed the research question and hypothesis, designed the study, and interpreted the results. T. T. B. and T. H. managed the research funding and infrastructure. J. S. wrote the manuscript. W. T. and J. S. performed the data analysis. T. T. B., T. H., D. S., P. T., B. Aissani, B. Aouizerat, M. V., M. H. K., D. G., K. M., M. C., M. S., A. A. A., M. K. A., and H. B. critically reviewed and revised the manuscript.
Disclaimer. The contents do not represent the views of the US Department of Veterans Affairs of the United States Government. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health.
Financial support. This work was supported by the WIHS, which is funded primarily by the National Institute of Allergy and Infectious Diseases, with additional co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute on Mental Health. Targeted supplemental funding for specific projects is also provided by the National Institute of Dental and Craniofacial Research, the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Deafness and other Communication Disorders, and the National Institutes of Health Office of Research on Women’s Health. WIHS data collection support was provided to the University of Alabama Birmingham-MS WIHS (grant number U01-AI-103401); Atlanta WIHS (grant number U01-AI-103408); Bronx WIHS (grant number U01-AI-035004); Brooklyn WIHS (grant number U01-AI-031834); Chicago WIHS (grant number U01-AI-034993); Metropolitan Washington WIHS (grant number U01-AI-034994); Miami WIHS (grant number U01-AI-103397); University of North Carolina WIHS (grant number U01-AI-103390); Connie Wofsy Women’s Human Immunodeficiency Virus Study, Northern California (grant number U01-AI-034989); WIHS Data Management and Analysis Center (grant number U01-AI-042590); Southern California WIHS (grant number U01-HD-032632; WIHS I–WIHS IV); University of California San Francisco Clinical & Translational Science Institute (CTSA) (grant number UL1-TR000004); Atlanta CTSA (grant number UL1-TR000454); University of North Carolina Center for AIDS Research (CFAR) (grant number P30-AI-050410); and University of Alabama-Birmingham CFAR (grant number P30-AI-027767). Analysis was supported by WIHS Data Management and Analysis Center (grant number U01-AI-04259 to T. T. B. and T. H.). This work was supported by the Tennessee Valley Healthcare System-Veterans Affairs Hospital.
Potential conflicts of interest. T. T. B. is supported in part by the National Institutes of Health/ National Institute of Allergy and Infectious Diseases (grant number K24AI120834) and has served as a consultant to Gilead Sciences, ViiV Healthcare, Merck, EMD-Serono, and Theratechnologies. J. S. is supported in part by the Johns Hopkins University Center for Acquired Immunodeficiency Syndrome Research (grant number P30AI094189). All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Tien PC, Schneider MF, Cox C, et al. Association of HIV infection with incident diabetes mellitus: impact of using hemoglobin A1C as a criterion for diabetes. J Acquir Immune Defic Syndr 2012; 61:334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ledergerber B, Furrer H, Rickenbach M, et al. ; Swiss Human Immunodeficiency Virus Cohort Study Factors associated with the incidence of type 2 diabetes mellitus in HIV-infected participants in the Swiss HIV Cohort Study. Clin Infect Dis 2007; 45:111–9. [DOI] [PubMed] [Google Scholar]
- 3. Butt AA, McGinnis K, Rodriguez-Barradas MC, et al. ; Veterans Aging Cohort Study HIV infection and the risk of diabetes mellitus. AIDS 2009; 23:1227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown TT, Cole SR, Li X, et al. Antiretroviral therapy and the prevalence and incidence of diabetes mellitus in the multicenter AIDS cohort study. Arch Intern Med 2005; 165:1179–84. [DOI] [PubMed] [Google Scholar]
- 5. Stanley TL, Grinspoon SK. Body composition and metabolic changes in HIV-infected patients. J Infect Dis 2012; 205(Suppl 3):S383–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grunfeld C, Delaney JA, Wanke C, et al. Preclinical atherosclerosis due to HIV infection: carotid intima-medial thickness measurements from the FRAM study. AIDS 2009; 23:1841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith CJ, Ryom L, Weber R, et al. ; The Data Collection on Adverse events of anti-HIV Drugs (D:A:D) Study Group. Trends in underlying causes of death in people with HIV from 1999 to 2011 (D:A:D): a multicohort collaboration. Lancet 2014; 384:241–8. [DOI] [PubMed] [Google Scholar]
- 8. Ernster L, Schatz G. Mitochondria: a historical review. J Cell Biol 1981; 91:227s–55s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sivitz WI, Yorek MA. Mitochondrial dysfunction in diabetes: from molecular mechanisms to functional significance and therapeutic opportunities. Antioxid Redox Signal 2010; 12:537–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kraja AT, Liu C, Fetterman JL, et al. Associations of mitochondrial and nuclear mitochondrial variants and genes with seven metabolic traits. Am J Hum Genet 2019; 104:112–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gómez-Durán A, Pacheu-Grau D, López-Gallardo E, et al. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet 2010; 19:3343–53. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Brown TT, Samuels DC, et al. The role of mitochondrial DNA variation in age-related decline in gait speed among older men living with human immunodeficiency virus. Clin Infect Dis 2018;67:778–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hulgan T, Haubrich R, Riddler SA, et al. European mitochondrial DNA haplogroups and metabolic changes during antiretroviral therapy in AIDS Clinical Trials Group Study A5142. AIDS 2011; 25:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hulgan T, Haas DW, Haines JL, et al. Mitochondrial haplogroups and peripheral neuropathy during antiretroviral therapy: an adult AIDS clinical trials group study. AIDS 2005; 19:1341–9. [DOI] [PubMed] [Google Scholar]
- 15. Canter JA, Robbins GK, Selph D, et al. ; Acquired Immunodeficiency Syndrome Clinical Trials Group Study 384 Team; New Work Concept Sheet 273 Team African mitochondrial DNA subhaplogroups and peripheral neuropathy during antiretroviral therapy. J Infect Dis 2010; 201:1703–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fuku N, Park KS, Yamada Y, et al. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet 2007; 80:407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kofler B, Mueller EE, Eder W, et al. Mitochondrial DNA haplogroup T is associated with coronary artery disease and diabetic retinopathy: a case control study. BMC Med Genet 2009; 10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chinnery PF, Mowbray C, Patel SK, et al. Mitochondrial DNA haplogroups and type 2 diabetes: a study of 897 cases and 1010 controls. J Med Genet 2007; 44:e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Micheloud D, Berenguer J, Guzmán-Fulgencio M, et al. European mitochondrial DNA haplogroups and metabolic disorders in HIV/HCV-coinfected patients on highly active antiretroviral therapy. J Acquir Immune Defic Syndr 2011; 58:371–8. [DOI] [PubMed] [Google Scholar]
- 20. Hulgan T, Stein JH, Cotter BR, et al. ; Acquired Immunodeficiency Syndrome Clinical Trials Group A5152s and Data Analysis Concept Sheet 252 Study Teams. Mitochondrial DNA variation and changes in adiponectin and endothelial function in HIV-infected adults after antiretroviral therapy initiation. AIDS Res Hum Retroviruses 2013; 29:1293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nasi M, Guaraldi G, Orlando G, et al. Mitochondrial DNA haplogroups and highly active antiretroviral therapy-related lipodystrophy. Clin Infect Dis 2008; 47:962–8. [DOI] [PubMed] [Google Scholar]
- 22. Barkan SE, Melnick SL, Preston-Martin S, et al. The Women’s Interagency HIV Study. WIHS collaborative study group. Epidemiology 1998; 9:117–25. [PubMed] [Google Scholar]
- 23. Adimora AA, Ramirez C, Benning L, et al. Cohort profile: the Women’s Interagency HIV Study (WIHS). Int J Epidemiol 2018; 47:393–394i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haffner SM, Kennedy E, Gonzalez C, Stern MP, Miettinen H. A prospective analysis of the HOMA model. The Mexico City diabetes study. Diabetes Care 1996; 19:1138–41. [DOI] [PubMed] [Google Scholar]
- 25. Shendre A, Wiener HW, Irvin MR, et al. Genome-wide admixture and association study of subclinical atherosclerosis in the Women’s Interagency HIV Study (WIHS). PLOS One 2017; 12:e0188725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloss‐Brandstätter A, Pacher D, Schönherr S, Weissensteiner H, Binna R, Specht G, Kronenberg F. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Human Mutat 2011; 32:25–32. [DOI] [PubMed] [Google Scholar]
- 27. van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009; 30:E386–94. [DOI] [PubMed] [Google Scholar]
- 28. Thiébaut R, Daucourt V, Mercié P, et al. Lipodystrophy, metabolic disorders, and human immunodeficiency virus infection: Aquitaine Cohort, France, 1999. Groupe d’Epidémiologie Clinique du Syndrome d’Immunodéficience Acquise en Aquitaine. Clin Infect Dis 2000; 31:1482–7. [DOI] [PubMed] [Google Scholar]
- 29. Justman JE, Benning L, Danoff A, et al. Protease inhibitor use and the incidence of diabetes mellitus in a large cohort of HIV-infected women. J Acquir Immune Defic Syndr 2003; 32:298–302. [DOI] [PubMed] [Google Scholar]
- 30. Mehta SH, Moore RD, Thomas DL, Chaisson RE, Sulkowski MS. The effect of HAART and HCV infection on the development of hyperglycemia among HIV-infected persons. J Acquir Immune Defic Syndr 2003; 33: 577–84. [DOI] [PubMed] [Google Scholar]
- 31. Lewis W, Copeland WC, Day BJ. Mitochondrial DNA depletion, oxidative stress, and mutation: mechanisms of dysfunction from nucleoside reverse transcriptase inhibitors. Lab Invest 2001; 81:777–90. [DOI] [PubMed] [Google Scholar]
- 32. McComsey GA, Daar ES, O’Riordan M, et al. Changes in fat mitochondrial DNA and function in subjects randomized to abacavir-lamivudine or tenofovir DF-emtricitabine with atazanavir-ritonavir or efavirenz: AIDS Clinical Trials Group study A5224s, substudy of A5202. J Infect Dis 2013; 207:604–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tranah GJ, Manini TM, Lohman KK, et al. Mitochondrial DNA variation in human metabolic rate and energy expenditure. Mitochondrion 2011; 11: 855–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alawad AO, Merghani TH, Ballal MA. Resting metabolic rate in obese diabetic and obese non-diabetic subjects and its relation to glycaemic control. BMC Res Notes 2013; 6:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bancks MP, Kershaw K, Carson AP, Gordon-Larsen P, Schreiner PJ, Carnethon MR. Association of modifiable risk factors in young adulthood with racial disparity in incident type 2 diabetes during middle adulthood. JAMA 2017; 318:2457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2010; 33(Suppl 1):S62–S69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.