Abstract
Aims
Our objective was to investigate the association of common variants in the coding region of advanced glycosylation end‐product specific receptor (RAGE) and the prognosis of heart failure (HF).
Methods and results
A total of 3394 HF patients were continuously enrolled from January 2009 to August 2018 with a median follow‐up of 20.4 months. Additionally, 2861 healthy subjects also participated in the study. By sequencing these two groups, we identified a common functional missense variant rs2070600 in the coding region of RAGE, which showed a significant association with the prognosis of HF [hazard ratio = 0.53, 95%, confidence interval (CI) = 0.30–0.94, P = 0.03], but no association with the risk of HF (odds ratio = 0.52, 95%, CI = 0.66–1.04, P = 0.106). A series of functional assays revealed that rs2070600‐A, but not ‐G allele, suppressed the expression of RAGE protein by facilitating the binding of miR‐125a‐3p. Furthermore, the RAGE messenger RNA levels of human peripheral blood lymphocytes were reduced in subjects with the rs2070600‐AA genotype compared with subjects with the rs2070600‐GG or ‐AG genotypes. Additionally, our Western blot results from human heart tissue showed increased RAGE expression in HF samples compared with that in healthy donors.
Conclusions
Our results demonstrate that the common missense variant rs2070600‐A allele is associated with a reduced risk of cardiovascular death and cardiac transplantation by facilitating the binding of miR‐125a‐3p.
Keywords: Heart failure, RAGE, Genetics, Prognosis, miR‐125a‐3p
Introduction
Heart failure (HF) is the terminal phase of many cardiovascular diseases, which are often driven by molecular events and genetic causes. Patients with HF have poor prognosis and poor therapeutic options. The optimal development of precision treatment relies on risk stratification and prognostic biomarker identification. Several genome‐wide association studies have identified hundreds of loci associated with the risk of cardiomyopathy 1 and coronary artery disease. 2 However, the link between specific genes and HF is largely undescribed. Advanced glycosylation end product receptor (RAGE, GenBank accession no. NC_000006) localizes on chromosome 6p21 and is a key regulator of endogenous inflammatory effects 3 and reactive oxygen species. 4 The advanced glycosylation end product/RAGE axis is associated with multiple diseases and complications in humans and animal models 5 , 6 and has served as a key therapeutic target 7 for the synthesis of small‐molecule inhibitors. 8 The RAGE gene encodes the advanced glycosylation end product receptor, which belongs to the immunoglobulin superfamily of cell surface receptors because of its special domain 9 that can bind multiple ligands 10 (e.g. advanced glycosylation end products, β‐sheets, S100, and HMGB1). This binding allows the receptors to act as nicotinamide adenine dinucleotide phosphate and nuclear factor‐κ‐gene binding 11 that mediate the pathophysiological process of cardiovascular diseases. 12 , 13 , 14 , 15 Polymorphisms and single‐nucleotide polymorphisms (SNPs) of RAGE have been implicated in disease development in previous studies. 16 Notably, a common nonsynonymous SNP, rs2070600, located in a coding region that causes a conversion at position 82 from glycine to serine (G82S), may be linked to rheumatoid arthritis, 17 cancer, 18 lung diseases, 19 diabetic nephropathy, 20 Alzheimer's disease, 21 and coronary artery disease. 22 However, the association of rs2070600 with HF is undefined, 23 , 24 and further research is needed in particular ethnic groups.
In this report, we hypothesized that the common missense variant of RAGE, rs2070600, is associated with HF outcomes and can be used to predict the prognosis of HF. We evaluated the association between RAGE risk variants and the prognosis of HF. Another case–control study was performed to assess the risk of HF. We then provided functional evidence that the common mutation at the chromosome 6p21.32 risk loci affects RAGE gene expression and investigated the underlying mechanisms at the genetic and cellular levels.
Methods
Study design and eligibility
This study was approved by the Ethics Committee of Tongji Hospital and written informed consent was obtained from all participants. The investigation conformed with the principles outlined in the Declaration of Helsinki. Details on sample recruitment, inclusion and exclusion criteria, data collection, and risk factor definition are described in our previous report. 25 The clinical characteristics of the study samples are shown in Table 1. The reference population for the resequencing efforts were 48 healthy, Han Chinese subjects recruited from a group of individuals undergoing routine health examinations at Tongji Hospital in Wuhan, Hubei province. The HF cohort, including 3394 HF patients, was enrolled from hospitalized patients in Tongji Hospital between January 2009 and August 2018. Finally, 3022 patients remained from the 3394 for prognosis analysis, as follow‐up and genotype was not completed in 200 and 172 patients, respectively. The primary endpoints were heart transplantation and cardiovascular death. The 2861 healthy, community‐based Han Chinese individuals were originally recruited from the general population in 2004, with ages ranging from 23 to 82 years. In addition, 232 patients without coronary artery disease were confirmed with coronary angiography and collected from January to August 2018. Details on inclusion criteria and data collection are provided in the online Supporting Information methods.
Table 1.
Baseline characteristics of the study samples
| Cohort | Re‐sequencing (n = 48) | Control population (n = 2861) | HF population (n = 3022) | Non‐CAD cases (n = 232) |
|---|---|---|---|---|
| Male, n (%) | 52 | 43.52 | 65.62 | 63.38 |
| Mean age (year) | 59.10 ± 10.10 | 58.60 ± 10.30 | 59.60 ± 14.10 | 59.73 ± 10.83 |
| TC (mmol/L) | 4.71 ± 0.34 | 4.93 ± 0.96 | 3.89 ± 1.16 | 3.95 ± 1.02 |
| TG (mmol/L) (median) | 1.25 ± 0.58 | 1.46 ± 1.00 | 1.50 ± 1.14 | 2.34 ± 3.94 |
| HDL (mmol/L) | 1.37 ± 0.37 | 1.46 ± 0.35 | 1.03 ± 0.57 | 1.18 ± 0.43 |
| LDL (mmol/L) | 2.40 ± 0.70 | 2.76 ± 0.79 | 2.36 ± 0.90 | 2.41 ± 0.85 |
| BUN (mmol/L) | / | / | 8.13 ± 5.67 | / |
| Cr (mmol/L) | / | / | 107.15 ± 135.41 | / |
| NT‐proBNP (pg/mL) | / | / | 1868(424,5924) | / |
| Blood pressure (mm Hg) | ||||
| Systolic | 130.15 ± 19.13 | 138.62 ± 24.43 | 131.15 ± 25.22 | 130.80 ± 20.87 |
| Diastolic | 82.35 ± 9.71 | 81.64 ± 13.13 | 80.30 ± 16.16 | 80.37 ± 13.38 |
| LVEF | / | / | 40.10 ± 12.94 | / |
| NYHA | / | / | 1533/952/537(II/III/IV) | / |
| Hypertension, n (%) | 0 | 504 (17.60) | 2372 (78.50) | 104(44.68) |
| Diabetes, n (%) | 0 | / | 913 (30.20) | 35(15.0) |
| Current/ex‐smoker, n (%) | 0 | / | 1122 (37.13) | 76(32.86) |
| Previous myocardial infarction, n (%) | 0 | / | 387(11.81) | / |
| Heart failure, n (%) | 0 | / | 3022 (100) | / |
| History of cerebrovascular disease, n (%) | 0 | / | 275(9.10) | / |
| Beta‐blocker user (%) | 0 | / | 1389 (46.0) | / |
BUN, blood urea nitrogen; CAD, coronary artery disease; Cr, creatinine; DBP, diastolic blood pressure; HDL‐C, high‐density lipoprotein cholesterol; HF, heart failure; LDL‐C, low‐density lipoprotein cholesterol; LVEF, left ventricular ejection fraction; NT‐proBNP, N terminal pro B type natriuretic peptide; NYHA, New York Heart Association; SBP, systolic blood pressure; TC, total cholesterol; TG, triglyceride.
Human tissue samples
The heart samples used in this study were obtained from five patients who underwent heart transplantation at Tongji Hospital and three healthy donors that suffered traffic accidents. Samples were frozen in liquid nitrogen and then stored at −80°C until use. The clinical characteristics of the study samples are summarized in Supporting Information, Table S1 . This study was approved by the Institutional Ethics Committee of Tongji Hospital and is in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from the study participants or their relatives.
Single‐nucleotide polymorphism selection and genotyping
DNA of human samples was extracted from peripheral leucocytes according to our previously published method. 26 Human peripheral lymphocytes were isolated using lymphocyte separation medium (Tian Jin Hao Yang Biological Manufacture, China, LTS10770125). The SNP rs2070600 was genotyped in our study using the TaqMan 5′‐nuclease assay on the TaqMan 7900HT Sequence Detection System (Applied Biosystems, Foster City, California) under the following conditions: 10 min at 95°C (enzyme activation), 40 cycles at 95°C for 15 s each, and 60°C for 1 min (annealing/extension). The endpoint read was performed for allelic discrimination after amplification. The genotyping procedure was consistent with our previous report. 27 Details regarding primers and probes are provided in Supporting Information, Table S2 .
Genetic variation screening
Sequence data were generated by Sanger sequencing of genomic DNA derived from 48 healthy individuals. Polymerase chain reaction (PCR) fragments covering the exons and introns of RAGE (consensus sequence NC_000006.11 GRCh37.p13) were screened using fluorescent dye‐terminator cycle, and products were analysed with an Applied Biosystems 3130xl Genetic Analyser (Applied Biosystems, Foster City, CA). The Chromas programme (Technelysium Pty. Ltd., Helensvale, Queensland, Australia) was used to identify putative polymorphisms that were then confirmed by two independent observers. All identified variants were confirmed by repeated sequencing. Details regarding primers are given in Supporting Information, Table S3 .
In silico analyses
Variants in high linkage disequilibrium (LD) with the variant rs2070600 were identified using HaploReg v4.1 (https://pubs.broadinstitute.org). The respective regions were analysed for potential transcription factor binding sites using JASPAR 2016 (http://jaspar2016.genereg.net/). MiRWalk2.0 was used to predict putative micro RNAs (miRNAs) that target the rs2070600 locus (http://zmf.umm.uni-heidelberg.de/). The influence of rs2070600 on local messenger RNA (mRNA) structure was predicted using UNAFold (http://unafold.rna.albany.edu/).
Plasmid construction, cell culture, and transient transfection
Expression plasmids of the predicted nine transcription factors (E2F6, MSC, ZNF263, ID4, TFAP2A, TFAP2C, EGR1, STAT3, and YTHDF1) were constructed with pcDNA3.1 through amplification of human cDNA derived from the above‐mentioned human heart tissue. According allele frequency of rs2070600 and ancestral allele in dbSNP, we defined the genotype of rs2070600‐GG as wildtype, ‐AA as mutation. Vectors expressing the RAGE wild‐type (rs2070600, ‐GG allele) or mutant (‐AA allele) type were constructed into pcDNA3.1 and flag‐pcDNA3.1 using cDNA derived from human peripheral lymphocytes with different rs2070600 genotypes. AC16 and HEK293T cells were used for the following functional assays. Cell culture and transient transfection procedures are described in detail in Supporting Information, Table S4 .
Western blot analysis
Lipofectamine™ 2000 transfection reagent (Invitrogen) was used to transfect miRNA and construct the cells. AC16 cells were transfected with 3.0 μg pcDNA3.1‐RAGE‐rs2070600‐G or ‐A plasmid and 100 nmol of chemically synthesized has‐miR‐125a‐3p (RIBOBIO Co., Ltd, Guangzhou, China), miRNA inhibitor(s), or miR‐negative control (RIBOBIO Co., Ltd, Guangzhou, China) (Supporting Information, Table S5 ). After 48 h, the cells were washed and homogenized with lysis solution (50 mM Tris‐Cl, pH 8.0; 150 mM NaCl; 0.02% sodium azide; 0.1% SDS; 1 μg/mL aprotinin; 1% Nonidet P‐40; and 0.5% sodium deoxycholate) containing protease inhibitors (100 μg/mL phenylmethylsulfonyl fluoride, 2 μg/mL aprotinin, and 2 μg/mL leupeptin). Samples were then centrifuged at 12 000 × g for 20 min at 4°C, and supernatants were collected. Cell lysates were resolved by 10% SDS‐polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. After blocking with 5% nonfat milk, blots were probed with RAGE antibody (ABclonal, China, A13264), anti‐FLAG antibody (Cell Signalling Technology, USA, 147935), and glyceraldehyde 3‐phosphate dehydrogenase antibody (Santa Cruz Biotechnology, USA, sc‐293,335). Bands were visualized using enhanced chemiluminescence reagents (Pierce Chemical, Rockford, IL) and quantified by densitometry.
Quantitative polymerase chain reaction
Total RNA was isolated from peripheral blood lymphocytes frozen tissues or cells by using TRIzol reagent kit (Thermo Fisher Scientific, 15,596,018) according to the manufacturer's instructions and 250–500 ng of total RNA was reverse‐transcribed using EasyScript First‐Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). RAGE and ACTB (or glyceraldehyde 3‐phosphate dehydrogenase) mRNA levels were measured using absolute quantification methods on the ABI 7900 Fast Real‐Time PCR System (Applied Biosystems Inc.). All experiments were performed in triplicate to avoid experimental error. Related primer sequences are provided in Supporting Information, Table S6 . The sorted chemically synthesized primers of miR‐125a‐3p for qPCR were obtained from RiboBio (Guangzhou, China). U6 was used to control endogenous miRNA.
Statistical analysis
Data were analysed using spss version 24.0 (SPSS, Inc., Chicago, Illinois) for Windows (Microsoft Corp., Redmond, WA). Linkage disequilibrium was calculated using Haploview version 4.1. The polymorphisms were tested for Hardy–Weinberg equilibrium using the χ2 test. We performed multivariate logistic regression analyses based on different genetic models with adjustment for traditional risk factors to test the association between SNPs and HF risk. The genetic models utilized were as follows: (i) additive model (AA vs. AG vs. GG); (ii) recessive model, AA vs. (AG + GG); and (iii) dominant model (AA + AG) vs. GG. Analysis of clinical prognosis of HF was performed using the Cox proportional hazards model. Significant differences were assessed by either one‐way ANOVA followed by Bonferroni's post hoc test or unpaired or paired two‐tailed Student's t‐test, where appropriate. All biostatistics calculations were performed using Prism (GraphPad). Data are expressed as mean ± SEM of n experiments. All probability values were two‐sided, and P < 0.05 was considered significant.
Results
DNA resequencing results
Through RAGE gene resequencing performed in 48 unrelated age‐matched and sex‐matched, healthy controls from the Han Chinese population, we identified 17 polymorphisms in RAGE. Of these, three were located in the exon region, and 14 were located in the intron region (Table 2 ). All 17 polymorphisms were in Hardy–Weinberg equilibrium in our resequencing population. The results were confirmed using 1000 Genomes Browser (https://www.internationalgenome.org/1000-genomes-browsers). Because the SNPs located in the coding region were more likely to affect the function of proteins by changing amino acids, we chose to evaluate the effect of common missense mutations in RAGE on the prognosis of HF. Rs2070600 in the RAGE coding region leads to the conversion of glycine to serine (G82S), as reported in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php).
Table 2.
Characteristics of region of advanced glycosylation end‐product variants identified by resequencing in 48 control subjects
| Gene position a | dbSNP ID b | HGVS c | Gene region | Maj > Min d | MAF |
|---|---|---|---|---|---|
| chr6:32184157 | rs3131300 | NC_000006.11:g.32151934A > G | Intron1 | C/T | 0.084 |
| chr6:32183666 | rs2070600 | NC_000006.11:g.32151443C > T | Exon3(G82S) | G/A | 0.236 |
| chr6:32183681 | rs80096349 | NC_000006.11:g.32151458G > A | Exon3 | A/G | 0.01 |
| chr6:32183445 | rs1035798 | NC_000006.11:g.32151222G > A | Intron3 | C/T | 0.145 |
| chr6:32183517 | rs2269422 | NC_000006.11:g.32151294 T > C | Intron3 | A/G | 0.052 |
| chr6:32182721 | rs17846798 | NC_000006.11:g.32150498G > A | Intron6 | C/T | 0.052 |
| chr6:32182783 | rs17846810 | NC_000006.11:g.32150560G > T | Intron6 | C/A | 0.01 |
| chr6:32182784 | rs17846809 | NC_000006.11:g.32150561G > A | Intron6 | C/T | 0.01 |
| chr6:32182519 | rs184003 | NC_000006.11:g.32150296C > A | Intron7 | G/T | 0.188 |
| chr6:32181979 | rs55640627 | NC_000006.11:g.32149756C > T | Intron8 | C/T | 0.021 |
| chr6:32182106 | rs204996 | NC_000006.11:g.32149883C > T | Intron8 | G/A | 0.02 |
| chr6:32181760 | rs3134941 | NC_000006.11:g.32149537C > G | Intron8 | G/C | 0.01 |
| chr6:32182039 | rs3134940 | NC_000006.11:g.32149816 T > C | Intron8 | A/G | 0.084 |
| chr6:32182024 | rs9391855 | NC_000006.11:g.32149801C > T | Intron8 | G/A | 0.236 |
| chr6:32181795 | rs2853807 | NC_000006.11:g.32149572G > A | Intron8 | C/T | 0.02 |
| chr6:32181483 | rs2071288 | NC_000006.11:g.32149260C > T | Intron9 | G/A | 0.052 |
| chr6:32181442 | rs143357175 | NC_000006.11:g.32149219G > A | Exon10 | C/T | 0.01 |
MAF, minor allele frequency.
Base pair position is based on NCBI GRCh38.
Polymorphisms are numbered relative to transcription start site.
With major allele given first followed by minor allele.
Region of advanced glycosylation end‐product specific receptor SNP rs2070600 G > A had a minor allele frequency (MAF) of 0.236 in the resequencing population (Table 1), 0.225 in HF patients, 0.221 in controls, and 0.233 in non‐coronary artery disease patients (Table 4 ). According to the dbSNP and 1000 Genomes Browser, we identified guanine ‘G’ coding G82 to be the major allele (wildtype) and adenine ‘A’ as the minor allele (mutation) throughout the report.
Table 4.
Association between rs2070600 variants and heart failure risk
| SNP | Group | Genotype | Allele frequency | Additive a | Dominant a | Recessive a | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | GA | GG | G | A | AdjustedP value | OR (95%CI) | AdjustedP value | OR (95%CI) | AdjustedP value | OR (95%CI) | ||
| rs2070600G > A | HF | 167 | 1054 | 1640 | 0.775 | 0.225 | 0.171 | 0.94 (0.86–1.03) | 0.358 | 0.95 (0.85–1.05) | 0.106 | 0.52 (0.66–1.04) |
| Control | 156 | 1091 | 1775 | 0.779 | 0.221 | |||||||
| Non‐CVD | 10 | 93 | 129 | 0.767 | 0.233 | |||||||
CVD, cardiovascular disease; HF, heart failure.
HF vs. control; odds ratios (ORs) and 95% confidence intervals (CIs) were obtained by logistic regression, with or without adjustment for sex, age, hypertension, diabetes, hyperlipidemia, and smoking status.
In silico predictions of functional single‐nucleotide polymorphisms
Among the 17 variants identified, 14 were located in the intronic region while only one common missense variant, rs2070600 (MAF > 0.05), was located in an exonic region. This SNP was in exon 3 and showed strong linkage disequilibrium with rs9391855, which is located in intron 8 (D' = 1, r2 = 1) (Supporting Information, Figure S1 ).
Subsequent bioinformatics analysis was performed to predict the potential functional variants of this gene. The identified variants were annotated using RegulomeDB (http://www.regulomedb.org/), PolyPhen‐2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (https://sift.bii.a-star.edu.sg/), and SNPs3D (http://biogps.org/plugin/321/snps3d/). The rs2070600 variant was found to likely affect function with a PolyPhen‐2 score of 1 (probably damaging), SIFT score of 0.03 (deleterious), and SNPs3D of −0.01 (probably damaging), which, together, strongly suggested functional damage to the RAGE protein (Supporting Information, Table S7 ). Furthermore, RNA structure prediction of ~250‐nts of RAGE mRNA flanking rs2070600 revealed a genotype‐specific structure of RAGE mRNA (Supporting Information, Figure S2 ).
Region of advanced glycosylation end‐product specific receptor rs2070600 variant affects the prognosis of heart failure
Among the 3022 HF patients, cardiovascular death or cardiac transplantation occurred in 295 patients (16.7%) in the GG genotype group (n = 1775), 169 patients (15.3%, n = 1091) in the AG genotype group, and 12 patients (7.7%) in the AA genotype group (n = 156) and beta blocker users in 1389 objectors (46%). Cox proportional hazards analysis showed that the rs2070600‐A allele was significantly associated with a decreased risk of cardiovascular death and cardiac transplantation in crude [hazard ratio (HR) = 0.49, 95% confidence interval (CI) = 0.28–0.87, P = 0.015] and multivariable‐adjusted (HR = 0.53, 95% CI = 0.30–0.94; P = 0.030) recessive genetic models (Table 3 ) (Figure 1A ). However, under dominant models, the rs2070600 genotype did not significantly influence the prognosis of HF patients in the crude model (HR = 0.84, 95% CI = 0.70–1.02, P = 0.074) or multivariable model (HR = 0.86, 95% CI = 0.71–1.04, P = 0.113) after adjusting for sex, age, hypertension, diabetes, hyperlipidaemia, smoking, and beta‐blocker use. In the additive models, the rs2070600‐AA homozygotes showed a higher event‐free survival rate compared with survival rate of rs2070600‐GG homozygotes in crude (AAvsGG; HR = 0.53, 95% CI = 0.30–0.92; P = 0.024), but not multivariable (HR = 0.59, 95% CI = 0.34–1.03; P = 0.064) models (Figure 1). After grouped by beta‐blocker use, the association between rs2070600 and the prognosis of HF showed no statistical difference in group without beta‐blocker use but a trend in group with beta‐blocker use in recessive model (Supporting Information, Table S8, Figure S11). Considering the fact that only 46% of the HF patients used beta‐blockers, the number of patients in group with beta‐blocker use may be too small to detect a statistically significant difference, which needed further investigation. In addition, we surveyed the association between the SNP and HF risk. We carried out a case–control study consisting of 3022 HF patients and 2861 control subjects. Our results showed that the rs2070600 did not significantly influence the susceptibility to HF in recessive model (AA vs. AG + GG, OR = 0.52, 95% CI = 0.66–1.04, P = 0.106) and any of other models (Table 4 ).
Table 3.
Association between rs2070600 variant and prognosis of chronic heart failure
| SNP rs ID | Analysis model | Dominant a | Recessive b | Additive c | |||
|---|---|---|---|---|---|---|---|
| (M > m) | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| rs2070600 (G > A) | Crude | 0.84 (0.70–1.02) | 0.074 | 0.49 (0.28–0.87) | 0.015 * | 0.53 (0.30–0.92) | 0.024 d , * |
| Adjusted | 0.86 (0.71–1.04) | 0.113 | 0.53 (0.45–1.14) | 0.030 * | 0.59 (0.34–1.03) | 0.064 d | |
| 0.91 (0.75–1.10) | 0.332 e | ||||||
M, major allele; m, minor allele; SNP, single nucleotide polymorphism.
Hazard ratio (HR) and 95% confidence intervals (95% CI) were obtained by Cox regression analysis, with or without adjustment for sex, age, hypertension, diabetes, hyperlipidaemia and smoking status, beta blocker use.
Dominant (GGvsAG+AA).
Recessive (AAvsAG+GG).
Additive (AAvsAGvsGG).
In additional model AA vs. GG.
AG vs. GG.
P < 0.05.
Figure 1.
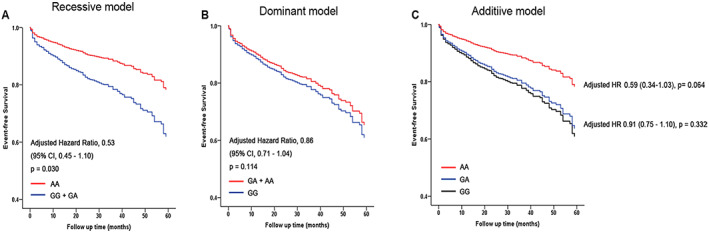
The association between genotypes of rs2070600 and the outcome of heart failure in three genetic models. (A–C) Survival analysis of different genotypes on the prognosis of heart failure using Cox proportional hazards analysis after adjusted for traditional risk factors (sex, age, hypertension, diabetes, hyperlipidemia, smoking status, and beta‐blocker treatment) in recessive (A), dominant (B), and additive model (C), respectively. Cox proportional hazards analysis showed the association of genotypes of rs2070600‐A allele with cardiovascular deaths or cardiac transplantation in recessive model.
Increased expression of RAGE in the failing heart
To explore the expression of RAGE in failing heart, we reanalysed the RNA sequencing data from GEO database (GSE57338). The expression of RAGE is higher in the HF group compared with the control group (HF VS control logFC = 0.052, P = 0.024, Supporting Information Figure S9). Furthermore, we detected RAGE expression in heart tissue samples from HF patients (five cases) and healthy donors (three controls) by western blot analysis. The results showed that the protein levels of RAGE were significantly higher in HF patients compared to levels in control subjects. To further confirm the association between the rs2070600 genotype and RAGE protein expression, the DNA was extracted and sequenced. As shown in Figure 2A, there were four major allele homozygotes (rs2070600‐GG) and one heterozygote in HF patient group compared with three major allele homozygotes in the healthy donor group. Western blot analysis revealed that RAGE expression increased by 48% in rs2070600 GG allele homozygote HF tissues (P = 0.041).
Figure 2.
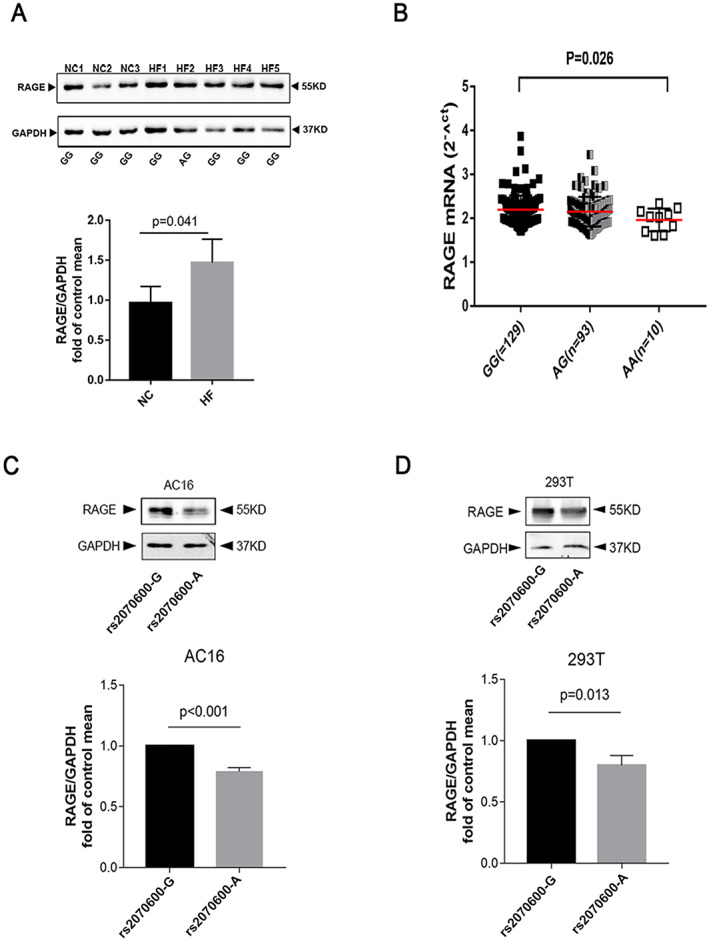
Rs2070600‐allele influences expression of region of advanced glycosylation end‐product specific receptor (RAGE), and the level of RAGE in failing heart. (A) Comparison of RAGE transcript levels in peripheral blood lymphocytes between rs2070600‐AA allele (n = 10), ‐AG (n = 93) and ‐GG allele carriers (n = 129). (B) Protein extracts of human cardiac tissue from patients with severe HF (n = 5) and control (non‐HF) hearts (n = 3) were normalized to GAPDH levels (GAPDH: glyceraldehyde 3‐phosphate dehydrogenase). Genotypes of the samples were described. (C–D) After transient transfection of pc3.1‐RAGE‐rs2070600‐GG and ‐AA, western blot showed the protein expression of ‐AA was significantly lower than ‐GG both in AC16 and 293 T cells. The data are presented as mean ± SD from three independent experiments.
In vivo effects of rs2070600 variant on messenger RNA levels
To further investigate the hypothesis that rs2070600 could influence gene expression, genotype‐dependent RAGE expression was assessed in RNA extracts of peripheral blood lymphocytes from 232 normal individuals. The rs2070600‐AA genotype showed significantly lower RAGE mRNA levels compared with the rs2070600‐GG genotype (Figure 2B).
To confirm the influence of rs2070600 on protein expression, plasmids harbouring G or A alleles of rs2070600 were transiently transfected into AC16 cells and HEK293T cells. Reduced RAGE expression was detected in AC16 cells transfected with rs2070600‐A allele compared with expression after transfection with rs2070600‐G allele (Figure 2C). This result was reiterated in 293 T cell lines (Figure 2D) and HUVEC cell lines (Supporting Information, Figure S3 ). To eliminate the possibility that rs2070600 could affect the binding affinity of the antibody (ABclonal, China, A13264), which contains a sequence corresponding to amino acids 41–340 of human AGER (NP_001127.1) (https://abclonal.com.cn/catalog/A13264), a pcDNA3.1 vector containing Flag‐pcDNA3.1‐RAGE‐rs2070600‐G or ‐A was transfected into AC16 and 293 T, and an anti‐FLAG antibody was used to detect the level of RAGE protein. The results showed that RAGE expression of the rs2070600‐A allele was lower than that of the ‐G allele, which was consistent with the above results (Supporting Information, Figure S6 ).
Micro RNAs modulate RAGE expression
To further investigate the mechanism by which rs2070600 could influence the expression of RAGE, bioinformatics analysis and cell experiments were performed. Bioinformatics analysis revealed that rs2070600 was located in the potential binding site of several transcription factors including E2F6, MSC, ZNF263, ID4, TFAP2A, TFAP2C, EGR1, STAT3, and YTHDF1. However, no significant changes in RAGE expression were observed in AC16 cells transfected with the above nine transcription factors (Supporting Information, Figure S4 ).
Considering that a common variant in the coding region could affect the binding affinity of microRNA, 28 we then focused on searching for the potential microRNA that could provide the interpretation. Seven candidate miRNAs were predicted to be influenced by rs2070600 using bioinformatics analysis (Supporting Information, Table S5 ).
To identify the functional miRNAs, the pcDNA3.1‐RAGE‐rs2070600‐A construct was co‐transfected with the predicted miRNAs into AC16 cells. As shown in Figure 3A, B, miR125a‐3p displayed efficient suppression of RAGE expression, while the other six miRNAs did not significantly decrease the expression of RAGE (Supporting Information, Figure S5 ). We further explored the effects of the association between miRNA‐125a‐3p overexpression and inhibition of RAGE expression. The PcDNA3.1‐RAGE‐rs2070600‐G or pcDNA3.1‐RAGE‐rs2070600‐A constructs were co‐transfected with either 100 nmol of has‐miR‐125a‐3p, has‐miR‐125a‐3p inhibitor(s), or miR‐negative control (RIBOBIO Co., Ltd, Guangzhou, China). Overexpression of miRNA‐125a‐3p downregulated RAGE expression level, while inhibition of miR‐125a‐3p significantly increased the expression of RAGE in AC16 cells transfected with pcDNA3.1‐RAGE‐rs2070600‐A. However, no effects of miRNA‐125a‐3p or inhibitor on RAGE expression were observed in AC16 cells transfected with pcDNA3.1‐RAGE‐rs2070600‐G (Figure 3B–C ). Additionally, the relative level of miR‐125a‐3p was detected in peripheral blood lymphocytes, and we found no difference among participants carrying the rs2070600‐GG, AG, or AA genotypes (Supporting Information, Figure S6 ). Furthermore, compared with health control, an increased tendency expression of mir‐125a‐3p in failing heart was found, but it was no statistical difference (Supporting Information, Figure S7A). To investigate how inflammatory stimuli influence the miR‐125a‐3p expressing in different cells types, we used 100 ng/L lipopolysaccharide to treat different cells (AC16, 293 T, HUVEC). We had a consistent result that lipopolysaccharide can induce mir‐125a‐3p high expression in intervention groups compared to controls. The details were supplied in Figure S7B–D . Additionally, the DNA sequences proximal to rs2070600 in AC16(A) and 293 T(B) cell lines were offered in Figure S8.
Figure 3.
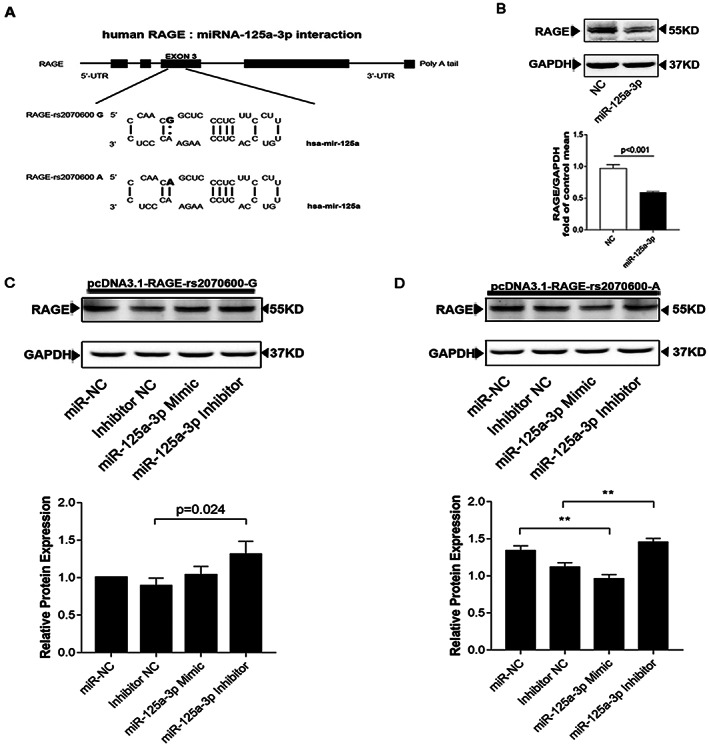
Mir‐125a‐3p directly targets the rs2070600‐A allele and mediates allelic expression. (A) Schema graph to illustrate the loci of rs2070600 G/A variant occurs at the miR‐125a‐3p binding site. Mir‐125a‐3p co‐action rs2070600‐A allele which in the coding region sequence of the gene to regulate the expression of RAGE. T base‐paired with U in the Watson–Crick mode (solid line), whereas allele G did not (shown without line). (B) miR‐125a‐3p negatively regulates the protein level of RAGE in AC16 analysed by Western blotting. (C–D) PcDNA3.1‐RAGE‐rs2070600‐G (C) or ‐rs2070600‐A (D) were co‐transfected with miR‐125a‐3p‐mimics, negative control miRNA (miR NC), miR‐125a‐3p‐inhibitor, or inhibitor negative control (Inhibitor NC) into AC16. Overexpression of MiR‐125a‐3p significantly reduced the level of RAGE in pcDNA3.1‐RAGE‐rs2070600‐A transfected cell, while the expression of RAGE increased after inhibition of MiR‐125a‐3p. No obvious differences were observed in AC16 transfected with pcDNA3.1‐RAGE‐rs2070600‐G.
Discussion
We identified a common SNP (rs2070600) in the coding region sequence (CDS) of RAGE significantly associated with the prognosis of HF in a prospective, observational, single‐centre study consisting of 3394 HF patients and 2861 control participants. The SNP affects the expression of RAGE through interacting with miR‐125a‐3p.
Chronic inflammation contributes to the progression of HF. 29 RAGE is a key mediator of inflammatory response. Under normal conditions, it plays a small role in normal physiological development and in healthy homeostasis. After acute infection or massive injury, RAGE is active and releases proinflammatory factors, 30 directly recruits leucocytes, and detects pathogen DNA. 31 RAGE aggravated the inflammatory response and tissue damage in HF following induction of amassing ligands. 32 Persistent activation of RAGE/nuclear factor‐κ‐gene binding modulates pro‐inflammatory cytokine expression, such as interleukin (IL)‐1, IL‐6, and tumor necrosis factor‐α, which leads to the activation of cardiomyocyte death pathways. 33
Plasma soluble RAGE levels were obviously higher in the rs2070600‐GG genotype of Korean subjects compared with the AA and AG genotypes. 34 As a protein biomarker, soluble RAGE was associated with all‐cause mortality in the Framingham Heart Study (n = 3523). 35 From the above studies, we could infer that the rs2070600 haplotype may be linked to the outcomes of cardiovascular diseases. Our study further uncovered the direct relationship between rs2070600 genotype and prognosis of HF. Based on our case–control study, rs2070600 was not associated with the risk of HF, which was in line with the Framingham offspring study 36 where no association was observed between rs2070600 and the incidence of cardiovascular disease in diabetic or non‐diabetic subjects (n = 1632). In brief, although the common missense variant (rs2070600) of RAGE does not cause HF, it may play an important role in the progression of HF.
We identified an exclusive missense mutation, rs2070600, in the RAGE CDS region that has a MAF of 0.232. Further bioinformatics analysis strongly suggested that rs2070600 is a functional variant. By querying the GTEX database (https://gtexportal.org), it was confirmed that rs2070600 is the expression quantitative trait loci. In a study exploring the association between rs2070600 and lung function, Miller et al. 19 showed that the major allele of rs2070600 is linked to higher FEV1 and FEV1/FVC in 1024 U.K. participants. Rs2070600 (G82S) is a non‐synonymous polymorphism that changes the acid sequence of the encoded protein from glycine to serine. A series of assays were carried out to investigate the underlying mechanisms. In vivo gene expression analysis of human lymphocytes revealed that the rs2070600‐AA genotype decreased RAGE expression compared with RAGE expression resultant from the rs2070600‐GG or rs2070600‐AG genotypes, which was consistent with the results from cell experiments that RAGE expression was lower in cells transfected with pcDNA3.1‐rs2070600‐A compared with the expression in cells transfected with pcDNA3.1‐rs2070600‐T allele. Taken together, we confirmed the predicted result of variant rs2070600 that the minor allele could decrease RAGE protein levels.
Multiple studies have shown that mRNAs can bind to CDSs and effectively inhibit translation. 37 , 38 The current study revealed a possible underlying mechanism by which miR‐125a‐3p directly targets the rs2070600‐A allele and decreases the expression of RAGE. Our result was consistent with a recent report by Fu et al. 39 that the characteristic of miRNAs that directly targeted the CDS were prone to extensive base‐pairing at the 3′ seed. However, the specific role of miR‐125a‐3p in the pathogenesis of HF should be investigated in future studies. To our knowledge, we have discovered the RAGE gene variant locus of a disease‐associated miR‐SNP interaction found within the CDS of RAGE. Furthermore, assessing whether rs2070600‐AA genotype risk is mingled by environmental factors or other common RAGE SNPs is necessary to explore the regulatory controls that impact RAGE expression.
Several limitations of our study should be mentioned. The first limitation of our study is that this is an observational study and that the positive association between RAGE variants and HF needs to be confirmed in prospective cohort studies. Second, the HF patients in our study were recruited mainly from Wuhan Tongji Hospital, and the results need to be validated in further multicentre studies including larger HF populations. Third, we cannot rule out the possibility that other variants in linkage disequilibrium with rs2070600 may account for the association with the prognosis of HF. Finally, other regulatory factors may be involved in the regulation of RAGE gene expression.
Conclusions
The genetic and functional evidence indicated that rs2070600 in the coding region of RAGE is associated with the prognosis of HF in the Han Chinese population. The rs2070600‐AA genotype shows a favourable prognosis for HF compared with the rs2070600‐G allele carriers, which is dependent on beta‐blocker use. A mechanistic study demonstrated that miR‐125a‐3p could directly target the rs2070600‐A allele to decrease RAGE expression. Targeting RAGE is an attractive way to decrease HF‐associated mortality and improve the prognosis of HF in the future.
Conflict of interest
None declared.
Funding
This work was supported by the grant and key project from National Key R&D Program of China (2017YFC0909400) and the National Natural Science Foundation of China (81770351 and 81630010).
Supporting information
Table S1: Clinical characteristics of Non‐HF and HF Samples
Table S2: Primers used for PCR and sequencing
Table S3: Sequence of probes and primers sets
Table S4. Primers used for PCR and plasmid construction
Table S5. Characteristics of Putative Allele‐Specific microRNAs
Table S6. Primers used for Quantitative Real‐Time PCR analysis
Table S7. Haploblock Structure of RAGE genetic variants and functional annotation of the variants
Table S8. Stratified analysis of beta blocker user for analysis the association rs2070600 with heart failure outcomes
Figure S1. Linkage disequilibrium (LD) structure and haplotype blocks of the RAGE gene. LD (D’) for identified polymorphisms in RAGE, as generated by Haploview version 4.1 from the genotype data of 48 random controls. Haplotype blocks derived from these genotypes using the solid spine LD setting are outlined in black.
Figure S2. The effect of rs2070600 genotype on local mRNA structure. RNA structure of ~250‐nts of RAGE mRNA flanking rs2070600 (red open‐circle) was predicted using UNAfold. Results revealed a genotype‐specific structure of RAGE mRNA. The table summarizes unfold microRNA targeting prediction results which indicate that rs2070600 A allele is energetically more favorable.
Figure S3. Rs2070600 genotype influences expression of RAGE in HUVEC cells. The rs2070600 mutant A allele displayed significantly reduced protein expression of RAGE in HUVEC cells. The data are presented as mean ± SD from three independent experiments.
Figure S4. Interaction between transcription factors and RAGE expression in AC16 cells. The predicted transcription factors didn't significantly influence the expression of RAGE. AC16 cells were transfected with pcDNA3.1 empty vector or pcDNA3.1‐TFAP2C construct, pcDNA3.1‐ZNF263 construct, pcDNA3.1‐YTFDH1 construct (A), or pcDNA3.1‐ID4 construct, pcDNA3.1‐E2F6 construct (B), or pcDNA3.1‐MSC construct (C), or pcDNA3.1‐TFAP2A construct (D), or pcDNA3.1‐STAT3 construct (E), or pcDNA3.1‐EGR1 construct (F). The data are presented as mean±SD from three independent experiments. N.S., no significant.
Figure S5. Interaction between miRNA and RAGE expression in AC16 cells. Six of the predicted miRNAs could not significantly suppress the expression of RAGE. AC16 cells were transfected pcDNA3.1‐RAGE‐rs2070600‐A 300ng with 100 nmol miR‐Negative Control (RIBOBIO Co., Ltd, Guangzhou, China) and other 8 miRNAs. The data are presented as mean ± SD from three independent experiments. NS, no significant.
Figure S6 Flag label plasmids of pcDNA‐rs2070600‐G(RAGE‐WT) and ‐rs2070600‐A (RAGE‐MU) transfected into AC16 and 293T cells, and the relative level of miR‐125a‐3p in peripheral blood lymphocytes.
Figure S7 Relative miR‐125a‐3p expressed of vivo and vitro (A) Relative miR‐125a‐3p expression in failing heart and health controls measured by real‐time PCR. Data are expressed as mean ± SEM. LPS 100ng/L induced miR‐125a‐3p expressed in vitro. Relative miR‐125a‐3p expression in AC16 (B), 293T(C) and HUVES(D).
Figure S8. DNA sequences proximal to rs2070600 in AC16(A) and 293T(B) cell lines.
Figure S9 Differential of RAGE/Endogenous RAGE ligands gene expression analysis in GSE57338
Figure S10 Relative RAGE expression in vitro normalize by GAPDH, RAGE mRNA expression levels was detected by qPCR, and GAPDH was used as endogenous control to mRNA. (A) AC16,(B)293T.
Figure S11 Effects of rs2070600 on the prognosis of HF patients grouped by β‐blocker use under different models. By Cox proportional hazards analysis after adjusted for traditional risk factors (sex, age, hypertension, diabetes, hyperlipidemia, smoking status), the link of rs2070600 with heart failure outcomes showed no statistical significance in group with and without β‐blocker use in recessive(A‐B), dominant(C‐D) and additive models(E‐F).
Acknowledgements
We thank the team of clinical follow‐up in the Department of Cardiovascular Medicine at Tongji Hospital for their excellent work. We also thank the clinicians in the Division of Cardiology for their tremendous help and efforts.
Li, S. , Hu, D. , Hu, S. , Sun, Y. , Zhang, Y. , Li, H. , Chen, Y. , Liu, H. , Cui, G. , and Wang, D. W. (2020) Association of rs2070600 in advanced glycosylation end‐product specific receptor with prognosis of heart failure. ESC Heart Failure, 7: 3561–3572. 10.1002/ehf2.12769.
Shiyang Li and Dong Hu equally contributed to this work.
References
- 1. Tayal U, Prasad S, Cook SA. Genetics and genomics of dilated cardiomyopathy and systolic heart failure. Genome Med 2017; 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bjorkegren JLM, Kovacic JC, Dudley JT, Schadt EE. Genome‐wide significant loci: how important are they? Systems genetics to understand heritability of coronary artery disease and other common complex disorders. J Am Coll Cardiol 2015; 65: 830–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chavakis T, Bierhaus A, Al‐Fakhri N, Schneider D, Witte S, Linn T, Nagashima M, Morser J, Arnold B, Preissner KT, Nawroth PP. The pattern recognition receptor (RAGE) is a counterreceptor for leukocyte integrins: a novel pathway for inflammatory cell recruitment. J Exp Med 2003; 198: 1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kang R, Tang D, Lotze MT, Zeh HJ 3rd. RAGE regulates autophagy and apoptosis following oxidative injury. Autophagy 2011; 7: 442–444. [DOI] [PubMed] [Google Scholar]
- 5. Cho MH, Castaldi PJ, Hersh CP, Hobbs BD, Barr RG, Tal‐Singer R, Bakke P, Gulsvik A, San Jose Estepar R, van Beek EJ, Coxson HO, Lynch DA, Washko GR, Laird NM, Crapo JD, Beaty TH, Silverman EK, Nett Genetics E, Investigators CO . A genome‐wide association study of emphysema and airway quantitative imaging phenotypes. Am J Respir Crit Care Med 2015; 192: 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldin A, Beckman JA, Schmidt AM, Creager MA. Advanced glycation end products: sparking the development of diabetic vascular injury. Circulation 2006; 114: 597–605. [DOI] [PubMed] [Google Scholar]
- 7. Koyama H, Yamamoto H, Nishizawa Y. RAGE and soluble RAGE: potential therapeutic targets for cardiovascular diseases. Mol Med 2007; 13: 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bongarzone S, Savickas V, Luzi F, Gee AD. Targeting the receptor for advanced glycation endproducts (RAGE): a medicinal chemistry perspective. J Med Chem 2017; 60: 7213–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sirois CM, Jin T, Miller AL, Bertheloot D, Nakamura H, Horvath GL, Mian A, Jiang J, Schrum J, Bossaller L, Pelka K, Garbi N, Brewah Y, Tian J, Chang C, Chowdhury PS, Sims GP, Kolbeck R, Coyle AJ, Humbles AA, Xiao TS, Latz E. RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. J Exp Med 2013; 210: 2447–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fritz G. RAGE: a single receptor fits multiple ligands. Trends Biochem Sci 2011; 36: 625–632. [DOI] [PubMed] [Google Scholar]
- 11. Wautier MP, Chappey O, Corda S, Stern DM, Schmidt AM, Wautier JL. Activation of NADPH oxidase by AGE links oxidant stress to altered gene expression via RAGE. Am J Physiol Endocrinol Metab 2001; 280: E685–E694. [DOI] [PubMed] [Google Scholar]
- 12. Pickering RJ, Tikellis C, Rosado CJ, Tsorotes D, Dimitropoulos A, Smith M, Huet O, Seeber RM, Abhayawardana R, Johnstone EK, Golledge J, Wang Y, Jandeleit‐Dahm KA, Cooper ME, Pfleger KD, Thomas MC. Transactivation of RAGE mediates angiotensin‐induced inflammation and atherogenesis. J Clin Invest 2019; 129: 406–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Volz HC, Laohachewin D, Seidel C, Lasitschka F, Keilbach K, Wienbrandt AR, Andrassy J, Bierhaus A, Kaya Z, Katus HA, Andrassy M. S100A8/A9 aggravates post‐ischemic heart failure through activation of RAGE‐dependent NF‐kappaB signaling. Basic Res Cardiol 2012; 107: 250. [DOI] [PubMed] [Google Scholar]
- 14. Kraakman MJ, Lee MK, Al‐Sharea A, Dragoljevic D, Barrett TJ, Montenont E, Basu D, Heywood S, Kammoun HL, Flynn M, Whillas A, Hanssen NM, Febbraio MA, Westein E, Fisher EA, Chin‐Dusting J, Cooper ME, Berger JS, Goldberg IJ, Nagareddy PR, Murphy AJ. Neutrophil‐derived S100 calcium‐binding proteins A8/A9 promote reticulated thrombocytosis and atherogenesis in diabetes. J Clin Invest 2017; 127: 2133–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andrassy M, Volz HC, Igwe JC, Funke B, Eichberger SN, Kaya Z, Buss S, Autschbach F, Pleger ST, Lukic IK, Bea F, Hardt SE, Humpert PM, Bianchi ME, Mairbaurl H, Nawroth PP, Remppis A, Katus HA, Bierhaus A. High‐mobility group box‐1 in ischemia‐reperfusion injury of the heart. Circulation 2008; 117: 3216–3226. [DOI] [PubMed] [Google Scholar]
- 16. Serveaux‐Dancer M, Jabaudon M, Creveaux I, Belville C, Blondonnet R, Gross C, Constantin JM, Blanchon L, Sapin V. Pathological implications of receptor for advanced glycation end‐product (AGER) gene polymorphism. Dis Markers 2019; 2019: 2067353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Steenvoorden MM, van der Helm‐van Mil AH, Stoeken G, Bank RA, Devries RR, Huizinga TW, Degroot J, Toes RE. The RAGE G82S polymorphism is not associated with rheumatoid arthritis independently of HLA‐DRB1*0401. Rheumatology (Oxford) 2006; 45: 488–490. [DOI] [PubMed] [Google Scholar]
- 18. Yue L, Zhang Q, He L, Zhang M, Dong J, Zhao D, Ma H, Pan H, Zheng L. Genetic predisposition of six well‐defined polymorphisms in HMGB1/RAGE pathway to breast cancer in a large Han Chinese population. J Cell Mol Med 2016; 20: 1966–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller S, Henry AP, Hodge E, Kheirallah AK, Billington CK, Rimington TL, Bhaker SK, Obeidat M, Melen E, Merid SK, Swan C, Gowland C, Nelson CP, Stewart CE, Bolton CE, Kilty I, Malarstig A, Parker SG, Moffatt MF, Wardlaw AJ, Hall IP, Sayers I. The Ser82 RAGE variant affects lung function and serum RAGE in smokers and sRAGE production in vitro. PLoS ONE 2016; 11: e0164041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lindholm E, Bakhtadze E, Sjogren M, Cilio CM, Agardh E, Groop L, Agardh CD. The −374 T/A polymorphism in the gene encoding RAGE is associated with diabetic nephropathy and retinopathy in type 1 diabetic patients. Diabetologia 2006; 49: 2745–2755. [DOI] [PubMed] [Google Scholar]
- 21. Deo P, Dhillon VS, Chua A, Thomas P, Fenech M. APOE epsilon4 carriers have a greater propensity to glycation and sRAGE which is further influenced by RAGE G82S polymorphism. J Gerontol A Biol Sci Med Sci 2019. Nov 2. pii: glz259. [DOI] [PubMed] [Google Scholar]
- 22. Ma WQ, Qu QR, Zhao Y, Liu NF. Association of RAGE gene Gly82Ser polymorphism with coronary artery disease and ischemic stroke: a systematic review and meta‐analysis. Medicine (Baltimore) 2016; 95: e5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Biros E, Moran CS, Norman PE, Hankey GJ, Yeap BB, Almeida OP, Flicker L, White R, Jones R, Golledge J. Association between the advanced glycosylation end product‐specific receptor gene and cardiovascular death in older men. PLoS ONE 2015; 10: e0134475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J, Zou L, Song Z, Lang X, Huang S, Lu F, Han L, Xu Z. Meta‐analysis of RAGE gene polymorphism and coronary heart disease risk. PLoS ONE 2012; 7: e50790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang J, Li C, Song Y, Fan X, You L, Tan L, Xiao L, Li Q, Ruan G, Hu S, Cui W, Li Z, Ni L, Chen C, Woo AY, Xiao RP, Wang DW. ADRB2 polymorphism Arg16Gly modifies the natural outcome of heart failure and dictates therapeutic response to beta‐blockers in patients with heart failure. Cell Discov 2018; 4: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ding H, Wu B, Wang H, Lu Z, Yan J, Wang X, Shaffer JR, Hui R, Wang DW. A novel loss‐of‐function DDAH1 promoter polymorphism is associated with increased susceptibility to thrombosis stroke and coronary heart disease. Circ Res 2010; 106: 1145–1152. [DOI] [PubMed] [Google Scholar]
- 27. Cui G, Li Z, Li R, Huang J, Wang H, Zhang L, Ding H, Wang DW. A functional variant in APOA5/A4/C3/A1 gene cluster contributes to elevated triglycerides and severity of CAD by interfering with microRNA 3201 binding efficiency. J Am Coll Cardiol 2014; 64: 267–277. [DOI] [PubMed] [Google Scholar]
- 28. Zhang X, Yoon JY, Morley M, McLendon JM, Mapuskar KA, Gutmann R, Mehdi H, Bloom HL, Dudley SC, Ellinor PT, Shalaby AA, Weiss R, Tang WHW, Moravec CS, Singh M, Taylor AL, Yancy CW, Feldman AM, McNamara DM, Irani K, Spitz DR, Breheny P, Margulies KB, London B, Boudreau RL. A common variant alters SCN5A‐miR‐24 interaction and associates with heart failure mortality. J Clin Invest 2018; 128: 1154–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buckley LF, Abbate A. Interleukin‐1 blockade in cardiovascular diseases: a clinical update. Eur Heart J 2018; 39: 2063–2069. [DOI] [PubMed] [Google Scholar]
- 30. Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, Neurath MF, Slattery T, Beach D, McClary J, Nagashima M, Morser J, Stern D, Schmidt AM. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell 1999; 97: 889–901. [DOI] [PubMed] [Google Scholar]
- 31. Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak‐Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll‐like receptor 9‐dependent activation by DNA‐containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol 2007; 8: 487–496. [DOI] [PubMed] [Google Scholar]
- 32. Volz HC, Kaya Z, Katus HA, Andrassy M. The role of HMGB1/RAGE in inflammatory cardiomyopathy. Semin Thromb Hemost 2010; 36: 185–194. [DOI] [PubMed] [Google Scholar]
- 33. Hamid T, Guo SZ, Kingery JR, Xiang X, Dawn B, Prabhu SD. Cardiomyocyte NF‐kappaB p65 promotes adverse remodelling, apoptosis, and endoplasmic reticulum stress in heart failure. Cardiovasc Res 2011; 89: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jang Y, Kim JY, Kang SM, Kim JS, Chae JS, Kim OY, Koh SJ, Lee HC, Ahn CW, Song YD, Lee JH. Association of the Gly82Ser polymorphism in the receptor for advanced glycation end products (RAGE) gene with circulating levels of soluble RAGE and inflammatory markers in nondiabetic and nonobese Koreans. Metabolism 2007; 56: 199–205. [DOI] [PubMed] [Google Scholar]
- 35. Ho JE, Lyass A, Courchesne P, Chen G, Liu C, Yin X, Hwang SJ, Massaro JM, Larson MG, Levy D. Protein biomarkers of cardiovascular disease and mortality in the community. J Am Heart Assoc 2018; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hofmann MA, Yang Q, Harja E, Kedia P, Gregersen PK, Cupples LA, Schmidt AM, Hudson BI. The RAGE Gly82Ser polymorphism is not associated with cardiovascular disease in the Framingham offspring study. Atherosclerosis 2005; 182: 301–305. [DOI] [PubMed] [Google Scholar]
- 37. Borel C, Gagnebin M, Gehrig C, Kriventseva EV, Zdobnov EM, Antonarakis SE. Mapping of small RNAs in the human ENCODE regions. Am J Hum Genet 2008; 82: 971–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hausser J, Syed AP, Bilen B, Zavolan M. Analysis of CDS‐located miRNA target sites suggests that they can effectively inhibit translation. Genome Res 2013; 23: 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang K, Zhang X, Cai Z, Zhou J, Cao R, Zhao Y, Chen Z, Wang D, Ruan W, Zhao Q, Liu G, Xue Y, Qin Y, Zhou B, Wu L, Nilsen T, Zhou Y, Fu XD. A novel class of microRNA‐recognition elements that function only within open reading frames. Nat Struct Mol Biol 2018; 25: 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Clinical characteristics of Non‐HF and HF Samples
Table S2: Primers used for PCR and sequencing
Table S3: Sequence of probes and primers sets
Table S4. Primers used for PCR and plasmid construction
Table S5. Characteristics of Putative Allele‐Specific microRNAs
Table S6. Primers used for Quantitative Real‐Time PCR analysis
Table S7. Haploblock Structure of RAGE genetic variants and functional annotation of the variants
Table S8. Stratified analysis of beta blocker user for analysis the association rs2070600 with heart failure outcomes
Figure S1. Linkage disequilibrium (LD) structure and haplotype blocks of the RAGE gene. LD (D’) for identified polymorphisms in RAGE, as generated by Haploview version 4.1 from the genotype data of 48 random controls. Haplotype blocks derived from these genotypes using the solid spine LD setting are outlined in black.
Figure S2. The effect of rs2070600 genotype on local mRNA structure. RNA structure of ~250‐nts of RAGE mRNA flanking rs2070600 (red open‐circle) was predicted using UNAfold. Results revealed a genotype‐specific structure of RAGE mRNA. The table summarizes unfold microRNA targeting prediction results which indicate that rs2070600 A allele is energetically more favorable.
Figure S3. Rs2070600 genotype influences expression of RAGE in HUVEC cells. The rs2070600 mutant A allele displayed significantly reduced protein expression of RAGE in HUVEC cells. The data are presented as mean ± SD from three independent experiments.
Figure S4. Interaction between transcription factors and RAGE expression in AC16 cells. The predicted transcription factors didn't significantly influence the expression of RAGE. AC16 cells were transfected with pcDNA3.1 empty vector or pcDNA3.1‐TFAP2C construct, pcDNA3.1‐ZNF263 construct, pcDNA3.1‐YTFDH1 construct (A), or pcDNA3.1‐ID4 construct, pcDNA3.1‐E2F6 construct (B), or pcDNA3.1‐MSC construct (C), or pcDNA3.1‐TFAP2A construct (D), or pcDNA3.1‐STAT3 construct (E), or pcDNA3.1‐EGR1 construct (F). The data are presented as mean±SD from three independent experiments. N.S., no significant.
Figure S5. Interaction between miRNA and RAGE expression in AC16 cells. Six of the predicted miRNAs could not significantly suppress the expression of RAGE. AC16 cells were transfected pcDNA3.1‐RAGE‐rs2070600‐A 300ng with 100 nmol miR‐Negative Control (RIBOBIO Co., Ltd, Guangzhou, China) and other 8 miRNAs. The data are presented as mean ± SD from three independent experiments. NS, no significant.
Figure S6 Flag label plasmids of pcDNA‐rs2070600‐G(RAGE‐WT) and ‐rs2070600‐A (RAGE‐MU) transfected into AC16 and 293T cells, and the relative level of miR‐125a‐3p in peripheral blood lymphocytes.
Figure S7 Relative miR‐125a‐3p expressed of vivo and vitro (A) Relative miR‐125a‐3p expression in failing heart and health controls measured by real‐time PCR. Data are expressed as mean ± SEM. LPS 100ng/L induced miR‐125a‐3p expressed in vitro. Relative miR‐125a‐3p expression in AC16 (B), 293T(C) and HUVES(D).
Figure S8. DNA sequences proximal to rs2070600 in AC16(A) and 293T(B) cell lines.
Figure S9 Differential of RAGE/Endogenous RAGE ligands gene expression analysis in GSE57338
Figure S10 Relative RAGE expression in vitro normalize by GAPDH, RAGE mRNA expression levels was detected by qPCR, and GAPDH was used as endogenous control to mRNA. (A) AC16,(B)293T.
Figure S11 Effects of rs2070600 on the prognosis of HF patients grouped by β‐blocker use under different models. By Cox proportional hazards analysis after adjusted for traditional risk factors (sex, age, hypertension, diabetes, hyperlipidemia, smoking status), the link of rs2070600 with heart failure outcomes showed no statistical significance in group with and without β‐blocker use in recessive(A‐B), dominant(C‐D) and additive models(E‐F).


