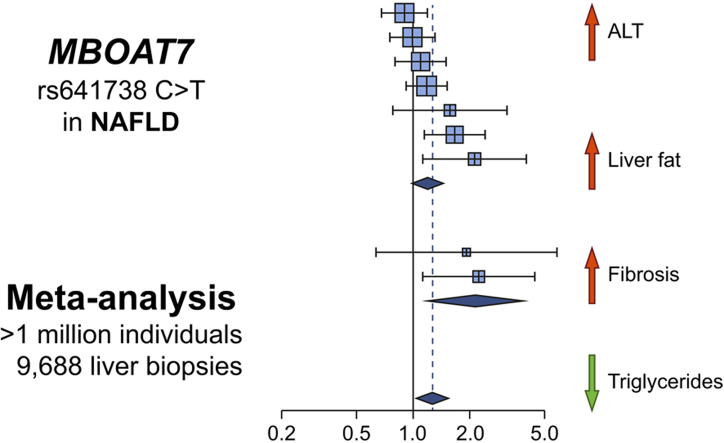
Abstract
Background & Aims
A common genetic variant near MBOAT7 (rs641738C>T) has been previously associated with hepatic fat and advanced histology in NAFLD; however, these findings have not been consistently replicated in the literature. We aimed to establish whether rs641738C>T is a risk factor across the spectrum of NAFLD and to characterise its role in the regulation of related metabolic phenotypes through a meta-analysis.
Methods
We performed a meta-analysis of studies with data on the association between rs641738C>T genotype and liver fat, NAFLD histology, and serum alanine aminotransferase (ALT), lipids or insulin. These included directly genotyped studies and population-level data from genome-wide association studies (GWAS). We performed a random effects meta-analysis using recessive, additive and dominant genetic models.
Results
Data from 1,066,175 participants (9,688 with liver biopsies) across 42 studies were included in the meta-analysis. rs641738C>T was associated with higher liver fat on CT/MRI (+0.03 standard deviations [95% CI 0.02–0.05], pz = 4.8×10–5) and diagnosis of NAFLD (odds ratio [OR] 1.17 [95% CI 1.05–1.3], pz = 0.003) in Caucasian adults. The variant was also positively associated with presence of advanced fibrosis (OR 1.22 [95% CI 1.03–1.45], pz = 0.021) in Caucasian adults using a recessive model of inheritance (CC + CT vs. TT). Meta-analysis of data from previous GWAS found the variant to be associated with higher ALT (pz = 0.002) and lower serum triglycerides (pz = 1.5×10–4). rs641738C>T was not associated with fasting insulin and no effect was observed in children with NAFLD.
Conclusions
Our study validates rs641738C>T near MBOAT7 as a risk factor for the presence and severity of NAFLD in individuals of European descent.
Lay summary
Fatty liver disease is a common condition where fat builds up in the liver, which can cause liver inflammation and scarring (including ‘cirrhosis’). It is closely linked to obesity and diabetes, but some genes are also thought to be important. We did this study to see whether one specific change (‘variant’) in one gene (‘MBOAT7’) was linked to fatty liver disease. We took data from over 40 published studies and found that this variant near MBOAT7 is linked to more severe fatty liver disease. This means that drugs designed to work on MBOAT7 could be useful for treating fatty liver disease.
Keywords: MBOAT7, Fibrosis, NAFLD, Triglyceride, Diabetes, ALSPAC
Graphical abstract
Highlights
-
•
Meta-analysis of 42 studies (>1 million participants) to assess the role of rs641738C>T near MBOAT7 in NAFLD.
-
•
rs641738C>T positively associated with liver fat, ALT, fibrosis and HCC.
-
•
rs641738C>T negatively associated with serum triglycerides.
-
•
Consistent associations found in studies of Caucasian populations only.
Introduction
Since the first genome-wide association study (GWAS) of liver fat,1 >20 genetic single nucleotide variants (SNVs) have been associated with NAFLD.2 These studies have deepened our understanding of the condition, its heritability and its relationship with cardiometabolic disease.
rs641738C>T near membrane-bound O-acyltransferase domain-containing 7 (MBOAT7) was initially identified as a genome-wide significant risk variant for alcohol-related cirrhosis [odds ratio (OR) = 1.35, p = 1.03×10–9],3 although this was not replicated in a more recent analysis.4 It has since been implicated in the pathogenesis of NAFLD,5 HCC,6 as well as in fibrosis development in chronic HBV and HCV,7,8 and primary sclerosing cholangitis.9 However, unlike variants in patatin-like phospholipase domain containing protein 3 (PNPLA3), transmembrane 6 superfamily member 2 (TM6SF2), and 17β-hydroxysteroid dehydrogenase type 13 (HSD17B13), it was not identified to have genome-wide significance for liver fat or serum alanine aminotransferase (ALT).1,10,11
Rs641738 is located a few hundred base pairs downstream of the 3′-untranslated region of MBOAT7, which belongs to a family of genes that encode specific acyl donors and acceptors.12 MBOAT7 encodes lysophosphatidylinositol acyltransferase 1 (LPIAT1), which contributes to the regulation of free arachidonic acid in cells.13,14 Rs641738C>T is associated with lower hepatic expression of MBOAT7 at both the mRNA15 and protein levels.5 Given its role in inflammatory lipid pathways, most mechanistic work relating to rs641738 has focussed on MBOAT7.16
In NAFLD, the rs641738C>T variant was first demonstrated to be associated with increased hepatic fat content and severity of fibrosis in individuals of European descent.5 Proton magnetic resonance spectroscopy data from 2,736 individuals showed a modest increase in hepatic fat in those with the TT-genotype (4.1%) compared with those with the CT- (3.6%) or CC-genotype (3.5%, p = 0.005). Follow-up studies of European subjects corroborated the initial findings, and suggested a role in development of HCC.17,18 However, these results were not replicated in adults of other ancestries5,[19], [20], [21] or in children.22
In addition, bi-allelic loss-of-function mutations in MBOAT7 cause autosomal recessive mental retardation 57 (Online Mendelian Inheritance in Man #617188) and no liver phenotype has been reported in these patients to date.14,23 However, rare likely pathogenic (coding) variants in MBOAT7 are associated with HCC in NAFLD.24
In summary, the association between rs641738C>T and hepatic fat content, as well as its effects on severity of NAFLD, remain unclear. Moreover, the broader metabolic effects of this SNV, including its association with markers of insulin resistance and dyslipidaemia, have not been assessed. Understanding the broader metabolic effects of rs641738C>T is important if MBOAT7 were to be investigated as a drug target in NAFLD.
Here, we conducted a large meta-analysis to determine whether rs641738C>T influences the development or stage of NAFLD and related traits.
Methods
Data sources and study selection
Two data sources were included in the meta-analysis: (i) studies that looked at the effect of the variant on traits of interest by genotyping the variant; and (ii) look-up from GWAS of traits of interest.
Studies were sourced through Medline, Embase, HuGe Navigator, Web of Science, bioRxiv and medRxiv. The search terms used were: ‘(MBOAT7 or membrane-bound-o-acyltransferase) or (rs641738 or rs626283) or (TMC4)’. In addition, HuGe Navigator Phenopedia was searched using terms related to liver disease (see Supplementary Methods). There were no restrictions on either date or language. The search was completed on July 28, 2020. Reference lists of publications were also reviewed.
A separate search was conducted for all potentially relevant GWAS through GWAS Catalogue,25 Phenoscanner,26 Type 2 diabetes knowledge portal27 and Cardiovascular disease knowledge portal28 (see Supplementary Methods).
After removal of duplicates, titles and abstracts were screened for eligibility independently by 2 authors (investigators), with inclusion/exclusion criteria applied to potentially eligible full texts.
HuGENet guidelines29 were followed throughout and MOOSE reporting guidelines30 were used. This study was prospectively registered on the PROSPERO Database of Systematic Reviews (CRD42018105507; www.crd.york.ac.uk/PROSPERO/display_record.php?ID=CRD42018105507).
Inclusion and exclusion criteria
Studies were included if genotyping of rs641738C>T [or rs626283G>C (R2 >0.98 in European and American populations31)/rs2576452C>T (R2 = 0.92 in Guzman et al.32), which are in strong linkage disequilibrium with rs641738C>T] was conducted and data on one of the outcomes of interest were reported. Narrative review articles, in vitro studies and investigations involving animals, fish and invertebrates were excluded. Studies that investigated liver disease of other aetiologies were also excluded. There was no restriction on ethnicity or ancestry. The types of study eligible for inclusion were case-control, cohort, GWAS, systematic reviews and meta-analyses. Preprint and abstract publications were not eligible for inclusion. Several studies reported on the same cohort (or patient sample) in more than 1 article. In these instances, data only from the larger of the overlapping cohorts were included in analyses. A full list of overlapping cohorts and articles is provided in Table S1.
Data collection
Details of the recruitment of controls and cases were obtained from each study and, where necessary, clarified by discussion with the authors of the study. In particular, it was noted when cases and controls were not recruited from the same population or clinics.
Hepatic steatosis or NAFLD (as diagnosis) was evaluated as a dichotomous variable where radiological (liver ultrasound, controlled attenuation parameter [CAP, with cut-off >248 dB/m], CT, MRI, or histological assessment were used. Hepatic fat content was collected as a continuous variable from CT, magnetic resonance spectroscopy (MRS), MRI, and proton density fat fraction (PDFF). Non-invasive assessment of hepatic fat content was also assessed using semiquantitative scoring in the Fenland cohort, as previously described,33 and using CAP.
Individual participant-level histology data were extracted according to the NASH Clinical Research Network scoring system34 and, where not otherwise diagnosed by a pathologist’s assessment, NASH was defined using the Fatty Liver Inhibition of Progression algorithm.35 The above data were collected for each genotype separately (CC, CT, and TT).
Participant demographics and characteristics meta-data were collected from each study, including sex, age, ethnicity, presence of type 2 diabetes mellitus (T2DM), and body mass index (BMI). Where possible, individual patient-level data were obtained.
The authors of 59 studies were contacted for additional data or clarification, of whom 49 replied. Data from 11 potentially relevant studies could not be included, which are listed in the Supplementary Methods.
Additional details regarding cohorts with genome-wide data, the Avon Longitudinal Study of Parents and Children (ALSPAC)[36], [37], [38] data extracted from the UK BioBank (UKBB), quality assessment and statistical analysis are found in the Supplementary Methods.
Results
Database searches identified 1,167 articles (Fig. S1), of which 44 articles were included: 42 primary studies (Tables S2–S4), 1 systematic review, and 1 meta-analysis (Table S5).
In total, 1,066,175 individuals (5,711 children) were included in the meta-analysis. Most studies were in adults (32/42, 76%) and in individuals from predominantly Caucasian populations (26/42, 62%). Of the 42 studies included, 14 (9,688 participants, including 584 children) reported data on liver histology.
Studies were generally of high quality, although, in 5 studies11,22,[39], [40], [41] (4 in adults and 1 in children), the control group was recruited from a different population or sample to the cases (Table S3).
One previous meta-analysis was included,42 which used data from 5 case-control studies to assess the effect of rs641738C>T on the diagnosis of NAFLD. The meta-analysis included 2,560 cases and 8,738 controls and found no evidence of an association between this variant and a diagnosis of NAFLD (Table S5). One previous systematic review43 found positive associations between rs641738C>T in adults of Caucasian, Hispanic, and Black descent, with limited data in children (Table S5).
Liver fat, NAFLD and severe steatosis in adults
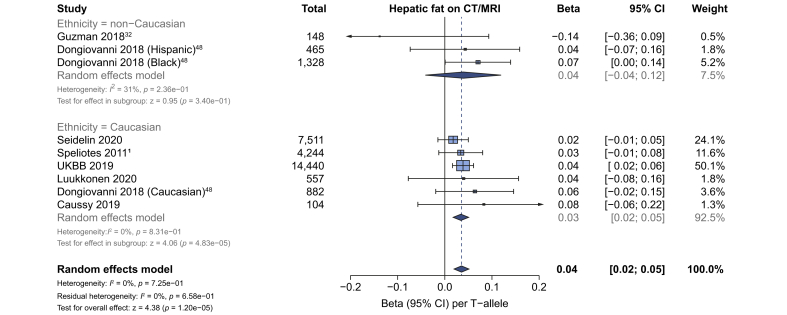
Seven studies (29,679 participants) reported data on hepatic fat as a continuous variable assayed by CT or MRI. On meta-analysis, rs641738C>T was associated with higher liver fat in studies in Caucasian populations using an additive model of inheritance, with a per T-allele change of β 0.034 (95% CI 0.018–0.051), pz = 4.8x10–5) standard deviations in inverse-normalised liver fat (Fig. 1), whereas no consistent effect was observed in non-Caucasian populations. A similar trend was observed using a dominant model of inheritance in studies of Caucasian populations: mean difference in hepatic fat +0.18% (95% CI 0.2–0.34; pz = 0.04; Table S6).
Fig. 1.
The effect of rs641738C>T on liver fat.
Data from 29,679916 individuals with CT, MRI or MRS liver fat. rs641738C>T was positively associated with liver fat in Caucasian populations (using an additive model of inheritance), where data represent SD change in normalised liver fat per T-allele. Meta-analysis was performed using random effects with DerSimonian-Laird method for estimation of tau2. Additional references are available in the Supplementary Data. MRS, magnetic resonance spectroscopy; UKBB, UK BioBank.
Given the difference in sensitivity and specificity of modalities used to assess liver fat, a subanalysis by modality of imaging was performed. No significant differences were observed between studies using CT, MRI, or MRS for quantification of liver fat (Fig. S2).
A similar trend was observed using CAP and semiquantitative ultrasound to assess steatosis severity in 12,224 adults (β 0.02 [95% CI -0.002–0.04], pz = 0.08; Fig. S3).
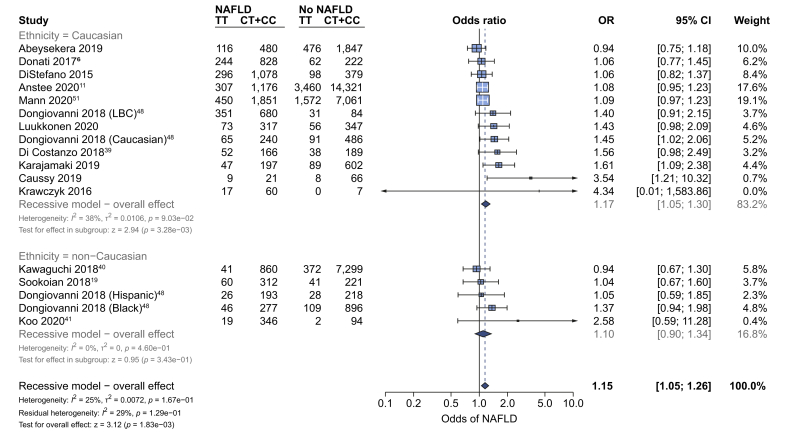
Data from a range of diverse modalities were used to assess the effect of this variant on the diagnosis of NAFLD, to reflect real-world diagnostic practice. rs641738C>T was associated with NAFLD as a trait (OR 1.15 [95% CI 1.05–1.26], pz = 0.002) using a recessive model of inheritance (Fig. 2) but not using additive or dominant models (Table S7). The effect was only observed in studies of Caucasian populations (OR 1.17 [95% CI 1.05–1.3], pz = 0.003). Subgroup analysis by modality of diagnosis found that the 95% CIs for all modalities overlapped, except for MRI-PDFF, which had only 1 study (Fig. S4). The association remained after excluding 4 studies in which there was a lack of similarity between cases and controls (OR 1.19 [95% CI 1.07–1.33], pz = 0.0017) using a recessive model of inheritance.
Fig. 2.
rs641738C>T is associated with higher odds of diagnosis of NAFLD.
Data from 52,17333,263 adults (11,3019,713 cases and 40,87223,550 controls) with radiologically or histologically defined steatosis for presence vs. absence of NAFLD using a recessive model of inheritance (CC + CT vs. TT). Meta-analysis was performed using random effects with DerSimonian-Laird method for estimation of tau2. Additional references are available in the Supplementary Data. LBC, Liver Biopsy Cohort; OR, odds ratio.
However, Egger's test suggested evidence of study distribution (publication) bias (p = 0.013) and when using the Trim and Fill method to account for this bias, the positive association remained but was attenuated (OR 1.11 [95% CI 1.01–1.23], pz = 0.037; Fig. S5).
In patients with NAFLD, data from 8 studies (6,206 participants) showed that rs641738C>T was not significantly associated with the presence of severe steatosis (S1-S2 vs. S3) on liver biopsy (OR 1.08 [95% CI 0.78–1.5], pz = 0.64; Table 1 and Fig. S6).
Table 1.
Summary of results in adults from meta-analyses for dichotomous outcomes.
| Outcome | Genetic model | Subanalysis | No. of studies | Heterogeneity |
Effect summary |
||
|---|---|---|---|---|---|---|---|
| I2 | pQ | OR (95% CI) | pZ | ||||
| NAFLD diagnosis (control vs. NAFLD) | Recessive | Overall | 17 | 0.25 | 0.17 | 1.15 (1.05–1.26) | 0.0018 |
| Non-Caucasian | 5 | 0 | 0.46 | 1.1 (0.9–1.34) | 0.343 | ||
| Caucasian | 12 | 0.38 | 0.09 | 1.17 (1.05–1.3) | 0.0033 | ||
| Severe steatosis (S1–S2 vs. S3) | Recessive | Overall | 8 | 0.67 | 0 | 1.08 (0.78–1.5) | 0.642 |
| Non-Caucasian | 1 | NA | NA | 1.11 (0.39–3.16) | 0.852 | ||
| Caucasian | 7 | 0.72 | 0 | 1.08 (0.76–1.54) | 0.676 | ||
| NASH (NAFL vs. NASH) | Recessive | Overall | 9 | 0.33 | 0.15 | 1.14 (0.96–1.36) | 0.128 |
| Non-Caucasian | 3 | 0 | 0.58 | 1.24 (0.81–1.9) | 0.324 | ||
| Caucasian | 6 | 0.53 | 0.06 | 1.14 (0.93–1.41) | 0.213 | ||
| Any fibrosis (F0 vs. F1–F4) | Recessive | Overall | 9 | 0.52 | 0.03 | 1.27 (1.04–1.54) | 0.0183 |
| Non-Caucasian | 2 | 0 | 0.82 | 2.14 (1.2–3.84) | 0.0105 | ||
| Caucasian | 7 | 0.51 | 0.06 | 1.19 (0.99–1.45) | 0.068 | ||
| Advanced fibrosis (F0–F2 vs. F3–F4) | Recessive | Overall | 8 | 0 | 0.65 | 1.2 (1.02–1.42) | 0.027 |
| Non-Caucasian | 2 | 0 | 0.64 | 0.96 (0.5–1.85) | 0.911 | ||
| Caucasian | 6 | 0 | 0.5 | 1.22 (1.03–1.45) | 0.0206 | ||
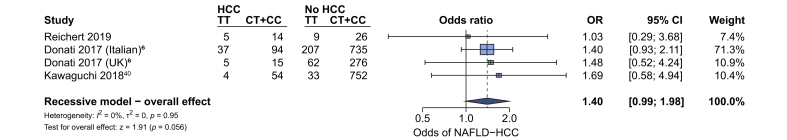
| HCC (NAFLD-HCC vs. NAFLD no-HCC) | Recessive | Overall | 4 | 0 | 0.95 | 1.4 (0.99–1.98) | 0.056 |
Meta-analyses were performed using random effects with subgroup analysis for Caucasian and non-Caucasian populations. Additive, recessive and dominant genetic models were tested for all outcomes. Results using a recessive model of inheritance (CC + CT vs. TT) are shown for all outcomes. Given the use of 3 genetic models, the critical p value for effect summary is pz <0.017. Full results (with all genetic models) are in Table S5. Meta-analyses were performed using random effects with DerSimonian-Laird method for estimation of tau2. OR, odds ratio.
Histological NASH in adults
Data from 9 studies (7,719 participants) found that rs641738C>T was not associated with the presence of NASH on biopsy in adults (OR 1.24 [95% 0.96–1.36], pz = 0.128; Fig. S7).
Fibrosis in adults
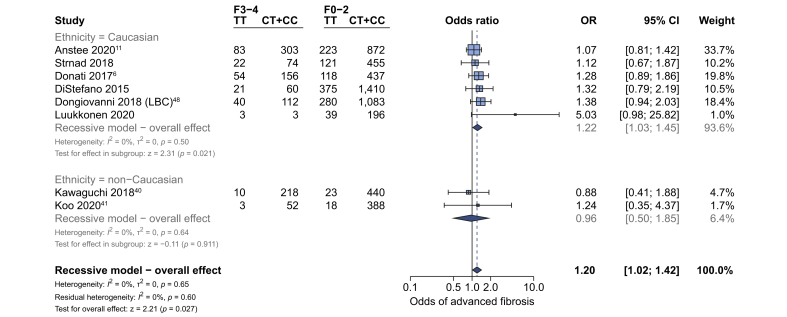
Liver biopsy data on the presence of advanced fibrosis were available from 8 studies (7,692 adults). Our primary outcome, presence of advanced fibrosis in adults (stage F0–F2 vs. stage F3–F4), showed a borderline positive association with rs641738C>T in Caucasian populations (OR 1.22 [95% 1.03–1.45], pz = 0.021; Fig. 3). In addition, 2 studies used International Statistical Classification of Diseases and Related Health Problems (ICD) codes in the UKBB cohort to identify individuals with NAFLD and advanced fibrosis or cirrhosis.44,45 Both found positive associations below genome-wide significance: for example, using an additive model of inheritance, Emdin et al. found the association between rs641738C>T and cirrhosis as β 1.22 (SE 0.06, p = 0.03), using an additive genetic model.44
Fig. 3.
The effect of rs641738C>T on the presence of advanced fibrosis in adult patients with NAFLD.
Data from 7,6926,211 adults (1,214828 cases and 6,4785,383 controls) with biopsy-proven NAFLD comparing advanced fibrosis (F3–F4) vs. F0–F2, using a recessive model of inheritance (CC + CT vs. TT). Meta-analysis was performed using random effects with DerSimonian-Laird method for estimation of tau2. Additional references are available in the Supplementary Data. LBC, Liver Biopsy Cohort; OR, odds ratio.
Data from 9 studies (8,389 participants) found that the presence of any fibrosis (F0 vs. F1–F4) was also borderline positively associated with rs641738C>T overall (OR 1.27 [95% 1.04–1.54], pz = 0.018) as well as in non-Caucasian populations as a subgroup (Fig. S8).
Development of HCC
Four cohorts (2,328 participants, 228 cases of NAFLD-HCC) reported on the development of HCC in patients with NAFLD. rs641738C>T was associated with increased odds of HCC in NAFLD only when using a dominant model (CC vs. CT + TT) of inheritance (OR 1.64 [95% CI 1.18–2.27], pz = 0.003, Fig. 4).
Fig. 4.
rs641738C>T is associated with higher odds of NAFLD-HCC.
Data from 2,328 adults with NAFLD assessing for the presence vs. absence of HCC, using a recessive model of inheritance (CC + CT vs. CT). Meta-analsysis was performed using random effects with DerSimonian-Laird method for estimation of tau2.
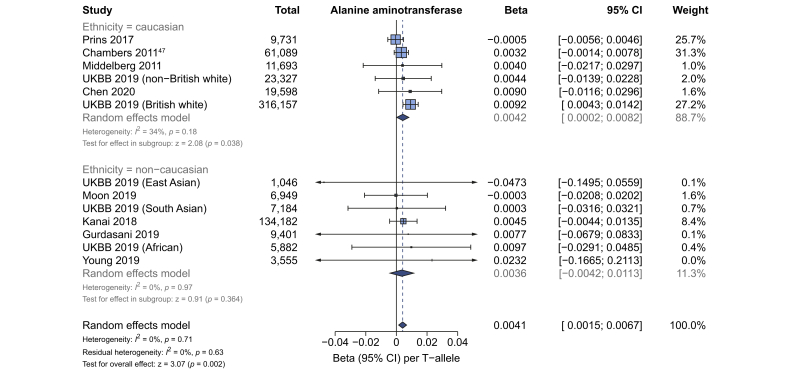
Effect on alanine aminotransferase
Data from GWAS using log-transformed ALT (609,794 participants) were available for meta-analysis to investigate the effect of rs641738C>T on ALT. The variant showed a positive association with ALT (β 0.004 [95% CI 0.002–0.007], pz = 0.002), which was observed in Caucasian populations but not in non-Caucasian populations on subanalysis (Fig. 5; Table S7).
Fig. 5.
rs641738C>T is positively associated with ALT in Caucasian populations in GWAS.
Meta-analysis using linear regression of GWAS summary statistics from 609,794 participants for the association between rs641738C>T and logarithmically transformed ALT. Meta-analysis was performed using random effects with DerSimonian-Laird method for estimation of tau2. Additional references are available in the Supplementary Data. GWAS, genome-wide association studies; UKBB, UK BioBank.
Additionally, in the UKBB cohort, rs641738C>T was associated with a small, but statistically significant (p = 2.0×10–8) increase in untransformed ALT: 0.18 IU/L higher ALT per T-allele in this variant (Table S8).
In the remaining cohort and case-control studies included in the meta-analysis (15,208 adults), rs641738C>T was not found to be significantly associated with a change in ALT; for example, the mean difference using a recessive model (CC + CT vs. TT) was +0.32 IU/L ([95% CI -0.06–0.7], pz = 0.08; Table S9) in Caucasian populations.
Effect on serum lipids and insulin
Data from GWAS using log-transformed serum triglycerides (850,241 participants) found that rs641738C>T was associated with lower triglycerides (β -0.01 [95% CI -0.018, -0.006], pz = 1.5×10–4), which was observed in Caucasian populations but not in non-Caucasian populations on subanalysis (Fig. S9). Similar findings were obtained from a meta-analysis of cohort and case-control studies, particularly when using an additive model (β -0.03 [95% CI -0.05, -0.01], pz = 0.00091; Table S9).
Data from GWAS (852,409 participants) found rs641738C>T to be positively associated with total cholesterol in Caucasian populations (β 0.007 [95% CI 0.003–0.01], pz = 2.1×10–4), which was not observed in non-Caucasian populations (Fig. S10). A borderline positive association was also observed between rs641738C>T and HDL cholesterol (β 0.009 [95% CI 0.001–0.02], pz = 0.02; Table S7). There was no effect on fasting insulin levels found in population-level GWAS (β 0.009 [95% CI -0.03–0.04], pz = 0.64; Table S7). However, a negative association was observed using data from cohort and case-control studies with a dominant genetic model (mean difference -1.4 pmol/L [95% CI -2.1, -0.65], pz = 0.004; Table S9).
Effect of rs641738C>T on paediatric NAFLD
Data from 10 studies (5,711 children) were used in the meta-analysis. rs641738C>T was not significantly associated with the diagnosis of NAFLD, liver fat content, stage of liver histology, or serum biochemistry in children (Table S10).
Meta-regression shows interaction between rs641738C>T and type 2 diabetes mellitus
Finally, we aimed to determine using meta-regression whether baseline participant characteristics influenced the association of rs641738C>T with histological outcomes. There was a negative association with the presence of T2DM and effect size for NASH vs. NAFL (β -1.8 [SE 0.65], p = 0.006; Fig. S11A). A similar negative trend with T2DM was observed for severe steatosis (S1–S2 vs. S3, β -2.6 [SE 1.5], p = 0.08) and presence of fibrosis (F0 vs. F1-4, β -1.5 [SE 0.8], p = 0.06; Table S11). In addition, the effect size for any fibrosis was greater in cohorts with an older mean age (β 0.05 [SE 0.02], p = 0.014; Fig. S11D).
Discussion
Identification of genetic variants associated with NAFLD has the potential to inform preclinical research and our understanding of hepatic metabolism. In this meta-analysis, we validated rs641738C>T near MBOAT7 as a risk factor for the full spectrum of NAFLD in Caucasian adults.
A 2-stage GWAS initially identified rs641738C>T as a genome-wide significant locus for alcohol-related cirrhosis.3 MBOAT7 was a potentially interesting target as an enzyme involved in (phospho)lipid metabolism, conceptually similar to other SNVs at GWAS significance in alcoholic and non-alcoholic liver disease, namely TM6SF2 and PNPLA3. Later studies found the variant to influence the full spectrum of fatty liver disease, from steatosis to NASH, to fibrosis, cirrhosis, and HCC.5,17 However, these associations have not been consistently replicated in the literature.19 We conducted a meta-analysis to firmly establish the association of rs641738C>T with the presence and severity of NAFLD and associated metabolic traits.
Main findings
We found that rs641738C>T was associated with higher liver fat content, higher ALT, and with higher odds of NAFLD diagnosis, fibrosis, and HCC, particularly in Caucasian adults and in the homozygous ‘TT’ genotype. The effect sizes of rs641738C>T reported here are small compared with those of PNPLA3 p.I148M and TM6SF2 p.E167K, the 2 strongest steatogenic variants.46 Also, the magnitude of change in ALT is small relative to that associated with variants in PNPLA3, HSD17B13, mitochondrial amidoxime reducing component 1 (MTARC1), and TM6SF2. This might account for the absence of this variant (or others near MBOAT7) from GWAS for NAFLD in the general population.1,10,11,45,47 The effect size (and associated p value) was too small to be identified as significant genome-wide. The marginal positive effect on hepatic triglyceride content suggests that this variant acts through alterations in the composition as well as quantity of hepatic lipid.17 This is consistent with preclinical data on lipotoxicity, where the composition of hepatic fats influences the development of NASH. By contrast, a recent Mendelian randomisation study using these variables as instruments to assess causality of fatty liver in determining fibrosis showed that the effect of steatosis highly correlates with fibrosis in all the genetic variables, indicating that quantity of lipid rather than quality might be more important.48 Functional studies are needed to understand the relationship between quality/quantity of fat and hepatotoxic/-protective mechanisms in causing progression of disease.
The function of this variant is still relatively poorly understood and there is conflicting evidence as to whether rs641738C>T is associated with changes in the hepatic expression of MBOAT7. Results from the GTEx Consortium showed a strong negative association with T-allele,15 which is supported by data from Schadt et al.49 MBOAT7 protein expression correlated with mRNA in liver biopsies from Mancina et al.,5 but this finding was not replicated by Sookoian et al.19 MBOAT7 encodes LPIAT1, a 6-transmembrane domain protein involved in acyl-chain remodelling of membranes that influence intracellular membrane composition and circulating phosphatidylinositols.50 Furthermore, recent metabolite profiling data implicated MBOAT7 as the causal gene for this SNV.51 Moreover, transmembrane channel-like 4 (TMC4) was found to have a low expression in the liver,5 which is consistent with no mechanistic data supporting its role in NAFLD.
The hypothesis that MBOAT7 is the causal gene underlying the association with liver disease at the locus is supported by the observation that mice deficient for MBOAT7 have altered hepatic concentrations of polyunsaturated phosphatidylinositol.50 Similarly, metabolite data from humans are strongly suggestive that rs641738C>T reduces MBOAT7 function.52 In addition, 2 independent groups found that loss of MBOAT7 (but not TMC4) increases the severity of NAFLD in mice fed a high-fat diet.53,54
These analyses suggest that rs641738C>T impacts the severity of NAFLD through a recessive model of inheritance, although some analyses using an additive genetic model were suggestive of a role (e.g. for liver fat and ALT). Other genetic variants are known to impact all-cause mortality in a recessive manner, notably variants that perturb homeostatic iron regulator protein (HFE).44 Further mechanistic work is required to understand the extent to which the haplo-insufficient state affects hepatocyte function.
We found no evidence of an effect of rs641738C>T on insulin resistance (the key driver of hepatic steatosis) as determined by unaltered fasting insulin concentrations. GWAS meta-analyses of T2DM have implicated p.I148M in PNPLA3 and p.E167K in TM6SF2 as significant risk loci (albeit with very modest effect sizes compared with their effects on liver disease)55 and Mendelian randomisation studies indicate a causal role in determining insulin resistance mediated by the degree of liver damage.48,56 Similarly, these 2 variants are associated with reduced risk of coronary artery disease; although our analysis did find lower serum triglycerides to be associated with this variant, it has not been associated with lower rates of cardiovascular disease.57 However, we did observe a negative association between effect size and prevalence of diabetes on meta-regression, suggesting that this variant has the greatest effect in less insulin-resistant individuals.
A strength of this meta-analysis is the large number of individuals with liver biopsy-derived phenotypic data as well as the use of population-based GWAS data. The larger number of included studies and participants is likely to account for the different conclusions reached in this study compared with the previous meta-analysis by Xia et al.42
Limitations and quality of evidence
An important practical consideration is the population frequency of this variant in different ethnicities. The mean allelic frequency of the effect (T) allele is highly variable: from 0.24 in East Asians compared with 0.53 in those of South Asian ancestry.58 Moreover, the majority of studies included in this meta-analysis used self-reported ethnicity, rather than genetic ancestry.
Although this analysis did include data from individuals of multiple ethnicities (and genetic ancestries), we only found evidence of an effect of this variant in Caucasian individuals. This is consistent with the initial discovery and it is likely that rs641738C>T is a proxy for the true causal variant. However, because of differences in patterns of linkage disequilibrium, we cannot exclude the possibility that a different nearby locus is associated with liver-related phenotypes in individuals of other genetic ancestries.
A limitation of using meta-analysis for a single variant is the lack of adjustment for population stratification. When further genome-wide data are available, a formal GWAS meta-analysis might be able to address this. We found significant differences between adult and paediatric histological analyses. Although there were fewer clinical events (e.g. with advanced fibrosis) in children, the analyses did not show a trend congruous with those in adults. Paediatric NAFLD has a different histological phenotype to that of adults (with prominent periportal inflammation) and, therefore, it is plausible that this is a true lack of association in children with NAFLD.
Data from multiple diagnostic or imaging modalities were combined in several analyses. Although we observed minimal heterogeneity between modalities, these techniques have differing accuracy for the diagnosis of steatosis, which has the potential to affect results. The subgroup analysis of hepatic fat by modality suggested a marginally greater effect size in studies using MRS, which is regarded as a highly sensitive technique. There is potential that, through the inclusion of other modalities (e.g. CT), we have underestimated the effect size associated with this variant.
The magnitude of effect observed across all associations was small compared with other well-established variants. The clinical relevance of rs738409C>G in PNPLA3 has been validated with hard end-points,59 but large cohorts will be required to prospectively demonstrate the clinical risk associated with this variant near MBOAT7.
Although there was minimal heterogeneity across included studies, there was evidence of publication bias, but the effect on diagnosis of NAFLD appeared to persist after attempting to account for this. Also of note, the numbers of individuals with NAFLD and HCC were comparatively low, limiting the power to assess for an association of this variant with non-cirrhotic HCC, as has been previously reported.6 The HCC analysis was also unique in only demonstrating an effect in the dominant, rather than recessive, model of inheritance. Further work in this area might improve the accuracy of effect estimates.
Conclusions
rs641738C>T near MBOAT7 is positively associated with liver fat, ALT and histological severity in Caucasian adults with NAFLD, but negatively associated with serum triglycerides and with relatively small effect sizes throughout. These data validate this locus as significant in the pathogenesis of NAFLD.
Abbreviations
ALSPAC, Avon Longitudinal Study of Parents and Children; BMI, body mass index; CAP, controlled attenuation parameter; GWAS, genome-wide association study; HFE, homeostatic iron regulator protein; HSD17B13, 17β-hydroxysteroid dehydrogenase type 13; LPIAT1, lysophosphatidylinositol acyltransferase 1; MBOAT7, membrane bound O-acyltransferase domain containing 7; MTARC1, mitochondrial amidoxime reducing component 1; OR, odds ratio; MRS, magnetic resonance spectroscopy; PDFF, proton density fat fraction; PNPLA3, patatin-like phospholipase domain containing protein 3; SNV, single nucleotide variant; T2DM, type 2 diabetes mellitus; TM6SF2, transmembrane 6 superfamily member 2; TMC4, transmembrane channel-like 4; UKBB, UK BioBank.
Financial support
J.P.M. is supported by a Wellcome Trust Fellowship (216329/Z/19/Z), a European Paediatric Research Society award and a Children's Liver Disease Foundation grant. The EU-PNAFLD is supported by an EASL Registry Grant. NIH grants: R01HD028016 (S.C.), R01DK111038 (S.C)., R01DK114504 (N.S.), DK091601 (J.K.D.) and UL1TR001105 (J.K.). Supported by the Nonalcoholic Steatohepatitis Clinical Research Network (NASH CRN) of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (U01DK061718, U01DK061728, U01DK061731, U01DK061732, U01DK061734, U01DK061737, U01DK061738, U01DK061730, and U01DK061713); the National Center for Advancing Translational Sciences (UL1TR000439, UL1TR000436, UL1TR000006, UL1TR000448, UL1TR000100, UL1TR000004, UL1TR000423, UL1TR000058, and UL1TR001881); and the NIDDK (DK063491 to the Southern California Diabetes Endocrinology Research Center). This study was supported by the German Federal Ministry for Education and Research (BmBF) through the Livers Systems Medicine (LiSyM) project. This work was supported by grants from the Swiss National Funds (SNF no. 310030_169196) and the Swiss Foundation for Alcohol Research (SSA) to F.S. This Raine Study was supported by the National Health and Medical Research Council of Australia (grant numbers 403981, 353514 and 572613). The UK Medical Research Council and Wellcome (grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. ALSPAC GWAS data was generated by Sample Logistics and Genotyping Facilities at Wellcome Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. A comprehensive list of grants funding is available on the ALSPAC website (www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf); this research was specifically funded by grants from MRC and Alcohol Research UK (MR/L022206/1) and NIH (5R01AA018333-05) to K.W.M.A. and M.H. L.V. was supported by MyFirst Grant AIRC n.16888, Ricerca Finalizzata Ministero della Salute RF-2016-02364358, Ricerca corrente Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico. German Federal Ministry of Education and Research (BMBF LiSyM 031L0051 to F.L.). P.L. is supported by grants from the Sigrid Jusélius Foundation and the Novo Nordisk Foundation. The Fenland study was funded by grants to the MRC Epidemiology Unit (MC UU12015/1, MC UU 12015/5). R.B. and M.K. are employees of and shareholders in Perspectum Diagnostics Ltd. C.A.P. is funded by a Wellcome Trust Clinical PhD Programme (206274/Z/17/Z). J.M.B. is supported by the Spanish Carlos III Health Institute (ISCIII; PI15/01132, PI18/01075, CIBERehd, and Miguel Servet Program CON14/00129) co-financed by Fondo Europeo de Desarrollo Regional (FEDER) and La Caixa Scientific Foundation (HR17-00601). Research from the GUARDIAN Study was supported DK085175 and DK118062, and phenotyping in IRASFS was supported by HL060944, HL061019, HL060919 and HL060894 IRASFS from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). H.Y. is funded by a Diabetes UK RD Lawrence fellowship (17/0005594). Q.M.A. and A.D. supported by the EPoS (Elucidating Pathways of Steatohepatitis) consortium funded by the Horizon 2020 Framework Program of the European Union under Grant Agreement 634413. Q.M.A., A.D., L.V. and A.G. are members of the LITMUS (Liver Investigation: Testing Biomarker Utility in Steatohepatitis) consortium funded by the European Union Innovative Medicines Initiative 2 (IMI2) Joint Undertaking under grant agreement 777377. Q.M.A. is a Newcastle NIHR Biomedical Research Center Investigator.
Authors' contributions
Study concept and design: J.P.M.; acquisition of data: all; analysis and interpretation of data: K.T., J.K., S.S., L.V., J.P.M.; drafting of the manuscript: K.T., J.P.M.; critical revision of the manuscript for important intellectual content: all authors; statistical analysis: K.T., J.K., S.S., L.V., J.P.M.; obtained funding: J.P.M., S.C., N.S., J.K.D., J.K., P.L., J.M.B., C.A.P., H.Y., K.W.M.A., L.A., Q.M.A., A.D.K., T.B., G.S.G., C.H., J.H., J.L., P.E.M., T.A.M., N.D.P., S.R., J.I.R., E.K.S., S.S., A.T.H., L.E.W., L.V., H.Y.J., K.A.Y.; study supervision: J.P.M.
Conflict of interest
C.E. reports receiving personal fees from Navitor Pharma and Novartis. The other authors declare no conflicts of interest that pertain to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
The authors are grateful to the Raine Study participants and their families, and to the Raine Study research staff for cohort coordination and data collection. The authors gratefully acknowledge the following institutes for providing funding for Core Management of the Raine Study: The University of Western Australia (UWA), Curtin University, the Raine Medical Research Foundation, the UWA Faculty of Medicine, Dentistry and Health Sciences, the Telethon Kids Institute, the Women and Infants Research Foundation (King Edward Memorial Hospital), and Edith Cowan University). We are extremely grateful to all the families who took part in the ALSPAC study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. This study was conducted using data from the Fenland study. The authors gratefully acknowledge the help of the MRC Epidemiology Unit Support Teams, including Field, Laboratory, and Data Management Teams. The authors are grateful to the members of the EU-PNAFLD Registry, including Anita Vreugdenhil, Anna Alisi, Piotr Socha, Wojciech Jańczyk, Ulrich Baumann, Sanjay Rajwal, Indra van Mourik, Florence Lacaille, Myriam Dabbas, Deirdre A. Kelly, and the late Valerio Nobili. The authors are also grateful to the members of the GOLD consortium: Gudny Eiriksdottir, Melissa E. Garcia, Vilmundur Gudnason, Tamara B. Harris, Lauren J. Kim, Lenore J. Launer, Michael A. Nalls, Albert V. Smith,Jeanne M. Clark, Ruben Hernaez, W. H. Linda Kao, Braxton D. Mitchell, Alan R. Shuldiner, Laura M. Yerges-Armstrong, Ingrid B. Borecki, J. Jeffrey Carr, Mary F. Feitosa, Jun Wu, Johannah L. Butler, Caroline S. Fox, Joel N. Hirschhorn, Udo Hoffmann, Shih-Jen Hwang, Joseph M. Massaro, Christopher J. O'Donnell, Cameron D. Palmer, Dushyant V. Sahani, Elizabeth K. Speliotes. We would also like to thank Naga Chalasani for his helpful comments. This research made use of the UK Biobank resource under project number 9914.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jhep.2020.08.027.
Contributor Information
Jake P. Mann, Email: jm2032@cam.ac.uk.
EU-PNAFLD Investigators:
Anita Vreugdenhil, Anna Alisi, Piotr Socha, Wojciech Jańczyk, Ulrich Baumann, Sanjay Rajwal, Indra van Mourik, Florence Lacaille, Myriam Dabbas, Deirdre A. Kelly, and Valerio Nobili
GOLD Consortium:
Gudny Eiriksdottir, Melissa E. Garcia, Vilmundur Gudnason, Tamara B. Harris, Lauren J. Kim, Lenore J. Launer, Michael A. Nalls, Albert V. Smith, Jeanne M. Clark, Ruben Hernaez, W.H. Linda Kao, Braxton D. Mitchell, Alan R. Shuldiner, Laura M. Yerges-Armstrong, Ingrid B. Borecki, J. Jeffrey Carr, Mary F. Feitosa, Jun Wu, Johannah L. Butler, Caroline S. Fox, Joel N. Hirschhorn, Udo Hoffmann, Shih-Jen Hwang, Joseph M. Massaro, Christopher J. O'Donnell, Cameron D. Palmer, Dushyant V. Sahani, and Elizabeth K. Speliotes
Supplementary data
References
- 1.Speliotes E.K., Yerges-armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romeo S., Sanyal A., Valenti L. Leveraging human genetics to identify potential new treatments for fatty liver disease. Cell Metab. 2020;31:35–45. doi: 10.1016/j.cmet.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Buch S., Stickel F., Trépo E., Way M., Herrmann A., Nischalke H.D. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47:1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 4.Innes H., Buch S., Hutchinson S., Guha I.N., Morling J.R., Barnes E. Genome-wide association study for alcohol-related cirrhosis identifies risk loci in MARC1 and HNRNPUL1. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.06.014. In press. [DOI] [PubMed] [Google Scholar]
- 5.Mancina R.M., Dongiovanni P., Petta S., Pingitore P., Meroni M., Rametta R. The MBOAT7-TMC4 Variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150:1219–1230. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donati B., Dongiovanni P., Romeo S., Meroni M., McCain M., Miele L. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci Rep. 2017;7:4492. doi: 10.1038/s41598-017-04991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thabet K., Asimakopoulos A., Shojaei M., Romero-Gomez M., Mangia A., Irving W.L. MBOAT7 rs641738 increases risk of liver inflammation and transition to fibrosis in chronic hepatitis C. Nat Commun. 2016;7:12757. doi: 10.1038/ncomms12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thabet K., Chan H.L.Y., Petta S., Mangia A., Berg T., Boonstra A. The membrane-bound O-acyltransferase domain-containing 7 variant rs641738 increases inflammation and fibrosis in chronic hepatitis B. Hepatology. 2017;65:1840–1850. doi: 10.1002/hep.29064. [DOI] [PubMed] [Google Scholar]
- 9.Freund C., Wahlers A., Begli N.H., Leopold Y., Klöters-Plachky P., Mehrabi A. The MBOAT7 rs641738 variant is associated with an improved outcome in primary sclerosing cholangitis. Clin Res Hepatol Gastroenterol. 2020 doi: 10.1016/j.clinre.2019.12.006. In press. [DOI] [PubMed] [Google Scholar]
- 10.Abul-Husn N.S., Cheng X., Li A.H., Xin Y., Schurmann C., Stevis P. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378:1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anstee Q.M., Darlay R., Cockell S., Meroni M., Govaere O., Tiniakos D. Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically-characterised cohort. J Hepatol. 2020;73:505–515. doi: 10.1016/j.jhep.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda S., Inoue T., Lee H.C., Kono N., Tanaka F., Gengyo-Ando K. Member of the membrane-bound O-acyltransferase (MBOAT) family encodes a lysophospholipid acyltransferase with broad substrate specificity. Genes Cells. 2008;13:879–888. doi: 10.1111/j.1365-2443.2008.01212.x. [DOI] [PubMed] [Google Scholar]
- 13.Gijón M.A., Riekhof W.R., Zarini S., Murphy R.C., Voelker D.R. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. 2008;283:30235–30245. doi: 10.1074/jbc.M806194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansen A., Rosti R.O., Musaev D., Sticca E., Harripaul R., Zaki M. Mutations in MBOAT7, encoding lysophosphatidylinositol acyltransferase I, lead to intellectual disability accompanied by epilepsy and autistic features. Am J Hum Genet. 2016;99:912–916. doi: 10.1016/j.ajhg.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meroni M., Dongiovanni P., Longo M., Carli F., Baselli G., Rametta R. Mboat7 down-regulation by hyper-insulinemia induces fat accumulation in hepatocytes. EBioMedicine. 2020;52:102658. doi: 10.1016/j.ebiom.2020.102658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luukkonen P.K., Zhou Y., Hyötyläinen T., Leivonen M., Arola J., Orho-Melander M. The MBOAT7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of non-alcoholic fatty liver disease in humans. J Hepatol. 2016;65:1263–1265. doi: 10.1016/j.jhep.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 18.Krawczyk M., Rau M., Schattenberg J.M., Bantel H., Pathil A., Demir M. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: a multicenter biopsy-based study. J Lipid Res. 2017;58:247–255. doi: 10.1194/jlr.P067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sookoian S., Flichman D., Garaycoechea M.E., Gazzi C., Martino J.S., Castaño G.O. Lack of evidence supporting a role of TMC4-rs641738 missense variant - MBOAT7- intergenic downstream variant - in the susceptibility to nonalcoholic fatty liver disease. Sci Rep. 2018;8:5097. doi: 10.1038/s41598-018-23453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koo B.K., Joo S.K., Kim D., Bae J.M., Park J.H., Kim J.H. Additive effects of PNPLA3 and TM6SF2 on the histological severity of non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2018;33:1277–1285. doi: 10.1111/jgh.14056. [DOI] [PubMed] [Google Scholar]
- 21.Umano G.R., Caprio S., Di Sessa A., Chalasani N., Dykas D.J., Pierpont B. The rs626283 variant in the MBOAT7 gene is associated with insulin resistance and fatty liver in Caucasian obese youth. Am J Gastroenterol. 2018;113:376–383. doi: 10.1038/ajg.2018.1. [DOI] [PubMed] [Google Scholar]
- 22.Hudert C.A., Selinski S., Rudolph B., Bläker H., Loddenkemper C., Thielhorn R. Genetic determinants of steatosis and fibrosis progression in paediatric non-alcoholic fatty liver disease. Liver Int. 2019;39:540–556. doi: 10.1111/liv.14006. [DOI] [PubMed] [Google Scholar]
- 23.Yalnızoǧlu D., Özgül R.K., Oǧuz K.K., Özer B., Yücel-Yılmaz D., Gürbüz B. Expanding the phenotype of phospholipid remodelling disease due to MBOAT7 gene defect. J Inherit Metab Dis. 2019;42:381–388. doi: 10.1002/jimd.12016. [DOI] [PubMed] [Google Scholar]
- 24.Pelusi S., Baselli G., Pietrelli A., Dongiovanni P., Donati B., McCain M.V. Rare pathogenic variants predispose to hepatocellular carcinoma in nonalcoholic fatty liver disease. Sci Rep. 2019;9:3682. doi: 10.1038/s41598-019-39998-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamat M.A., Blackshaw J.A., Young R., Surendran P., Burgess S., Danesh J. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. 2019;35:4851–4853. doi: 10.1093/bioinformatics/btz469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Type 2 diabetes knowledge portal. www.type2diabetesgenetics.org/
- 28.Cardiovascular Disease Knowledge Portal. www.broadcvdi.org/
- 29.Little J., Higgins J., editors. The HuGENetTM HuGE Review Handbook. Centers for Disease Control and Prevention; Atlanta: 2006. [Google Scholar]
- 30.Stroup D.F., Berlin J.A., Morton S.C., Olkin I., Williamson G.D., Rennie D. Meta-analysis of observational studies. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 31.Arnold M., Raffler J., Pfeufer A., Suhre K., Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics. 2015;31:1334–1336. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guzman C.B., Duvvuru S., Akkari A., Bhatnagar P., Battioui C., Foster W. Coding variants in PNPLA3 and TM6SF2 are risk factors for hepatic steatosis and elevated serum alanine aminotransferases caused by a glucagon receptor antagonist. Hepatol Commun. 2018;2:561–570. doi: 10.1002/hep4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Lucia Rolfe E., Brage S., Sleigh A., Finucane F., Griffin S.J., Wareham N.J. Validity of ultrasonography to assess hepatic steatosis compared to magnetic resonance spectroscopy as a criterion method in older adults. PLoS One. 2018;13:87–99. doi: 10.1371/journal.pone.0207923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 35.Bedossa P., Burt A.A., Gouw A.H.A., Lackner C., Schirmacher P., Terracciano L. Utility and appropriateness of the fatty liver inhibition of progression (FLIP) algorithm and steatosis, activity, and fibrosis (SAF) score in the evaluation of biopsies of nonalcoholic fatty liver disease. Hepatology. 2014;60:565–575. doi: 10.1002/hep.27173. [DOI] [PubMed] [Google Scholar]
- 36.Boyd A., Golding J., Macleod J., Lawlor D.A., Fraser A., Henderson J. Cohort profile: the “children of the 90s”–the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Northstone K., Lewcock M., Groom A., Boyd A., Macleod J., Timpson N. The Avon Longitudinal Study of Parents and Children (ALSPAC): an update on the enrolled sample of index children in 2019. Wellcome Open Res. 2019;4:51. doi: 10.12688/wellcomeopenres.15132.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Costanzo A., Belardinilli F., Bailetti D., Sponziello M., D'Erasmo L., Polimeni L. Evaluation of polygenic determinants of non-alcoholic fatty liver Disease (NAFLD) by a candidate genes resequencing strategy. Sci Rep. 2018;8:1–10. doi: 10.1038/s41598-018-21939-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawaguchi T., Shima T., Mizuno M., Mitsumoto Y., Umemura A., Kanbara Y. Risk estimation model for nonalcoholic fatty liver disease in the Japanese using multiple genetic markers. PLoS One. 2018;13:1–16. doi: 10.1371/journal.pone.0185490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koo B.K., Joo S.K., Kim D., Lee S., Bae J.M., Park J.H. Development and validation of a scoring system, based on genetic and clinical factors, to determine risk of steatohepatitis in Asian patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2020;18:2592–2599. doi: 10.1016/j.cgh.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 42.Xia Y., Huang C.X., Li G.Y., Chen K.H., Han L., Tang L. Meta-analysis of the association between MBOAT7 rs641738, TM6SF2 rs58542926 and nonalcoholic fatty liver disease susceptibility. Clin Res Hepatol Gastroenterol. 2019;43:1–9. doi: 10.1016/j.clinre.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Ismaiel A., Dumitrascu D.L. Genetic predisposition in metabolic-dysfunction-associated fatty liver disease and cardiovascular outcomes: systematic review. Eur J Clin Invest. 2020:e13331. doi: 10.1111/eci.13331. [DOI] [PubMed] [Google Scholar]
- 44.Emdin C.A., Haas M.E., Khera A.V., Aragam K., Chaffin M., Klarin D. A missense variant in Mitochondrial Amidoxime Reducing Component 1 gene and protection against liver disease. PLoS Genet. 2020;16:e1008629. doi: 10.1371/journal.pgen.1008629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen V.L., Chen Y., Du X., Handelman S.K., Speliotes E.K. Genetic variants that associate with cirrhosis have pleiotropic effects on human traits. Liver Int. 2020;40:405–415. doi: 10.1111/liv.14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sookoian S., Pirola C.J., Valenti L., Davidson N.O. Genetic pathways in nonalcoholic fatty liver disease: insights from systems biology. Hepatology. 2020;72:330–346. doi: 10.1002/hep.31229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chambers J.C., Zhang W., Sehmi J., Li X., Wass M.N., Van Der Harst P. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dongiovanni P., Stender S., Pietrelli A., Mancina R.M., Cespiati A., Petta S. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J Intern Med. 2018;283:356–370. doi: 10.1111/joim.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:1020–1032. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee H.-C., Inoue T., Sasaki J., Kubo T., Matsuda S., Nakasaki Y. LPIAT1 regulates arachidonic acid content in phosphatidylinositol and is required for cortical lamination in mice. Mol Biol Cell. 2012;23:4689–4700. doi: 10.1091/mbc.E12-09-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mann J.P., Pietzner M., Wittemans L.B., De Lucia Rolfe E., Nicola D., Imamura F. Insights into genetic variants associated with NASH-fibrosis from metabolite profiling. Hum Mol Genet. 2020:ddaa162. doi: 10.1093/hmg/ddaa162. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shin S.-Y., Fauman E.B., Petersen A.-K., Krumsiek J., Santos R., Huang J. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helsley R.N., Varadharajan V., Brown A.L., Gromovsky A.D., Schugar R.C., Ramachandiran I. Obesity-linked suppression of membrane-bound O-acyltransferase 7 (MBOAT7) drives non-alcoholic fatty liver disease. eLife. 2019;8:e49882. doi: 10.7554/eLife.49882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka Y., Shimanaka Y., Caddeo A., Kubo T., Mao Y., Kubota T. LPIAT1/MBOAT7 depletion increases triglyceride synthesis fueled by high phosphatidylinositol turnover. Gut. 2020 doi: 10.1136/gutjnl-2020-320646. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parisinos C.A., Wilman H.R., Thomas E.L., Kelly M., Nicholls R.C., McGonigle J. Genome-wide and Mendelian randomisation studies of liver MRI yield insights into the pathogenesis of steatohepatitis. J Hepatol. 2020;73:241–251. doi: 10.1016/j.jhep.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brouwers M.C.G.J., Simons N., Stehouwer C.D.A., Koek G.H., Schaper N.C., Isaacs A. Relationship between nonalcoholic fatty liver disease susceptibility genes and coronary artery disease. Hepatol Commun. 2019;3:587–596. doi: 10.1002/hep4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grimaudo S., Pipitone R.M., Pennisi G., Celsa C., Cammà C., Di Marco V. Association between PNPLA3 rs738409 C>G variant and liver-related outcomes in patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2020;18:935–944. doi: 10.1016/j.cgh.2019.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.