Abstract
PARP inhibitor monotherapies are effective to treat patients with breast, ovary, prostate, and pancreatic cancer with BRCA1 mutations, but not to the much more frequent BRCA wild-type cancers. Searching for strategies that would extend the use of PARP inhibitors to BRCA1-proficient tumors, we found that the stability of BRCA1 protein following ionizing radiation (IR) is maintained by postphosphorylational prolyl-isomerization adjacent to Ser1191 of BRCA1, catalyzed by prolyl-isomerase Pin1. Extinction of Pin1 decreased homologous recombination (HR) to the level of BRCA1-deficient cells. Pin1 stabilizes BRCA1 by preventing ubiquitination of Lys1037 of BRCA1. Loss of Pin1, or introduction of a BRCA1-mutant refractory to Pin1 binding, decreased the ability of BRCA1 to localize to repair foci and augmented IR-induced DNA damage. In vitro growth of HR-proficient breast, prostate, and pancreatic cancer cells were modestly repressed by olaparib or Pin1 inhibition using all-trans retinoic acid (ATRA), while combination treatment resulted in near-complete block of cell proliferation. In MDA-MB-231 xenografts and triple-negative breast cancer patient-derived xenografts, either loss of Pin1 or ATRA treatment reduced BRCA1 expression and sensitized breast tumors to olaparib. Together, our study reveals that Pin1 inhibition, with clinical widely used ATRA, acts as an effective HR disrupter that sensitizes BRCA1-proficient tumors to PARP inhibition.
Significance:
PARP inhibitors have been limited to treat homologous recombination–deficient tumors. All-trans retinoic acid, by inhibiting Pin1 and destabilizing BRCA1, extends benefit of PARP inhibitors to patients with homologous recombination–proficient tumors.
Introduction
Like DNA replication and RNA transcription, the DNA damage repair response requires the tight temporal and spatial coordination of protein complexes that sense DNA damage and conduct its repair. In eukaryotic cells, these proteins are frequently activated by serine/ threonine phosphorylation signaling cascades. Phosphorylated serine/threonine residues adjacent to a proline are targets for phospho-specific prolyl-isomerization, a postphosphorylational modification that regulates protein folding, function, and stability. This postphosphorylational modification of signaling molecules, carried out by the prolyl-isomerase Pin1, is thought to coordinate mitosis, and can be aberrantly activated in cancer (1, 2). Pin1 binds its phosphorylated target proteins via the N-terminal WW-domain, and then isomerizes the adjacent peptidyl-prolyl bond catalyzed by its C-terminal PPIase domain.
The consequences of this postphosphorylational modification can be pleiotropic and depend on a cell's gene expression and mutational profile. In general, Pin1 stabilizes and augments the function of proteins that promote cell-cycle progression, such as cyclin D1 (3, 4), c-Jun (5), Raf-1 (6), NF-κB (7), β-catenin (8), and TopoIIα (9). In mouse models, Pin1 overexpression promotes breast cancer formation (5, 10) while Pin1 ablation prevents breast (10) or prostate cancer (11). Pin1 is also a positive regulator of tumor suppressors P53 (12–14) and p73 (15, 16), although, in cancer, stabilization by Pin1 can enhance the oncogenic activity of mutant, pathogenic P53 (17). In DNA double-strand break (DSB) repair, Pin1 regulates CtIP polyubiquitination and turnover (18). However, an involvement of Pin1 in homologous recombination (HR) has so far not been shown.
The BRCA1 protein is phosphorylated in response to DNA damage, and phosphorylation is a requirement for the DNA damage repair function of BRCA1 (19, 20). Phosphorylation of BRCA1 occurs mostly on serine residues (19) in response to ionizing radiation (IR) and upon progression through the cell cycle. Kinases that phosphorylate BRCA1 include CDK1, 2, casein kinase 2 and DNA damage-responsive kinases such as ATM, ATR, hCds1, and AKT (19–24). Upon phosphorylation, BRCA1 localizes to distinct nuclear DNA repair foci (25–27). Without functional BRCA1, cells incur genomic instability and loss of cell-cycle checkpoint control (28). The nuclear focus formation appears to directly correlate with the ability of BRCA1 to promote DNA damage repair (28). Loss of functional BRCA1 predisposes to the development of breast or ovarian cancer. Moreover, loss of functional BRCA1 is predictive of response to treatments that augment DNA damage repair defects: patients with breast, ovary, prostate, and pancreatic cancer with BRCA mutations are sensitive to therapeutic PARP inhibition as the defect in HR results in defective replication stress response, which is further challenged by PARP inhibition leading to insurmountable DNA damage (29–31). This “synthetic lethality” of PARP inhibitors in HR-deficient tumors has prompted research into strategies that would render BRCA1-proficient tumors BRCA1 deficiency and hence sensitize to PARP inhibition (32, 33). Given Pin1's abundant expression in breast, prostate, pancreatic cancer, etc. (34, 35), the recent discovery that all-trans retinoic acid (ATRA) can inhibit Pin1's function (36, 37), and the presence of several S/P motifs in BRCA1 that could serve as Pin1 targets, we hypothesized that Pin1 might regulate the stability or function of BRCA1 and that pharmacologic Pin1 inhibition might enhance PARP inhibitor sensitivity of BRCA1-proficient cancers.
Materials and Methods
Drugs
ATRA (Sigma) was dissolved in DMSO at 150 mmol/L and diluted in a working solution of 1:1 PBS and 1 mol/L NaOH, and given in 200 mL at a concentration of 1.5 mg/kg intraperitoneally once a day. Olaparib (Selleckchem) was used as described previously (33).
Cells and cell culture
MCF10A cells from ATCC were cultured in DMEM/F-12 with 5% horse serum, 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, 100 ng/mL cholera toxin, and 10 μg/mL insulin, as described previously (38). MCF7, T47D, MDA-MB-231, AU565, and HEK293 from ATCC were maintained in DMEM with 10% FBS. SUM149 were maintained in Ham's F-12 medium supplemented with 5% bovine serum (FBS) with insulin (5 μg/mL) and hydrocortisone (2 μg/mL).
For the radiation treatment, the dosage was chosen according to the experiments and cells that were used. MCF10A cells were treated with 5 Gy for visualization of DNA repair foci by immunofluorescence assay. Breast cancer cells were treated with 10 Gy for BRCA1 degradation assay. HEK297 were treated with 20Gy for BRCA1 degradation and ubiquitination assay.
Cell viability and luciferase assays
For cell viability assays, cells were seeded at a density of 1,000 (MDA-MB-231) and 2,000 (MCF7) cells/well in 96-well plates. Treatment with ATAR or olaparib began from the second day. Cell viability was determined using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer's instructions, and absorption was read using a Wallac 3 plate reader.
Colony formation assay
Cells were seeded at a density of 500 (MDA-MB-231), 1,000 (MCF7), 1,000 (T47D), 500 (MCF10A), 1,000 (SUM149) cells/well in 6-well plates. The other day, cells were irradiated or treated with olaparib. The survival clones were measured 6 to 7 days after seeding.
Plasmid transfection and RNAi experimentation
HA-BRCA1, Flag-Pin1, or Myc-tagged BRCA1 fragments plasmid (39) in pcDNA3.1 were used for transfection with Lipofectamine 2000 (Invitrogen). Cells were switched to growth medium 8 hours after transfection. Point mutations of BRCA1 and Pin1 were generated on the basis of pcDNA3.1 HA-BRCA1 and Flag-Pin1 plasmids using the Site-Directed Mutagenesis Kit (Agilent). ShPin1 cells were generated as described previously (40). Briefly, the target sequence of Pin1 shRNA is 5'-CCACCGTCACACAGTATTTAT-3', which targets to Pin1 3'UTR (40). The control shRNA sequence is 5'-UAAGGCUAUGAAGAGAUAC-3'. Following lentiviral infection, cells were selected using puromycin.
Immunoblotting
Primary mAbs against β-actin (1:10,000, Sigma), HA tag (1:2,000, Cell Signaling Technology), Flag tag (1:2,000, Cell Signaling Technology), Myc tag (1:1,000, 9E10, Santa Cruz Biotechnology), and BRCA1 antibody (1:500, MS110, Calbiochem), mouse anti-Pin1 (1:3,000; ref. 41) were used. Standard procedures were used for immunoblotting.
Cycloheximide chase assay
Cells were transfected with HA-BRCA1 and 48 hours later subjected to irradiation and then switched to complete medium with 100 μg/mL cycloheximide. Cell lysates were prepared at indicated time points. Western bands of HA BRCA1 and β-actin were quantified using Image J Software.
Coimmunoprecipitation assay
Transient Flag-Pin1 or HA-BRCA1-expressing cells were irradiated and lysed with RIPA buffer 2 hours after irradiation. The cell lysate was precleared with PA or PG beads and then incubated with anti-Flag or anti-HA antibody for 2 hours at 4°C. Following precipitation, the pellets were washed three times with lysis buffer and then analyzed by immuoblotting to determine the precipitated BRCA1 or Pin1.
Pin1 GST-pulldown assay
HA-BRCA1 or Myc-tagged BRCA1 fragments were transfected into HEC293 cells. 48 hours later, cells were irradiated and lysed for GST-Pin1 pull-down (42). The lysates were precleared with glutathione beads, and then incubated with a GST-Pin1 fusion protein immobilized on glutathione agarose beads for 2 hours at 4°C. Following precipitation, the pellets were washed three times with lysis buffer and then analyzed by Western blotting to determine the BRCA1 amount that binds to Pin1.
In vitro ubiquitination assay
In vitro ubiquitination assay was performed as described previously (43). Briefly, HEK293 cells were transfected with HA-BRCA1. 48 hours later, they were pretreated with MG132 (20 μg/mL) for 3 hours andthen subjected toirradiation. 6 hours after irradiation, cells were lysed and incubated with Ni-NTA-agarose beads for 3 hours at 4°C and then precipitated. The precipitate was washed three times and the reactions stopped with SDS sample buffer prior to SDS-PAGE for immunoblotting.
Immunofluorescence
MCF10A and MCF7 cells were irradiated and then fixed at indicated time points for immunofluorescence, stained with antibodies against BRCA1 (1:50, MS110), Pin1 (1:100), and 53BP1 (1:50, Cell Signaling Technology) followed by fluorescence secondary antibody reaction and examined with a Zeiss Axiovert 200M fluorescence microscope.
Confocal fluorescence microscopy
MCF10A or MCF7 cells transfected with HA-BRCA1, were irradiated and then were fixed as indicated time fixed. After fixation and blocking, cells were stained with anti-BRCA1 (1: 50, MS110) or anti-HA antibodies (1:50, Cell Signaling Technology), γ-H2AX (1:50, bioLegend) over night at 4°C. Fluorescence-labeled secondary antibodies were used followed by three times washing with PBS/Triton X-100 (0.1%). Slides were then covered with DAPI (4',6-diamidino-2-phenylindole) containing fluormount and subjected to confocal fluorescence microscopy (Zeiss LSM 510 Inverted Live-Cell Confocal System).
IHC
Pin1 (1:200; ref. 40), Ki67 (BD Biosciences), and BRCA1 (1:100 MS110) staining were performed according to standard protocols on formalin-fixed and paraffin-embedded tumors. Immunolabeling was visualized with a mixture of DAB solution (Vector Laboratories), followed by counterstaining with hematoxylin.
TUNEL assay
Cell apoptosis was detected by TUNEL assay in formalin-fixed and paraffin-embedded sections using Cell Death Detection Kit (Roche). Briefly, the sections were dewaxed, rehydrated, and treated with Proteinase K, followed by adding the TUNEL reaction mixture. The staining of sections was visualized by Leica fluorescence microscopy.
Animal experimentation
All studies involving mice were approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center and Sun Yat-Sen University (Guangzhou, China), and performed in accordance with the relevant protocols. For xenograft treatments, 1.5 × 106 of MDA-MB-231 cells expressing shcontrol or shPin1 with/without adding back exogenous wild-type (WT) Pin1 or mutant Pin1 (S67E) were injected into the mammary fat pads of 6-week-old BALB/c nude mice (Jackson Laboratories). Two weeks later, when tumor growth was visible, mice were randomized to receive olaparib or control treatments. For intraperitoneal injection, vehicle or olaparib (50 mg kg−1 d−1) were administered 5 weeks. For ATRA and olaparib combination treatments, 1.5 × 106 of MDA-MB-231 cells were injected into fat pads of 6-week-old BALB/c nude mice (Jackson Laboratories).
For patient-derived xenograft (PDX) implantation, three fresh BRCA1-proficient triple-negative breast cancer (TNBC) specimens were collected from tumor resection of patients at Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University (Guangzhou, China) between 2017 and 2018. All samples were collected with written informed consent from patients according to the internal review and ethics boards of Sun Yat-Sen Memorial Hospital (Guangzhou, China). Detailed procedure is described previously (44). Briefly, 6-week-old NSG female mice were anaesthetized by isoflurance. Tumors were minced into 1 mm3 sized fragments and imbedded directly into the mammary fat pads to get the first generation of the PDX. Once the PDX of first generation reached diameter of 1 cm, they were harvested and then minced into 1 mm3 sized fragments and imbedded directly into the mammary fat pads to get the second generation of PDX for treatments. For the treatment experiment, PDXs derived from each patient were transplanted into four mice and then randomized into four groups. Once tumors were established, mice were randomized to ATRA, olaparib, combination, or control treatments. For intraperitoneal injection, placebo, olaparib (50 mg kg−1d−1), ATRA (1.5 mg kg−1d−1), or combination of olaparib (50 mg/kg/d) and ATRA (1.5 mg/kg/d) were administered 5 weeks. Tumor sizes were recorded by a caliper and tumor volumes were calculated using formula L × W2 × 0.5, where L and W represent length and width, respectively. Mice were sacrificed after treatment of 5 weeks. Not any animal was excluded during the experiments.
For the characterization of PDXs, the DNA of PDX were subjected to next-generation sequencing of the HR DNA repair panel (AmoyDx), which included 32 genes (AR/ATM/ATR/BARD1/ BRAF/BRCA1/BRCA2/BRIP1/CDH1/CDK12/CHEK1/CHEK2/ERBB2/ ESR1/FANCA/FANCL/HDAC2/HOXB13/KRAS/MRE11/NBN/ NRAS/PALB2/PIK3CA/PPP2R2A/PTEN/RAD51B/RAD51C/RAD51D/ RAD54L/STK11/TP53). All coding region and junction region of exon and intron were sequenced by Illumina NextSeq500. The depth of sequencing was ≥1,000 × average coverage across the target region. Raw data were analyzed by AmoyDx ANDAS Data Analyzer. Genetic variants were filtered using MAF < 1% in 1,000 Genomes Project as a cutoff.
Statistical analysis
The in vitro data were presented as mean SD ± of three independent experiments. All statistical analyses were performed using SPSS 16.0 statistical software package (SPSS). Student t test and one-way ANOVA were used to compare cell viability, colony formation, and tumor volume with different treatments. In all cases, *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Results
Pin1 localizes to DNA damage repair foci
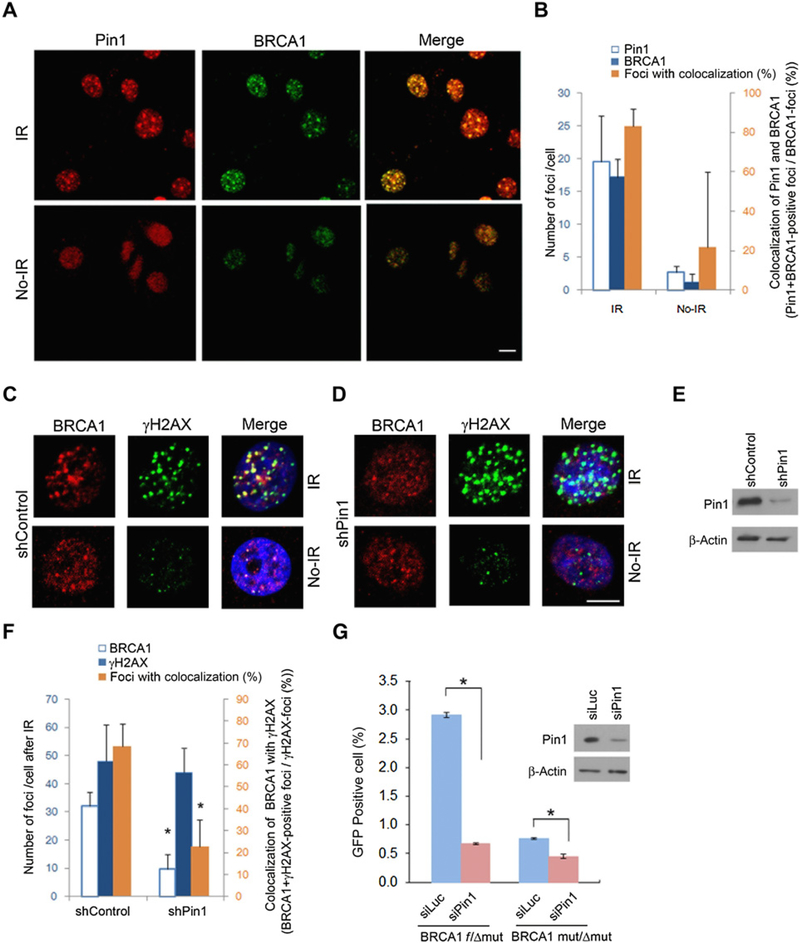
IR treatment is a common approach to induce DNA DBS (19). To examine the subcellular distribution of Pin1 in response to DBS damage, we applied 5Gy IR to treat mammary epithelial cells (MCF10A) or breast cancer cells (MCF7) with WT BRCA1 (WT BRCA1), which will form DNA repair foci in response to IR. The recruitment of BRCA1 (Fig. 1A; Supplementary Fig. S1A) or the transcriptional coactivator 53BP1 (Supplementary Fig. S1B) to discrete nuclear foci is a hallmark of DNA damage repair (45). We found that Pin1 colocalized to these repair foci after IR (Fig. 1A and B). To uncover a potential function of Pin1 for DNA damage repair, we examined the effects of Pin1 ablation, achieved by RNA interference and verified by immunoblotting or immunofluorescence for Pin1 (Supplementary Fig. S1A and S1B). Upon irradiation, the formation of foci that stained positive for 53BP1 or BRCA1 was greatly reduced when Pin1 expression was silenced in MCF7 or MCF10A cells (Supplementary Fig. S1A and S1B). The number of BRCA1 foci number per cell and the colocalization of BRCA1 foci with γH2AX foci after IR was reduced in Pin1-silenced cells (Fig. 1C–F), indicating a potential role for Pin1 in the formation of DNA damage repair foci, and the process of HR.
Figure 1.
Pin1 and BRCA1 colocalize in DNA damage repair foci and Pin1 ablation disrupts HR. A, BRCA1-proficient MCF10A cells were irradiated (5 Gy) and fixed 2 hours later for immunofluorescence with Pin1 and BRCA1. B, Quantification of data in A. Bars correspond to means ± SD of experimental triplicates. C and D, MCF10A cells were transduced with either shRNA control or shPin1, irradiated followed by immunostaining for BRCA1 and γH2AX 2 hours later. BRCA1 and γH2AX foci after irradiation in the presence (C) or absence of Pin1 (D). E, Immunoblotting of Pin1. F, Quantification of γH2AX and BRCA1 foci. Bars represent means ± SD of experimental triplicate experiments. Five cells were counted per experiment. G, Silencing Pin1 disrupts HR as assessed by short tract conversion in ES cells of the BRCA1f/Δmut genotype (left), similar to what is seen in ES cells of the BRCA1mut/Δmut genotype (right). Bars, means ± SD of experimental triplicates. *, P < 0.05.
To examine the effects of Pin1 in HR, we employed a functional reporter assay to measure HR (46). As the majority of postsynaptic HR events entails short track gene conversions (STGC), we used STGC as determined by repair of a GFP reporter gene as a read-out for HR in embryonic stem cells (ES cells). These mouse ES cells are of a BRCA1f/Δmut genotype, that is, they are heterozygous for WT BRCA1 and HR-proficient (Fig. 1G, left), or they incur loss of BRCA1 function, and HR deficiency (loss of exons 22–24 corresponding to loss of the BRCT domain in BRCA1; Fig. 1G, right). When transduced with Cre-recombinase, DSBs were induced in the cells and then cells undergo HR. Consistent with HR proficiency, about 3% of cells underwent successful HR when DSBs were induced inthe BRCA1f/Δmut cells. However, when Pin1 was silenced, the fraction of cells with HR decreased to 0.67% (Fig. 1G, left), similar to the rate of HR in cells in which both alleles are BRCA1 mutant (Fig. 1G, right). These observations suggest that Pin1 depletion is as effective as loss of BRCA1 in reducing HR and raise the possibility that Pin1 regulates BRCA1 function.
Pin1 binds to BRCA1 in a DNA damage–dependent manner
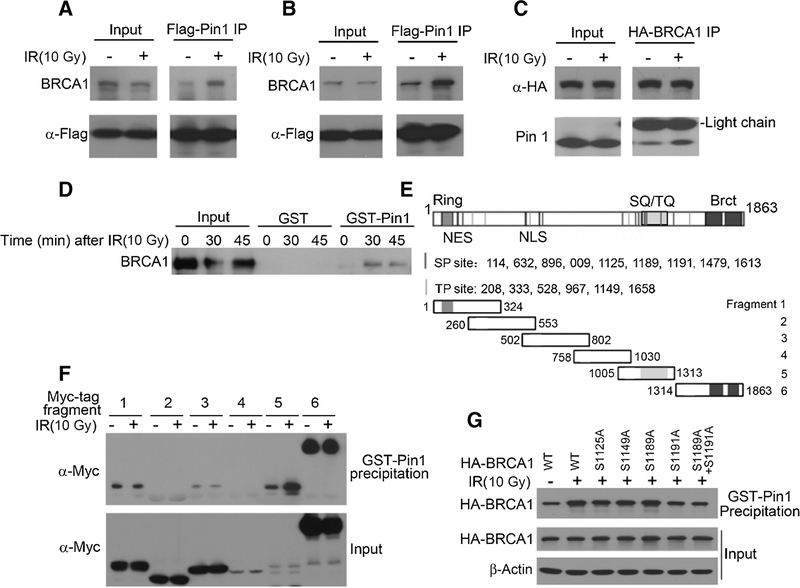
As Pin1 colocalized with BRCA1 in response to DNA damage and its depletion reduced HR and the formation of DNA damage repair foci, we examined whether Pin1 physically interacted with BRCA1 and at which site. We hypothesized that hyperphosphorylation of BRCA1 induced by IR (21) might provide phosphorylated Ser/Thr-Pro motifs that would serve as binding sites for Pin1. Flag-tagged Pin1 expressed in MCF7 or T47D breast cancer cells, which are BRCA1-proficient, precipitated with endogenous BRCA1 even when cells were not treated with IR. Precipitation of BRCA1 with Flag-tagged Pin1 increased within an hour after IR (Fig. 2A and B; Supplementary Fig. S2A and S2B). In the reciprocal experiment, HA-tagged BRCA1 expressed in HEK-293 cells was found to precipitate endogenous Pin1 at baseline and substantially more after IR (Fig. 2C; Supplementary Fig. S2C). As a third approach to examine the Pin1-BRCA1 interaction, we used the GST-pulldown technique to directly examine the interaction of Pin1 with BRCA1 in MCF7 cells. We found a weak interaction at baseline. However, as early as 30 minutes to 45 minutes after IR, GST-Pin1 pulled down BRCA1 (Fig. 2D; Supplementary Fig. S2D), indicating a DNA damage–dependent interaction of Pin1 with BRCA1.
Figure 2.
Serine 1191 is the phosphorylation site that serves as a docking site for Pin1 in BRCA1 upon DNA damage. Coimmunoprecipitation (IP) of endogenous BRCA1 and Flag-tagged Pin1 in MCF7 (A) and T47D (B). Cells were irradiated one hour prior to cell lysis and IP. C, Coimmunoprecipitation of endogenous Pin1 with HA-tagged BRCA1 transfected into HEK293 cells. Cells were pretreated as in A and B. D, GST-pulldown of endogenous BRCA1 with GST-Pin1 in MCF7 cells. MCF7 cells were irradiated as indicated. E, The BRCA1 ORF has nine candidate SP-motifs and six candidate TP-motifs. Sketch depicts Myc-tagged fragments that were created to determine Pin1-binding sites. F, GST-Pin1 pulldown of Myc-tagged BRCA1 fragments as indicated in E with GST-Pin1 in HEK293 cells. Cells were transfected with BRCA1 fragments and irradiated 48 hours later, followed by lysis 1 hour after irradiation. G, GST-Pin1 pulldown of S→A mutations in candidate SP-motifs in fragment 5. HEK293 cells were transfected with the respective fragments, irradiated, and lysed 1 hour later for GST-Pin1 pulldown.
Pin1 binds to phosphorylated serine 1191 of BRCA1 after radiation
BRCA1 has 15 Ser/Thr-Pro motifs, which are candidate sites for Pin1-mediated prolyl-isomerization (Fig. 2E). To address at which site Pin1 binds to BRCA1, we tested truncation fragments of BRCA1 for their ability to bind to Pin1 after radiation (Fig. 2E). We found that the N-terminal, C-terminal, and the middle fragments (AA 502–802 and AA1005–1313) are weakly bound to Pin1 in the absence of IR (Fig 2F; Supplementary Fig. S2E). However, the binding of the fragment AA1005–1313 with Pin1 was enhanced by IR (Fig. 2F; Supplementary Fig. S2E), suggesting that one of the Ser/Thr-Pro motifs in this fragment was phosphorylated in response to IR and was the binding site for Pin1 after IR. AA1005–1313 contains the SQ/TQ domain of BRCA1 and 4 Ser/Thr-Pro motifs (Ser1125, Thr1149, Ser1189, and Ser1191) that can potentially bind to Pin1. Among these sites, Ser1189 is phosphorylated by ATM and CDK1 (19, 24) and Ser1191 is phosphorylated by CDK1 after DNA damage (24). Both phosphorylation sites are required for BRCA1 to form DNA damage repair foci (24). We mutated putative phosphorylation sites in S/T-P motifs in BRCA1 and examined the ability of GST-Pin1 to pull down these mutants. As expected, binding of Pin1 to WT BRCA1 increased after IR, and this response was eliminated in a mutant at Serine 1191 (S1191A; Fig 2G; Supplementary Fig. S2F). We conclude that Ser1191, shown to be phosphorylated by CDK1, is the phosphorylation site that allows for binding of Pin1 to isomerize Pro1192.
Pin1 stabilizes BRCA1 after DNA damage
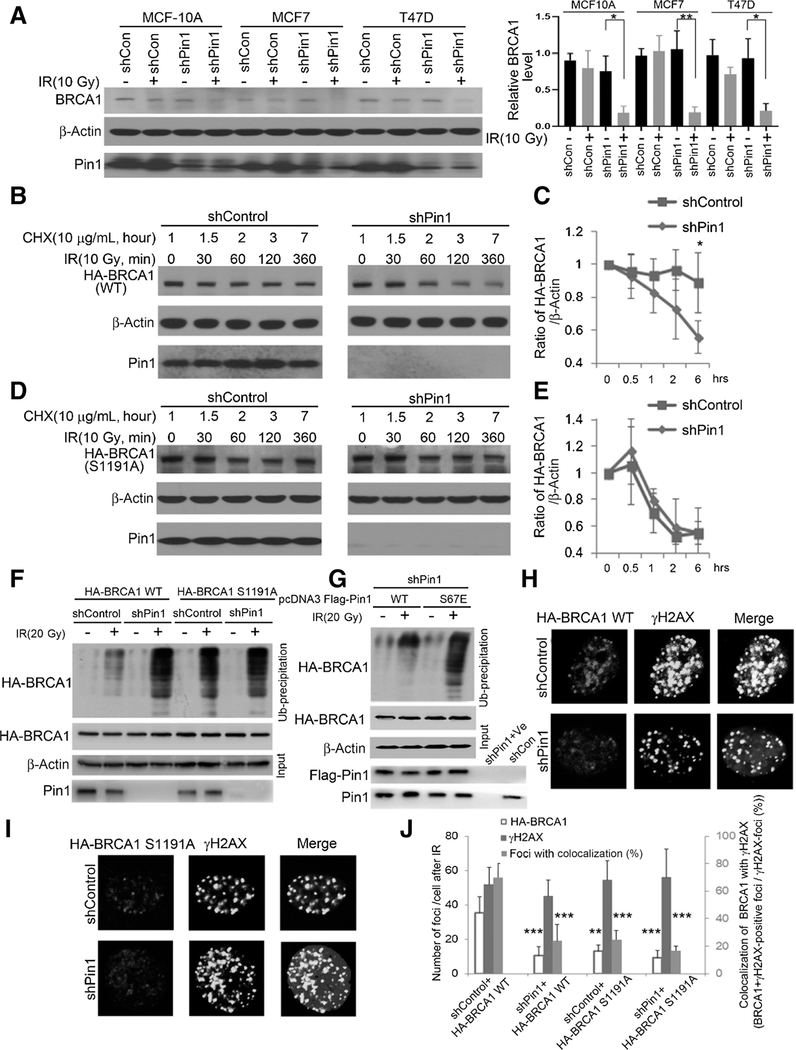
DNA damage–induced BRCA1 phosphorylation also promotes its ubiquitination and degradation (47). To understand how Pin1-mediated prolyl-isomerization affected BRCA1 and given that Pin1 is known to regulate protein stability of tumor suppressor genes (48), we examined the stability of BRCA1 in response to Pin1knockdown. While baseline BRCA1 expression was not affected by Pin1 shRNA (Fig. 3A; Supplementary Fig. S2G–S2I), Pin1 loss led to a decrease in BRCA1 expression after irradiation in mammary epithelial cells and breast cancer cells (Fig. 3A). A cycloheximide run-off assay showed that Pin1 knockdown in MCF7 cells decreased the half-life of BRCA1 to about 6 hours (Fig. 3B and C). In contrast, the S1191A BRCA1 mutant had a short half-life (<6 hours) regardless of the absence or presence of Pin1 (Fig. 3D and E). These data suggest that Pin1 facilitates the stabilization of BRCA1 after IR-induced phosphorylation of Ser1191 through prolyl-isomerization.
Figure 3.
Pin1-catalyzed prolyl-isomerization is required for radiation-induced stabilization of BRCA1. A, MCF10A, MCF7, and T47D cells were transduced with shRNA against Pin1 or control for 48 hours, then irradiated and lysis 5 hours later. The protein level of BRCA1 was detected by immunoblotting and quantified as ratio relative to β-actin (right). B, MCF7 cells were transfected with HA-BRCA1 and subjected to a cycloheximide (CHX) run-off assay in the absence (right) or presence (left) of Pin1. C, Quantification of HA-BRCA1 stability in B of biological triplicates (the vertical axis starting at 0.4). D, MCF7 cells were transfected with the HA-BRCA1 (S1191A) mutant, identified as Pin1-binding site, and subjected to a cycloheximide run-off assay in the absence (right) or presence (left) of Pin1. E, Quantification of HA-BRCA1 (S1191A) stability in D of biological triplicates (the vertical axis starting at 0.4). F, BRCA1 ubiquitination after irradiation is enhanced in the absence of Pin1 and after extinction of the Serine 1191 phosphorylation site. ShPin1 or ShControl HEK293 cells were transfected with HA-BRCA1 (WT or S1191A) for 48 hours, pretreated with MG132 3 hours prior to irradiation and lysed 6 hours after irradiation. G, The WT, but not the catalytically inactive Pin1 mutant (S67E), reduces BRCA1 ubiquitination. ShPin1 HEK293 cells were transfected with HA-BRCA1 and Flag-Pin1 (WT or S67E mutant) for 48 hours, pretreated with MG132 3 hours prior to irradiation, and lysed 6 hours after irradiation. Absence of Pin1 (H and J) mimics mutation of Serine 1191 (I and J) with regards to DNA repair foci formation. MCF7 cells were transfected with HA-BRCA1 (WT or S1191A) for 48 hours and then irradiated. Immunofluorescence was performed 5 hours after IR. *, P < 0.05; **, P < 0.01; ***, P < 0.001
BRCA1 expression increases in S and G2–M when it is recruited to DNA repair foci (49). Thereafter, BRCA1 undergoes ubiquitination and degradation (50). Consequently, we observed ubiquitination of BRCA1 after DNA damage, which greatly increased upon Pin1 knockdown (Fig. 3F). Consistent with its shortened half-life irrespective of Pin1 (Fig. 3D and E), ubiquitination of the S1191A BRCA1 mutant in response to radiation was greatly increased and did not respond to extinction of Pin1 (Fig. 3F). Moreover, in the shPin1 cells, when adding back exogenous Pin1, only the WT, but not the catalytically inactive Pin1 mutant (S67E), reduced BRCA1 ubiquitination (Fig. 3G). The S1191A-BRCA1 mutant was also not efficiently recruited into the DNA damage repair foci regardless of the absence or presence of Pin1, as detected by the colocalization with γH2AX (Fig. 3H–J). These observations indicate that binding of Pin1 to the Ser1191-Pro motif of BRCA1 catalyzes the S1191/P1192 prolyl-isomerization and serves to stabilize BRCA1, allowing for its recruitment to DNA damage repair foci.
Pin1 stabilizes BRCA1 by preventing ubiquitination of Lysine 1037 of BRCA1
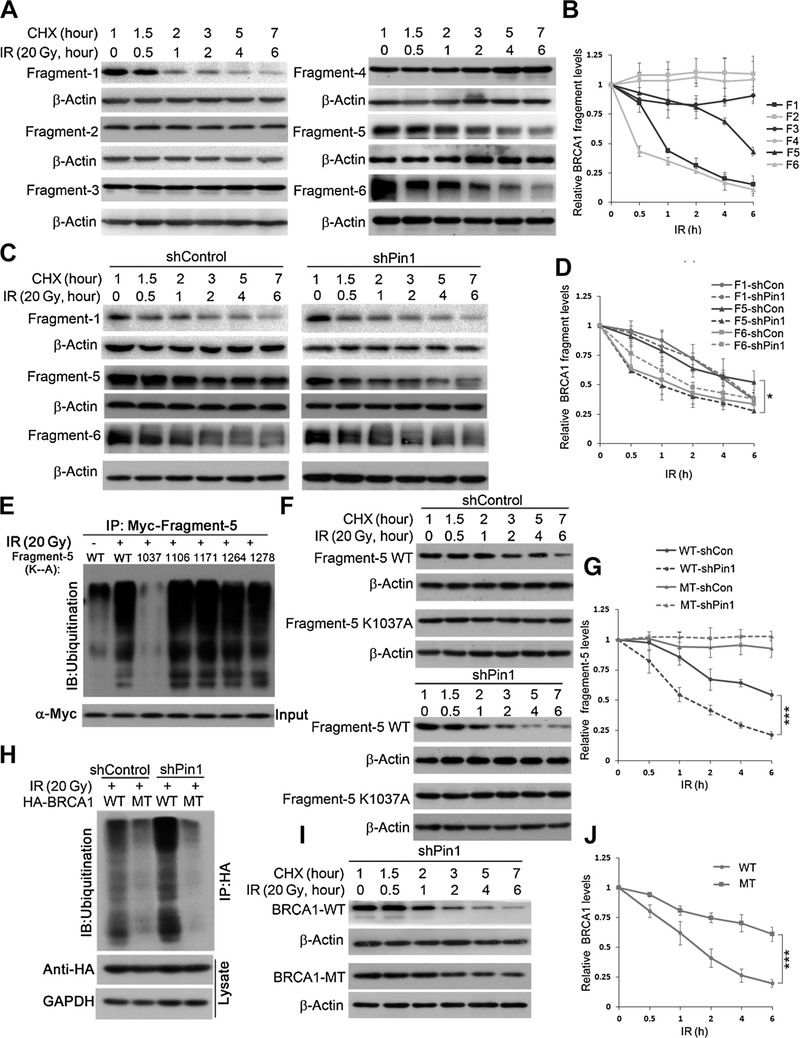
We next determined which Lysine residue is the target for ubiquitin-mediated degradation of BRCA1 after a genotoxic event. Toward this end, BRCA1 fragments were subjected to a protein decay assay after irradiation. While fragments 1, 5, and 6 were degraded after irradiation (Fig. 4A and B), only degradation of fragment 5 was Pin1-dependent, that is, selectively accelerated in the absence of Pin1 (Fig. 4C and D). To identify the lysine site(s) of BRCA1 for Pin1-dependent ubiquitination, the Ubpred algorithm (http://www.ubpred.org) was used for prediction. Five candidate lysine sites were identified and substituted with alanine (Fig. 4E). In vivo ubiquitination assays in Pin1 knockdown HEK293 cells after irradiation showed that the K1037A mutant largely abolished the ubiquitination of fragment 5 (Fig. 4E). Consistently, when the K1037A mutant was subjected to the protein degradation assay, it was significantly more stable than the WT fragment 5 irrespective of the presence or absence of Pin1 (Fig. 4F and G). To confirm the K1037 was the ubiquitination site for the stability of BRCA1 in vivo, the K1037A mutant of full-length HA-BRCA1 was generated. We found that the ubiquitination of K1037A-mutant BRCA1 was significantly reduced, irrespective of the absence or presence of Pin1 (Fig. 4H). In addition, the K1037A mutant of HA-BRCA1 was degraded significantly slower than the WT BRCA1 in the Pin1 knockdown HEK293 cells (Fig. 4I and J). The WT or K1037A mutant of HA-BRCA1 was also transfected into MCF7 cells. The K1037A-BRCA1 mutant was efficiently recruited into the DNA damage repair foci regardless of the absence or presence of Pin1 (Supplementary Fig. S3A–S3C). In summary, we showed that Pin1 prevented the ubiquitination of BRCA1 on K1037, thus enhancing the stability of BRCA1 stability in the process of DNA damage repair.
Figure 4.
Lysine 1037 of BRCA1 is the ubiquitination site that mediates BRCA1 degradation upon DNA damage in Pin1 knockdown cells. A, Myc-labeled BRCA1 fragments were transfected into HEK293 cells and subjected to acylcoheximide(CHX)run-off assay following irradiation treatment. B, Quantification of BRCA1 fragment stability in A inbiological triplicates. C, Myc-labeled BRCA1 fragments 1, 5, and 6 were transfected into HEK293 cells, respectively, and subjected to a cycloheximide run-off assay in the absence (right) or presence (left) of Pin1. Fragment 5 degraded faster in absence of Pin1 than in the presence of Pin1. D, Quantification of BRCA1 fragment stability in C in biological triplicates. E, Ubiquitination of BRCA1 fragment 5 after irradiation is suppressed in the K1037A mutant in the absence of Pin1. ShPin1 HEK293 cells were transfected with Myc-tagged fragment 5 (WT or indicated mutants) for 48 hours, pretreated with MG132 3 hours prior to irradiation, and lysed 6 hours after irradiation. F and G, Stability of Myc-tagged WT and mutant (K1037A) fragment 5. Myc-BRCA1 fragments were transfected into HEK293 cells and subjected to a cycloheximide run-off assay in the presence (top) or absence (bottom) of Pin1. G, Quantification of BRCA1 fragment 5 stability in F in biological triplicates. H, HA-BRCA1 ubiquitination after irradiation in the absence of Pin1. Shcontrol or shPin1 HEK293 cells were transfected with HA-BRCA1 (WT or K1037A mutant) for 48 hours, pretreated with MG132 3 hours prior to irradiation, and lysed 6 hours after irradiation. I, The mutant BRCA1 decaysslower than the WT in the absence of Pin1. WT or mutant (K1037A) HA-BRCA1 were transfected into shPin1 HEK293 cells and subjected to a cycloheximide run-off assay. J, Quantification of HA-BRCA1 stability in I in biological triplicates. *, P < 0.05; ***, P < 0.001
Pin1 inhibition sensitizes breast cells to radiation and PARP inhibition
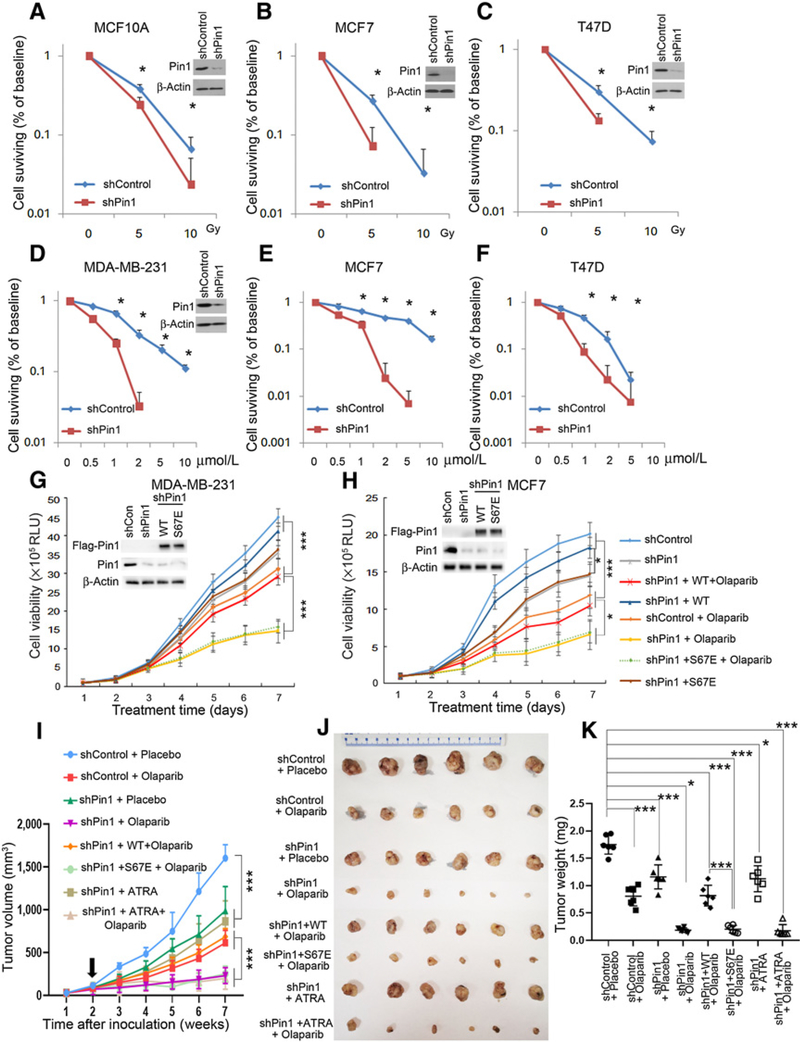
Our finding that the phospho-specific prolyl-isomerizase Pin1 stabilizes BRCA1 and promotes its function in DNA damage repair foci raises the possibility that Pin1 extinction can destabilize BRCA1, disrupt DNA damage repair via HR and hence lead to radiosensitization. We found that Pin1 inhibition by means of shRNA reduced cell survival after IR in long-term colony assays (Fig. 5AC). In MCF10A cells (BRCA1- and P53-proficient), Pin1 inhibition decreased colony formation at 5Gy IR by about 35% (Fig. 5A), in MCF7 (BRCA1- and P53-proficient) breast cancer cells colony formation was reduced by about 55% by Pin1 extinction (Fig. 4B). In T47D cells (BRCA1-proficient and P53-deficient), focus formation after irradiation was reduced by about 74% by Pin1 extinction (Fig. 5C). On the other hand, in SUM149 cells, BRCA1 mutant (2288delT) and Pin1 knockdown did not further decrease colony formation efficiency (Supplementary Fig. S4A), indicating that loss of Pin1 and loss of BRCA1 were redundant.
Figure 5.
Extinction of Pin1 sensitizes BRCA1-WT breast cancer cells to DNA damage. Colony formation assays were performed with shPin1 or control cells treated with IR(A–C) or olaparib (D–F). MCF10A (A), MCF7 (B and E), T47D (C and F), MDA-MB-231 (D). G and H, Effect of combining Pin1 knockdown with olaparib in suppressing the viability of BRCA1-proficient cells. Stable shPin1 or shControl MDA-MB-231 (G) and MCF7 (H) cells were treated with olaparib (10 μmol/L), or transfected with either WT or catalytically inactive Pin1 (S67E). Viability of shPin1 or control cells was examined in response to olaparib or ATRA in MDA-MB-231 (G) or MCF7 (H). I–K, Pin1 depletion sensitizes BRCA1-proficient breast tumors to PARP inhibitor, but not to ATRA treatment. MDA-MB-231 cells with stable expression of shControl, shPin1, or shPin1 plus Pin1 (WT or S67E; 1.5 million/mouse) were implanted into fat pads of nude mice and treatments were started 2 weeks after inoculation. Treatments were continued for 5 weeks when animals were euthanized. Tumor growth curve is shown in I. The harvest tumor was measured (J) and weighted (K). *, P < 0.05; ***, P < 0.001
PARP inhibitors induce DNA single-strand breaks, which will convert to double-strand breaks upon cellular replication, therefore activating BRCA1 for HR repair (51). Thus, BRCA-negative cells or HR-deficient cells are highly susceptible to PARP inhibitors (52, 53). We showed that Pin1 depletion increased the BRCA1 degradation under IR treatment. Here we analyzed BRCA1 stability after PARP inhibitor olaparib treatment in BRCA1-proficient cells. BRCA1 degraded faster in Pin1 knockdown cells than in the control MCF7 cells (Supplementary Fig. S4B and S4C), which suggested that PARP inhibition could be synergistically lethal with loss of Pin1. Thus, we examined the effects of Pin1 depletion on PARP inhibitor sensitivity in WT BRCA1 breast cancer cells MDA-MB-231 (triple-negative), MCF7, and T47D (ER+). We found that breast cancer cells with WT BRCA1, irrespective of triple-negative or ER positive, showed sharply reduced colony formation in the presence of the olaparib after Pin1 depletion (Fig. 5D–F), while the BRCA1-mutant SUM149 were not affected by Pin1 ablation (Supplementary Fig. S4D). In parallel, cell viability assays showed that the growth of MDA-MB-231 and MCF7 cells was moderately delayed by Pin1 knockdown or olaparib monotherapy, while olaparib treatment in Pin1 knockdown cells resulted in near-complete block of cell proliferation (Fig. 5G and H). Furthermore, when adding exogenous Pin1 back inshPin1 MDA-MB-231 and MCF7 cells, only the WT, but not the catalytically inactive Pin1 mutant (S67E) could rescue the synergistic inhibitory effect of Pin1 RNAi with olaparib (Fig. 5G and H).
To measure the effects of Pin1 depletion on responses to PARP inhibitors in vivo, xenografts of MDA-MB-231 cells expressing control or Pin1 shRNA with/without adding back exogenous WT Pin1 or mutant Pin1 (S67E) were established in nu/nu mice. 2 weeks after tumor cell inoculation, the mice were commenced on vehicle control or olaparib for 5 weeks. Pin1 knockdown or olaparib monotherapy moderately delayed growth of xenografts. However, when Pin1 knockdown was combined with olaparib, tumor growth was substantially delayed (Fig. 5I and J). When exogenous WT Pin1 was added back to the shPin1 cells, olaparib treatment only moderately delayed tumor growth, whereas the mutant Pin1 (S67E) did not have such effect (Fig. 5I and J). The mean tumor volume of Pin1-depleted xenografts was about 30% smaller than shcontrol tumors (P < 0.01), confirming previous reports that Pin1 ablation suppresses breast cancer growth (10). The mean tumor volume and weight were significantly reduced when Pin1 depletion was combined with olaparib (Fig. 5I–K). As expected, Pin1 depletion led to reduced levels of BRCA1 (Supplementary Fig. S5A and S5B), which could be restored by adding back exogenous WT Pin1 but not mutant Pin1 (S67E). The combination of Pin1 depletion and olaparib effectively lowered the proliferation of tumor cells, as assessed by Ki67 staining, and increased tumor cell apoptosis, as detected by TUNEL assay (Supplementary Fig. S5A, S5C, and S5D).
ATRA is an HR-disrupting agent
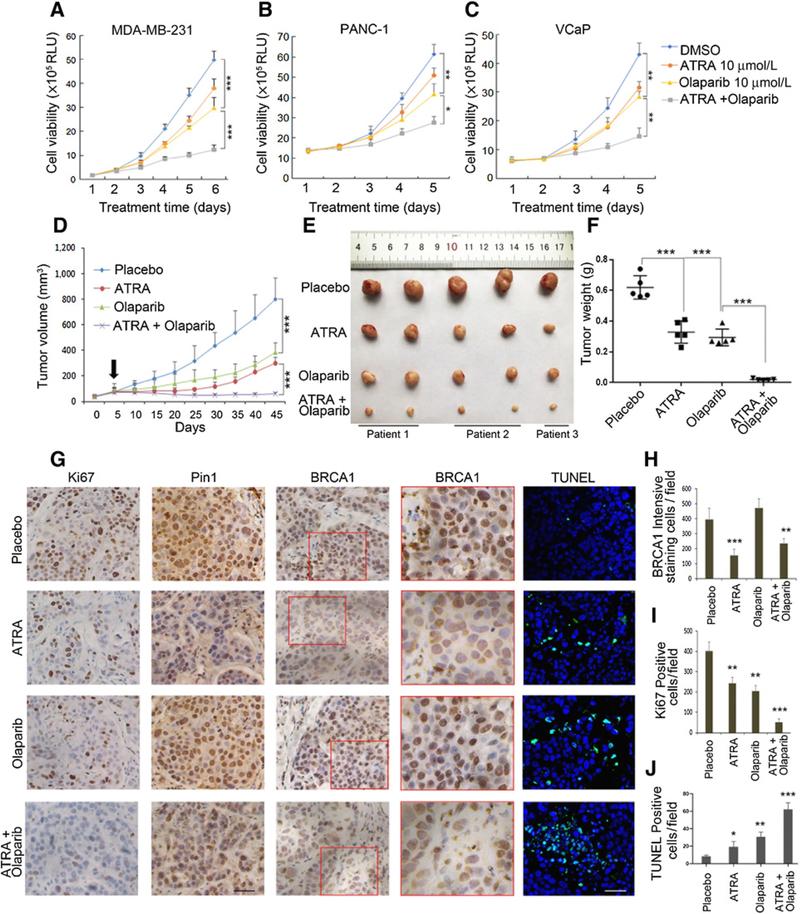
ATRA has been reported to be a Pin1 inhibitor, leading to Pin1 degradation (36, 37), hence we expected treatments with ATRA to result in similar effects as genetic Pin1 ablation. We found that either ATRA or olaparib delayed cell growth, while the combination of ATRA and olaparib stalled cell growth synergistically, not only in MDA-MB-231 and MCF7 cells, but also in BRCA1/2-WT pancreatic cancer cell PANC-1 and prostate cancer cell VCaP (Fig. 6A–C; Supplementary Fig. S5E). These in vitro data suggest that ATRA, through Pin1 degradation, acts as a HR-disrupting agent and hence might sensitize BRCA1-WT tumors to PARP inhibition in vivo. To test this hypothesis, we treated mice-bearing MDA-MB-231 xenografts with single-agent ATRA, olaparib, or their combination for 5 weeks. We found that the combination of ATRA and olaparib significantly delayed tumor growth (Supplementary Fig. S6A–S6C). As expected, ATRA led to reduced levels of Pin1 and BRCA1 (Supplementary Fig. S6D and S6E), and the combination of ATRA with olaparib effectively lowered the proliferative rate of tumor growth and increased apoptotic rate as assessed by Ki67 staining and TUNEL assay, respectively (Supplementary Fig. S6D, S6F, and S6G). Thus, inhibiting Pin1 by ATRA shows similar effect as shRNA Pin1 in sensitizing BRCA1-WT xenografts to olaparib treatment (Supplementary Fig. S5). In addition, ATAR treatment in shPin1 xenografts did not significantly delay tumor growth further (Fig. 5I–K). When ATAR was combined with shPin1 and olaparib, the inhibitory effect to the tumor was similarly as strong as shPin1 combined with olaparib (Fig. 5I–K). These results strengthened that ATRA action was mediated through Pin1.
Figure 6.
ATRA is as effective as Pin1 knockdown in regards to sensitizing tumors to PARP inhibitor treatments. A–C, Cell proliferation assays under the treatment of ATRA (10 μmol/L) and olaparib using MDA-MB-231 (A), PANC-1 (B), and VCaP (C) cells. D–F, ATRA treatments sensitize BRCA1-proficient PDX from TNBCs to PARP inhibitor. PDX from TNBC tissues were implanted into the mammary fat pads of NSG mice. Treatments as indicated were started about 2 to 3 weeks after inoculation when the tumor volume reached around 100 mm3 (D). Arrow, the initial time point of treatment. Treatments were continued for 35 days when animals were euthanized and tumors were measured (E) and weighed (F). G–J, ATRA treatments mimic Pin1 ablation with regards to extinction of BRCA1 expression, and the combination of ATRA and olaparib has antiproliferative effects. IHC of Ki67, Pin1, and BRCA1, as well as TUNEL assay in representative tumors at treatment endpoint. G, Red boxed area in BRCA1 is shown in a higher magnification on the right. H, Quantification of BRCA1 staining. I, Quantification of Ki67 staining. J, Quantification of TUNEL staining. For in vivo treatments, tissues from three patients of TNBCs were used to generate PDX. Totally, 20 PDX-bearing animals were used (n = 5 per group). Scale bars, 100 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001
The therapeutic effect of the combination of ATRA with olaparib was further tested in PDX of TNBCs, which did not contain BRCA1/2 mutations, or alterations of core genes in HR DNA repair pathways (Supplementary Table S1 and S2). The tumors from 3 patients were propagated anddivided into fourgroups andtreatedwhen the volumes reached approximately100 mm3. The combination of ATRA and olaparib significantly halted tumor growth (Fig. 6D–F), while single-agent treatment only slowed down the growth of these PDX moderately (Fig. 6D–F). As expected, ATRA led to reduced levels of Pin1 and BRCA1 (Fig. 6G and H). The combination of ATRA and olaparib effectively lowered the proliferative rate of tumor growth and increased apoptotic rate as assessed by Ki67 and TUNEL staining, respectively (Fig. 6G, I, and J). On the other hand, the mouse weights of each group were not significantly different, suggesting that the combination does not have significant systemic toxicity (Supplementary Fig. S6H and S6I).
Discussion
It has previously been reported that Pin1 deficiency causes a defect in nonhomologous end joining (NHEJ) because in the absence of Pin1, the endonuclease CtIP is stabilized, leading to more efficient DSB end resection (18). Here we show that Pin1 not only controls the balance between DSB end resection and NHEJ, but also regulates HR directly by protecting BRCA1 from degradation. DNA DSB repair via NHEJ or HR has evolved in eukaryotic cells starting with budding yeast, simultaneously with the emergence of the phospho-specific prolyl-isomerase Pin1. While NHEJ can proceed at any point during the cell cycle, HR is employed almost exclusively during S and G2 phases when a homologous sister chromatid is available. As Pin1 serves to coordinate mitotic progression, it is consistent that a DNA damage repair mechanism operating specifically in the S- and G2-phases of the cell cycle, that is, HR would be included in the regulatory network coordinated by Pin1. BRCA1 protein levels and its phosphorylation continuously accumulate, starting at S-phase and culminating at mid-G2, coinciding with peak Pin1 expression, phosphorylation, and nuclear localization (40). Here we show that Pin1-mediated phospho-specific prolyl-isomerization on S1191/P1192 is needed for the maintenance of BRCA1 in DNA damage repair foci. We have showed that the fragment 5 of BRCA1 contains the S/T-P site that is in charge of the increased binding with Pin1 upon DNA damage. By point mutation, we revealed that S1191/P1192 is the only site in fragment 5 of BRCA1 that binds with Pin1 upon DNA damage (Fig. 2). Thus, even Pin1 can bind to other S/T-P sites of BRCA1, those sites would not be involved in the regulation of Pin1 to BRCA1 under DNA damage. The S1191A mutation of BCRA1 abolished the interaction of Pin1 with S1191/P1192 and therefore failed to isomerize S1191/P1192 and induce K1037 ubiquitination. In the shPin1 cells, when adding back exogenous Pin1, only the WT, but not the catalytically inactive Pin1 mutant (S67E), reduced BRCA1 ubiquitination. These data supported that Pin1 catalyzed the S1191A/P1192 phospho-specific prolyl-isomerization and then stabilized BRCA1 by preventing its ubiquitination at K1037 in BRCA1, thus extending its availability in DNA damage repair foci.
Interestingly, Pin1 was recently also found to interact with BRCA1 at Ser114, promoting the BRCA1-BARD1 interaction (54). The study of BRCA1-BARD1 interaction supported that protecting replication fork and retaining HR proficiency are separate functions of BRCA1 (54). This is concordant with our data that Pin1-mediated stabilization of BRCA1 in facilitating HR after DNA damage was specifically dependent on Pin1-BRCA1 S1191/P1192 interaction, which was separated from the BARD1-BRCA1 S114 interaction for protecting replication fork. Notably, we found that the N-terminal fragment of BRCA1, including Serine 114 as well as the C-terminal fragment 6 that includes the BRCT domains, also underwent degradation, which was, however, not Pin1-sensitive (Fig. 4B). These findings are consistent with Daza-Martin and colleagues, who have shown that prolyl-isomerization on S114 is required for BRCA1-BARD1 heterodimerization (54) and they support a pivotal role for Pin1 in maintaining BRCA1-mediated genomic stability, which is likely achieved by both, an increased stability of BRCA1 and the enablement of its interactions with its heterodimeric partners. Thus, both, the stability and the function of BRCA1 during DNA damage repair or replication can be regulated through prolyl-isomerization on multiple sites by Pin1, analogous to how P53 stability and function are regulated by Pin1-mediated prolyl-isomerization on multiple S/P sites in cell-cycle checkpoint control (12, 13). Our findings add to the complexity of the effects of phospho-specific prolyl-isomerization that generally promotes and facilitates mitotic progression. This includes the stabilization of proproliferative signaling molecules, and also molecules that safeguard against errors during mitosis such as the checkpoint control proteins P53, P63, or P73 (15, 55, 56).
PARP inhibition is to some extent a “stress test” for HR as inhibition of PARP1 leads to greater reliance on HR (57, 58). PARP inhibitors have emerged as a safe and effective agent to treat BRCA-mutant breast, ovary, prostate, and pancreatic cancers with HR defects (29–31). However, gBRCAwt cancers are less likely to respond, leading to efforts to sensitize these tumors by combining PARP inhibitors with HR disrupters (59, 60). ATRA has been used clinically to treat acute promyelocytic leukemia through induction of differentiation with few side effects. Our data here raise the possibility that ATRA, or an ATRA derivative, could act as an effective HR disrupter that sensitizes tumors to PARP inhibition. Given Pin1's abundant expression in multiple cancer types, the combination of ATRA and olaparib may have broad applicability to target HR-proficient tumors, besides the breast cancer. Moreover, effective Pin1 inhibition might convey additional therapeutic benefits such as disruption of the tumor's cyclinD1 signaling axis and extinction of a dominant mutant P53, found in the majority of TNBCs, all contributing to its efficacy in epithelial cancers.
Supplementary Material
Acknowledgments
This work was supported by grants from the Natural Science Foundation of China (81572890, 81672738, 81730077, 81772613, 81872370), Guangdong Science and Technology Department (2017B030314026), Science and Technology Program of Guangzhou (201704020095), the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2016ZT06S252), Elite Young Scholars Program of Sun Yat-Sen Memorial Hospital (Y201701), and grants from SU2C-AACR-DT0209, from Mary Kay Ash Foundation, Ovarian Cancer Research Foundation, Breast Cancer Alliance, Breast Cancer Research Foundation (BCRF), and Merck & Co. of USA.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
X.Z. Zhou and K.P. Lu have an ownership interest in patents and patent applications owned by Beth Israel Deaconess Medical Center. G.M. Wulf reports receiving a commercial research grant from Merck & Co., Inc. (A11071). No potential conflicts of interests were disclosed by the other authors.
Footnotes
Disclosure of Potential Conflicts of Interest
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol 2005;7:435–41. [DOI] [PubMed] [Google Scholar]
- 2.Yeh ES, Means AR. PIN1, the cell cycle and cancer. Nat Rev Cancer 2007;7:381–8. [DOI] [PubMed] [Google Scholar]
- 3.Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, Fujimori F, et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci U S A 2002;99:1335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Wang S, Zhu T, Zhou J, Xu Q, Lu Y, et al. Pin1 contributes to cervical tumorigenesis by regulating cyclin D1 expression. Oncol Rep 2006;16:491–6. [PubMed] [Google Scholar]
- 5.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, et al. Pin1 is overexpressed in breast cancer and cooperates with ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J 2001;20:3459–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell 2005;17: 215–24. [DOI] [PubMed] [Google Scholar]
- 7.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, et al. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell 2003;12:1413–26. [DOI] [PubMed] [Google Scholar]
- 8.Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol 2001;3:793–801. [DOI] [PubMed] [Google Scholar]
- 9.Xu YX, Manley JL. The prolyl isomerase pin1 functions in mitotic chromosome condensation. Mol Cell 2007;26:287–300. [DOI] [PubMed] [Google Scholar]
- 10.Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J 2004; 23:3397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryo A, Uemura H, Ishiguro H, Saitoh T, Yamaguchi A, Perrem K, et al. Stable suppression of tumorigenicity by Pin1-targeted RNA interference in prostate cancer. Clin Cancer Res 2005;11:7523–31. [DOI] [PubMed] [Google Scholar]
- 12.Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G, et al. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 2002;419: 849–53. [DOI] [PubMed] [Google Scholar]
- 13.Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, et al. The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 2002;419:853–7. [DOI] [PubMed] [Google Scholar]
- 14.Wulf GM, Liou YC, Ryo A, Lee SW, Lu KP. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J Biol Chem 2002;277:47976–9. [DOI] [PubMed] [Google Scholar]
- 15.Mantovani F, Piazza S, Gostissa M, Strano S, Zacchi P, Mantovani R, et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol Cell 2004; 14:625–36. [DOI] [PubMed] [Google Scholar]
- 16.Oberst A, Rossi M, Salomoni P, Pandolfi PP, Oren M, Melino G, et al. Regulation of the p73 protein stability and degradation. Biochem Biophys Res Commun 2005;331:707–12. [DOI] [PubMed] [Google Scholar]
- 17.Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, et al. A Pin1/ mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 2011;20: 79–91. [DOI] [PubMed] [Google Scholar]
- 18.Steger M, Murina O, Huhn D, Ferretti LP, Walser R, Hanggi K, et al. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol Cell 2013;50:333–43. [DOI] [PubMed] [Google Scholar]
- 19.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 1999;286:1162–6. [DOI] [PubMed] [Google Scholar]
- 20.Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 2000;404:201–4. [DOI] [PubMed] [Google Scholar]
- 21.Gatei M, Scott SP, Filippovitch I, Soronika N, Lavin MF, Weber B, et al. Role for ATM in DNA damage-induced phosphorylation of BRCA1. Cancer Res 2000;60: 3299–304. [PubMed] [Google Scholar]
- 22.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, et al. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev 2000;14:2989–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J. Ataxia telangiectasia-related protein is involved in the phosphorylation of BRCA1 following deoxyribonucleic acid damage. Cancer Res 2000;60:5037–9. [PubMed] [Google Scholar]
- 24.Johnson N, Cai D, Kennedy RD, Pathania S, Arora M, Li YC, et al. Cdk1 participates in BRCA1-dependent S phase checkpoint control in response to DNA damage. Mol Cell 2009;35:327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 1997;88:265–75. [DOI] [PubMed] [Google Scholar]
- 26.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 1997;90:425–35. [DOI] [PubMed] [Google Scholar]
- 27.Scully R, Anderson SF, Chao DM, Wei W, Ye L, Young RA, et al. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci U S A 1997;94:5605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 2002;108:171–82. [DOI] [PubMed] [Google Scholar]
- 29.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009;361:123–34. [DOI] [PubMed] [Google Scholar]
- 30.Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 2019;381:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015;373: 1697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010;376:235–44. [DOI] [PubMed] [Google Scholar]
- 33.Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmana J, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov 2012;2:1048–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou XZ, Lu KP. The isomerase PIN1 controls numerous cancer-driving pathways and is a unique drug target. Nat Rev Cancer 2016;16:463–78. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Xu X, Wen X, Xu S, Wang L, Lu W, et al. Targeting PIN1 exerts potent antitumor activity in pancreatic ductal carcinoma via inhibiting tumor metastasis. Cancer Sci 2019;110:2442–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei S, Kozono S, Kats L, Nechama M, Li W, Guarnerio J, et al. Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat Med 2015;21:457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozono S, Lin YM, Seo HS, Pinch B, Lian X, Qiu C, et al. Arsenic targets Pin1 and cooperates with retinoic acid to inhibit cancer-driving pathways and tumor-initiating cells. Nat Commun 2018;9:3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003;30:256–68. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 1998;2:317–28. [DOI] [PubMed] [Google Scholar]
- 40.Luo ML, Gong C, Chen CH, Hu H, Huang P, Zheng M, et al. The Rab2A GTPase promotes breast cancer stem cells and tumorigenesis via Erk signaling activation. Cell Rep 2015;11:111–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo ML, Gong C, Chen CH, Lee DY, Hu H, Huang P, et al. Prolyl isomerase Pin1 acts downstream of miR200c to promote cancer stem-like cell traits in breast cancer. Cancer Res 2014;74:3603–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S, et al. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol Cell 2011;42:147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell 2010; 18:147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10(+)GPR77(+) cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell 2018;172:841–56. [DOI] [PubMed] [Google Scholar]
- 45.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. lap53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 2000;151:1381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandramouly G, Kwok A, Huang B, Willis NA, Xie A, Scully R. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat Commun 2013;4:2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parameswaran B, Chiang HC, Lu Y, Coates J, Deng CX, Baer R, et al. Damage-induced BRCA1 phosphorylation by Chk2 contributes to the timing of end resection. Cell Cycle 2015;14:437–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci 2011;36: 501–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mok MT, Henderson BR. A comparison of BRCA1 nuclear localization with 14 DNA damage response proteins and domains: identification of specific differences between BRCA1 and 53BP1 at DNA damage-induced foci. Cell Signal 2010;22:47–56. [DOI] [PubMed] [Google Scholar]
- 50.Choudhury AD, Xu H, Baer R. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J Biol Chem 2004;279:33909–18. [DOI] [PubMed] [Google Scholar]
- 51.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21. [DOI] [PubMed] [Google Scholar]
- 52.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 2006;66: 8109–15. [DOI] [PubMed] [Google Scholar]
- 53.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol 2008;26:3785–90. [DOI] [PubMed] [Google Scholar]
- 54.Daza-Martin M, Starowicz K, Jamshad M, Tye S, Ronson GE, MacKay HL, et al. Isomerization of BRCA1-BARD1 promotes replication fork protection. Nature 2019;571:521–27. [DOI] [PubMed] [Google Scholar]
- 55.Li C, Chang DL, Yang Z, Qi J, Liu R, He H, et al. Pin1 modulates p63alpha protein stability in regulation of cell survival, proliferation and tumor formation. Cell Death Dis 2013;4:e943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin CH, Li HY, Lee YC, Calkins MJ, Lee KH, Yang CN, et al. Landscape of Pin1 in the cell cycle. Exp Biol Med 2015;240:403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuzminov A. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci U S A 2001;98:8241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays 2001;23:447–55. [DOI] [PubMed] [Google Scholar]
- 59.Juvekar A, Hu H, Yadegarynia S, Lyssiotis CA, Ullas S, Lien EC, et al. Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proc Natl Acad Sci U S A 2016;113:E4338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson SF, Cruz C, Greifenberg AK, Dust S, Stover DG, Chi D, et al. CDK12 inhibition reverses de novo and acquired PARP inhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Rep 2016;17: 2367–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.