Abstract
Purpose of review:
this historical perspective reviews how work of Eric H. Davidson was a catalyst and exemplar for explaining hematopoietic cell fate determination through gene regulation.
Recent findings:
Researchers studying blood and immune cells pioneered many of the early mechanistic investigations of mammalian gene regulatory processes. These efforts included the characterisation of complex gene regulatory sequences exemplified by the globin and T/B cell receptor gene loci, as well as the identification of many key regulatory transcription factors through the fine mapping of chromosome translocation breakpoints in leukaemia patients. As the repertoire of known regulators expanded, assembly into gene regulatory network models became increasingly important, not only to account for the truism that regulatory genes do not function in isolation, but also to devise new ways of extracting biologically meaningful insights from ever more complex information. Here we explore how Eric H. Davidson’s pioneering studies of gene regulatory network control in non-vertebrate model organisms have had an important and lasting impact on research into blood and immune cell development.
Summary:
The intellectual framework developed by Davidson continues to contribute to hematopoietic research, and his insistence on demonstrating logic and causality still challenges the frontier of research today.
Keywords: Gene regulatory network, Causal modeling, Transcription factor, Cis-regulatory element
Introduction
Cellular decision making underpins early development and adult haematopoiesis
The haematopoietic system has long served as a paradigm of how a hierarchically organised differentiation system might mediate the long-term maintenance of adult tissues that are characterized by a high turn-over. Haematopoietic stem cells (HSCs) at the top of this hierarchy serve as an emergency reservoir as well as making a small but steady contribution to the more rapidly proliferating downstream progenitor populations. At the molecular level, this hierarchy is underpinned by cellular decision making processes, which are balanced to ensure that multipotent progenitors give rise to the appropriate numbers of downstream mature cells (for review see [1]).
Cellular decision making also lies at the heart of early development, where, following fertilization of the egg, cellular diversity is rapidly generated through a process of rapid cell division accompanied by the establishment of distinct gene expression programs. Such alterations in gene expression critically depend on the deployment of lineage-specific transcription factors (TFs), or more typically small combinations of such TFs. Of note, TFs of, for example, the Gata, Ets and bHLH families play important roles in early development as well as adult haematopoiesis [2,3]. It is important however to recognize that there are important differences between early embryonic development and adult tissue maintenance. For example, multipotent cells in the embryo such as epiblast or neuromesodermal progenitors only exist for a specified time window, after which they will have turned over into downstream progeny. In contrast, adult stem cells self-renew as well as generate differentiated derivatives.
The building blocks for regulatory network models
A network model may be defined simply as a model of a set of data, whereby the network model provides a flexible way of representing the individual objects as well as their relationships. The objects in such models are commonly referred to as nodes, and their relationships as edges. Network models are commonly used to interpret and simulate highly complex systems. The question therefore arises what specific entities could form the nodes for network models that would prove useful to advance our understanding of biology. Together with Roy Britten, Eric H. Davidson published a visionary paper in 1969, presenting a theoretical framework for how distinct sequences in the genome may communicate with each other through sequence-specific trans-acting regulators, then envisioned as mobile RNA molecules [4]. The interactions (or edges in network terminology) between the DNA sequences and mobile regulators would thus define the inner workings of a network capable of decoding the regulatory blueprint present within the genome, and thus ensure cell type and condition-specific gene expression programs. As emphasized by Davidson over the following decades, this kind of gene regulatory network model did not simply aim to represent correlations, but rather was intended to show causal mechanisms that either drive system state change or maintain system stability [5,6].
Following on from the 1969 theory paper, research across many laboratories identified DNA-binding proteins as key mediators of converting DNA-encoded regulatory information into cell type-specific gene expression programs. Many of these so-called transcription factor (TF) proteins were first identified by haematopoiesis researchers, either by mapping the breakpoints of recurrent chromosome translocations in leukemia patients, or through biochemical fractionation of proteins binding to regulatory sequences of the haemoglobin or immunoglobulin genes (e.g. [7–9]).
While the identification of each of these individual TFs undoubtedly represented a major advance, another step-change was needed to integrate DNA regulatory sequences and TF proteins into regulatory network models. At a practical level, this required the vision to embrace long-term experiments to systematically analyse all the individual components of the machinery that drive expression of individual genes. Here again, Eric Davidson and co-workers led the way, using sea urchin development as an experimentally tractable model system, with the overall goal of decoding the hardwiring of metazoan development through comprehensive characterisation of the organization and function of genomic regulatory systems [10–12]. Below we outline some of the broadly relevant messages that came from these pioneering studies.
Insights from regulatory network models of sea urchin development: the sea urchin system
Davidson developed gene regulatory network models aiming for full explanation of the highly ordered process, reproducible in space and time, through which a complex embryo develops from a single fertilized egg [11,13,14]. The sea urchin embryo was a particularly clear system in which to attempt this, because future tissue “territories” of the free-living larva become distinct within the first 6 cell divisions, prior to any cell migration. The embryo partitions into ≥10 different territories before gastrulation [15,16], each distinguished by different combinations of newly-expressed TFs. Thus, in this system, the entire diversification of embryonic regions can be transformed into the question of how regulatory genes get activated in the correct combinations, in the correct places, times, and orders.
The gene regulatory network models developed by Davidson and colleagues explained how genomically encoded regulatory systems could: create multiple cell types from one cell; produce complex tissues in an invariant geometry from a single cell; and convert transient signals into more lasting stable states with predictable timing, entirely through the ability of TFs to regulate each other’s expression. The foundation for this analysis was the nature of cis-regulatory elements that control gene expression. Network explanations were made possible by the fact that key genes in most systems are regulated simultaneously by multiple “upstream” factors, often with distinct positive or negative roles (early examples: [17,18]). Rule sets for expression of a given gene in some domain are physically embodied in genomic cis-regulatory sequences that comprise enhancers, i.e., clustered, specific binding sites for the factors that need to work coordinately, while the same gene may be expressed under different conditions by using a different cis-regulatory element. The sea urchin system is an exceptionally tractable one for molecular embryology, and this enabled the Davidson team to isolate cis-regulatory elements linked to specific network genes, demonstrating that they drive reporter gene expression in specific spatial and temporal patterns in the whole embryo, and proving that their activities depend on direct DNA binding from multiple TFs [17,19,20].
Network logic embodied in molecular biology
Combinatoriality:
Because cis-regulation responds to multiple inputs, any given gene in the network can respond to logical combinations (AND, OR, ANDNOT) of existing TFs [21,22]. This logic is crucial for explaining how complexity increases as cells divide in an embryo. For example, it means that a gene may require both an inherited factor, which defines its “lineage” criterion for expression, and an independent signal-dependent factor that is activated only if the cell is adjacent to a particular neighbour. As embryonic cells divide in a stereotyped pattern, this causes predictable divergence of the gene expression and fate of one daughter cell from the other.
Distinct rules mediated by distinct cis-regulatory elements:
The same gene can be regulated by different cis-regulatory elements, not only to enable expression in different cell types under different control, but also enabling the gene to be activated initially by one set of conditions and then maintained in the same cell lineage by different conditions. Specific early examples came from the sea urchin system [23,24], as well as from Drosophila blastoderm and wing discs [25,26]. For example, a cis-element switch enabled a gene first activated by a transient signal to be maintained afterwards, independent of the initial signal, by direct or indirect positive autoregulatory feedback. Therefore, not only mapping of factor binding sites in one “minimal” regulatory element, but rather defining the whole system of regulatory elements for a gene, is needed to account for that gene’s developmental domain(s) of expression.
Robustness, not parsimony:
Certain sets of regulatory genes are co-expressed in a sustained way in the sea urchin embryo to stabilize a developmental state [27,28]. Factors reinforce each other’s expression by direct positive cross-regulation, using redundant “additive OR” logic. This cross-regulation was shown to have a deep evolutionary history [27,28]. Although certain individual connections were direct in one species but indirect in another, the gene set as a whole remained positively cross-regulating across long evolutionary times [29]. These gene network circuits decisively refute the idea that evolution selects mainly for parsimony. Instead, these recursive, redundant system architectures for locking down developmental states are selected for highly reproducible performance and avoidance of failure [30].
Repression:
A dominant feature of the embryonic gene network model in the sea urchin was the prominence of transcriptional repression, which was recognized early as the central mechanism for setting boundaries between programs active in different territories [24]. Davidson and colleagues identified multiple examples of sequence-specific, regulated repression, sometimes under switch-like modulation by signalling pathways [11,15,16]. To date, evidence from the sea urchin and Drosophila embryo systems still provides clearer insights into the molecular basis of gene-specific repression than most available mammalian data.
Network architecture, not master regulators:
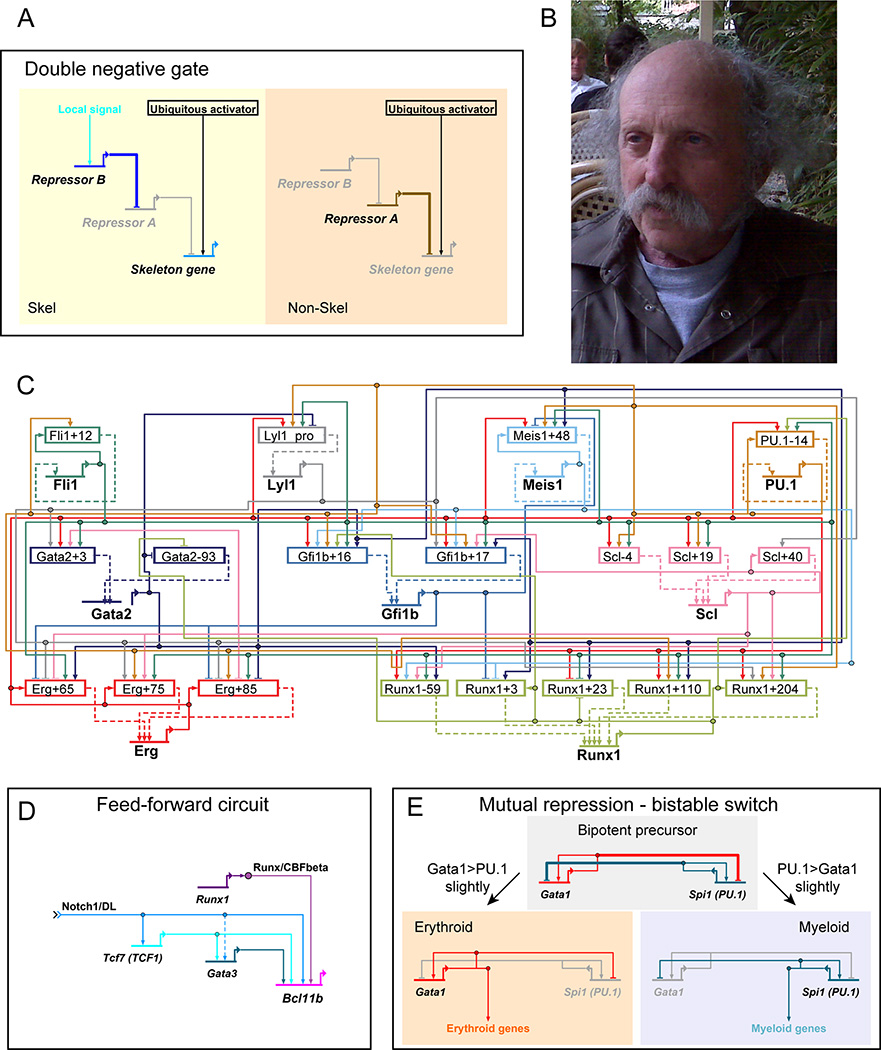
Importantly, gene regulatory network architecture can cause a factor’s regulatory impact to appear to be the reverse of its biochemical activity. The sea urchin system illustrated that repression relationships could be nested to create a “double negative gate”. This circuit enables a ubiquitous activator to turn on a complex program only in one embryonic cell lineage [31,32]. Whereas this program is silenced in most of the embryo by a first repressor, within the selected region a second repressor blocks expression of the first repressor, allowing the ubiquitous activator to trigger the program (Fig. 1A). Thus, through the double-negative gate, network architecture can produce a pattern of gene expression that does not match the expression patterns of any of its direct activators. This is a caveat for purely correlation-based network inference.
Figure 1.
Gene regulatory network circuits in context. (A) Double negative gate, used for sea urchin embryonic skeleton specification [31]. Skel: in skeletogenic precursors. Non-Skel: in rest of embryo. Horizontal lines with bent arrows: genes. Arrows: positive regulation. Bar-end lines: negative regulation. Grayed out: inactive/non-expressed. (B) Eric Davidson, 2010, in Berlin. (C) Core regulatory network for hematopoietic stem and progenitor cells [60], with inputs to genes transduced via discrete cis-regulatory elements (rectangles). Note dense positive cross-regulation. (D) Feed-forward circuit, example from pro-T cell lineage commitment [68]. (E) Mutual repression-based bistable circuit, GATA1 vs. PU.1 [69].
Information processing by the regulatory genome in hematopoiesis
It became clear from the 1990’s that hematopoiesis could only be explained by the actions of hematopoietic TFs if one could account for the orderly developmental control of expression of the factors themselves. Davidson (Fig. 1B) strongly encouraged the enterprise of mapping the regulatory sequences that controlled the genes encoding these factors. Successful research in several groups, prior to whole-genome mapping, identified stem and progenitor-cell associated regulatory elements for SCL (Tal1) [33–36], Gfi1 [37], Gata2 [38–42], Runx1 [43] and Lmo2 [44], regulatory elements with erythroid-associated activity for Gata1 [45], and elements important for regulation of PU.1 (Spi1) in distinct myeloid and lymphoid contexts [46–55]. Each of these genes was shown to respond to inputs delivered via distinct cis-regulatory elements in different developmental contexts, confirming the generality of rules previously established in the invertebrate embryos. While these mapping studies based on classic, functionally-monitored methods may not have been comprehensive, they yielded strong insights about the regulatory switches that determine expression patterns for pivotal genes.
Examples of regulatory models of haematopoiesis inspired by Eric Davidson
Because of the rapid progress characterising key hematopoietic TFs, it became clear early that hematopoietic programmes must be determined by regulators combinatorially [56]. Very few hematopoietic TFs are strictly lineage-specific in expression; almost all play roles in multiple hematopoietic lineages. Furthermore, while different cell lineages may express different members of a given TF family, family members generally share DNA-binding specificity. Thus, from the “viewpoint” of the DNA sequence, an isolated GATA site might equally mediate control by GATA1 in erythroid or mast cells, by GATA2 in stem cells, or by GATA3 in T cells. In reality, lineage specificity must emerge from the unique combinations of TFs expressed in different cell types. The mobilization and impacts of these different combinations require explanation by a gene regulatory network.
The Davidson group itself focused primarily on logic models of gene regulatory networks, with implicit [11], or explicit timing [13]. The most directly comparable hematopoietic regulatory network models may well be a series of models developed by Roger Patient’s group, with the overall aim of encapsulating key processes that regulate early developmental haematopoiesis in Xenopus (summarized in [57]). Starting from a gene regulatory network logic model relating the known regulators of erythroid development [58], the Patient group collected and integrated spatiotemporal expression and signalling data coupled with comprehensive perturbation screens in Xenopus embryos. The resulting models do not reach the same degree of temporal or whole organism coverage achieved with the sea urchin work, and direct cis-regulatory information did not constitute a major component of the Xenopus work. Nevertheless, interesting parallels emerged including early repression and stepwise cascades of combinatorial TF interplay during the process of establishing and then stabilising cell fates [57].
Differentiation of hematopoietic progenitors from an embryonic hemangioblast or hemogenic endothelium must begin with a robust regulatory “launching pad”. Pimanda et al. recognized that in early multipotent hematopoietic progenitors, the cis-regulatory elements of several of the key regulatory genes share target sequences for the TFs encoded by the other genes in the set [38]. In a mutually supportive triad, GATA2, SCL, and Fli1 maintained each other’s expression through direct positive cross-regulation. Further work revealed that this triad was at the core of a larger network, also including Lmo2, Lyl1, Runx1, and Erg, in a core heptad, and three other factors. These frequently bound together to the same regulatory elements, including those controlling their own expression [59,60], in a mechanism strongly evocative of the densely interconnected cross-regulatory circuits seen in certain conserved embryonic gene regulatory networks [28]. A comprehensive model of this network was established, anchored in molecular biology, by testing each factor-bound cis-regulatory element for functional hematopoietic activity, determining the roles of binding sites within those elements by mutation, and then using the results to construct a dynamic Bayesian network model to account for the cells’ regulatory state [60](Fig. 1C). The strong performance of this model in predicting experimental observations showed the feasibility of predictive gene network modelling in this system, and also revealed substantial robustness to withstand single factor perturbations reminiscent of the robustness of sea urchin regulatory networks.
The Graf and Thieffry groups extended a related approach to pathways for hematopoietic cell identity change, using TF binding maps in B and myeloid cells and regulatory perturbation effects on B-cell and myeloid transcriptomes to explore these divergent developmental pathways. They constructed an asynchronous Boolean regulatory network model, which performed well in accounting for cytokine-driven lymphomyeloid differentiation, requirements for different TFs in setting distinct fates, and the ability of C/EBP family TFs to transdifferentiate B lineage cells to myeloid fates [61].
Component subcircuits in hematopoietic differentiation
Many gene regulatory network-centered analyses of hematopoiesis have focused on identifying roles of particular network subcircuits, rather than seeking comprehensive predictiveness. For example, driving the transitions in several lineages are feed-forward circuits, which are used in many biological contexts [62] besides developmental gene regulatory networks. Examples are the E2A-EBF1-Pax5 circuitry promoting B-cell specification (rev. by [63]); the GATA1-FOG1 (Zfpm1) and GATA1-KLF1-FLI1 circuitry that operate during megakaryocyte-erythroid differentiation [64–66]; and the Notch signalling-TCF1-GATA-3-Bcl11b cascade driving early T-cell lineage commitment (rev. by [67,68])(Fig. 1D). Notably, in these developmental contexts, feed-forward circuits have also generally been found to drive an output repressing a lineage alternative as well as an output activating the favoured programme.
Hematopoietic lineage choice by bipotent precursors is often described as an example of bistable switch architecture based on balanced mutual repression, and driven to irreversibility by the positive autoregulation of each of the “contestants”. This simple choice-making circuit architecture promotes dichotomous irreversible outcomes, with stochastic timing, based on equilibrium solutions to simple ordinary differential equations with very few variables. Thus, such a model has been popular to explain the instability of bipotent developmental intermediates and to explain how the fate decisions of single cells become irreversible. Mutual repression-based switches have been invoked for the PU.1-GATA1 opposition in myeloid vs. erythroid lineage choice [69–71](Fig. 1E), and for a Gfi1-Egr/Nab opposition circuit that converts modestly different PU.1 and C/EBPα ratios into dichotomous granulocyte vs. macrophage outcomes [72]. More recently, using single-cell transcriptome analysis, fluorescent TF expression reporters, and regulatory gene perturbation, mutual Gfi1-Irf8 opposition has been shown to be perhaps even more central for the granulocyte-macrophage lineage choice [73].
The prevalence of mutual repression subcircuit models for hematopoietic lineage choice contrasts with the circuits described by Davidson in sea urchin embryos, where such subcircuits were rare. If this difference is real, it could reflect a difference between the deterministic timeclock of a developing embryo and the stochasticity of choices governed by bistable mutual repression. Interestingly, on closer inspection, the mutual repression-based switch models are simpler than the hematopoietic reality. For example, if the PU.1-GATA1 antagonism were absolute, mast cells, basophils, and eosinophils would not exist. Indeed, several lines of evidence increasingly indicate more conditionality, and thus more regulatory participants, in the PU.1—GATA1 relationship than a simple stochastic winner-take-all opposition [74–77], reflecting the fact that even single cells in hematopoiesis face more than two potential choices.
Competing feed-forward mechanisms are often embedded within a more complex architecture together with bistable switch circuits, as proposed in the Gfi1/Egr-Nab based granulocyte-macrophage choice model [72]. For example, in later T lymphocyte development, cells use embedded feed-forward and cross-inhibition circuit motifs for the CD4+ helper vs. CD8+ killer decision within the thymus and the TH1 vs. TH2 effector subtype decision in the periphery [78–81]. These are irreversible but non-deterministically programmed developmental choices regulated by environmental signals, which are clearly advantageous for hematopoietic functions in adult organisms.
Challenges and caveats: the importance of underlying biological differences
Despite the powerful influence of Davidson’s approaches, there are differences between the network models needed to account for non-vertebrate embryogenesis and for hematopoietic systems in postnatal mammals. These are noted briefly here; for a more detailed review, see [82]. Key differences concern dose dependence and timing.
As Davidson’s group built increasingly complete models of the sea urchin embryo developmental gene regulatory network, their theoretical work increasingly stressed the Boolean (digital) quality of the network behaviour. A temporally synchronized Boolean model by Peter et al., modeling regulatory genes simply as being “off” or “on”, already seemingly approached a complete, temporally accurate account of the generation and patterning of both mesoderm and endoderm in sea urchin embryos [13]. However, the underlying set of assumptions was problematic for parallels with hematopoiesis, since hematopoietic TFs have repeatedly been found to work on their targets in level-sensitive ways.
The developmental genetics field has long exploited the near-equivalence between heterozygous mutant embryos and wildtype control embryos, but in mammalian hematopoiesis many regulatory loci are haploinsufficient, e.g. [83–86]. Such twofold gene dosage differences affect the distribution of fate decisions among nominally equivalent starting cells or the kinetics of progression from progenitor to differentiated state. PU.1, although required in myeloid and lymphoid programmes alike, has strongly differential effects on these programmes depending on its expression level [87–89]. Furthermore, elevated expression of transcription factors in hematopoiesis does not simply flip a physiological bistable switch. Some TF gain-of-function perturbations drive cells into an unanticipated alternative fate: e.g., overly high levels of the T-cell TF, GATA-3, drive T-cell precursors to generate mast cells instead [90]. These relationships are understandable based on incoherent (self-antagonizing) feedforward circuits, common in systems biology [62], but are not readily represented in classic topological gene network models (for a workaround applicable to some cases, see [91]).
Step timing in embryonic networks, which change state as soon as new TFs or signal-dependent modifications of TFs appear [13,92], seems very different from that in networks governing hematopoietic differentiation, which drive programmes that change slowly over many days. In postnatal hematopoietic precursors, even after experimental introduction or knockout of a TF, it often takes more than a day for changes in specific target gene expression to become measurable. This makes direct, indirect, and feed-forward dependent effects hard to disentangle. The slow response may be due in part to epigenetic state inertia, as discussed elsewhere [82,93]. However, it is also possible that hematopoietic gene regulatory networks themselves are more highly buffered against state changes. For example, in mutual repression-based hematopoietic choices, cells take days to select their fate [72], a marked contrast from the embryonic boundaries that are established by repression within minutes or hours.
Conclusions
In haematopoiesis research as well as many other fields, the use of the term “regulatory network” far exceeds the actual number of studies that include genuine experiments and/or analyses that would be required to gain new insights at the regulatory network scale. At least two relevant take-home messages seem to follow from this observation. Firstly, there is broad recognition that regulatory network analysis is a powerful approach to advance from a descriptive to a mechanistic understanding of biological processes. Secondly, actual regulatory network studies are not a small add-on to an existing body of work, but instead represent a substantial undertaking that is not for the faint-hearted.
Notably, many of the principles that underlie cellular decision making processes apply broadly across biological systems. Hematopoiesis researchers therefore have been able to benefit from pioneering studies such as the work by Davidson and co-workers on regulatory network control of sea urchin development. Against the backdrop of a scientific environment characterized by increasing specialization, there is an important lesson here on the enormous benefits that come from cross-fertilization and crossing artificial boundaries between diverse fields, with important implications for researchers as well as research funders, across all fields of hematopoiesis research and beyond.
Key points:
Gene regulatory networks explain programs of developmental change
Nodes in gene regulatory networks respond to combinatorial inputs from transcription factors via cis-regulatory elements
Causal gene regulatory network models must be based on systematic perturbation tests and molecular evidence
Lessons about developmental system behavior based on embryology lead to powerful insights about hematopoiesis
Acknowledgements
We thank the late Eric H. Davidson for many critical, stimulating discussions.
Financial support and sponsorship
Relevant support for EVR was from USPHS grants R01HL119102 and R01HD100039 and the Ruddock Professorship at Caltech. Research in the BG laboratory is supported by Wellcome, MRC, Cancer Research UK, NIH-NIDDK and Blood Cancer UK.
Conflicts of Interest
EVR is a member of the Scientific Advisory Board for Century Therapeutics and has been a consultant for A2 Biotherapeutics. BG has been a consultant for Autolus, and the BG lab has received funding from Astra Zeneca, Novo Nordisk and Autolus.
REFERENCES
*References: Highly notable references across the time frame of this historical perspective.
- [1].Laurenti E, Göttgens B: From haematopoietic stem cells to complex differentiation landscapes. Nature 2018, 553:418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Menegatti S, de Kruijf M, Garcia-Alegria E, Lacaud G, Kouskoff V: Transcriptional control of blood cell emergence. FEBS Lett 2019, 593:3304–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Göttgens B: Regulatory network control of blood stem cells. Blood 2015, 125:2614–2620. [DOI] [PubMed] [Google Scholar]
- [4].Britten RJ, Davidson EH: Gene regulation for higher cells: a theory. Science 1969, 165:349–357.* Multiple genes can be expressed coordinately in complex organisms even though they are not arranged in operons as in bacteria. To explain this, Britten and Davidson proposed that certain special genes in higher organisms could encode trans-acting regulators that would act on multiple loci in parallel (gene battery) via shared cis-regulatory sequences. While the identities of the regulators were not correctly predicted, the logic was highly influential.
- [5].Levine M, Davidson EH: Gene regulatory networks for development. Proc Natl Acad Sci U S A 2005, 102:4936–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Davidson EH: The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic Press; 2006. [Google Scholar]
- [7].Zon LI, Tsai SF, Burgess S, Matsudaira P, Bruns GA, Orkin SH: The major human erythroid DNA-binding protein (GF-1): primary sequence and localization of the gene to the X chromosome. Proc Natl Acad Sci U S A 1990, 87:668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Murre C, McCaw PS, Baltimore D: A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 1989, 56:777–783. [DOI] [PubMed] [Google Scholar]
- [9].Begley CG, Aplan PD, Davey MP, Nakahara K, Tchorz K, Kurtzberg J, Hershfield MS, Haynes BF, Cohen DI, Waldmann TA, et al. : Chromosomal translocation in a human leukemic stem-cell line disrupts the T-cell antigen receptor delta-chain diversity region and results in a previously unreported fusion transcript. Proc Natl Acad Sci U S A 1989, 86:2031–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Arnone MI, Davidson EH: The hardwiring of development: organization and function of genomic regulatory systems. Development 1997, 124:1851–1864. [DOI] [PubMed] [Google Scholar]
- [11].Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. : A genomic regulatory network for development. Science 2002, 295:1669–1678.* This paper provided the first comprehensive gene regulatory network model for development of multiple embryonic tissues from a fertilized egg, based on “bottom up” empirical data about cross-regulation of >40 regulatory genes. Not a correlation-based cluster model, every link in this network represented an experimentally demonstrated causal relationship. This model was revised, refined and extended until Davidson’s death.
- [12].Peter IS, Davidson EH: Genomic Control Process: Development and Evolution. San Diego, CA: Academic Press, Elsevier; 2015. [Google Scholar]
- [13].Peter IS, Faure E, Davidson EH: Predictive computation of genomic logic processing functions in embryonic development. Proc Natl Acad Sci U S A 2012, 109:16434–16442.* This paper formally tested how close the gene network for sea urchin development came to completeness, by constructing a Boolean model to simulate the known network operations to test whether it could account for observed regulatory dynamics embryo-wide. Vector equations were based on perturbation data and cis regulatory mapping. Explicitly realistic step time clocks and intra-embryonic cell proximities were included in the successful model.
- [14].Oliveri P, Davidson EH: Gene regulatory network controlling embryonic specification in the sea urchin. Curr Opin Genet Dev 2004, 14:351–360. [DOI] [PubMed] [Google Scholar]
- [15].Li E, Cui M, Peter IS, Davidson EH: Encoding regulatory state boundaries in the pregastrular oral ectoderm of the sea urchin embryo. Proc Natl Acad Sci U S A 2014, 111:E906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Peter IS, Davidson EH: A gene regulatory network controlling the embryonic specification of endoderm. Nature 2011, 474:635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yuh CH, Bolouri H, Davidson EH: Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science 1998, 279:1896–1902.* This paper rigorously showed how distinct components of the cis-regulatory system of a single gene contributed to generate the observed dynamic gene regulation pattern. The functions demonstrated included not only positive regulation but also distinctive kinetics of expression mediated by different elements, spatially restricted negative regulation, and importantly inter-modular interactions leading to non-additive composite outputs.
- [18].Small S, Blair A, Levine M: Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J 1992, 11:4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kirchhamer CV, Davidson EH: Spatial and temporal information processing in the sea urchin embryo: modular and intramodular organization of the CyIIIa gene cis-regulatory system. Development 1996, 122:333–348. [DOI] [PubMed] [Google Scholar]
- [20].Yuh CH, Davidson EH: Modular cis-regulatory organization of Endo16, a gut-specific gene of the sea urchin embryo. Development 1996, 122:1069–1082. [DOI] [PubMed] [Google Scholar]
- [21].Istrail S, De-Leon SB, Davidson EH: The regulatory genome and the computer. Dev Biol 2007, 310:187–195. [DOI] [PubMed] [Google Scholar]
- [22].Peter IS, Davidson EH: Modularity and design principles in the sea urchin embryo gene regulatory network. FEBS Lett 2009, 583:3948–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yuh CH, Bolouri H, Davidson EH: Cis-regulatory logic in the endo16 gene: switching from a specification to a differentiation mode of control. Development 2001, 128:617–629. [DOI] [PubMed] [Google Scholar]
- [24].Kirchhamer CV, Yuh CH, Davidson EH: Modular cis-regulatory organization of developmentally expressed genes: two genes transcribed territorially in the sea urchin embryo, and additional examples. Proc Natl Acad Sci U S A 1996, 93:9322–9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Small S, Arnosti DN, Levine M: Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development 1993, 119:762–772. [PubMed] [Google Scholar]
- [26].Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB: Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature 1996, 382:133–138. [DOI] [PubMed] [Google Scholar]
- [27].Hinman VF, Nguyen AT, Cameron RA, Davidson EH: Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci U S A 2003, 100:13356–13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Davidson EH, Erwin DH: Gene regulatory networks and the evolution of animal body plans. Science 2006, 311:796–800. [DOI] [PubMed] [Google Scholar]
- [29].Hinman VF, Davidson EH: Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci U S A 2007, 104:19404–19409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Oliveri P, Davidson EH: Development. Built to run, not fail. Science 2007, 315:1510–1511. [DOI] [PubMed] [Google Scholar]
- [31].Oliveri P, Tu Q, Davidson EH: Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci U S A 2008, 105:5955–5962.* This paper showed that the first cell type to acquire its lineage identity in a sea urchin embryo is specified not by any dedicated, lineage-specific positive regulator, but rather by the operation of a “double negative gate”, a spatially restricted, causal logic circuit based on repression within the sea urchin embryo gene regulatory network.
- [32].Revilla-i-Domingo R, Oliveri P, Davidson EH: A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci U S A 2007, 104:12383–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Göttgens B, Broccardo C, Sanchez MJ, Deveaux S, Murphy G, Göthert JR, Kotsopoulou E, Kinston S, Delaney L, Piltz S, et al. : The scl +18/19 stem cell enhancer is not required for hematopoiesis: identification of a 5’ bifunctional hematopoietic-endothelial enhancer bound by Fli-1 and Elf-1. Mol Cell Biol 2004, 24:1870–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Göttgens B, Nastos A, Kinston S, Piltz S, Delabesse EC, Stanley M, Sanchez MJ, Ciau-Uitz A, Patient R, Green AR: Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 2002, 21:3039–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Barton LM, Göttgens B, Gering M, Gilbert JG, Grafham D, Rogers J, Bentley D, Patient R, Green AR: Regulation of the stem cell leukemia (SCL) gene: a tale of two fishes. Proc.Natl.Acad.Sci.U.S.A 2001, 98:6747–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Göttgens B, McLaughlin F, Bockamp EO, Fordham JL, Begley CG, Kosmopoulos K, Elefanty AG, Green AR: Transcription of the SCL gene in erythroid and CD34 positive primitive myeloid cells is controlled by a complex network of lineage-restricted chromatin-dependent and chromatin-independent regulatory elements. Oncogene 1997, 1997:2419–2418.* Here, the complex developmental expression pattern of the stem- and erythroid-cell regulatory gene SCL was shown to reflect action of multiple discrete cis-regulatory elements. These were mapped by DNase-hypersensitivity and shown to have distinct cell type-dependent regulatory activities in reporter assays.
- [37].Wilson NK, Timms RT, Kinston SJ, Cheng YH, Oram SH, Landry JR, Mullender J, Ottersbach K, Gottgens B: Gfi1 expression is controlled by five distinct regulatory regions spread over 100 kilobases, with Scl/Tal1, Gata2, PU.1, Erg, Meis1, and Runx1 acting as upstream regulators in early hematopoietic cells. Mol.Cell Biol. 2010, 30:3853–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pimanda JE, Ottersbach K, Knezevic K, Kinston S, Chan WY, Wilson NK, Landry JR, Wood AD, Kolb-Kokocinski A, Green AR, et al. : Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc.Natl.Acad.Sci.U.S.A 2007, 104:17692–17697.* This paper provided the first evidence that the hematopoietic stem/progenitor state is stabilized by a “kernel”-like circuit of regulatory genes whose products directly stimulate each other’s expression. The circuit model was based on identification of GATA2, FLI1 and SCL as three transcription factors that directly bind together at key cis-regulatory DNA elements previously shown to be functionally important for each of the genes.
- [39].Robert-Moreno A, Espinosa L, de la Pompa JL, Bigas A: RBPjκ-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development 2005, 132:1117–1126. [DOI] [PubMed] [Google Scholar]
- [40].Khandekar M, Suzuki N, Lewton J, Yamamoto M, Engel JD: Multiple, distant Gata2 enhancers specify temporally and tissue-specific patterning in the developing urogenital system. Mol Cell Biol 2004, 24:10263–10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *[41].Snow JW, Trowbridge JJ, Fujiwara T, Emambokus NE, Grass JA, Orkin SH, Bresnick EH: A single cis element maintains repression of the key developmental regulator Gata2. PLoS Genet 2010, 6:e1001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Martowicz ML, Grass JA, Boyer ME, Guend H, Bresnick EH: Dynamic GATA factor interplay at a multicomponent regulatory region of the GATA-2 locus. J Biol Chem 2005, 280:1724–1732.* References *41 and *42 demonstrate that a key event in erythroid differentiation, the downregulation of Gata2 expression, depends on a gene regulatory network negative feedback circuit that transforms a key cis-regulatory element of Gata2 from a positive to a negative function. This results from a “GATA switch”, as activating GATA2 is displaced from its autoregulatory binding site by repressive GATA1.
- [43].Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng JF, Prabhakar S, Rubin EM, Li PS, Sloane-Stanley J, Kong AS, et al. : Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 2007, 110:4188–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Landry JR, Bonadies N, Kinston S, Knezevic K, Wilson NK, Oram SH, Janes M, Piltz S, Hammett M, Carter J, et al. : Expression of the leukemia oncogene Lmo2 is controlled by an array of tissue-specific elements dispersed over 100 kb and bound by Tal1/Lmo2, Ets, and Gata factors. Blood 2009, 113:5783–5792. [DOI] [PubMed] [Google Scholar]
- [45].Kobayashi M, Nishikawa K, Yamamoto M: Hematopoietic regulatory domain of gata1 gene is positively regulated by GATA1 protein in zebrafish embryos. Development 2001, 128:2341–2350. [DOI] [PubMed] [Google Scholar]
- [46].Leddin M, Perrod C, Hoogenkamp M, Ghani S, Assi S, Heinz S, Wilson NK, Follows G, Schönheit J, Vockentanz L, et al. : Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood 2011, 117:2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher A, et al. : Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 2009, 114:299–309.* This paper used high-resolution DNase hypersensitivity mapping, DNA methylation mapping, and DamID-based analysis of transient Runx1 binding to resolve the kinetics of the cascade from Runx activation, to PU.1 activation, to activation of a PU.1 target gene. The results showed a potent activity of Runx1 as a hit-and-run chromatin opening factor required to initiate gene expression but not to maintain expression or chromatin accessibility.
- [48].Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, et al. : Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet 2006, 38:27–37. [DOI] [PubMed] [Google Scholar]
- [49].Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H, Akashi K, Moreau-Gachelin F, Li Y, Zhang P, et al. : Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol.Cell Biol. 2005, 25:2832–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li Y, Okuno Y, Zhang P, Radomska HS, Chen HM, Iwasaki H, Akashi K, Klemsz MJ, McKercher SR, Maki RA, et al. : Regulation of the PU.1 gene by distal elements. Blood 2001, 98:2958–2965. [DOI] [PubMed] [Google Scholar]
- [51].Hoogenkamp M, Krysinska H, Ingram R, Huang G, Barlow R, Clarke D, Ebralidze A, Zhang P, Tagoh H, Cockerill PN, et al. : The Pu.1 locus is differentially regulated at the level of chromatin structure and noncoding transcription by alternate mechanisms at distinct developmental stages of hematopoiesis. Mol Cell Biol 2007, 27:7425–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chou ST, Khandros E, Bailey LC, Nichols KE, Vakoc CR, Yao Y, Huang Z, Crispino JD, Hardison RC, Blobel GA, et al. : Graded repression of PU.1/Sfpi1 gene transcription by GATA factors regulates hematopoietic cell fate. Blood 2009, 114:983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zarnegar MA, Rothenberg EV: Ikaros represses and activates PU.1 cell-type-specifically through the multifunctional Sfpi1 URE and a myeloid specific enhancer. Oncogene 2012, 31:4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zarnegar MA, Chen J, Rothenberg EV: Cell-type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol Cell Biol 2010, 30:4922–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, et al. : PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet 2008, 40:51–60. [DOI] [PubMed] [Google Scholar]
- [56].Rothenberg EV, Anderson MK: Elements of transcription factor network design for T-lineage specification. Devel.Biol 2002, 246:29–44. [DOI] [PubMed] [Google Scholar]
- [57].Ciau-Uitz A, Patient R: Gene Regulatory Networks Governing the Generation and Regeneration of Blood. J Comput Biol 2019, 26:719–725.* This review summarizes the gene regulatory network relationships assembled by Roger Patient and colleagues based on years of detailed characterization of the intercellular signals and responding regulatory genes that mediate emergence of hematopoietic progenitor cells in amphibian embryos. This classically-established model system has advantages for experimental embryology, revealing roles of signaling factors more easily than in mammals.
- [58].Swiers G, Patient R, Loose M: Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev Biol 2006, 294:525–540. [DOI] [PubMed] [Google Scholar]
- [59].Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. : Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 2010, 7:532–544.* This paper showed the importance of tracking multiple transcription factors by parallel ChIP-seq analyses. The results substantiated the direct cross-regulation of a core circuit of hematopoietic regulatory genes, as their products bound together at known cis-regulatory elements. However, the frequency of highly clustered binding also suggested that transcription factors affect each other’s site choices and may not “read” DNA autonomously.
- [60].Schutte J, Wang H, Antoniou S, Jarratt A, Wilson NK, Riepsaame J, Calero-Nieto FJ, Moignard V, Basilico S, Kinston SJ, et al. : An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. eLife 2016, 5:e11469.* This paper crystallized the molecular biology and logic relationships between the recursively wired central regulatory genes introduced in refs. *38 and *59 and assembled them into a formal computational model. Exploiting detailed cis-regulatory element activity profiles and site mutagenesis data, the dynamic Bayesian network model gave realistically graded expression predictions, capturing the responses of the real network to many perturbations.
- [61].Collombet S, van Oevelen C, Sardina Ortega JL, Abou-Jaoude W, Di Stefano B, Thomas-Chollier M, Graf T, Thieffry D: Logical modeling of lymphoid and myeloid cell specification and transdifferentiation. Proc Natl Acad Sci U S A 2017, 114:5792–5799.* This paper modeled the multifactor gene regulatory network underlying the pathway of B lineage cell conversion to myeloid cells by forced expression of C/EBP transcription factors. Using ChIP-seq and experimental perturbation data to define network edges, a discrete-valued logical model was used to simulate outcomes of sustained or pulsed C/EBP exposures. In silico perturbations identified factors within the system affecting outcomes.
- [62].Alon U: Network motifs: theory and experimental approaches. Nat Rev Genet 2007, 8:450–461. [DOI] [PubMed] [Google Scholar]
- [63].Boller S, Grosschedl R: The regulatory network of B-cell differentiation: a focused view of early B-cell factor 1 function. Immunol Rev 2014, 261:102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mancini E, Sanjuan-Pla A, Luciani L, Moore S, Grover A, Zay A, Rasmussen KD, Luc S, Bilbao D, O’Carroll D, et al. : FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors. EMBO J 2012, 31:351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S: GATA switches as developmental drivers. J Biol Chem 2010, 285:31087–31093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Palii CG, Cheng Q, Gillespie MA, Shannon P, Mazurczyk M, Napolitani G, Price ND, Ranish JA, Morrissey E, Higgs DR, et al. : Single-Cell Proteomics Reveal that Quantitative Changes in Co-expressed Lineage-Specific Transcription Factors Determine Cell Fate. Cell Stem Cell 2019, 24:812–820 e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Rothenberg EV: Programming for T-lymphocyte fates: modularity and mechanisms. Genes Dev 2019, 33:1117–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rothenberg EV: Dynamic control of the T-cell specification gene regulatory network. Curr Opin Syst Biol 2019, 18:62–76.* This review summarized the cross-regulatory relationships, logic, and distinctive regulatory gene cohorts that have been found to guide T-cell precursors through the transition from multipotency to commitment. Key points were PU.1--Notch opposition, roles of E proteins and Bcl11b, and use of partly overlapping factor sets to program related but divergent fates. Overlap is facilitated by incomplete repression and dosage-sensitive action at many nodes.
- [69].Huang S, Guo YP, May G, Enver T: Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol 2007, 305:695–713.* This influential systems-biology study provided a formal dynamic model of GATA1 vs. PU.1 antagonism driving erythromyeloid divergence, but with the capacity to sustain a metastable, uncommitted intermediate state as well. The ability of GATA1 and PU.1 each to positively regulate themselves was found to be important to account for the observed bipotent precursor cells which express target genes of both GATA1 and PU.1.
- [70].Chickarmane V, Enver T, Peterson C: Computational modeling of the hematopoietic erythroid-myeloid switch reveals insights into cooperativity, priming, and irreversibility. PLoS Comput Biol 2009, 5:e1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cantor AB, Orkin SH: Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 2002, 21:3368–3376. [DOI] [PubMed] [Google Scholar]
- [72].Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H: Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 2006, 126:755–766. [DOI] [PubMed] [Google Scholar]
- [73].Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL: Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 2016, 537:698–702.* Single cell RNA-seq and supervised clustering with a well-curated set of genes were used to create a framework for analysis of granulocyte vs. macrophage fate. Fluorescent reporter alleles of key regulators Gfi1 and Irf8 were used to define the cellular pathways of lineage decision. Perturbations in the single-cell gene expression trajectory when either Gfi1 or Irf8 was mutated revealed the gene regulatory network that actually controls the lineage choice.
- [74].Wheat JC, Sella Y, Willcockson M, Skoultchi AI, Bergman A, Singer RH, Steidl U: Single-molecule imaging of transcription dynamics in somatic stem cells. Nature 2020, 583:431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Monteiro R, Pouget C, Patient R: The gata1/pu.1 lineage fate paradigm varies between blood populations and is modulated by tif1γ. EMBO J. 2011, 30:1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hoppe PS, Schwarzfischer M, Loeffler D, Kokkaliaris KD, Hilsenbeck O, Moritz N, Endele M, Filipczyk A, Gambardella A, Ahmed N, et al. : Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature 2016, 535:299–302. [DOI] [PubMed] [Google Scholar]
- [77].Gillespie MA, Palii CG, Sanchez-Taltavull D, Shannon P, Longabaugh WJR, Downes DJ, Sivaraman K, Espinoza HM, Hughes JR, Price ND, et al. : Absolute Quantification of Transcription Factors Reveals Principles of Gene Regulation in Erythropoiesis. Mol Cell 2020, 78:960–974 e911.* Most gene regulatory network models infer regulatory factor expression levels by RNA expression, but where action is dose-dependent, actual protein levels are important and could be different. This study used calibrated CYTOF-measured transcription factor levels in single cells to generate a protein level-based gene regulatory network model of erythroid specification. Some key cross-regulatory relationships proved to be context dependent.
- [78].Ebihara T, Seo W, Taniuchi I: Roles of RUNX Complexes in Immune Cell Development. Adv Exp Med Biol 2017, 962:395–413. [DOI] [PubMed] [Google Scholar]
- [79].Zhu J, Yamane H, Paul WE: Differentiation of effector CD4 T cell populations. Annu.Rev.Immunol 2010, 28:445–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R: Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4+ T cells. Nat.Immunol 2008, 9:1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE: GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 2006, 16:3–10. [DOI] [PubMed] [Google Scholar]
- [82].Rothenberg EV: Encounters across networks: Windows into principles of genomic regulation. Mar Genomics 2019, 44:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Prasad MA, Ungerbäck J, Ahsberg J, Somasundaram R, Strid T, Larsson M, Mansson R, De Paepe A, Lilljebjorn H, Fioretos T, et al. : Ebf1 heterozygosity results in increased DNA damage in pro-B cells and their synergistic transformation by Pax5 haploinsufficiency. Blood 2015, 125:4052–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH, Enver T, Vyas P, Scadden DT: Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood 2005, 106:477–484. [DOI] [PubMed] [Google Scholar]
- [85].Sun W, Downing JR: Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood 2004, 104:3565–3572. [DOI] [PubMed] [Google Scholar]
- [86].Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC: Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPα ratio and granulocyte colony-stimulating factor. Nat.Immunol 2003, 4:1029–1036. [DOI] [PubMed] [Google Scholar]
- [87].DeKoter RP, Singh H: Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science 2000, 288:1439–1441. [DOI] [PubMed] [Google Scholar]
- [88].Carotta S, Wu L, Nutt SL: Surprising new roles for PU.1 in the adaptive immune response. Immunol Rev 2010, 238:63–75. [DOI] [PubMed] [Google Scholar]
- [89].Kueh HY, Champhekar A, Nutt SL, Elowitz MB, Rothenberg EV: Positive feedback between PU.1 and the cell cycle controls myeloid differentiation. Science 2013, 341:670–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Taghon T, Yui MA, Rothenberg EV: Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol 2007, 8:845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Cacace E, Collombet S, Thieffry D: Logical modeling of cell fate specification-Application to T cell commitment. Curr Top Dev Biol 2020, 139:205–238. [DOI] [PubMed] [Google Scholar]
- [92].Clark E, Akam M: Odd-paired controls frequency doubling in Drosophila segmentation by altering the pair-rule gene regulatory network. Elife 2016, 5:e18215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rothenberg EV: Causal Gene Regulatory Network Modeling and Genomics: Second-Generation Challenges. J Comput Biol 2019, 26:703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]