Abstract
Zebrafish have a fifty-year history as a model organism for studying vertebrate developmental biology and more recently have emerged as a powerful model system for studying vertebrate microbiome assembly, dynamics, and function. In this Review, we discuss the strengths of the zebrafish model for both observational and manipulative microbiome studies, and we highlight some of the important insights gleaned from zebrafish gut microbiome research.
Introduction
Starting in the late 1960’s, George Streisinger at the University of Oregon established the zebrafish (Danio rerio) as a model organism to study complex vertebrate biology using genetic approaches1. The organism’s high fecundity, manageable laboratory husbandry, rapid early development, and accessible embryology all made the prospect of forward genetic screens feasible, although this project was vastly more ambitious than Streisinger’s previous genetic studies with bacteriophage. Fifty years later, the zebrafish model has exceeded Streisinger’s expansive vision. Enabled by pioneering approaches in advanced microscopy, chemical screening, and genome engineering, the zebrafish is now a premier model organism for studying a broad array of topics in biology and biomedicine.
One particularly fruitful area of zebrafish research is the study of vertebrate-associated microbial communities or microbiomes2. This research field takes advantage of the high fecundity and easy laboratory husbandry of zebrafish to perform large, well-powered, and well-controlled observational studies of zebrafish microbiomes. In parallel, the organisms’ accessible embryology allows researchers to perform manipulative studies in which embryos are sterilized and then grown microbe-free or in the presence of defined microbes. The ease of embryo sterilization eliminates the husbandry burden of maintaining colonies of sterile zebrafish across generations. A brief overview of the basic experimental paradigms and models available for studying zebrafish microbiome in observational and manipulative studies is presented in Fig.1.
Fig. 1|. Experimental paradigms for studying zebrafish microbiome assembly, dynamics, and function.
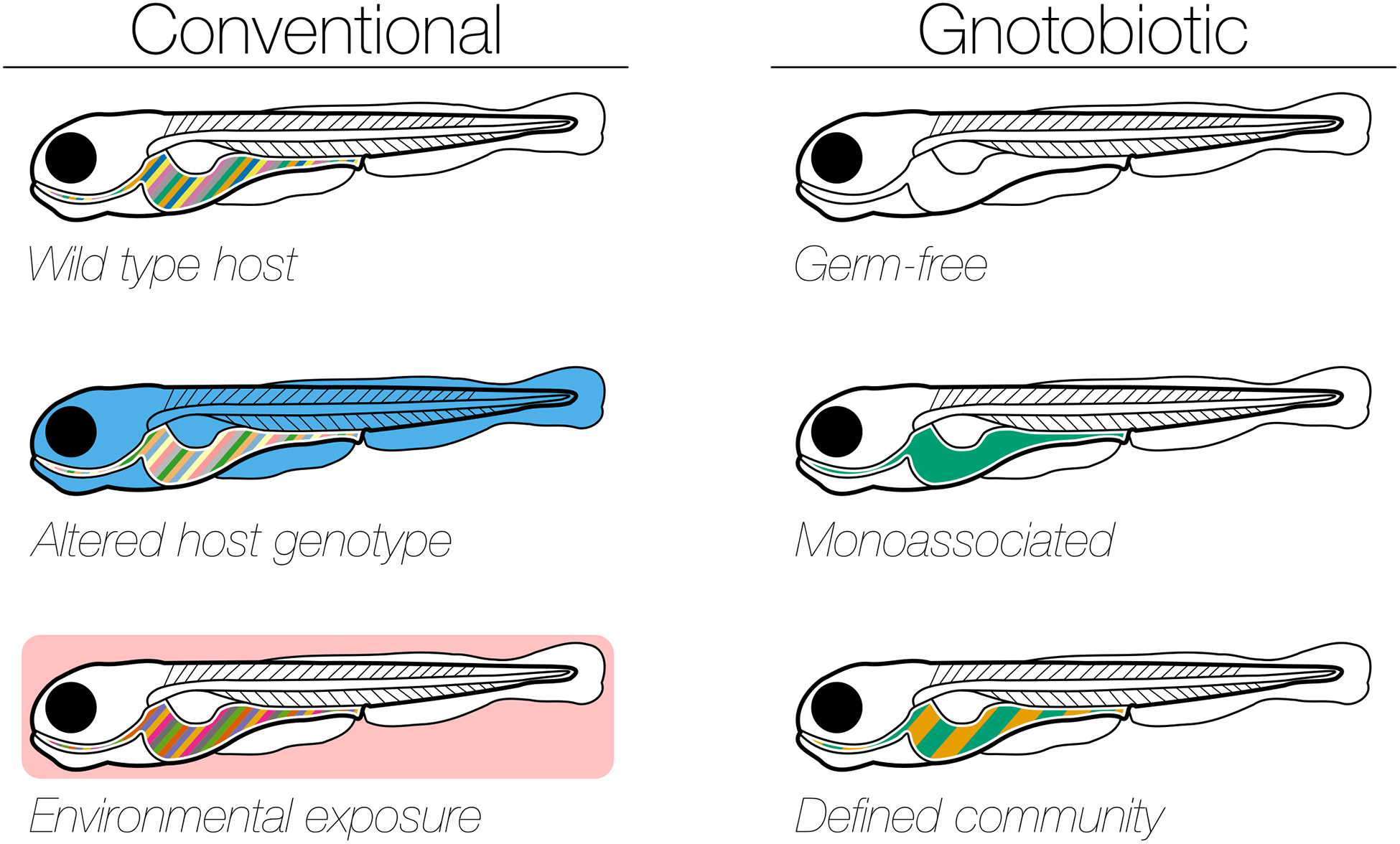
A brief overview of the basic experimental paradigms in zebrafish microbiome studies. The list is not exhaustive, and these paradigms are not mutually exclusive (more than one paradigm can be utilized in a single experiment). a, Experiments that utilize conventionally raised zebrafish. Firstly, observational studies of the microbiome (represented as colored stripes in the larval fish gut) can be made using wild type zebrafish raised according to standardized protocols. Secondly, the roles of specific host genetic factors in the assembly of the gut microbiome can be tested by utilizing mutant zebrafish lines (blue larval fish). Thirdly, the roles of external factors (such as environmental chemicals or contact with other hosts) in the assembly of the gut microbiome can be tested by altering the environment in which the zebrafish are raised (pink box). b, Experiments that utilize gnotobiotically raised zebrafish. Firstly, germ-free zebrafish (for example, fish that remain unexposed to microbes after hatching), can be utilized to test the requirements for the microbiome in host development and physiology. Secondly, monoassociations (fish with an all green gut) can be utilized to test the sufficiency for a specific microbial taxon in host development and physiology. Thirdly, defined communities (fish with striped green and orange gut), can be utilized to test the sufficiency of specific consortia of microbes in host development and physiology. This paradigm can also be utilized to test the properties of microbial colonization dynamics for a particular set of microbes. Whereas conventional microbiome analyses can take place at any part of the zebrafish life cycle, the gnotobiotic study designs are currently limited to larval and young juvenile fish.
Observational studies give researchers insights into how host–microbe systems function as a whole, and allow them to generate hypotheses about the specific mechanisms responsible for the observed phenomena. Such studies, for example, have addressed how the zebrafish intestinal microbiome changes across development or in response to environmental perturbations3,4. Manipulative studies allow researchers to rigorously test mechanisms of host–microbiome interactions using models of varying degrees of complexity. For example, researchers have shown that specific developmental deficits in zebrafish grown in the absence of microbes can be restored upon addition of particular bacterial proteins5,6.
In this Review, we explore how investigations using gnotobiotic zebrafish have revealed specific requirements for microbial associations during normal development and uncovered specific microbes and microbial products that promote these processes. These manipulative experiments complement studies of the drivers of zebrafish microbiome composition across the animals’ life history and in diverse environmental settings. Reoccurring themes of particularly impactful microbial constituents and environmental drivers indicate the ecological relevance of laboratory-based experiments and validate the utility of the zebrafish model to reveal generalizable insights into host-microbe interactions.
Investigations with gnotobiotic zebrafish
Microbiome manipulations in zebrafish.
The earliest zebrafish microbiome studies focused on the larval stage of zebrafish development7,8, which parallels the neonatal period of mammalian development. Whereas embryogenesis progresses within the essentially sterile environment of the egg, larval life, which begins at hatching, is marked by the animal’s first encounter with environmental microbes. Key to the success of zebrafish microbiome studies was the establishment of relatively simple and low-cost protocols for deriving sterile or “germ-free” zebrafish embryos through surface sterilization of the eggshell or chorion9. Such germ-free animals could then be used for gnotobiology (“known biology”) investigations in which animals are either maintained germ-free or associated with defined microbial strains or consortia (Fig 1b).
During the first days of larval life, the zebrafish is endowed with a yolk that serves as an internal nutrient source, thereby simplifying its husbandry requirements. Rearing germ-free zebrafish to later developmental stages introduces new husbandry challenges such as providing nutritionally adequate, sterile food and maintaining water quality in the absence of denitrifying bacteria. These husbandry constraints have limited the studies of older germ-free zebrafish to date. The focus of germ-free zebrafish studies on larval stages provides a fortuitous complement to germ-free research in mammals, for which phenotypic characterization of neonates can be logistically challenging before weaning. However, studies that compare the requirements for microbiota in mouse and zebrafish for processes such as maturation of the adaptive immune system or elaboration of complex social behaviors, have been challenging because in zebrafish, these processes occur at later developmental stages, which are inaccessible with current gnotobiotic husbandry methods10,11. One approach to study the influence of microbiome on adult behavior has been to test whether addition of exogenous bacteria to conventionally reared adult fish can alter specific traits. For example, two studies have reported changes in adult zebrafish behavior upon introducing human probiotic strains Lactobacillus plantarum12 and Lactobacillus rhamnosus13 to tanks of adult zebrafish, although the studies did not establish whether the responses were specific to these bacteria or whether the probiotic strains colonized the zebrafish. Capitalizing on the high-throughput behavioral phenotyping assays developed for zebrafish14, Phelps et al.15 in 2017 determined that microbial colonization of zebrafish larvae by 9 days post-fertilization (dpf) is necessary to elicit normal locomotor activity. Integrating the emerging toolkits for behaviorally phenotyping of zebrafish and manipulating their microbiomes holds great potential for clarifying the zebrafish microbiome’s contribution to behavior. However, more work is needed to determine whether discoveries that emerge from such investigations are translatable to humans. The different methods for interrogating the zebrafish microbiome that are applicable across all life stages, or specific to larva or older stages are summarized in Table 1.
Table 1|.
Methods available for interrogating the zebrafish microbiome at each life stage.
| Larva and adult stages | Larva stage | Adult stage |
|---|---|---|
|
|
|
Defining developmental requirements for microbiota.
Experiments comparing germ-free (Fig. 1b) with conventionally raised (Fig. 1a) larval zebrafish have enabled the identification of specific requirements for microbiota in vertebrate development. For example, germ-free larvae have decreased rates of intestinal epithelial cell proliferation and fewer intestinal secretory cells relative to absorptive enterocytes, all phenotypes that are reversed upon restoration of the microbiota7,8,16–18. The inflammatory tone of the intestine is diminished in the germ-free state, with fewer resident neutrophils and reduced NF-κB activation19. The physiology of the germ-free intestine is also altered compared to conventional zebrafish, with reduced absorption of macromolecules including proteins and lipids7,8,20,21. The influence of the microbiota extends to extra-intestinal tissues as well. For example, the pancreas of germ-free larvae has fewer insulin-producing β cells than the pancreas of conventionally reared larvae5. Behavioral differences between germ-free and conventionally reared zebrafish larvae have also been reported, with germ-free fish exhibiting increased locomotion15,22 and, in one report, decreased thigmotaxis (zebrafish with thigmotactic behavior show a preference for the perimeter of the dish)22, which has been interpreted as a measure of anxiety. Many of these altered responses in germ-free zebrafish mirror findings in other model systems such as mouse and fruit flies23, suggesting a conservation of animal responses to resident microbiota during development.
Identifying microbiota members sufficient for promoting specific aspects of development.
The early zebrafish microbiome studies provided the first descriptions and culture isolations of zebrafish-associated bacteria for gnotobiotic experiments7,8. These studies revealed that the larval intestine is dominated by fast-growing and metabolically versatile Gammaproteobacteria, including representatives of the Aeromonas, Pseudomonas, and Vibrio genera. Members of these bacterial groups are relatively amenable to laboratory culture and genetic manipulation24. With bacterial isolates in hand, researchers could perform the first studies of gnotobiotic zebrafish (host–microbe systems in which all microbiota members are known) by introducing specific microorganisms (monoassociations) or defined consortia into germ-free zebrafish larvae (Fig. 1b). The use of gnotobiotic zebrafish allowed researchers to identify specific bacteria or bacterial products that are sufficient to reverse germ-free phenotypes. For example, different defects in lipid absorption by enterocytes could be restored by distinct bacteria: a representative of Exiguobacterium increased lipid droplet number whereas other Chryseobacterium and Pseudomonas species promoted lipid droplet growth21. Increased locomotor activity in germ-free zebrafish was found to be reversed by multiple bacteria including zebrafish-derived Aeromonas and Vibrio species15 and a human probiotic strain Lactobacillus plantarum22, although the latter study did not establish whether the added bacteria colonized the zebrafish. A study to determine the microbiota requirement for normal pancreatic β-cell numbers using monoassociations with a panel of zebrafish-derived bacteria showed that β-cell mass could be restored by certain Aeromonas and Shewanella strains but not by other bacterial isolates5. This finding led to the identification of β-cell Expansion Factor A (BefA), a secreted bacterial protein produced specifically by those strains, which is sufficient to induce expansion of the β-cell population when added to germ-free larvae5.
Investigating microbiota colonization dynamics.
Another feature of the larval zebrafish that has accelerated microbiome research is its high optical clarity, which allows for the visual tracking of individual cells and specific cell types. In conjunction with gnotobiotic methods and light-sheet microscopy, this optical clarity has proven to be useful in studying real-time dynamics of fluorescent protein expressed by microbiome members24,25. For example, light-sheet microscopy of monoassociated larval zebrafish has revealed growth dynamics of a common microbiome constituent, Aeromonas, at much finer temporal and spatial resolutions than achievable using culturing- or sequencing-based methods26. Similar approaches have also revealed mechanisms for intermicrobial competition. For example, when the same Aeromonas species mentioned above is co-introduced into the zebrafish with a Vibrio species, which is also commonly found in the zebrafish microbiome, the Vibrio species consistently excludes the Aeromonas species27. Light-sheet microscopy revealed that this competitive advantage is conferred to the Vibrio species by its ability to avoid peristaltic expulsion from the gut. Whereas the Vibrio species is highly motile and exhibits primarily planktonic growth, the Aeromonas species aggregates into non-motile clusters, which are regularly expelled, resulting in stochastic Aeromonas population crashes. In these experiments, the planktonic Vibrio species was found to dominate in the proximal intestine, whereas the aggregated Aeromonas population that was more susceptible to gut motility was distributed more distally27. Further investigations revealed that when the planktonic Vibrio is treated with low dose antibiotics28 or genetically engineered to be amotile29, it becomes more aggregated and more easily expelled from the intestine. A broader survey on the spatial organization of phylogenetically diverse bacterial residents of the zebrafish intestine found this relationship between aggregation and distal displacement to be generalizable, indicating that the physical properties of bacterial populations contribute to microbiome biogeography30. With their optical clarity and gnotobiotic amenability, zebrafish larvae provide opportunities to study microbiome cellular dynamics that few other vertebrate models can offer.
Bacterial genetic requirements for host colonization.
In addition to enabling live imaging of bacterial dynamics via the introduction of fluorescent protein encoding genes, the genetic tractability of zebrafish microbiome members has made possible the interrogation of bacterial genetic requirements for host associations. For example, Stephens et al.31 in 2015 used a negative selection transposon mutagenesis screen on two species of bacteria commonly found in the zebrafish microbiome. This method allowed the researchers to track specific subpopulations of each species, as well as identify particular genes necessary for the successful colonization of the larval zebrafish intestine in the presence or absence of other competing bacterial populations. In doing so, they found that intraspecies priority effects strongly determined whether a specific subpopulation could colonize, and they identified genes involved in chemotaxis and motility as being important for successful colonization in all situations. In 2016, Bae et al.32 used a chemical mutagenesis and high-throughput sequencing approach to identify genes required for bacterial motility in Exiguobacterium acetylicum, a zebrafish bacterial isolate that was recalcitrant to traditional genetic mutagenesis methods; they also showed that Exiguobacterium motility was important for host colonization.
Exploring host selection through microbiota transplantations.
In addition to understanding how resident microbes interact with each other and their hosts, the gnotobiotic zebrafish model allows researchers to examine the ways in which a host selects for a specific consortium of microbes. For example, Rawls et al.33 in 2006 transplanted conventional mouse microbiomes into germ-free zebrafish, and vice versa. In doing so, they found that while the lineages present in the transplanted community were more similar to the community of origin, the relative abundances of these lineages more closely resembled a typical microbiome of the new host. This gnotobiotic framework therefore revealed meaningful differences in the selection imposed on the microbiome by two important model organisms. A technical limitation to these studies is that microbiota transplantations into zebrafish are done by addition of samples into the aqueous media, thus introducing a second selection for microbial survival in the water column and reducing the efficiency of transmission.
Further explorations of host selection on microbiota assembly have been enabled by the plethora of available mutant zebrafish lines and the ease of genome engineering in this model organism (Fig. 1a). Studies comparing conventionally raised wild-type and mutant zebrafish have provided insights into the mechanisms underlying host selection such as the innate34 and adaptive35 branches of the immune system, as discussed below.
Drivers of zebrafish gut microbiome composition
In addition to enabling manipulative gnotobiotic studies, the zebrafish offers the possibility of doing large-scale, controlled, and well-powered observational studies of microbiome composition under different conditions. To date, the majority of these studies have investigated the gut microbiome through characterizations of dissected intestines or collected fecal material. Future investigations to characterize the microbial communities colonizing other tissues such as the skin or the gills will be of great interest.
Comparison of zebrafish microbiome across laboratories and wild-caught fish.
One of the limitations of any model organism is that the laboratory environment in which it is maintained very rarely reflects the natural environment in which its wild conspecifics live. Thus, extrapolating results from the laboratory to the “real world” can be a tenuous exercise. However, observational studies have provided evidence that while laboratory zebrafish do not have microbiomes identical to their wild counterparts, the microbiome of zebrafish from these different habitats show strong similarities in terms of taxonomic composition36. Indeed, a Terminal restriction fragment length polymorphism (TRFLP)-based comparison of zebrafish microbiome samples collected from facilities across the United States, as well as samples collected from fish caught in the species’ native environment—flood-plains of the Indian subcontinent37—revealed that gut microbiome composition varies across facilities, and that the microbiota of many facility-managed fish are more similar to those of wild-caught fish than fish grown in other facilities36. A pyrosequencing-based investigation of zebrafish grown in two geographically disparate facilities, as well as wild-caught zebrafish, revealed the presence of 21 operational taxonomic units (OTUs; a proxy for microbial species) that are common to all three fish populations36. These OTUs included members of the genera Aeromonas, Shewanella, Pseudomonas, Stenotrophomonas, Vibrio, Burkholderia, Diaphorobacter, Cetobacterium, Streptococcus, Bacillus, Cloacibacterium, and Propionibacterium. These findings suggest that these microbial taxa are especially capable of dispersing into the zebrafish gut or growing within the gut environment. Similar taxa were found in subsequent Illumina-based profiling of zebrafish intestinal bacteria from the same facilities3,38. Additional investigations of the zebrafish gut microbiome that utilized fish from facilities distinct from those analyzed in this aforementioned study accordingly observed the same taxa in these distinct cohorts of fish4. These results collectively demonstrate that a subcomponent of the zebrafish gut microbiome associates with the host regardless of husbandry practices or geographic location and that facility-managed zebrafish microbiomes can model those of wild zebrafish.
At high levels of taxonomic classification (bacterial phylum), there are similarities in composition across a broad range of teleost species. For example, the microbiomes of many fish contain a high relative abundance of Proteobacteria and Fusobacteria39. An analysis of clone libraries generated from gut microbes carried by zebrafish or several other wild teleost fish species (Acanthurus nigricans, Lutjanus bohar, Chlorurus sordidus, Chaenocephalus aceratus, Notothenia coriiceps, Takifugu niphobles, and Pelteobagrus fulvidraco) similarly revealed that Gammaproteobacteria and Fusobacteria consistently appear in the guts of these diverse fishes36. On the other hand, certain bacterial taxa are carried by select fish hosts consistent with co-evolutionary processes, although diet and trophic level effects likely contribute to these patterns of gut microbiome composition40. Collectively, these findings indicate that the zebrafish is likely to be an ecologically relevant model organism, as teleosts account for the greatest number of vertebrate species on the planet. But, as with all models, researchers should consider the experimental context of the model’s use (developmental stage and diet) to ensure effective interpretation of their results.
The role of development and diet in zebrafish microbiome assembly.
The ability to rear zebrafish through their entire life cycle in the laboratory provides the opportunity to survey their microbiomes across their lifespan and under different dietary regimens. To date, the largest developmental study was conducted in 2016 on a cohort of sibling zebrafish reared under standard laboratory conditions in replicate tanks, with a subset of the fish’s digestive tracts sampled at seven time points across the lifespan3. This longitudinal observational study demonstrated that zebrafish microbiomes change across the lifespan in both composition and diversity3. For example, α-diversity (the total number of different taxa, sometimes weighted by abundance) of the zebrafish microbiomes decreased across time, with the mean OTU richness of 4 dpf larvae being >400 taxa, whereas that of adults was closer to 100. In parallel, the β-diversity (the pairwise differences in composition) of the zebrafish microbiome increased through developmental time: when larval fish were compared to other larval fish, the microbiomes were taxonomically similar, but when adult fish were compared to other adult fish, a diversification and individualization in the taxonomic composition of the microbiomes could be seen. The study also showed that these changes in diversity are marked by major shifts in the relative abundances of a handful of bacterial groups. For example, while the metabolically versatile Gammaproteobacteria are dominant throughout the lifespan, the Fusobacteria, which are typically obligate anaerobes, expanded to be a major group in adults3. One interpretation of this pattern is that as the digestive tract and associated bacterial communities mature, the environment becomes anoxic, opens up a niche for obligate anaerobes.
Concurrently with developmental changes across zebrafish lifespan, the animals also undergo dietary shifts in the laboratory setting. As larvae shift from endogenous egg yolk to exogenous feeding, typically they will receive both commercial fish food and live feeds (such as paramecia or rotifers at larval stages, and Artemia (brine shrimp) at juvenile and adult stages). In the longitudinal study on the composition of the zebrafish intestinal microbial community across development, microbiome shifts occurred concurrently with changes in diet, including a bloom of Alphaproteobacteria in juveniles, species that were also abundant in the Artemia introduced at that time3. In a similar 2015 longitudinal study in which diets were controlled and restricted to commercial feeds of defined composition, Wong et al.38 found that there were significant differences between the gut microbiomes of fish fed high- versus low-fat diets, and that the significance of these differences depended on host age. Interestingly, some of the patterns observed with the longitudinal study in which fish were fed standard lab diets with live feed were not observed in the study of fish on artificial diets, such as a reduction in α-diversity over time. However, the fish developed more slowly on the artificial feeds and did not reach adulthood by the end of the study. In 2012, Semova et al.21 reported significant differences in the microbiome of unfed versus fed larval zebrafish at 6 dpf, additionally highlighting the interconnections between diet and animal development. Furthermore, they found that taxa within the Firmicutes phylum altered the cellular mechanism by which the intestinal epithelium absorbed lipids from the diet, providing evidence that the types of microbes cultivated by the host can have direct effects on host physiology through diet. A subsequent study from this group found that high fat diet suppressed the sensory functions and induced ER stress in the enteroendocrine cells of the intestinal epithelium20. This response required the presence of the microbiota and could be mediated by monoassociation with a specific Acinetobacter strain. Relatedly, fish fed high amounts of palmitic acid carried microbiome assemblages that were linked to ER stress and liver damage41, and gut microbiota was found to influence how dietary nucleotide composition affects zebrafish metabolism42. These observations are important to consider when selecting dietary formulations for zebrafish experiments, especially given that diet frequently varies across zebrafish facilities and often is not optimized for a particular experimental context43.
The progression of the zebrafish microbiome community composition across development appears to reflect changes in the ecological processes underpinning the community assembly44. For example, the early larval microbiome seems to be driven primarily by neutral processes, that is, random sampling of taxa from a source pool and stochastic loss and replacement of individual microbial taxa. At 4 dpf, neutral processes can account for >80% of the variance in the distribution of microbiome taxa across a zebrafish population. Through developmental time, however, the relative contributions of neutral processes diminish and selective processes become more important. By 380 dpf, the same neutral model can only account for <40% of the variance in the distribution of microbiome taxa.
The role of immunity in shaping the zebrafish microbiomes.
The changes in maturing zebrafish microbiomes, with increased individualization and signatures of host selection, suggest a role for the host immune system in microbiome assembly. The immune system in vertebrates consists of two branches: innate immunity, which responds to microbial-associated patterns conserved across a broad swath of taxa, and adaptive immunity, which responds to specific (as narrow as strain-level) molecular patterns. Studies utilizing mutant zebrafish lines lacking either the innate34 or adaptive35 branch of the immune system, have uncovered the potential of the immune system as a whole to impose selection on the intestinal microbiome (Fig. 1a). For example, when myd88–/– zebrafish lacking innate immunity and wild-type zebrafish were maintained in solitary housing, a stark difference in the composition of the microbiome could be observed between genotypes. Similarly, when rag1–/— zebrafish lacking adaptive immunity and wild-type zebrafish populations were maintained separately, the β-diversity of the microbiome between individuals was greater for wild-type zebrafish than knockout mutants, indicating that the microbiomes of knockout individuals were more homogenous than those of wild type individuals. These results illustrate the capacity of the immune system to select or, at the very least, act as an ecological filter that influences which bacteria taxa ultimately comprise the gut microbiome.
Both of these studies also highlighted the importance of microbial transmission between individual fish in microbiome assembly, even in the context of immune selection. While significant differences were observed in microbiome composition between wild-type and zebrafish lacking innate immunity when they were maintained in solitary housing, housing multiple individuals in the same tank (separated by genotype or mixed) dramatically reduced these differences. Likewise, housing wild-type and zebrafish lacking adaptive immunity separately resulted insignificant differences in the β-diversity within each genotype. However, when the two genotypes were co-housed, no significant differences between genotypes were observed. These experiments demonstrate the substantial role of the ecological context in determining how host factors or any potential factors influence the assembly of the intestinal microbiome, and suggest the importance of considering parameters such as host interactions networks to study microbiome assembly in other systems such as humans45.
Effects of toxins and pathogens on the zebrafish microbiome.
In addition to direct host-selection mechanisms, exogenous factors can also influence the microbiome. For example, environmental chemicals, which can directly affect the host, can also alter the composition of the intestinal microbiome (Fig. 1a). Zebrafish are a well-established model for understanding how environmental chemicals impact vertebrate physiology and a vast array of studies have similarly used zebrafish to ascertain how these chemicals affect the vertebrate gut microbiome. For example, in 2013, Merrifield et al.46 fed metal-nanoparticle contaminated food to adult zebrafish and found that these particles had antimicrobial effects on some putatively beneficial strains of gut bacteria (such as Cetobacterium somerae)46. Exposure to endocrine-disrupting agents, such as bisphenol A (BPA), perturbed the gut microbiome of both juvenile47 and adult48 zebrafish. Taking advantage of the ex-utero development of zebrafish, Catron et al.47 demonstrated in 2019 that embryonic exposure to various BPA-derivative chemicals differentially affects the composition of the juvenile gut microbiome. Moreover, the authors observed dosage-dependent effects of the chemical on the gut microbiome, suggesting that the microbiome’s sensitivity to a chemical depends on the chemical’s environmental concentration. Another study explored the effect of low dose ciprofloxacin on monocultures of bacteria in the larval zebrafish gut and showed, using live imaging, that this antibiotic treatment dramatically decreased bacterial population by enhancing bacterial aggregation and clearance from the gut28. In another study on the effects of antibiotics on the gut microbiome, the administration of triclosan-laden food to adult zebrafish, significantly altered the composition of the gut microbiome4. The study took advantage of the zebrafish model to screen a large number of individuals, allowing the identification of microbial taxa that were especially sensitive to the drug’s exposure, including OTUs identified as members of the Enterobacteriaceae family; these microbial taxa might serve as indicators of exposure. Moreover, the large sample sizes facilitated the determination of how the microbe–microbe interaction networks in the gut vary as a function of triclosan exposure. In particular, while many OTUs in the gut displayed evidence of resistance to triclosan, their patterns of covariation with other conserved members of the microbiome were significantly altered. This observation indicates that not only does exposure to triclosan affect the gut microbiome’s composition, but it also affects the inter-organismal dynamics among microbiota. A 2018 study demonstrated that feeding fish Lactobacillus plantarum can attenuate the toxic effects of chronic triclosan exposure, including the effect of the drug on the gut microbiome40. Collectively, these studies exemplify the importance of understanding the interaction between environmental chemicals and gut microbiota and the potential of zebrafish to serve as a relatively high-throughput model organism to discover these interactions.
In addition to environmental chemicals, pathogens can invade the host and cause alterations to the microbiome as well. For example, introduction of the nematode intestinal parasite, Pseudocapillaria tomentosa, resulted not only in physiological changes to the host, but also to compositional changes in the gut microbiome49. The composition of the microbiome was different between exposed and unexposed zebrafish, and was also significantly associated with each stage (egg, larva, adult) of the nematode life cycle as well as parasite burden. Moreover, the composition of the gut microbiome was linked to histopathological outcomes of exposure, which indicates that the microbiome might potentially mediate exposure outcomes or serve as a passive indicator of exposure effects. Indeed, a machine-learning classifier trained on these microbiomics data accurately predicted an individual’s exposure state, suggesting that the microbiome might contain useful information for zebrafish facilities that seek to monitor infectious agents49. Therefore, the zebrafish model allows for a systems-level analysis of infection. That is, in addition to the direct effect of the pathogen on the host, indirect effects of the pathogen on the microbiome that contribute to disease can also be assessed.
Zebrafish microbiome studies for discovering new pathogens and probiotics.
Not only can microbiome profiling be used to reveal how known pathogens perturb resident microbial communities, it can also identify new bacterial agents that initiate or drive disease progression. One such example came from standard laboratory surveillance of sentinel zebrafish in facilities, which revealed a high incidence of intestinal hyperplasia and tumors in elderly fish50. Subsequent studies established that this intestinal pathology was transmissible, and further showed that it tracked with a particular Mycoplasma species51. The same Mycoplasma species was also found to be enriched in fish infected with Pseudocapillaria tomentosa presenting with intestinal histopathological lesions49, suggesting that this organism has a tropism for damaged intestinal tissue or possibly a capacity to accelerate intestinal disease progression.
Another example of microbiome profiling to uncover candidate bacterial etiologies of disease comes from a zebrafish model of Hirschsprung’s disease, in which the enteric nervous system fails to develop, resulting in impaired gut motility and often enterocolitis. Zebrafish lacking the Hirschsprung’s disease gene sox10 develop spontaneous intestinal inflammation and perturbed intestinal microbiomes52. Correlating the extent of inflammation with alterations in microbiomes across individual sox10 mutant zebrafish larvae revealed that expansion of specific Vibrio taxa and reduction of specific Escherichia taxa correlated with higher levels of inflammation. Subsequent experiments demonstrated that addition of a Vibrio isolate was sufficient to exacerbate intestinal inflammation in the conventional sox10 mutant fish, whereas an E. coli isolate, used as a probiotic in humans, could ameliorate inflammation52. Related study designs exposed germ-free fish to various Aeromonas strains to uncover putative genetic mechanisms underlying motile aeromonad septicemia53. These studies demonstrate the utility of zebrafish for discovering new bacteria with pathogenic and beneficial potentials in vertebrate intestines.
Conclusions and future directions
As accumulating evidence demonstrates the important contribution of microbiota to animal development, physiology, and behavior, the scope of microbiome research continues to expand. We consider that the zebrafish model is an important player in this expansion. The model offers several advantages that facilitate mechanistic studies of the microbiome using gnotobiology, and microbiome profiling of large population sizes with controlled environmental exposures and housing configurations, thereby allowing researchers to span the many scales across which animals interact with their resident microbes, from molecules to populations.
Acknowledgements
This work was supported in part by grants from the National Institutes of Health, under [P01GM125576] to K.G., the National Institutes of Environmental Health Sciences [1R01ES030226] to T.J.S. K.S. was supported in part by the NIEHS Integrated Regional Training Program in Environmental Health Sciences grant (PI Robyn L Tanguay, T32-ES007060-38).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Grunwald DJ & Eisen JS Headwaters of the zebrafish — emergence of a new model vertebrate. Nat. Rev. Genet 3, 717–724 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Burns AR & Guillemin K The scales of the zebrafish: host–microbiota interactions from proteins to populations. Curr. Opin. Microbiol 38, 137–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephens WZ et al. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 10, 644–654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaulke CA, Barton CL, Proffitt S, Tanguay RL & Sharpton TJ Triclosan Exposure Is Associated with Rapid Restructuring of the Microbiome in Adult Zebrafish. PLoS One 11, e0154632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill JH, Franzosa EA, Huttenhower C & Guillemin K A conserved bacterial protein induces pancreatic beta cell expansion during zebrafish development. Elife 5, 1–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rolig AS et al. A bacterial immunomodulatory protein with lipocalin-like domains facilitates host–bacteria mutualism in larval zebrafish. Elife 7, 1–26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawls JF, Samuel BS & Gordon JI Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci 101, 4596–4601 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates JM et al. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol 297, 374–386 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Melancon E et al. Best practices for germ-free derivation and gnotobiotic zebrafish husbandry in Methods in Cell Biology 138, 61–100 (Elsevier Ltd, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis KL, Del Cid N & Traver D Perspectives on antigen presenting cells in zebrafish. Dev. Comp. Immunol 46, 63–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stednitz SJ et al. Forebrain Control of Behaviorally Driven Social Orienting in Zebrafish. Curr. Biol 28, 2445–2451.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis DJ et al. Lactobacillus plantarum attenuates anxiety-related behavior and protects against stress-induced dysbiosis in adult zebrafish. Sci. Rep 6, 33726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrelli L et al. Probiotic modulation of the microbiota-gut-brain axis and behaviour in zebrafish. Sci. Rep 6, 30046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truong L et al. Multidimensional In Vivo Hazard Assessment Using Zebrafish. Toxicol. Sci 137, 212–233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phelps D et al. Microbial colonization is required for normal neurobehavioral development in zebrafish. Sci. Rep 7, 11244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bates JM, Akerlund J, Mittge E & Guillemin K Intestinal Alkaline Phosphatase Detoxifies Lipopolysaccharide and Prevents Inflammation in Zebrafish in Response to the Gut Microbiota. Cell Host Microbe 2, 371–382 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheesman SE, Neal JT, Mittge E, Seredick BM & Guillemin K Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc. Natl. Acad. Sci 108, 4570–4577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Troll JV et al. Microbiota promote secretory cell determination in the intestinal epithelium by modulating host Notch signaling. Development 145, dev155317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murdoch CC & Rawls JF Commensal Microbiota Regulate Vertebrate Innate Immunity-Insights From the Zebrafish. Front. Immunol 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye L et al. High fat diet induces microbiota-dependent silencing of enteroendocrine cells. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semova I et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–88 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis DJ, Bryda EC, Gillespie CH & Ericsson AC Microbial modulation of behavior and stress responses in zebrafish larvae. Behav. Brain Res 311, 219–227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosch TCG, Guillemin K & McFall-Ngai M Evolutionary ‘Experiments’ in Symbiosis: The Study of Model Animals Provides Insights into the Mechanisms Underlying the Diversity of Host-Microbe Interactions. Bioessays 41, e1800256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiles TJ et al. Modernized Tools for Streamlined Genetic Manipulation and Comparative Study of Wild and Diverse Proteobacterial Lineages. MBio 9, 1–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taormina MJ et al. Investigating Bacterial-Animal Symbioses with Light Sheet Microscopy. Biol. Bull 223, 7–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jemielita M et al. Spatial and Temporal Features of the Growth of a Bacterial Species Colonizing the Zebrafish Gut. MBio 5, e01751–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiles TJ et al. Host Gut Motility Promotes Competitive Exclusion within a Model Intestinal Microbiota. PLOS Biol. 14, e1002517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlomann BH, Wiles TJ, Wall ES, Guillemin K & Parthasarathy R Sublethal antibiotics collapse gut bacterial populations by enhancing aggregation and expulsion. Proc. Natl. Acad. Sci. U. S. A 116, 21392–21400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiles TJ et al. Swimming motility of a gut bacterial symbiont promotes resistance to intestinal expulsion and enhances inflammation. PLOS Biol. 18, e3000661 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlomann BH, Wiles TJ, Wall ES, Guillemin K & Parthasarathy R Bacterial Cohesion Predicts Spatial Distribution in the Larval Zebrafish Intestine. Biophys. J 115, 2271–2277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens WZ et al. Identification of Population Bottlenecks and Colonization Factors during Assembly of Bacterial Communities within the Zebrafish Intestine. MBio 6, e01163–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae S, Mueller O, Wong S, Rawls JF & Valdivia RH Genomic sequencing-based mutational enrichment analysis identifies motility genes in a genetically intractable gut microbe. Proc. Natl. Acad. Sci 113, 14127–14132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawls JF, Mahowald MA, Ley RE & Gordon JI Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-free Recipients Reveal Host Habitat Selection. Cell 127, 423–433 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burns AR et al. Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc. Natl. Acad. Sci 114, 11181–11186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stagaman K, Burns AR, Guillemin K & Bohannan BJ The role of adaptive immunity as an ecological filter on the gut microbiota in zebrafish. ISME J. 11, 1630–1639 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roeselers G et al. Evidence for a core gut microbiota in the zebrafish. ISME J. 5, 1595–1608 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spence R, Gerlach G, Lawrence C & Smith C The behaviour and ecology of the zebrafish, Danio rerio. Biol. Rev 83, 13–34 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Wong S et al. Ontogenetic Differences in Dietary Fat Influence Microbiota Assembly in the Zebrafish Gut. MBio 6, e00687–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Givens C, Ransom B, Bano N & Hollibaugh J Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar. Ecol. Prog. Ser 518, 209–223 (2015). [Google Scholar]
- 40.Egerton S, Culloty S, Whooley J, Stanton C & Ross RP The Gut Microbiota of Marine Fish. Front. Microbiol 9, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding Q et al. The hepatotoxicity of palmitic acid in zebrafish involves the intestinal microbiota. J. Nutr 148, 1217–1228 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Guo X et al. The Growth-Promoting Effect of Dietary Nucleotides in Fish Is Associated with an Intestinal Microbiota-Mediated Reduction in Energy Expenditure. J. Nutr 147, 781–788 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Teame T et al. The use of zebrafish (Danio rerio) as biomedical models. Anim. Front 9, 68–77 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burns AR et al. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 10, 655–664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brito IL & Alm EJ Tracking strains in the microbiome: Insights from metagenomics and models. Front. Microbiol 7, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merrifield DL et al. Ingestion of metal-nanoparticle contaminated food disrupts endogenous microbiota in zebrafish (Danio rerio). Environ. Pollut 174, 157–163 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Catron TR et al. Host Developmental Toxicity of BPA and BPA Alternatives Is Inversely Related to Microbiota Disruption in Zebrafish. Toxicol. Sci 167, 468–483 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Liu Y et al. Influence of Endogenous and Exogenous Estrogenic Endocrine on Intestinal Microbiota in Zebrafish. PLoS One 11, e0163895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaulke CA et al. A longitudinal assessment of host-microbe-parasite interactions resolves the zebrafish gut microbiome’s link to Pseudocapillaria tomentosa infection and pathology. Microbiome 7, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paquette CE et al. A Retrospective Study of the Prevalence and Classification of Intestinal Neoplasia in Zebrafish ( Danio Rerio ). Zebrafish 10, 228–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burns AR et al. Transmission of a common intestinal neoplasm in zebrafish by cohabitation. J. Fish Dis 41, 569–579 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rolig AS et al. The enteric nervous system promotes intestinal health by constraining microbiota composition. PLoS Biol. 15, 1–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ran C et al. Aeromonas veronii and aerolysin are important for the pathogenesis of motile aeromonad septicemia in cyprinid fish. Environ. Microbiol 20, 3442–3456 (2018). [DOI] [PubMed] [Google Scholar]


