The identification of nine different genotypes among members of G. vaginalis allowed us to distinguish an uneven distribution of virulence-associated genetic traits within this taxon and thus suggest the potential occurrence of putative pathogen and commensal G. vaginalis strains. These findings, coupled with metagenomics microbial profiling of human vaginal microbiota, permitted us to get insights into the distribution of the genotypes among the human population, highlighting the presence of different structural communities in terms of G. vaginalis genotypes.
KEYWORDS: Gardnerella vaginalis, phylogenomics, metagenomics, Bifidobacteriaceae
ABSTRACT
Gardnerella vaginalis is described as a common anaerobic vaginal bacterium whose presence may correlate with vaginal dysbiotic conditions. In the current study, we performed phylogenomic analyses of 72 G. vaginalis genome sequences, revealing noteworthy genome differences underlying a polyphyletic organization of this taxon. Particularly, the genomic survey revealed that this species may actually include nine distinct genotypes (GGtype1 to GGtype9). Furthermore, the observed link between sialidase and phylogenomic grouping provided clues of a connection between virulence potential and the evolutionary history of this microbial taxon. Specifically, based on the outcomes of these in silico analyses, GGtype3, GGtype7, GGtype8, and GGtype9 appear to have virulence potential since they exhibited the sialidase gene in their genomes. Notably, the analysis of 34 publicly available vaginal metagenomic samples allowed us to trace the distribution of the nine G. vaginalis genotypes identified in this study among the human population, highlighting how differences in genetic makeup could be related to specific ecological properties. Furthermore, comparative genomic analyses provided details about the G. vaginalis pan- and core genome contents, including putative genetic elements involved in the adaptation to the ecological niche as well as many putative virulence factors. Among these putative virulence factors, particularly noteworthy genes identified were the gene encoding cholesterol-dependent cytolysin (CDC) toxin vaginolysin and genes related to microbial biofilm formation, iron uptake, adhesion to the vaginal epithelium, as well as macrolide antibiotic resistance.
IMPORTANCE The identification of nine different genotypes among members of G. vaginalis allowed us to distinguish an uneven distribution of virulence-associated genetic traits within this taxon and thus suggest the potential occurrence of putative pathogen and commensal G. vaginalis strains. These findings, coupled with metagenomics microbial profiling of human vaginal microbiota, permitted us to get insights into the distribution of the genotypes among the human population, highlighting the presence of different structural communities in terms of G. vaginalis genotypes.
INTRODUCTION
The human female reproductive tract harbors trillions of bacteria that play an important role in the health of women (1). In particular, human vaginal microbiota are believed to exert a preventive action against several diseases, such as bacterial vaginosis (BV), sexually transmitted diseases (STDs), and urinary tract infections (2–5). In this context, members of the Lactobacillus genus are generally dominant in the vaginal microenvironment of healthy women and exploit their beneficial role(s) through lactic acid production that keeps low pH and provide protection to the host against pathogenic bacteria (6–8).
In recent years, the composition of women’s vaginal microbiota has been investigated by means of next-generation DNA sequencing techniques, revealing that vaginal bacterial communities, i.e., vaginal microbiota, can be classified from three to nine ecotypes according to their specific microbial composition (9). In this context, it has been proposed that the vaginal microbiota of asymptomatic women from four ethnic groups could be clustered into five community-state types (CSTs) (9). Notably, CST I, CST II, CST III, and CST V were dominated by various species of Lactobacillus, i.e., Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus iners, and Lactobacillus jensenii, respectively, while CST IV was mainly constituted by obligated anaerobic bacteria, also including members of the Prevotella, Atopobium, and Gardnerella genera. Among the latter genus, a single species has been so far described, i.e., Gardnerella vaginalis, represented by Gram-positive, anaerobic, non-spore-forming bacteria commonly identified in the vaginal environment (10).
Great interest revolves around G. vaginalis since this microorganism was frequently detected as a dominant microorganism in chronic and acute BV incidence (11), which is an aberrant condition characterized by a shift of the vaginal microbiota composition (Lactobacillus dominated) toward a more diversified microbial community (12). It has been demonstrated that G. vaginalis cells possess the ability to adhere to the vaginal epithelium and develop a characteristic microbial biofilm (13, 14), enabling it to colonize the vaginal tract efficiently. In addition, G. vaginalis can produce other virulence factors, such as sialidase, which has been strongly linked with microbial biofilm production (15, 16), and cholesterol-dependent cytolysin (CDC) family toxin vaginolysin (17). However, it has also been observed that presence of G. vaginalis in the vaginal microbiota does not always imply BV (18). For this reason, several efforts were made to highlight which genomic differences could discriminate pathogenic from commensal strains (11, 19, 20). Nevertheless, the role of G. vaginalis in the pathogenesis of BV is still far from being fully understood.
Since its discovery in 1955, G. vaginalis was named Haemophilus vaginalis (10), and later, it was designated Corynebacterium vaginale (21). Afterward, taxonomic studies confirmed the need to introduce the new Gardnerella genus, also showing its taxonomic relatedness to the Bifidobacterium genus (22, 23). To date, G. vaginalis is taxonomically placed within the Bifidobacteriaceae family, and it is considered the only species of the Gardnerella genus. However, several studies have reported the existence of genetic heterogeneity among the various members of this genus (24–26).
Here, we carried out an exhaustive comparative genome analysis based on 72 publicly available genomic sequences of G. vaginalis, aiming to investigate the genomic variability of this taxon. Moreover, phylogenomics analyses were carried out to highlight the phylogenetic relationships of G. vaginalis with the other members of the Bifidobacteriaceae family. Finally, the screening of 34 publicly available vaginal shotgun metagenomic data sets allowed us to investigate the distribution of the here-identified G. vaginalis genotypes among the human population.
RESULTS AND DISCUSSION
General genome features of Gardnerella vaginalis.
In order to perform an exhaustive comparative genomic analysis of the G. vaginalis species, all the publicly available genome sequences of this taxon were retrieved from the NCBI database (Table 1). Notably, chromosomes of G. vaginalis used in this work were carefully selected, resulting in one of the largest high-quality databases developed to date, encompassing 72 G. vaginalis genomes (see Materials and Methods). The predicted average genomic GC content was 41.8%, a lower value than the other members of the Bifidobacteriaceae family (60.2% for the bifidobacterial strains and 52.9% for other genera of the Bifidobacteriaceae family) (27). Interestingly, the GC content showed low variability among analyzed strains, except for the CMW7778B chromosome, which deviates from the genomes of the other strains, with a GC content of 38%. As shown by previous studies, these findings allowed researchers to suggest that the adaptation to a limited niche complexity, along with a constant environmental temperature, may have affected the GC content of G. vaginalis genomes (28). In fact, G. vaginalis strains have been isolated so far only from the human urogenital tract, thus showing a restricted ecological niche whose temperature is maintained to be almost constant. In contrast, members of the Bifidobacterium genus that colonize a wide variety of ecological niches, including the gut of homeothermic and heterothermic animals, exhibited a higher GC content level (27).
TABLE 1.
General genome features of G. vaginalis
| Gardnerella vaginalis strain | ENA assembly no. | Genome status | Genome size (Mb) | GC content (%) | No. of CDS | No. of rRNA loci | No. of tRNA genes | Virulence gene(s) | Isolation source | BioProject accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| 5-1 | GCA_000176495.1 | Draft | 1.6728 | 42.0 | 1,273 | 1 | 45 | vly | Vagina | PRJNA40895 |
| 41V | GCA_000165635.2 | Draft | 1.6594 | 41.3 | 1,277 | 1 | 45 | vly | Vagina | PRJNA53893 |
| PSS_7772B | GCA_001546485.1 | Draft | 1.5967 | 42.9 | 1,169 | 1 | 44 | vly | Urine | PRJNA272100 |
| KA00225 | GCA_002896555.1 | Draft | 1.6700 | 40.8 | 1,187 | 2 | 45 | vly | Vagina | PRJNA338962 |
| 101 | GCA_000165615.2 | Draft | 1.5275 | 43.4 | 1,163 | 2 | 45 | vly | NAa | PRJNA53359 |
| CMW7778B | GCA_001563665.1 | Draft | 1.6026 | 38.0 | 1,150 | 1 | 44 | vly | Vagina | PRJNA272122 |
| N165 | GCA_003408785.1 | Draft | 1.7116 | 41.4 | 1,344 | 2 | 44 | vly, sld | Vaginal mucus | PRJNA310104 |
| 1400E | GCA_000263495.1 | Draft | 1.7163 | 41.2 | 1,331 | 3 | 44 | vly | Vagina | PRJNA42445 |
| 1500E | GCA_000263595.1 | Draft | 1.5482 | 43.0 | 1,157 | 3 | 45 | vly | Vagina | PRJNA42447 |
| 55152 | GCA_000263475.1 | Draft | 1.6432 | 41.3 | 1,244 | 1 | 45 | vly | Vagina | PRJNA42443 |
| GED7760B | GCA_001546455.1 | Draft | 1.4892 | 43.3 | 1,123 | 1 | 45 | sld | Vagina | PRJNA272108 |
| UGent 09.07 | GCA_003397665.1 | Draft | 1.7238 | 41.1 | 1,293 | 1 | 47 | vly | Vagina | PRJNA474758 |
| 00703C2mash | GCA_000263515.1 | Draft | 1.5467 | 42.3 | 1,185 | 2 | 45 | vly | Vagina | PRJNA42451 |
| 49145 | GCA_003034925.1 | Draft | 1.7014 | 41.2 | 1,325 | 1 | 45 | vly, sld | Vagina | PRJNA437230 |
| ATCC 49145 | GCA_001913835.1 | Draft | 1.7069 | 41.2 | 1,361 | 2 | 45 | vly, sld | Vagina | PRJNA342481 |
| GED7275B | GCA_001546445.1 | Draft | 1.5079 | 42.5 | 1,139 | 1 | 45 | vly | Vagina | PRJNA272096 |
| UMB0061 | GCA_002861165.1 | Draft | 1.7422 | 41.2 | 1,387 | 2 | 45 | vly, sld | Catheter | PRJNA316969 |
| 0288E | GCA_000263555.1 | Draft | 1.7088 | 41.2 | 1,338 | 1 | 45 | vly | Vagina | PRJNA42437 |
| 284V | GCA_000263435.1 | Draft | 1.6508 | 41.2 | 1,280 | 2 | 45 | vly | Vagina | PRJNA42431 |
| 00703Bmash | GCA_000263615.1 | Draft | 1.5661 | 42.3 | 1,227 | 1 | 45 | vly, sld | Vagina | PRJNA42449 |
| JCM 11026 | GCA_004336685.1 | Draft | 1.6571 | 41.3 | 1,225 | 1 | 45 | vly, sld | Vagina | PRJNA524873 |
| 6420B | GCA_000263575.1 | Draft | 1.4936 | 42.2 | 1,122 | 1 | 45 | vly | Vagina | PRJNA42441 |
| UMB0032B | GCA_002862005.1 | Draft | 1.7451 | 41.2 | 1,382 | 1 | 45 | vly, sld | Catheter | PRJNA316969 |
| 315-A | GCA_000214315.2 | Draft | 1.6533 | 41.4 | 1,298 | 1 | 45 | vly, sld | Vaginal | PRJNA52049 |
| NR010 | GCA_003408845.1 | Draft | 1.6227 | 45.5 | 1,181 | 3 | 45 | vly, sld | Vaginal mucus | PRJNA310104 |
| UMB0833 | GCA_002861885.1 | Draft | 1.6203 | 42.1 | 1,273 | 3 | 45 | sld | Catheter | PRJNA316969 |
| 6119V5 | GCA_000263655.1 | Draft | 1.4996 | 43.3 | 1,117 | 1 | 45 | vly | Vagina | PRJNA42455 |
| 3549624 | GCA_001049785.1 | Draft | 1.7323 | 41.4 | 1,298 | 2 | 45 | vly, sld | Vagina | PRJNA288563 |
| 00703Dmash | GCA_000263635.1 | Draft | 1.4908 | 43.4 | 1,121 | 1 | 45 | vly | Vagina | PRJNA42453 |
| 14018c | GCA_004336715.1 | Draft | 1.6578 | 41.3 | 1,232 | 1 | 45 | vly, sld | NA | PRJNA524879 |
| UMB0032A | GCA_002862015.1 | Draft | 1.7455 | 41.2 | 1,383 | 2 | 45 | vly, sld | Catheter | PRJNA316969 |
| UMB0770 | GCA_002861945.1 | Draft | 1.6960 | 41.2 | 1,323 | 1 | 45 | vly, sld | Catheter | PRJNA316969 |
| UMB0775 | GCA_002861925.1 | Draft | 1.7436 | 41.2 | 1,397 | 2 | 45 | vly, sld | Catheter | PRJNA316969 |
| GS 9838-1 | GCA_003397705.1 | Draft | 1.6221 | 41.9 | 1,231 | 2 | 45 | vly | Vagina | PRJNA474758 |
| UMB1686 | GCA_002884775.1 | Draft | 1.5106 | 43.3 | 1,124 | 2 | 45 | vly, sld | Catheter | PRJNA316969 |
| DNF01149 | GCA_002894105.1 | Draft | 1.7247 | 41.2 | 1,362 | 3 | 45 | vly, sld | Vagina | PRJNA338971 |
| N101 | GCA_003369895.1 | Draft | 1.5430 | 42.4 | 1,205 | 1 | 45 | vly, sld | Vaginal swab | PRJNA265097 |
| UMB0233 | GCA_002862045.1 | Draft | 1.6424 | 41.2 | 1,292 | 1 | 45 | vly, sld | Catheter | PRJNA316969 |
| 14019_MetR | GCA_001278345.1 | Draft | 1.6611 | 41.3 | 1,317 | 1 | 45 | vly, sld | NA | PRJNA294071 |
| W11 | GCA_003369875.1 | Draft | 1.5667 | 42.3 | 1,213 | 1 | 45 | sld | Vaginal swab | PRJNA265103 |
| N95 | GCA_003369965.1 | Draft | 1.5225 | 42.4 | 1,183 | 2 | 45 | vly, sld | Vaginal swab | PRJNA265092 |
| UMB0682 | GCA_002862065.1 | Draft | 1.6013 | 42.1 | 1,234 | 1 | 45 | vly | Catheter | PRJNA316969 |
| N153 | GCA_003369935.1 | Draft | 1.5418 | 42.4 | 1,167 | 1 | 45 | vly, sld | Vaginal swab | PRJNA265102 |
| N72 | GCA_003408815.1 | Draft | 1.6429 | 41.9 | 1,249 | 1 | 45 | vly | Vaginal mucus | PRJNA310104 |
| N160 | GCA_003408775.1 | Draft | 1.5097 | 43.3 | 1,119 | 3 | 43 | vly, sld | Vaginal mucus | PRJNA310104 |
| UGent 18.01 | GCA_003397585.1 | Draft | 1.5143 | 42.5 | 1,144 | 1 | 45 | sld | Vagina | PRJNA474758 |
| UMB0768 | GCA_002884835.1 | Draft | 1.6748 | 41.3 | 1,319 | 2 | 45 | vly, sld | Catheter | PRJNA316969 |
| UMB1642 | GCA_002884795.1 | Draft | 1.6288 | 41.8 | 1,233 | 2 | 45 | vly | Catheter | PRJNA316969 |
| UMB0264 | GCA_002884875.1 | Draft | 1.5151 | 42.3 | 1,155 | 2 | 45 | vly | Catheter | PRJNA316969 |
| UMB0913 | GCA_002861145.1 | Draft | 1.5136 | 42.1 | 1,140 | 1 | 45 | vly | Catheter | PRJNA316969 |
| UMB0170 | GCA_002884855.1 | Draft | 1.5147 | 42.3 | 1,153 | 2 | 45 | vly | Catheter | PRJNA316969 |
| UMB0912 | GCA_002861125.1 | Draft | 1.5138 | 42.1 | 1,140 | 1 | 45 | vly | Catheter | PRJNA316969 |
| UMB0830 | GCA_002861905.1 | Draft | 1.5592 | 42.3 | 1,224 | 1 | 45 | vly, sld | Catheter | PRJNA316969 |
| 75712 | GCA_000263535.1 | Draft | 1.6730 | 41.3 | 1,302 | 2 | 45 | vly | Vagina | PRJNA42435 |
| UMB0386 | GCA_002861965.1 | Draft | 1.6757 | 41.2 | 1,323 | 2 | 45 | vly, sld | Catheter | PRJNA316969 |
| UGent 25.49 | GCA_003397605.1 | Draft | 1.6586 | 41.2 | 1,246 | 2 | 45 | vly, sld | Vagina | PRJNA474758 |
| GS 10234 | GCA_003397745.1 | Draft | 1.5890 | 41.9 | 1,181 | 2 | 45 | vly | Vagina | PRJNA474758 |
| UGent 09.48 | GCA_003397635.1 | Draft | 1.4709 | 42.2 | 1,095 | 1 | 45 | vly | Vagina | PRJNA474758 |
| UGent 21.28 | GCA_003397615.1 | Draft | 1.5479 | 42.5 | 1,189 | 1 | 45 | sld | Vagina | PRJNA474758 |
| UMB0298 | GCA_002861975.1 | Draft | 1.6760 | 41.2 | 1,319 | 2 | 45 | vly, sld | Catheter | PRJNA316969 |
| ATCC 14018 | GCA_003397685.1 | Draft | 1.6620 | 41.3 | 1,248 | 1 | 45 | vly, sld | Vagina | PRJNA474758 |
| FDAARGOS_296 | GCA_002206225.2 | Draft | 1.7710 | 41.3 | 1,375 | 2 | 45 | vly, sld | NA | PRJNA231221 |
| N144 | GCA_003408835.1 | Draft | 1.5824 | 42.3 | 1,217 | 1 | 45 | vly, sld | Vaginal mucus | PRJNA310104 |
| GH015 | GCA_003408745.1 | Draft | 1.5756 | 41.0 | 1,173 | 1 | 43 | vly, sld | Vaginal mucus | PRJNA310104 |
| JCM 11026 | GCA_001042655.1 | Complete | 1.6674 | 41.3 | 1,244 | 2 | 45 | vly, sld | Vagina | PRJDB63 |
| NCTC10287 | GCA_900637625.1 | Complete | 1.6674 | 41.4 | 1,252 | 2 | 45 | vly, sld | Vagina | PRJEB6403 |
| GV37 | GCA_001953155.1 | Complete | 1.7467 | 41.8 | 1,359 | 2 | 45 | vly | Blood culture | PRJNA360037 |
| HMP9231 | GCA_000213955.1 | Complete | 1.7265 | 41.2 | 1,354 | 2 | 45 | vly | Endometrium | PRJNA51067 |
| FDAARGOS_568 | GCA_003812765.1 | Complete | 1.7166 | 41.3 | 1,368 | 2 | 45 | vly, sld | NA | PRJNA231221 |
| ATCC 14019 | GCA_000159155.2 | Complete | 1.6674 | 41.4 | 1,209 | 2 | 45 | vly, sld | Vaginal | PRJNA31473 |
| 409-05 | GCA_000025205.1 | Complete | 1.6176 | 42.0 | 1,237 | 2 | 45 | vly | Vaginal | PRJNA31001 |
| UGent 06.41 | GCA_003293675.1 | Complete | 1.5635 | 42.1 | 1,162 | 2 | 45 | vly | Vagina | PRJNA474758 |
NA, not available.
G. vaginalis genome sequences considered in this study ranged in size from 1.47 Mb (UGent 09.48) to 1.77 Mb (FDAARGOS_296), with an average of 1,241 coding DNA sequences (CDS). Furthermore, these genomes had between 1 and 3 rRNA loci, and the number of tRNA genes ranged from 44 to 47. These data results were consistent with those of the genomes of nonbifidobacterial taxa of the Bifidobacteriaceae family, exhibiting averages of 1,502 CDS and 2.6 rRNA operons per genome and numbers of tRNA genes ranging from 45 to 48. Specifically, a statistical comparison between G. vaginalis chromosomes and nonbifidobacterial genomes showed that, within this latter group, the average numbers of CDS, rRNA operons, and tRNA genes were increased by 17.41% (P < 0.05), 40.10% (P < 0.05), and 2.53% (P < 0.05), respectively. Moreover, the analogous comparison with members of the Bifidobacterium genus showed averages of 1,865 CDS and 3.2 rRNA loci and a tRNA content ranging from 40 to 79, revealing that within the bifidobacterial group, the averages of these numbers were increased by 33.45% (P < 0.05), 50.37% (P < 0.05), and 14.99% (P < 0.05), respectively (27). Based on the statistical comparisons of the number of CDS, rRNA, and tRNA, it seems at first that the G. vaginalis species has undergone a selective pressure similar to nonbifidobacterial members of the Bifidobacteriaceae family rather than members of the Bifidobacterium genus. As mentioned above, G. vaginalis was correlated with BV incidence; nevertheless, it was also often found in healthy vaginal microbiota. It was supposed that certain lineages or species of Gardnerella are natural commensals and others can act as pathogens, triggering cases of symptomatic vaginal dysbiosis (29). To evaluate this hypothesis, we assessed the distribution of the two most studied and described genes that participate in the pathogenesis mechanism driven by G. vaginalis, i.e., those encoding the pore-forming CDC toxin vaginolysin (vly) and sialidase (sld), also known as neuraminidase (15, 17). Results showed that the genomes of 67 strains contained the vly gene, whereas more than half of the total number of Gardnerella chromosomes (40 genomes) were shown to encode a sialidase enzyme (Table 1). Notably, the sialidase enzymatic activity can reduce the protective vaginal mucosal layer, facilitating bacterial adhesion to the vaginal epithelium and subsequent microbial biofilm development, thus increasing the infectious capabilities of G. vaginalis strains (15).
The evaluation of the possible presence of mobile elements within G. vaginalis chromosomes, followed by investigations of the genomic regions adjacent to both their ends, allowed us to assess the occurrence of eight putative virulence genes in eight G. vaginalis genomes. Each of these protein-encoding genes contained a domain resembling the coding region for a virulence-related protein belonging to a member of the Streptococcus genus and was found alongside a putative genomic prophage island (Fig. S1 in the supplemental material). Furthermore, 15 strains of G. vaginalis contained noteworthy genes placed tightly adjacent to transposases predicted to belong to members of the IS256 and IS3 families. These genes included a sequence encoding a RelE/RelB toxin-antitoxin system that is thought to exert toxic effects on both bacterial and eukaryotic cell types (30), as well as a ribosomal protection protein (TetM) conferring tetracycline resistance (31) and a collagen-binding protein (Fig. S1). These characteristics may reflect how prophage-like sequences and insertion sequence (IS) elements can be responsible for genomic duplications, deletions, and rearrangements, contributing to the genetic makeup and biodiversity of this bacterial taxon (32).
Pan-genome and core genome of the G. vaginalis species.
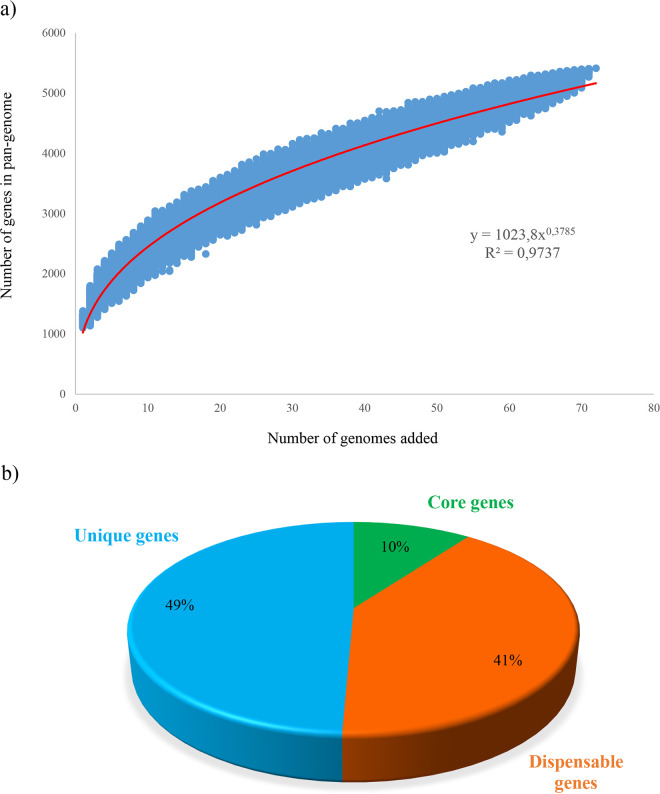
Previous comparative genomic studies involving much smaller numbers of G. vaginalis genome sequences highlighted significant genomic differences between the chromosomes of this species (19, 24, 33). In this context, pan-genome reconstruction can contribute to deciphering the evolutionary dynamics, i.e., selection pressure of beneficial genes, as well as species- and genus-level differences in overall gene content (34). In order to explore genetic differences, the genomes of 72 G. vaginalis strains were submitted to gene reannotation and subsequently analyzed from a pan-genome perspective, also unveiling their core genome and unique gene sequences. The pan-genome size of G. vaginalis has been shown to consist of 5,071 clusters of orthologous groups (COGs), and plotting it on a logarithmic scale as a function of the total amount of involved genomes revealed that the power trend line had not yet reached a plateau (Fig. 1). More precisely, adding a new G. vaginalis genome is predicted to add about 38 or 39 new genes to the G. vaginalis pan-genome.
FIG 1.
G. vaginalis pan-genome. (a) Pan-genome represented as a variation in size of the gene pool resulting from the sequential addition of the 72 G. vaginalis genomes. (b) Pie chart of the number of core genes (green), dispensable genes (orange), and unique genes (light blue) of G. vaginalis.
As previously mentioned, the pan-genome analysis allowed the evaluation of the core genome, defined as the set of gene families shared by all the organisms (34). In this comparison, a total repertoire of 514 COGs (10.1%) has been identified as a constituent of the core genome of G. vaginalis. Previous pan-genome analysis, including 60 Bifidobacterium pseudolongum genomes with an average genome size of 2.01 Mb, revealed a pan-genome consisting of 6,172 COGs corresponding to a core genome of 1,069 COGs (17.3%) (35). Likewise, a pan-genome curve based on 33 Bifidobacterium longum genomes with an average genome size of 2.35 Mb showed a pan-genome consisting of about 6,000 COGs and a core genome formed by 1,145 COGs (about 19%) (36). This evidence suggests that the G. vaginalis core genome could be considered smaller than that of other species belonging to the closely related Bifidobacterium genus.
Furthermore, through pan-genome analysis, we also identified the truly unique genes (TUGs) of Gardnerella, which ranged from 7 for the strain UMB0386 to 143 for KA00225. These findings showed that this species displays a modestly sized core genome corresponding to a relatively sizeable dispensable genome, i.e., the subset of genes shared by two or more strains (Fig. 1). Afterward, in silico analysis employing the eggNOG database allowed us to investigate the functional annotation of core genes. Excluding 14.9% that have no function, the large part of the encoded proteins belonging to the core proteome of G. vaginalis was related to essential cell maintenance, including translation (16.2%), carbohydrates, amino acids, and nucleotide metabolic processes (7.3%, 6.8%, and 6.6%, respectively) as well as inorganic ion transport (6.4%) (Fig. 1).
In addition, to get insights into specific genes supporting the adaptation of G. vaginalis to the vaginal environment, the genes belonging exclusively to the core genome of this species were further analyzed. A collection of 379 COGs, constituting the specific core genome of G. vaginalis, were obtained from the total amount of 514 COGs following the exclusion of COGs shared with other members of the Bifidobacteriaceae family (see Materials and Methods). This set of genes was evaluated from a functional annotation perspective. Such analysis revealed the ubiquitous presence of genes encoding C69-family dipeptidase, previously recognized as responsible for collagen molecule degradation (37), and a pullulanase, which seems to allow the efficient utilization of glycogen, i.e., the primary available carbon source in the vaginal lumen (38). The mere presence of the latter genes within the G. vaginalis chromosomes cannot demonstrate that these genes are still under selective pressure. Thus, further investigations are requested to confirm their activity and functionality. However, their presence in the genomes of G. vaginalis may represent a clue to genetic adaptation to the vaginal environment of this species.
The screening of G. vaginalis genomes revealed the presence of several common features related to virulence, i.e., cytotoxicity/hemolysis mechanisms, biofilm production, iron uptake, adhesion to the epithelium, and antimicrobial resistance. Specifically, the ability of G. vaginalis to adhere to the vaginal wall is mediated by genes encoding type IV Flp pili (Table S3). At the same time, the subsequent biofilm development seems to be related to type I glycosyltransferase, also involving sortase enzyme activity, which was detected in each G. vaginalis genome analyzed as well (39). Furthermore, G. vaginalis genomes contained genes associated with toxicity, including a CDC toxin, vaginolysin, highly conserved among G. vaginalis strains (17), and a serralysin characterized in Serratia marcescens annotated as serralysin (40). Finally, within the core G. vaginalis genes, seven genes encoding putative drug resistance proteins were found, including two genes that are predicted to confer resistance to macrolide antibiotics, one major facilitator superfamily (MFS) transporter, as well as four unknown multidrug efflux systems.
Phylogenomic analysis of G. vaginalis taxon.
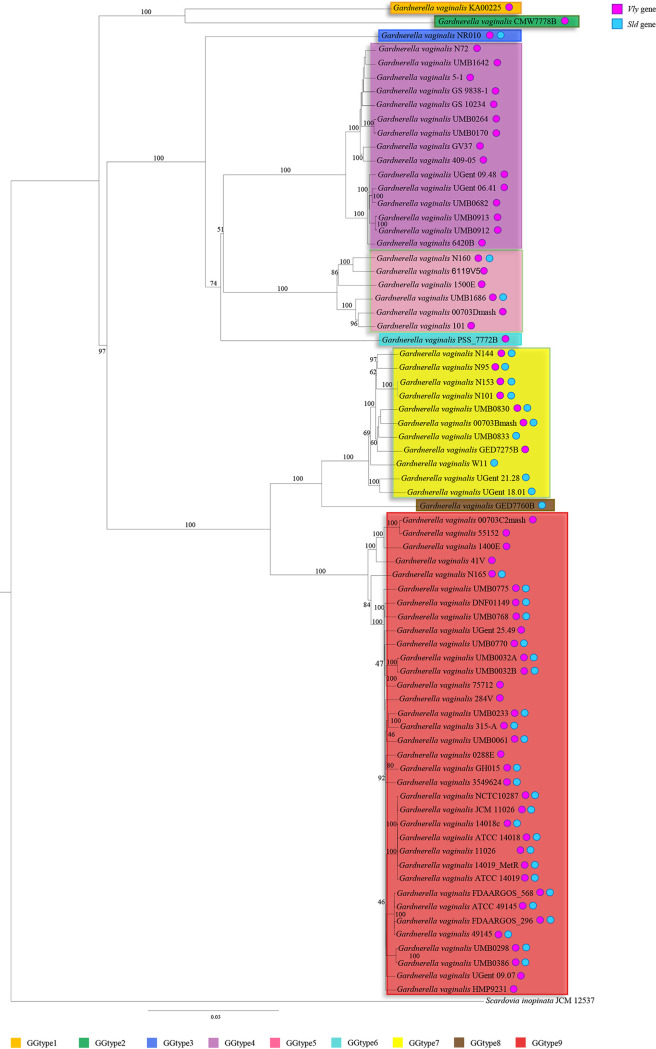
As previously mentioned, the Gardnerella genus is currently considered to be composed of just one species, G. vaginalis (41). Over time, since its discovery, G. vaginalis was renamed repeatedly. This complicated taxonomic classification history provides an idea of the difficult challenge faced due to considerable diversity within this species. In recent years, phylogenetic analysis based on comparison of chaperonin-60 (cpn60) sequences identified four subgroups within 112 G. vaginalis isolates (42). In contrast, analyses employing the bacterial 16S rRNA gene sequences did not give reliable support for a species-level resolution. To date, clear species identification events and the resultant presence of different species within the Gardnerella genus remain undiscovered. In this context, genomic comparisons represent a powerful in silico approach to highlight genomic differences between G. vaginalis strains, also contributing to its taxonomic classification. To infer the possible existence of phylogenomic-based clades within this species, the set of genes representing the core genome of G. vaginalis species was employed to perform a phylogenomic comparison. Specifically, we computed a phylogenetic tree based on the concatenation of 334 amino acid sequences (Fig. 2). Selected orthologous sequences were collected for previous genome comparison of all the chromosomes of 72 strains of G. vaginalis, together with the genome sequences of Scardovia inopinata JCM 12537 as a representative outgroup (Fig. 2). Remarkably, the resulting tree showed that most of the 72 G. vaginalis strains were grouped in two main clusters sharing the same phylogenetic branch. Moreover, within each cluster, it was possible to identify two additional groups, suggesting the existence of four putative different Gardnerella taxa (Fig. 2). Interestingly, G. vaginalis KA00225 and G. vaginalis CMW7778B were placed on separate branches with respect to other G. vaginalis strains, highlighting a polyphyletic evolutionary history of this species.
FIG 2.
Phylogenomic tree of G. vaginalis. A proteomic tree was constructed based on the concatenation of 334 G. vaginalis core genes identified in the pan-genome analysis of the 72 G. vaginalis strains. The tree was built by the neighbor-joining method, and bootstrap percentages above 50 are shown at node points, based on 1,000 replicates. Phylogenetic clusters of different genotypes are highlighted in different colors. Colored circles represent the occurrence of the vly (dark pink) and sld (light blue) genes in the corresponding G. vaginalis genomes.
In order to further explore the genomic differences among members of the G. vaginalis taxon, the pairwise percent average nucleotide identity (ANI) was assessed, resulting in values ranging from 99.9% to 81.5% (Table S4). Notably, previous studies employing ANI analysis to taxonomically distinct species of the Bifidobacteriaceae family identified an ideal ANI threshold value of 94% (27, 43). The analysis of ANI values among members of G. vaginalis revealed that the collected genome sequences fall into four main groups, within which ANI values were found above the species-level cutoff threshold of 94%. Conversely, genomes belonging to G. vaginalis strains GED7760B, PSS_7772B, CMW7778B, KA00225, and NR010 exhibited ANI values lower than 92% against each analyzed strain, highlighting another five putative different species of the Gardnerella genus (Table S4). These findings, together with data generated from the phylogenetic tree reconstruction, strongly support the existence of an extensive level of genomic variability between G. vaginalis strains and would cast doubt on the presence of a single species within this genus. Specifically, the calculation of ANI values allowed us to identify nine Gardnerella genotypes (GGtype1 to GGtype9), corresponding to putative different Gardnerella taxa (Fig. 2). Moreover, combining the genomic information related to the identified G. vaginalis virulence factors, we observed a heterogeneous distribution across the phylogenomic tree. In particular, the sialidase gene was detected almost exclusively in the genome sequences of G. vaginalis strains belonging to GGtype7 and GGtype9, together with the strains GED7760B and NR010, representative of GGtype8 and GGtype3, respectively (Fig. 2), suggesting these may be virulent genotypes. Conversely, the genomes of GGtype1, GGtype2, GGtype4, GGtype5, and GGtype6 appeared to lack the sialidase gene, revealing that these may be less virulent. Previous findings supported the existence of G. vaginalis strains that can provoke severe damage to the vaginal integrity through their ability to develop microbial biofilm and others that are linked with an asymptomatic medical condition (19). Thus, our results highlighted how G. vaginalis strains encoding sialidase might be phylogenetically related, reinforcing the notion of a putative subdivision in potentially pathogenic and commensal strains (Fig. 2).
Assessing the prevalence and the abundance of G. vaginalis genotypes among the human population.
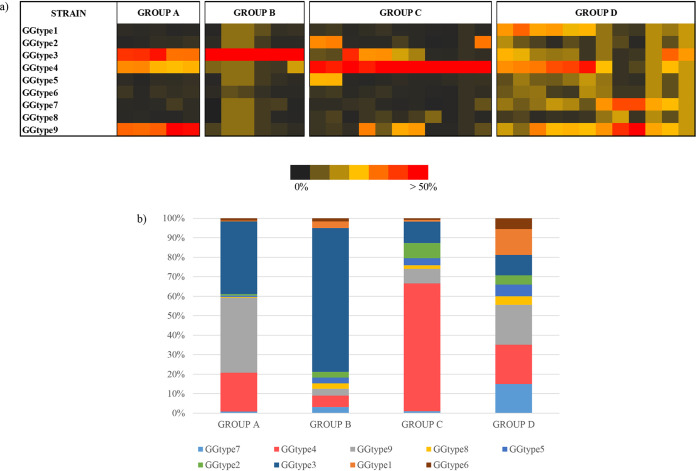
Our genome-based analyses demonstrated that G. vaginalis species consists of separate subgroups. In light of the above findings, we assessed the composition of the vaginal microbiota of 175 women, aiming to investigate the prevalence and the distribution of the nine G. vaginalis genotypes among the human population. Specifically, a preliminary survey was performed to evaluate the overall vaginal microbiota composition of the collected 175 vaginal samples, displaying an abundance of G. vaginalis taxon above 5% in 20% of the samples (see Materials and Methods). Samples that did not reach such threshold were discarded, resulting in a final collection of 34 metagenomic data sets, showing an abundance of G. vaginalis genomic reads ranging from 6.01% to 86.41%. Notably, six of the collected metagenomic data sets were obtained from vaginal samples of healthy pregnant women. In contrast, for the vast majority of the remaining 28 samples, it was not possible to get enough information regarding the health conditions of the subjects since the corresponding metadata were not available. Thereafter, these metagenomic data sets were assayed for the presence of the nine genotypes of G. vaginalis identified above, employing genome sequences belonging to strains KA00225, CMW7778B, NR010, UMB0264, 6119V5, PSS_7772B, 00703Bmash, GED7760B, and FDAARGOS_568 as representatives of each genotype. The minimum coverage of each gene was calculated based on the metagenomics reads with at least 99% full-length identity (see Materials and Methods). As displayed in Fig. 3, considering the uneven distribution and abundance of the nine G. vaginalis genotypes, it was possible to delineate four groups overall within the collected vaginal samples. In particular, G. vaginalis communities with a predominance of a single genotype were identified within 16 metagenomic data sets. More specifically, the latter showed a predominance of GGtype4 in group C (n = 10) and GGtype3 in group B (n = 6), with an average percentage of metagenomic reads of 66.03% and 74.91%, respectively. Moreover, group A (n = 5) was mainly constituted by a combination of the latter two genotypes together with GGtype9 (Fig. 3). Interestingly, GGtype9 and GGtype3 were identified as putative virulence genotypes; thus, their presence in the vaginal microbiota could be linked with possible adverse health effects (Fig. 2). Conversely, GGtype4, which was found dominant in group C, seems to have less virulence potential since it does not contain the sialidase gene.
FIG 3.
Metagenomic abundance of the different G. vaginalis genotypes. (a) Prevalence and distribution of the nine G. vaginalis genotypes observed in the 34 metagenome vaginal samples. (b) Average abundance of reads of each G. vaginalis genotype in the individuated four groups.
These findings allowed us to observe that different genotypes can be found within the female population, postulating their different impact on the vaginal environment. Moreover, these results may be consistent with the notion that G. vaginalis species can lead to a symptomatic unbalanced state of the vaginal microbiota in some instances and behave as natural commensal in others (18). Nevertheless, the involvement of specific genotypes in the development of significant clinical conditions should be investigated more in-depth in future metagenomic analyses that also include BV-positive vaginal microbiota samples.
Notably, all the metagenomic data sets from pregnant women fall in the same group (group D), characterized by the absence of a single predominant genotype. In fact, our results showed that the vaginal microbiota of pregnant women harbors a greater G. vaginalis biodiversity than that typical of nonpregnant women. It is known that during the late gestational period, the microbiome undergoes significant strain-level variation, and the physiological state of pregnancy may have an impact also on the structure of G. vaginalis communities (44).
Phylogenomic evaluation of G. vaginalis within Bifidobacteriaceae.
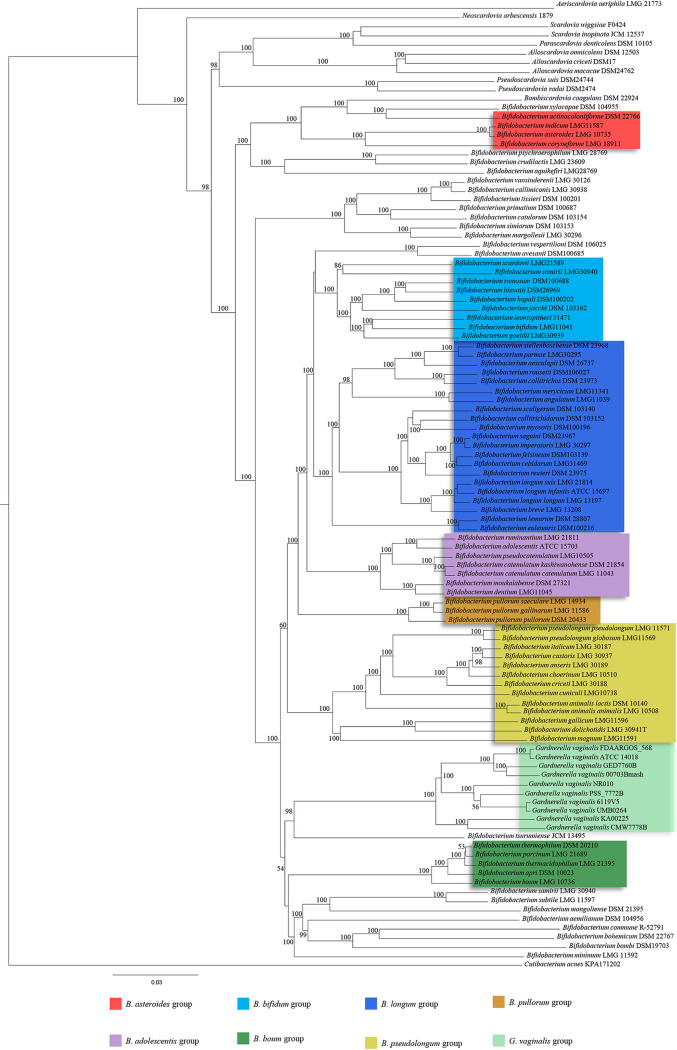
To date, G. vaginalis is considered a member of the Bifidobacteriaceae family since close relationships among this species and Bifidobacterium spp., based on 16S rRNA gene sequencing, were observed (45). Aiming to investigate the positioning of G. vaginalis within the Bifidobacteriaceae family, we performed a further phylogenetic analysis, including one representative strain for each genotype of G. vaginalis identified above by means of ANI values calculation, i.e., strains KA00225, CMW7778B, NR010, UMB0264, 6119V5, PSS_7772B, 00703Bmash, GED7760B, and FDAARGOS_568. These strains, together with the 96 type strains of the Bifidobacteriaceae family (bifidobacterial as well as nonbifidobacterial taxa) and Cutibacterium acnes KPA171202 as an outgroup, were employed to perform a comparative genomics analysis aimed to identify ubiquitously conserved protein sequences. The concatenation of 91 amino acid sequences shared between all considered genomes was used to construct the phylogenetic tree of the Bifidobacteriaceae family (Fig. 4). This analysis showed that most of the nodes were supported by 100% of the bootstrap values, validating the reliability of the phylogenetic tracing and robustness of the results. In accordance with a previous study, the obtained tree showed that Bifidobacterium spp. are separated from nonbifidobacterial taxa, belonging to the genera Scardovia, Parascardovia, and Alloscardovia (27). Furthermore, these latter represent the deepest branches of the Bifidobacteriaceae family tree and therefore evidenced a very early separation in the evolution of this family. Focusing on the G. vaginalis genotypes, this phylogenetic investigation highlighted its evolutionary positioning within the Bifidobacterium genus. More specifically, Bifidobacterium tsurumiense was identified as the phylogenetically closest-related taxon to G. vaginalis. Furthermore, the type strain G. vaginalis ATCC 14019 exhibits tight phylogenetical grouping with strains belonging to GGtype9, consistently with our ANI-based findings (see above). Interestingly, the nine G. vaginalis strains representative of the many related putative species are grouped, giving rise to a new cluster located alongside the previously described Bifidobacterium boum group (46). Overall, these findings clearly showed that employing a robust phylogenomic based approach, the G. vaginalis species resulted in being identified along with some currently classified Bifidobacterium species, thus suggesting the need for a reevaluation of the currently known taxonomy of the Bifidobacterium genus.
FIG 4.
Phylogenomic tree of the Bifidobacteriaceae family. The proteomic tree is based on the concatenation of 91 core genes shared by members of the Bifidobacteriaceae family. The tree was constructed by the neighbor-joining method, and bootstrap percentages above 50 are shown at node points, based on 1,000 replicates. Phylogenetic groups are highlighted in different colors. The G. vaginalis cluster is highlighted in light green.
In conclusion, the high degree of genetic heterogeneity observed among members of Gardnerella vaginalis has been investigated, suggesting inaccuracy in the current taxonomic classification that consists of a single species within the Gardnerella genus. In this study, through an exhaustive phylogenomic and comparative genomic analysis employing 72 publicly available G. vaginalis genome sequences, we identified nine different Gardnerella genotypes (GGtype1 to GGtype9). Notably, within the Bifidobacteriaceae family, G. vaginalis is phylogenetically located alongside the Bifidobacterium boum group (46), casting doubt on its current taxonomic classification due to the relatedness with other bifidobacterial species. Furthermore, the characterization of the pan-genome of G. vaginalis allowed us to obtain insights into the adaptation mechanisms to the vaginal environment. Our data showed that genes encoding collagen and glycogen utilization functions were ubiquitous genetic elements, while virulence-associated ones exhibited an uneven distribution among genotypes. Notably, among G. vaginalis genes encoding virulence factor, sialidase is especially noteworthy due to its involvement in the degradation of the vaginal mucosal layer as well as microbial biofilm formation (15). The latter gene was identified between members of four genotypes, i.e., GGtype3, GGtype7, GGtype8, as well as GGtype9, allowing to discern those genotypes with the highest putative virulence capability and potentially linked with major adverse health outcomes. Interestingly, the microbial profiling of the vaginal microbiota of 34 women allowed us to identify GGtype3 and GGtype9 as sialidase positive, as well as GGtype4, which conversely lacks the sialidase gene, as the most abundant genotypes among the human population. These findings are in line with previous studies since both pathogenic and commensal G. vaginalis strains have been previously described (11).
MATERIALS AND METHODS
Gardnerella vaginalis and Bifidobacteriaceae genome sequences.
Genome sequences of G. vaginalis strains were retrieved from the National Center for Biotechnology Information (NCBI) public database, resulting in 107 available genomes. Moreover, incomplete genomes (genome size less than 1.4 Mb) as well as genome sequences that exhibited low sequencing quality (genome coverage lower than 30× or containing unspecified nucleotide bases in conformity to IUPAC nomenclature), were discarded. Furthermore, a comparison of the G. vaginalis genome sequences was performed to evaluate the average nucleotide identity (ANI) values for each genome with respect to the genome of G. vaginalis ATCC 14018, which is the type strain of this species. Based on this analysis, there were inconsistencies in the predicted taxonomy of two strains belonging to the Lactobacillus genus, i.e., G. vaginalis UMB0388 and G. vaginalis MGYG-HGUT-00021. Finally, collected high-quality genome sequences of 72 G. vaginalis (Table 1) were compared to each other. Additional genomic and phylogenomic analyses were performed employing 96 type strains of the Bifidobacteriaceae family retrieved from the NCBI database, including 84 bifidobacterial genome sequences and 12 nonbifidobacterial genome sequences (27, 46) (Table S1 in the supplemental material).
Genome annotation.
In order to obtain comparable quality standards for the analyzed genomes, the 72 G. vaginalis genome sequences retrieved from the NCBI database were submitted to annotation employing the MEGAnnotator pipeline (47). Protein-encoding open reading frames (ORFs) were predicted using Prodigal (48). tRNA genes were detected using tRNAscan-SE v1.4 (49), while rRNA genes were identified using RNAmmer v1.2 (50). Outcomes of the gene-finder program were combined with data from RAPSearch2 analysis (Reduced Alphabet based Protein similarity Search) (51) of a nonredundant protein database provided by the NCBI and hidden Markov model profile (HMM) search (http://hmmer.org/) in the manually curated Pfam-A protein family database (52). Results were examined by Artemis (53), which was used for validating predicted genes and, where required, for genome manual editing consisting of removal or addition of coding regions as well as a redefinition of gene starts.
Virulence gene identification.
In order to perform a screening among genomes of the 72 G. vaginalis strains, amino acid sequences of nonredundant WP accessions, i.e., a CDC vaginolysin and 26 exo-alpha-sialidases, were retrieved from the Identical Protein Groups (IPG) resource of the NCBI database (https://www.ncbi.nlm.nih.gov/ipg/). Then, putative CDC vaginolysin and sialidase genes were identified through BLASTP analysis (E value cutoff of 1E−5) (54). A subsequent manual inspection of the resulting aligned proteins based on their amino acid sequence identity (greater than 42%), combined with the alignment length (more than 500 amino acids), allowed us to discard false positives from the prediction (Table 1). In addition, careful scrutiny of G. vaginalis core genes and the genomic regions adjacent to both ends of the identified mobile elements allowed us to discover further virulence traits. Such outcomes were subsequentially validated through the cross-examination of the virulence factor database (VFDB) (55).
Prophages and IS element identification.
The 72 G. vaginalis genomes were screened for prophage-associated genes using a custom database employing BLASTP analysis (54) (E value cutoff of 1E−5). The custom database was assembled through previously bifidoprophage-validated sequences retrieved from 60 bifidoprophages previously described (56). Then, a manual examination of the DNA region surrounding a putative prophage-encoding gene was performed, allowing the identification of complete prophage-like sequences (Table S2). Moreover, the same G. vaginalis genomes were also screened for the presence of IS elements (57) through the IS Finder online tool (https://isfinder.biotoul.fr/) (Table S2).
G. vaginalis pan-genome analysis.
A pan-genome calculation employing 72 genomes of G. vaginalis was performed using the PGAP (pan-genome analysis pipeline) (58). Predicted ORFs were organized into functional clusters employing the GF (gene family) method, which consists of a similarity search between each protein pair through BLAST analysis (cutoff E value of 1 × 10−10 and 50% identity over at least 80% of both protein sequences). Following this, a clustering in protein families of orthologous genes was performed using MCL (graph theory-based Markov clustering algorithm) (59). A pan-genome profile was built using an optimized algorithm integrated into PGAP software, based on a presence/absence matrix that included all protein families of orthologous genes identified in the analyzed genomes. Subsequently, the unique protein families for each of 72 G. vaginalis genomes were identified. Protein families shared between all genomes allowed us to build the core genome of the G. vaginalis species, defined by selecting the families that contained at least one protein member for each genome. A different pan- and core- genome analysis was performed on the 96 Bifidobacteriaceae type strains as described above, including G. vaginalis ATCC 14018, identifying 135 COGs belonging to the core genome of this family. Afterward, in order to obtain the core genes of G. vaginalis that were not shared with other members of the Bifidobacteriaceae family, the 135 gene sequences attributed to G. vaginalis ATCC 14018 were used to remove the corresponding COGs from the core genome of G. vaginalis species.
Phylogenomic comparison between G. vaginalis strains and their positioning within the Bifidobacteriaceae family.
In order to assess genome differences between G. vaginalis strains, a phylogenetic comparison involving the 72 genome sequences retrieved from NCBI was performed. For this purpose, the concatenated core genome sequences were aligned using MAFFT (60), and the resulting phylogenetic tree was constructed using the neighbor-joining method in Clustal W v2.1 (61). A visual core genome tree was developed using FigTree software (http://tree.bio.ed.ac.uk/software/figtree/). A value for the average nucleotide identity (ANI) was calculated for each genome pair using the fastANI software (62).
A further phylogenomic analysis, aiming to evaluate the phylogenetic position of G. vaginalis within the family Bifidobacteriaceae, was executed on 72 G. vaginalis genome sequences together with 96 Bifidobacteriaceae type strains as described above.
Whole-genome sequencing data collection and analysis.
The publicly available vaginal metagenomic data sets were retrieved from NCBI (BioProject accession no. PRJEB24147, PRJNA352475, PRJNA361427, PRJNA576566, and PRJNA379120). Specifically, we selected Illumina whole-genome shotgun (WGS) sequencing data concerning vaginal samples from midvagina and cervix swabs of fertile pregnant, as well as nonpregnant, women. The resulting 175 vaginal metagenomic data sets were analyzed through a shallow shotgun metagenomics approach (63), allowing us to achieve high taxonomic resolution at the species level. In order to reconstruct the microbiota composition of vaginal samples, the fastq files of the paired-end reads were used as input for the genome assemblies through the METAnnotatorX pipeline (64). The SPAdes software was used for de novo assembly of each genome sequence (65). To assess the distribution of the different G. vaginalis genotypes among the human population, the samples showing a relative abundance of this species below 5% were discarded. In fact, it was observed that below this threshold level, the number of G. vaginalis reads within samples was not enough to ensure a reasonable mapping accuracy of genotypes (see below). Afterward, the genome sequences belonging to the nine strains representative of as many G. vaginalis genotypes identified in this study were aligned with WGS reads. Metagenomics data sets were filtered by use of the fastq-mcf script (https://expressionanalysis.github.io/ea-utils/) (minimum mean quality score, 20; window size, 5 bp; quality threshold, 25; and minimum length, 80 bp) to obtain high-quality reads. Collected reads were aligned against the human genome using the Burrows-Wheeler Aligner program (66) (BWA-MEM algorithm with trigger reseeding, 1.5; minimum seed length, 19; matching score, 1; mismatch penalty, 4; gap open penalty, 6; and gap extension penalty, 1) and processed with the SAMtools software package (67), aiming to remove human reads. The final mapping against the genome sequences of the G. vaginalis genotypes was performed using Bowtie 2 (68) through multiple-hit mapping and “very-sensitive” policy. The mapping was performed using a minimum score threshold function (–score-min C,−13,0) to limit reads of arbitrary length to two mismatches and retain those matches with at least 99% full-length identity. HTSeq software (69) (running in union mode) was employed to calculate read counts corresponding to the G. vaginalis genotypes.
Supplementary Material
ACKNOWLEDGMENTS
We thank GenProbio Srl for the financial support of the Laboratory of Probiogenomics.
Part of this research was conducted using the High Performance Computing (HPC) facility of the University of Parma.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bradford LL, Ravel J. 2017. The vaginal mycobiome: a contemporary perspective on fungi in women's health and diseases. Virulence 8:342–351. doi: 10.1080/21505594.2016.1237332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taha TE, Hoover DR, Dallabetta GA, Kumwenda NI, Mtimavalye LA, Yang LP, Liomba GN, Broadhead RL, Chiphangwi JD, Miotti PG. 1998. Bacterial vaginosis and disturbances of vaginal flora: association with increased acquisition of HIV. AIDS 12:1699–1706. doi: 10.1097/00002030-199813000-00019. [DOI] [PubMed] [Google Scholar]
- 3.Sobel JD. 1999. Is there a protective role for vaginal flora? Curr Infect Dis Rep 1:379–383. doi: 10.1007/s11908-999-0045-z. [DOI] [PubMed] [Google Scholar]
- 4.Wiesenfeld HC, Hillier SL, Krohn MA, Landers DV, Sweet RL. 2003. Bacterial vaginosis is a strong predictor of Neisseria gonorrhoeae and Chlamydia trachomatis infection. Clin Infect Dis 36:663–668. doi: 10.1086/367658. [DOI] [PubMed] [Google Scholar]
- 5.Skarin A, Sylwan J. 1986. Vaginal lactobacilli inhibiting growth of Gardnerella vaginalis, Mobiluncus and other bacterial species cultured from vaginal content of women with bacterial vaginosis. Acta Pathol Microbiol Immunol Scand B 94:399–403. doi: 10.1111/j.1699-0463.1986.tb03074.x. [DOI] [PubMed] [Google Scholar]
- 6.Nam H, Whang K, Lee Y. 2007. Analysis of vaginal lactic acid producing bacteria in healthy women. J Microbiol 45:515–520. [PubMed] [Google Scholar]
- 7.Oh HY, Kim BS, Seo SS, Kong JS, Lee JK, Park SY, Hong KM, Kim HK, Kim MK. 2015. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect 21:674.e1–674.e9. doi: 10.1016/j.cmi.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 8.Lee JE, Lee S, Lee H, Song YM, Lee K, Han MJ, Sung J, Ko G. 2013. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 8:e63514. doi: 10.1371/journal.pone.0063514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner HL, Dukes CD. 1955. Haemophilus vaginalis vaginitis: a newly defined specific infection previously classified “nonspecific” vaginitis. Am J Obstet Gynecol 69:962–976. doi: 10.1016/0002-9378(55)90095-8. [DOI] [PubMed] [Google Scholar]
- 11.Aroutcheva AA, Simoes JA, Behbakht K, Faro S. 2001. Gardnerella vaginalis isolated from patients with bacterial vaginosis and from patients with healthy vaginal ecosystems. Clin Infect Dis 33:1022–1027. doi: 10.1086/323030. [DOI] [PubMed] [Google Scholar]
- 12.Ravel J, Brotman RM, Gajer P, Ma B, Nandy M, Fadrosh DW, Sakamoto J, Koenig SS, Fu L, Zhou X, Hickey RJ, Schwebke JR, Forney LJ. 2013. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 1:29. doi: 10.1186/2049-2618-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy L, Jespers V, Dahchour N, Mwambarangwe L, Musengamana V, Vaneechoutte M, Crucitti T. 2015. Unravelling the bacterial vaginosis-associated biofilm: a multiplex Gardnerella vaginalis and Atopobium vaginae fluorescence in situ hybridization assay using peptide nucleic acid probes. PLoS One 10:e0136658. doi: 10.1371/journal.pone.0136658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsang A, Bradbury JM. 1981. Separation and properties of prestalk and prespore cells of Dictyostelium discoideum. Exp Cell Res 132:433–441. doi: 10.1016/0014-4827(81)90118-X. [DOI] [PubMed] [Google Scholar]
- 15.Hardy L, Jespers V, Van den Bulck M, Buyze J, Mwambarangwe L, Musengamana V, Vaneechoutte M, Crucitti T. 2017. The presence of the putative Gardnerella vaginalis sialidase A gene in vaginal specimens is associated with bacterial vaginosis biofilm. PLoS One 12:e0172522. doi: 10.1371/journal.pone.0172522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soong G, Muir A, Gomez MI, Waks J, Reddy B, Planet P, Singh PK, Kaneko Y, Kanetko Y, Wolfgang MC, Hsiao Y-S, Tong L, Prince A. 2006. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J Clin Invest 116:2297–2305. doi: 10.1172/JCI27920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelber SE, Aguilar JL, Lewis KL, Ratner AJ. 2008. Functional and phylogenetic characterization of vaginolysin, the human-specific cytolysin from Gardnerella vaginalis. J Bacteriol 190:3896–3903. doi: 10.1128/JB.01965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janulaitiene M, Paliulyte V, Grinceviciene S, Zakareviciene J, Vladisauskiene A, Marcinkute A, Pleckaityte M. 2017. Prevalence and distribution of Gardnerella vaginalis subgroups in women with and without bacterial vaginosis. BMC Infect Dis 17:394. doi: 10.1186/s12879-017-2501-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornejo OE, Hickey RJ, Suzuki H, Forney LJ. 2018. Focusing the diversity of Gardnerella vaginalis through the lens of ecotypes. Evol Appl 11:312–324. doi: 10.1111/eva.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwich MD Jr, Alves JM, Buck GA, Strauss JF III, Patterson JL, Oki AT, Girerd PH, Jefferson KK. 2010. Drawing the line between commensal and pathogenic Gardnerella vaginalis through genome analysis and virulence studies. BMC Genomics 11:375. doi: 10.1186/1471-2164-11-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zinnemann K, Turner GC. 1963. The taxonomic position of “Haemophilus vaginalis” [Corynebacterium vaginale]. J Pathol 85:213–219. doi: 10.1002/path.1700850120. [DOI] [Google Scholar]
- 22.Pickett J. 1980. Transfer of Haemophilus vaginalis Gardner and Dukes to a new genus, Gardnerella: G. vaginalis (Gardner and Dukes) comb. nov. Int J Syst Evol Microbiol 30. doi: 10.1099/00207713-30-1-170. [DOI] [Google Scholar]
- 23.Piot P, van Dyck E, Goodfellow M, Falkow S. 1980. A taxonomic study of Gardnerella vaginalis (Haemophilus vaginalis) Gardner and Dukes 1955. J Gen Microbiol 119:373–396. doi: 10.1099/00221287-119-2-373. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed A, Earl J, Retchless A, Hillier SL, Rabe LK, Cherpes TL, Powell E, Janto B, Eutsey R, Hiller NL, Boissy R, Dahlgren ME, Hall BG, Costerton JW, Post JC, Hu FZ, Ehrlich GD. 2012. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J Bacteriol 194:3922–3937. doi: 10.1128/JB.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pleckaityte M, Janulaitiene M, Lasickiene R, Zvirbliene A. 2012. Genetic and biochemical diversity of Gardnerella vaginalis strains isolated from women with bacterial vaginosis. FEMS Immunol Med Microbiol 65:69–77. doi: 10.1111/j.1574-695X.2012.00940.x. [DOI] [PubMed] [Google Scholar]
- 26.Vaneechoutte M, Guschin A, Van Simaey L, Gansemans Y, Van Nieuwerburgh F, Cools P. 2019. Emended description of Gardnerella vaginalis and description of Gardnerella leopoldii sp. nov., Gardnerella piotii sp. nov. and Gardnerella swidsinskii sp. nov., with delineation of 13 genomic species within the genus Gardnerella. Int J Syst Evol Microbiol 69:679–687. doi: 10.1099/ijsem.0.003200. [DOI] [PubMed] [Google Scholar]
- 27.Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, Mangifesta M, Ferrario C, Modesto M, Mattarelli P, Jiří K, van Sinderen D, Ventura M. 2017. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18:568. doi: 10.1186/s12864-017-3955-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutta C, Paul S. 2012. Microbial lifestyle and genome signatures. Curr Genomics 13:153–162. doi: 10.2174/138920212799860698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schellenberg JJ, Patterson MH, Hill JE. 2017. Gardnerella vaginalis diversity and ecology in relation to vaginal symptoms. Res Microbiol 168:837–844. doi: 10.1016/j.resmic.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto TA, Gerdes K, Tunnacliffe A. 2002. Bacterial toxin RelE induces apoptosis in human cells. FEBS Lett 519:191–194. doi: 10.1016/S0014-5793(02)02764-3. [DOI] [PubMed] [Google Scholar]
- 31.Grossman TH. 2016. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med 6:a025387. doi: 10.1101/cshperspect.a025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Touchon M, Rocha EP. 2007. Causes of insertion sequences abundance in prokaryotic genomes. Mol Biol Evol 24:969–981. doi: 10.1093/molbev/msm014. [DOI] [PubMed] [Google Scholar]
- 33.Santiago GL, Deschaght P, El Aila N, Kiama TN, Verstraelen H, Jefferson KK, Temmerman M, Vaneechoutte M. 2011. Gardnerella vaginalis comprises three distinct genotypes of which only two produce sialidase. Am J Obstet Gynecol 204:450 e1-7. doi: 10.1016/j.ajog.2010.12.061. [DOI] [PubMed] [Google Scholar]
- 34.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. 2005. The microbial pan-genome. Curr Opin Genet Dev 15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Lugli GA, Duranti S, Albert K, Mancabelli L, Napoli S, Viappiani A, Anzalone R, Longhi G, Milani C, Turroni F, Alessandri G, Sela DA, van Sinderen D, Ventura M. 2019. Unveiling genomic diversity among members of the species Bifidobacterium pseudolongum, a widely distributed gut commensal of the animal kingdom. Appl Environ Microbiol 85:e03065-18. doi: 10.1128/AEM.03065-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Callaghan A, Bottacini F, O’Connell Motherway M, van Sinderen D. 2015. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genomics 16:832. doi: 10.1186/s12864-015-1968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakamoto T, Otokawa T, Kono R, Shigeri Y, Watanabe K. 2013. A C69-family cysteine dipeptidase from Lactobacillus farciminis JCM1097 possesses strong Gly-Pro hydrolytic activity. J Biochem 154:419–427. doi: 10.1093/jb/mvt069. [DOI] [PubMed] [Google Scholar]
- 38.van der Veer C, Hertzberger RY, Bruisten SM, Tytgat HLP, Swanenburg J, de Kat Angelino-Bart A, Schuren F, Molenaar D, Reid G, de Vries H, Kort R. 2019. Comparative genomics of human Lactobacillus crispatus isolates reveals genes for glycosylation and glycogen degradation: implications for in vivo dominance of the vaginal microbiota. Microbiome 7:49. doi: 10.1186/s40168-019-0667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeoman CJ, Yildirim S, Thomas SM, Durkin AS, Torralba M, Sutton G, Buhay CJ, Ding Y, Dugan-Rocha SP, Muzny DM, Qin X, Gibbs RA, Leigh SR, Stumpf R, White BA, Highlander SK, Nelson KE, Wilson BA. 2010. Comparative genomics of Gardnerella vaginalis strains reveals substantial differences in metabolic and virulence potential. PLoS One 5:e12411. doi: 10.1371/journal.pone.0012411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Morrison AJ, Thibodeau PH. 2015. Interdomain contacts and the stability of serralysin protease from Serratia marcescens. PLoS One 10:e0138419. doi: 10.1371/journal.pone.0138419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castro J, Jefferson KK, Cerca N. 2020. Genetic heterogeneity and taxonomic diversity among Gardnerella species. Trends Microbiol 28:202–211. doi: 10.1016/j.tim.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Schellenberg JJ, Paramel Jayaprakash T, Withana Gamage N, Patterson MH, Vaneechoutte M, Hill JE. 2016. Gardnerella vaginalis subgroups defined by cpn60 sequencing and sialidase activity in isolates from Canada, Belgium and Kenya. PLoS One 11:e0146510. doi: 10.1371/journal.pone.0146510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A, Mancabelli L, Mangifesta M, Taminiau B, Delcenserie V, van Sinderen D, Ventura M. 2014. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol 80:6383–6394. doi: 10.1128/AEM.02004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goltsman DSA, Sun CL, Proctor DM, DiGiulio DB, Robaczewska A, Thomas BC, Shaw GM, Stevenson DK, Holmes SP, Banfield JF, Relman DA. 2018. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res 28:1467–1480. doi: 10.1101/gr.236000.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leblond-Bourget N, Philippe H, Mangin I, Decaris B. 1996. 16S rRNA and 16S to 23S internal transcribed spacer sequence analyses reveal inter- and intraspecific Bifidobacterium phylogeny. Int J Syst Bacteriol 46:102–111. doi: 10.1099/00207713-46-1-102. [DOI] [PubMed] [Google Scholar]
- 46.Lugli GA, Milani C, Duranti S, Mancabelli L, Mangifesta M, Turroni F, Viappiani A, van Sinderen D, Ventura M. 2018. Tracking the taxonomy of the genus Bifidobacterium based on a phylogenomic approach. Appl Environ Microbiol 84:e02249-17. doi: 10.1128/aem.02249-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 48.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao Y, Tang H, Ye Y. 2012. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 55.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lugli GA, Milani C, Turroni F, Tremblay D, Ferrario C, Mancabelli L, Duranti S, Ward DV, Ossiprandi MC, Moineau S, van Sinderen D, Ventura M. 2016. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol 18:2196–2213. doi: 10.1111/1462-2920.13154. [DOI] [PubMed] [Google Scholar]
- 57.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–6. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 62.Jain C, Rodriguez RL, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hillmann B, Al-Ghalith GA, Shields-Cutler RR, Zhu Q, Gohl DM, Beckman KB, Knight R, Knights D. 2018. Evaluating the information content of shallow shotgun metagenomics. mSystems 3:e00069-18. doi: 10.1128/mSystems.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Milani C, Casey E, Lugli GA, Moore R, Kaczorowska J, Feehily C, Mangifesta M, Mancabelli L, Duranti S, Turroni F, Bottacini F, Mahony J, Cotter PD, McAuliffe FM, van Sinderen D, Ventura M. 2018. Tracing mother-infant transmission of bacteriophages by means of a novel analytical tool for shotgun metagenomic datasets: METAnnotatorX. Microbiome 6:145. doi: 10.1186/s40168-018-0527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anders S, Pyl PT, Huber W. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.