Third-generation cephalosporins (3GCs) are critically important antibacterials, and 3GC resistance (3GC-R) threatens human health, particularly in the context of opportunistic pathogens such as Escherichia coli. There is some evidence for zoonotic transmission of 3GC-R E. coli through food, but little work has been done examining possible transmission via interaction of people with the local near-farm environment. We characterized acquired 3GC-R E. coli found on dairy farms in a geographically restricted region of the United Kingdom and compared these with E. coli from people living in the same region, collected in parallel. While there is strong evidence for recent farm-to-farm transmission of 3GC-R strains and plasmids—including one epidemic plasmid that has a remarkable capacity to be transmitted—there was no evidence that 3GC-R E. coli found on study farms had a significant impact on circulating 3GC-R E. coli strains or plasmids in the local human population.
KEYWORDS: antibiotic resistance, phylogenetic analysis, plasmid analysis, zoonotic infections
ABSTRACT
Third-generation cephalosporin resistance (3GC-R) in Escherichia coli is a rising problem in human and farmed-animal populations. We conducted whole-genome sequencing analysis of 138 representative 3GC-R isolates previously collected from dairy farms in southwest England and confirmed by PCR to carry acquired 3GC-R genes. This analysis identified blaCTX-M (131 isolates encoding CTX-M-1, -14, -15, -and 32 and the novel variant CTX-M-214), blaCMY-2 (6 isolates), and blaDHA-1 (1 isolate). A highly conserved plasmid was identified in 73 isolates, representing 27 E. coli sequence types. This novel ∼220-kb IncHI2 plasmid carrying blaCTX-M-32 was sequenced to closure and designated pMOO-32. It was found experimentally to be stable in cattle and human transconjugant E. coli even in the absence of selective pressure and was found by multiplex PCR to be present on 26 study farms representing a remarkable range of transmission over 1,500 square kilometers. However, the plasmid was not found among human urinary E. coli isolates we recently characterized from people living in the same geographical location, collected in parallel with farm sampling. There were close relatives of two blaCTX-M plasmids circulating among eight human and two cattle isolates, and a closely related blaCMY-2 plasmid was found in one cattle and one human isolate. However, phylogenetic evidence of recent sharing of 3GC-R strains between farms and humans in the same region was not found.
IMPORTANCE Third-generation cephalosporins (3GCs) are critically important antibacterials, and 3GC resistance (3GC-R) threatens human health, particularly in the context of opportunistic pathogens such as Escherichia coli. There is some evidence for zoonotic transmission of 3GC-R E. coli through food, but little work has been done examining possible transmission via interaction of people with the local near-farm environment. We characterized acquired 3GC-R E. coli found on dairy farms in a geographically restricted region of the United Kingdom and compared these with E. coli from people living in the same region, collected in parallel. While there is strong evidence for recent farm-to-farm transmission of 3GC-R strains and plasmids—including one epidemic plasmid that has a remarkable capacity to be transmitted—there was no evidence that 3GC-R E. coli found on study farms had a significant impact on circulating 3GC-R E. coli strains or plasmids in the local human population.
INTRODUCTION
Third-generation cephalosporin-resistant (3GC-R) Escherichia coli isolates have been increasingly reported in both animal and human populations and are considered pathogens of major concern for humans (1, 2). 3GCs such as cefotaxime and ceftazidime have been listed by the World Health Organization (WHO) as “highest-priority critically important antimicrobials” (HP-CIAs) because of their importance for human health (3). Resistance to 3GCs in E. coli can be caused by several mechanisms but is primarily attributed to the acquisition of extended-spectrum β-lactamases (ESBLs) or plasmid-mediated AmpC β-lactamases (pAmpCs) (4). Plasmids encoding ESBLs and pAmpCs frequently harbor additional resistance genes and so can present a significant therapeutic challenge (5). In recent years, the promotion and implementation of the One Health approach in antimicrobial resistance by the WHO have emphasized the importance of surveillance in both animal and human populations and have highlighted gaps in this knowledge (6). It has been well established in numerous global studies that in humans, certain E. coli lineages (e.g., blaCTX-M-encoding ST131) play a major role in the dissemination of ESBL genes; however, such a depth of information does not exist for isolates from animal populations (2). Human-associated pandemic lineages have been reported in animal populations, albeit to a much lesser extent than in human populations (7).
In humans, blaCTX-M variants are the globally dominant ESBL type, with some variants exhibiting geographical associations (e.g., blaCTX-M-15 in Europe and North America and blaCTX-M-14 in Asia) (2). Transmission of ESBLs occurs largely through horizontal gene transfer, with conjugative IncF plasmids being reported as the dominant vehicles for blaCTX-M genes (8, 9). Previous studies using typing methods including whole-genome sequencing (WGS) have suggested transmission of both strains and ESBL plasmids across animal and human populations (10, 11). Epidemic plasmids have been reported across different host populations and in multiple countries (12). For example, one epidemic plasmid type—pCT, encoding blaCTX-M-14—was identified in cattle and human E. coli isolates in England and found to exist in human isolates from several countries across 3 continents (12).
Antimicrobial use in food animals may provide selective pressure for resistance genes and plasmids which could theoretically be spread to humans (13). However, recent reports suggest that such transmission is limited, at least in the United Kingdom (14). In dairy farming, antibiotics are used both therapeutically in the treatment of common infections, such as mastitis, and preventatively. For example, in so-called dry cow therapy, an antibacterial preparation is inserted into a cow’s udder between lactations to prevent mastitis (15). A survey of dairy farms in England and Wales in 2012 revealed that the fourth-generation cephalosporin (4GC) cefquinome (another HP-CIA) was the most used dry cow therapy treatment (16). By 2017, however, only 5.3% of total dry cow therapy active ingredients were HP-CIAs. Indeed, there has been a significant decline in the use of HP-CIAs on dairy farms in the United Kingdom in recent years (17).
Recently, we reported a survey of 53 dairy farms located in southwest England in which we investigated the prevalence of 3GC-R E. coli (18). PCR analysis confirmed that 648 of 1,226 such isolates (52.7%) carried blaCTX-M and 13 (1.1%) carried a pAmpC gene. The remaining 566 (46.2%) isolates did not carry any putative acquired 3GC-R gene, and a subsequent analysis confirmed that hyperproduction of the chromosomally encoded AmpC β-lactamase was the mechanism of 3GC-R in these isolates (19).
The study reported here aimed to characterize, using WGS, representative blaCTX-M- and pAmpC-positive E. coli isolates collected on dairy farms during our earlier surveillance study (18). Furthermore, our aim was to compare these isolates at the strain and plasmid-encoded 3GC-R gene levels with blaCTX-M- and pAmpC-positive urinary E. coli isolates (20) collected from humans living in the same 50- by 50-km region that was the location of the majority of dairy farms under study.
RESULTS AND DISCUSSION
WGS analysis of E. coli carrying acquired 3GC-R genes from dairy farms.
A total of 138 representative isolates, PCR positive for blaCTX-M or pAmpC genes (18) and chosen to give coverage of all 42 farms positive for acquired 3GC-R genes, were subjected to WGS (Table 1). blaCTX-M-32, encoding a group 1 enzyme first described in a human clinical isolate in 2004 (21), was the most common 3GC-R gene identified and was found in 79/138 sequenced isolates encompassing 27 E. coli sequence types (STs) from 25 farms. Other 3GC-R genes identified were blaCTX-M-14 (18 isolates; 6 STs from 9 farms), blaCTX-M-1 (16 isolates; 8 STs from 6 farms), blaCTX-M-15 (16 isolates; 5 STs from 10 farms), blaCMY-2 (6 isolates; 3 STs from 3 farms), and blaCTX-M-214 (3 isolates; 2 STs from 3 farms); in addition, one isolate harbored blaDHA-1 and one isolate had both blaCTX-M-1 and blaCTX-M-14. CTX-M-214 is a novel CTX-M-9 variant, first identified in this study, which differs from CTX-M-9 by a single amino acid, Ala112Thr. In all three isolates carrying blaCTX-M-214, the gene was identified on a contig which also contained an IncI-ST26 plasmid replicon as well as aadA2, sul1, and dfrA16.
TABLE 1.
Characteristics of 3GC-R isolates (138 isolates from 42 farms) subjected to WGSa
| Parameter | Abundance of acquired 3GC-R mechanisms identified by WGS and no. of STs/farms involved |
||||||
|---|---|---|---|---|---|---|---|
| blaCTX-M-32 | blaCTX-M-1 | blaCTX-M-14 | blaCTX-M-15 | blaCTX-M-214 | blaDHA-1 | blaCMY-2 | |
| No. of isolates | 79 | 16a | 18a | 16 | 3 | 1 | 6 |
| No. of STs/no. of farms | 27/25 | 8/6 | 6/9 | 5/10 | 2/3 | 1/1 | 3/3 |
One isolate harbored both blaCTX-M-1 and blaCTX-M-14.
Identification and characterization of pMOO-32.
Following observations of the high prevalence of blaCTX-M-32, a search for common plasmid replicon types was conducted which revealed an IncHI2-ST2 replicon in almost all the sequenced blaCTX-M-32-positive isolates. Conjugation into E. coli DH5α was attempted using blaCTX-M-32-positive farm isolate DK as the donor (Table 2). Transconjugants were identified at high frequency, approximately 10−4 per donor. One successful transconjugant was sent for WGS employing both long- and short-read methods to sequence the plasmid to closure. pMOO-32 is a 226,022-bp conjugative plasmid belonging to the ST2-IncHI2 incompatibility group, harboring repHI2 and repHI2A replication genes. It contains 245 putative open reading frames (ORFs) and has a GC content of 45.5% (Fig. 1). pMOO-32 carries the antimicrobial resistance genes blaCTX-M-32, strA, strB, aph(6)-Ic, aph(3′)-IIa, and tet(B), as well as genes encoding resistance to the heavy metal compound tellurite (terABCDEFWXYZ) and a HipAB type II toxin-antitoxin system, along with a second partial system (higB toxin gene). blaCTX-M-32 is present downstream of an ISECp1 element within which there is an ISKpn26 insertion in the opposite orientation (Fig. 2). The same genetic environment was also observed in 4 blaCTX-M-32-positive but IncHI2 plasmid-negative ST10 isolates collected from 2 farms. There were 2 additional IncHI2 plasmid-negative ST765 isolates, both from the same farm, that carried blaCTX-M-32 where the immediate genetic environment differed by a truncation in ISEcp1. In these 6 IncHI2 plasmid-negative isolates, blaCTX-M-32 is carried on an IncFII plasmid not previously reported.
TABLE 2.
Disc susceptibility testing of E. coli DK and pMOO-32 transconjugants of various E. coli recipient strainsa
| Antimicrobial agent | Zone diameter (mm) and resistance category |
||||||
|---|---|---|---|---|---|---|---|
| E. coli DK | E. coli DH5α | E. coli DH5α TR | E. coli HG | E. coli HG TR | E. coli HC4 | E. coli HC4 TR | |
| AMP | <6 (R) | 30 (S) | <6 (R) | <6 (R) | <6 (R) | 20 (S) | <6 (R) |
| CTX | 10 (R) | 45 (S) | 18 (I) | 35 (S) | 10 (R) | 34 (S) | 13 (R) |
| CAZ | 20 (I) | 45 (S) | 30 (S) | 34 (S) | 20 (I) | 32 (S) | 23 (S) |
| FEP | 19 (R) | 45 (S) | 28 (S) | 28 (S) | 18 (R) | 35 (S) | 24 (I) |
| ETP | 30 (S) | 45 (S) | 44 (S) | 34 (S) | 33 (S) | 36 (S) | 36 (S) |
| ATM | 15 (R) | 45 (S) | 24 (I) | 34 (S) | 15 (R) | 35 (S) | 19 (R) |
| STRb | <6 (R) | 25 (S) | 10 (R) | <6 (R) | <6 (R) | 13 (R) | <6 (R) |
| TOB | 17 (S) | 28 (S) | 26 (S) | <6 (R) | <6 (R) | 18 (S) | 19 (S) |
| NEOb | 8 (R) | 20 (S) | 12 (R) | <6 (R) | <6 (R) | 14 (I) | 8 (R) |
| TETb | <6 (R) | 36 (S) | <6 (R) | <6 (R) | <6 (R) | 30 (S) | <6 (R) |
AMP, ampicillin; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; ETP, ertapenem; ATM, aztreonam; STR, streptomycin; TOB, tobramycin; NEO, neomycin; TET, tetracycline; TR, pMOO-32 transconjugant. Susceptible (S), intermediate (I), and resistant (R) were determined as defined by CLSI criteria (resistant results are highlighted in bold).
Streptomycin and neomycin sensitivities were determined using tobramycin EUCAST interpretation guidelines, and tetracycline sensitivities were determined according to guidelines for Yersinia enterocolitica.
FIG 1.
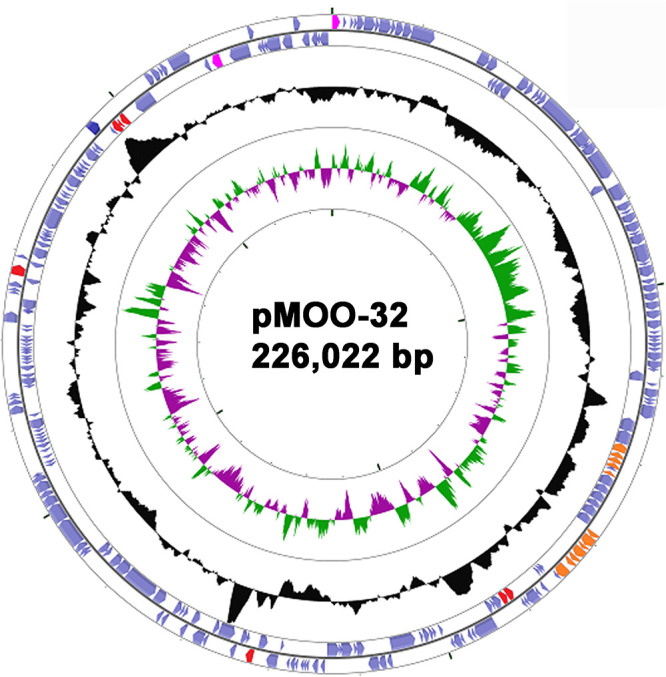
Plasmid pMOO-32, created using CGView (39). The outer two rings represent predicted coding sequences, facing clockwise (outermost) and counterclockwise, respectively. Those predicted to encode antimicrobial resistance proteins are shown in red, plasmid replicon functions are in pink, and telluride resistance proteins are in orange. GC content is in black, above or below 50%, and GC skew is illustrated in the central ring in green (biased toward GC) or magenta (biased toward AT).
FIG 2.
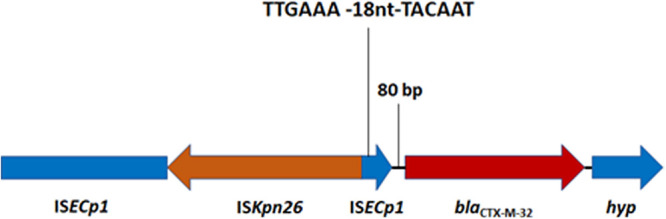
Genetic environment of blaCTX-M-32 in pMOO-32 and other IncHI2-positive, blaCTX-M-32-harboring isolates.
Conjugation attempts using the pMOO-32-positive farm isolate DK as donor into a 3GC-susceptible (3GC-S) cattle ST88 E. coli isolated from one of the study farms (18) as well as into a 3GC-S human urinary ST1193 E. coli isolate (20) were both successful (Table 2). ST1193 was selected as a recently described fluoroquinolone-resistant global clone, often implicated as a cause of human infections (22), while ST88 was selected as a particularly prevalent ST among cattle isolates (23). Antimicrobial disc testing showed that the pMOO-32-carrying donor was, as expected from the genotype, resistant to ampicillin, cefotaxime, cefepime, aztreonam, streptomycin, neomycin, and tetracycline. The cattle ST88 and human ST1193 transconjugants were, in addition to their starting wild-type resistance profile, resistant to cefotaxime, cefepime, and aztreonam. These results (Table 2) are indicative of the functionality of the blaCTX-M-32 gene harbored by pMOO-32.
Epidemiology of pMOO-32-like plasmids on farms.
Using the complete nucleotide sequence of pMOO-32 as a reference, sequencing reads from all 73 isolates positive for blaCTX-M-32 and the IncHI2 replicon were mapped. While there are limitations in using short-read sequencing data in this way, the plasmids in these 73 isolates exhibited 94 to 100% identity to the reference sequence. Any differences could be attributed to a loss or gain of mobile genetic elements, but no major rearrangements of the plasmid backbone or changes to resistance gene content were observed. The isolates carrying pMOO-32-like plasmids comprised 27 E. coli STs. On 10 farms, pMOO-32-like plasmids were found in isolates of more than one ST. The most frequently identified STs were ST69 and ST10, found in 18 isolates from 7 farms and 6 isolates from 4 farms, respectively. Therefore, we conclude that pMOO-32-like plasmids are dominant in this study area, a result of horizontal and clonal transmission.
We next designed a multiplex PCR, based on the pMOO-32 sequence and used it to screen all group 1 blaCTX-M-PCR-positive isolates from our study farms (18) not previously subjected to WGS. The aim was to test them for the presence of pMOO-32-like plasmids. It was found that 26/53 (49.1%) farms within our study area tested positive for the presence of a pMOO-32-like plasmid by this test. The origins and geographical reach of pMOO-32 remain to be established, but all positive farms were located in a 40- by 40-km subregion of the wider study area, where 26/33 farms were positive, suggesting that the plasmid remained geographically confined at the time of sample collection.
We hypothesized that the observed dominance of pMOO-32-like plasmids could be a consequence of the HipAB-type II toxin-antitoxin system encoded by the plasmid, leading to persistence even in the absence of antibacterial selective pressure. In support of this, we found that pMOO-32 was stably maintained over 10 days of passaging in the absence of antibiotic pressure. All tested farm isolates, their respective DH5α transconjugants, and the human and cattle E. coli isolate transconjugants were found to have retained the plasmid.
No evidence for recent sharing of 3GC-R E. coli and limited evidence for recent sharing of 3GC-R E. coli plasmids between humans and dairy farms.
The ability of pMOO-32 to readily transfer into human urinary E. coli ST1193 (Table 2) and be maintained in the absence of antimicrobial pressure indicates the zoonotic potential of this plasmid. We next aimed to see if there was evidence of recent sharing of 3GC-R isolates or plasmids, including pMOO-32, between dairy farms and humans living in the same geographical region as the farms. To do this, we compared farm isolates WGS data (described above) with human urinary E. coli WGS data collected in parallel within the same 50- by 50-km geographical region (20). Since 10 dairy farms under study here lie outside the 50- by 50-km region (18), isolates from these farms were excluded.
Core genome phylogenetic analysis was carried out using WGS data from 112 dairy farm and 212 human urinary E. coli isolates carrying acquired 3GC-R genes collected within the target region. This analysis (Fig. 3) revealed only four STs, including examples of both farm and human isolates. In no case was the single nucleotide polymorphism (SNP) difference between any pair of human and farm isolate core genomes suggestive of recent transmission. ST10 was the closest (≥205 SNPs different between human and cattle isolates) (Fig. 3, inset); the others were ST540 (929 SNPs different), ST58 (≥1,388 SNPs different), and ST69 (≥831 SNPs different). In contrast, there was clear evidence of recent farm-to-farm transmission of isolates from multiple STs (e.g., ST10 and ST69, for which, in both cases, there was a 3-SNP minimum distance between pairs of isolates representing two farms) (Fig. 3). AmpC-hyperproducing E. coli isolates from these same farms showed a similar pattern: no evidence of strain sharing between dairy farms and humans but strong evidence for recent farm-to-farm transmission (19). We conclude, therefore, that 3GC-R isolates from farms do not readily circulate among the local human population and cause urinary tract infection (UTI).
FIG 3.
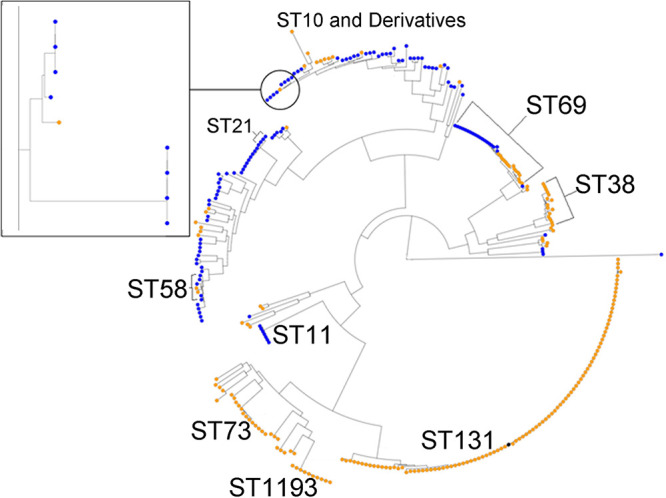
Phylogenetic analysis of E. coli from dairy farms and human UTI collected in parallel in a 50- by 50-km region. Human isolates are in orange; cattle isolates are in blue. The reference ST131 isolate is in black. Certain key STs are highlighted, particularly STs with representatives from human and cattle isolates: ST21 (ST540), ST69, ST58, and ST10. The inset shows a more detailed analysis of ST10 isolates, which represents the closest relationship between a human and a cattle isolate: 205 SNPs different across the core genome.
We next considered transmission of plasmid-mediated 3GC-R between farm and human E. coli isolates (i.e., those that that have caused UTI). We found that 37/107 blaCTX-M-positive E. coli isolates from farm samples within our 50- by 50-km study region carried blaCTX-M variants also seen among 189 blaCTX-M-positive urinary E. coli cultured from people living in the same region during the same time period (20). None of the human isolates carried blaCTX-M-32 or any plasmid related to pMOO-32. By filtering sequenced isolates by their blaCTX-M variants and plasmid replicon types, plasmids that shared high degrees of sequence identity in farm and human isolates were identified. There were three different blaCTX-M variant/plasmid replicon type combinations found in both farm and human isolates. For each, the longest blaCTX-M-containing contig was used to identify the finished NCBI sequence with the highest identity, which was used as a reference for read mapping-based comparisons.
One IncI1-ST3 plasmid, found in an ST345 farm isolate, harbored blaCTX-M-1. Mapping of sequencing reads showed that this plasmid exhibited 100% coverage and 97.5% sequence identity to an unpublished ∼106-kb IncI1-ST3 plasmid (pTC_N40607; GenBank accession no. CP007651) found in E. coli obtained from meat and cattle isolates in the United States. Six human urinary E. coli isolates representing ST23, ST127, ST131, ST141, and ST2015 harbored blaCTX-M-1 on a plasmid that exhibited 99.4 to 100% coverage and 96.4 to 98.7% identity when sequencing reads were mapped to pTC_N40607.
Another plasmid type—again obtained from a single farm isolate, in this case belonging to ST58—exhibited 100% coverage and 98.5% identity by read mapping to a published IncK plasmid, pCT (GenBank accession no. NC_014477). pCT is ∼94 kb and harbors blaCTX-M-14. pCT-like plasmids have been reported in both human and veterinary E. coli isolates across three continents (12, 23). Among human urinary E. coli isolates found in this study, two also carried pCT-like plasmids. Both isolates were the pandemic clone ST131, and their pCT-like plasmids exhibited 96.4 or 97.2% identity and 100% coverage to pCT.
In terms of plasmids carrying pAmpC genes, we identified a blaCMY-2-bearing plasmid, p96 (GenBank accession no. CP023370). This is an ∼96 kb IncI1 plasmid derived from an E. coli isolate that caused urinary tract infections in dogs in Scotland (24). Mapping analyses showed that one of seven blaCMY-2-carrying human urinary isolates (ST80) showed 100% coverage and 99.9% identity to p96. An almost identical plasmid was found in one farm isolate (ST1480) that showed 99.6% coverage and 98.9% identity to p96. As with the two blaCTX-M plasmids, this blaCMY-2 plasmid appears to be globally widespread. BLAST analysis found blaCMY-2 plasmids with >95% coverage and >98% identity to p96 in E. coli isolates from humans in Taiwan (25) and chicken meat in the United States (GenBank accession no. CP048295).
Concluding remarks.
Overall, this analysis found no evidence of recent, direct sharing of E. coli between farms and the local human population that have resulted in UTI. Three farm E. coli isolates carried a blaCTX-M or blaCMY-2 plasmid almost identical to one of three plasmids found in urinary E. coli in the local human population. However, these three plasmids are known to be widely disseminated in humans and animals across several continents. Furthermore, no human-farm plasmid pair shared 100% identity. So, while there is some general evidence of plasmids circulating between human and animal populations, as reported in a number of recent studies in Western countries (14, 26, 27), the overall level of overlap between United Kingdom dairy farm and human 3GC-R E. coli identified in this study was very small and not suggestive of any novel or recent zoonotic transmission events. In contrast, both recent and sustained farm-to-farm transmission of 3GC-R E. coli, and particularly of a newly identified epidemic plasmid, pMOO-32, across many different E. coli STs and dairy farms was clearly seen. Identifying the vectors for this transmission will inform interventions that might facilitate a more rapid reduction in 3GC-R E. coli on dairy farms.
MATERIALS AND METHODS
Bacterial isolates, identification, and susceptibility testing.
Details of farm sample collection and microbiological analysis were recently reported (18). In brief, samples of fecally contaminated sites were collected using sterile overshoes on 53 dairy farms located in southwest England between January 2017 and December 2018. Samples were plated on TBX agar (Sigma-Aldrich, Poole, United Kingdom) containing 16 mg liter−1 cephalexin. Up to 5 E. coli colonies per cefalexin plate were replated on TBX agar containing 2 mg liter−1 cefotaxime to confirm resistance. The presence of acquired 3GC-R genes (blaCTX-M, blaSHV, blaCMY-2, and blaDHA-1) was confirmed by PCR. Disc susceptibility testing was performed and interpreted according to EUCAST guidelines (28).
Conjugation.
Conjugation experiments were performed using rifampin-resistant (Rifr) E. coli DH5α with both human and cattle E. coli isolates as the recipients (Table 3). Briefly, 1 ml each of overnight broth cultures of donor and recipient cells were mixed in a 3:1 ratio before centrifugation and resuspension in 50 μl of phosphate-buffered saline (PBS). Aliquots (5 μl) were spotted onto LB agar (Oxoid, Basingstoke, United Kingdom) plates and incubated at 37°C for 6 h. Growth was collected and resuspended in 100 μl of PBS before being plated on MacConkey agar (Oxoid) plates containing either 32 mg liter−1 rifampin (for Rifr E. coli DH5α) or 0.5 mg liter−1 ciprofloxacin (for strains HC4 and HG) and 2 mg liter−1 cefotaxime. Transconjugant colonies were screened by PCR.
TABLE 3.
Characteristics of E. coli strains used in conjugation experiments
| Isolate | Source/host | ST | Resistance genes | Use |
|---|---|---|---|---|
| DK | Cattle | 155 | strA, strB, aph(6)-Ic, aph(3′)-IIa, tet(B), blaCTX-M-32 | Donor |
| HC4 | Human | 1193 | aadA5, dfrA17, mdf(A), sul1 | Recipient |
| HG | Cattle | 88 | aph(6)-Id, ant(2ʺ)-Ia, aph(3′)-Ia, aadA24, aph(3ʺ)-Ib, sul1, sul2, tet(A), tet(B), blaTEM-1, blaOXA-1, catA1, floR | Recipient |
WGS and analyses and pMOO-32 PCR.
Representative isolates were selected for WGS based on resistance phenotype, β-lactamase gene carriage, and farm of isolation, as defined previously (18). WGS was performed by MicrobesNG (https://microbesng.uk/) on a HiSeq 2500 instrument (Illumina, San Diego, CA, USA) using 2 × 250-bp paired-end reads. Reads were trimmed using Trimmomatic (29) and assembled into contigs using SPAdes 3.13.0 (30) (http://cab.spbu.ru/software/spades/). Resistance genes, plasmid replicon types, and sequence types (according to the Achtman scheme [31]) were assigned using ResFinder (32), PlasmidFinder (33), and MLST 2.0 on the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/) platform. Enhanced genome sequencing (combining Illumina and MinION reads) was performed by MicrobesNG on one transconjugant, and reads were assembled using Unicycler (34). Contigs were annotated using Prokka 1.2 (35).
Reads were aligned to plasmid reference sequences using the progressive Mauve alignment software (36), CLC Genomics Workbench 12 (Qiagen, Manchester, United Kingdom), or BWA (37) and SAMtools (38), with variant positions being called using BCFtools (38). pMOO-32 was visualized using the CGView server (39) (http://stothard.afns.ualberta.ca/cgview_server/). The most appropriate plasmid reference for each analysis was identified using BLASTn with the largest blaCTX-M-containing contig relevant to each analysis.
A multiplex PCR, targeting five size-distinguishable regions of pMOO-32, was designed to indicate the presence of pMOO-32-like plasmids (Table 4).
TABLE 4.
Primers used for the pMOO-32 multiplex PCR
| Primer | Sequence (5′–3′) | Product size (bp) | Target |
|---|---|---|---|
| aph(3′)-lla_F | TGGCTACCCGTGATATTGCT | 642 | aph(3′)-lla/aph(6)-Ic junction |
| aph(6)-lc_R | CTGGCGGACGGGAAGTATC | ||
| HI2A_F | AGCCTTTCTCACGGTAGCAT | 526 | HI2 repA |
| HI2A_R | TTCAATTGTCGGTGAGCGTC | ||
| TraI_F | CGGGAAAAGCTGCACTCAAT | 396 | traI |
| TraI_R | AAGACTTTGTGAGCTTGGCG | ||
| TetB_F | TTCAGCGCAATTGATAGGCC | 285 | tetB |
| TetB_R | ATCCCACCACCAGCCAATAA | ||
| CTX-M-32_F | TTAGGAAGTGTGCCGCTGTA | 180 | blaCTX-M-32 |
| CTX-M-32_R | CACGGCCATCACTTTACTGG |
Plasmid stability assay.
Three representative pMOO-32 PCR-positive isolates, obtained from different farms, and their transconjugant counterparts were subjected to 10 days of serial passaging on nonselective LB agar. Each day, a single colony was picked and restreaked. After 10 days, one colony from each plate was screened for the presence of pMOO-32 by PCR.
Phylogenetic analysis.
Sequence alignment and phylogenetic analysis were carried out using the Bioconda environment (40) on the Cloud Infrastructure for Microbial Bioinformatics (CLIMB) (41). The reference sequence was E. coli ST131 isolate EC958 complete genome (accession no. HG941718). Sequences were first aligned to a closed reference sequence and analyzed for SNP differences, while omitting insertion and deletion elements, using the Snippy alignment program. Alignment was then focused on regions of the genome found across all isolates, the core genome, using the Snippy-core program, thus eliminating the complicating factors of insertions and deletions (https://github.com/tseemann/snippy). Aligned sequences were then used to construct a maximum-likelihood phylogenetic tree using RAxML, utilizing the GTRCAT model of rate heterogeneity and the software’s autoMR and rapid bootstrap to find the best-scoring maximum-likelihood tree and including tree branch lengths, defined as the number of base substitutions per site compared (42, 43). Finally, phylogenetic trees were illustrated using the web-based Microreact program (44).
Data availability.
Newly determined sequence data have been deposited in GenBank under accession numbers MH121688 (CTX-M-214) and MK169211 (pMOO-32).
ACKNOWLEDGMENTS
Genome sequencing was provided by MicrobesNG (http://www.microbesng.uk). We thank all the farmers who participated in this study.
This work was funded by grant NE/N01961X/1 to M.B.A., K.K.R., and T.A.C. and grant BB/T004592/1 to K.K.R. and M.B.A. from the Antimicrobial Resistance Cross Council Initiative supported by the seven United Kingdom research councils. W.W.Y.L. is supported by a scholarship from the Medical Research Foundation National PhD Training Program in Antimicrobial Resistance Research (MRF-145-0004-TPG-AVISO).
We declare no conflict of interests. Farming businesses who permitted access to collect the isolates studied here were not involved in the design of this study or in data analysis and were not involved in drafting the manuscript for publication.
Conceived the Study: K.K.R., M.B.A. Collection of Data: J.F., N.N., O.M., K.M., supervised by T.A.C., M.B.A. Cleaning and Analysis of Data: J.F., O.M., W.W.Y.L., H.S., V.C.G., supervised by K.K.R., M.B.A. Initial Drafting of Manuscript: J.F., M.B.A. Corrected and Approved Manuscript: all authors.
REFERENCES
- 1.Department of Health. 2012. ESBLs—a threat to human and animal health? Report by the Joint Working Group of DARC and ARHAI. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/215180/dh_132534.pdf.
- 2.Bevan ER, Jones AM, Hawkey PM. 2017. Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. 2016. Critically important antimicrobials for human medicine: ranking of medically important antimicrobials for risk management of antimicrobial resistance due to non-human use. 5th revision. http://apps.who.int/iris/bitstream/handle/10665/255027/9789241512220-eng.pdf;jsessionid=7E5054076A3F8ED22FA8C41A51687950?sequence=1.
- 4.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta K, Bhadelia N. 2014. Management of urinary tract infections from multidrug-resistant organisms. Infect Dis Clin North Am 28:49–59. doi: 10.1016/j.idc.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization. 2016. Antimicrobial resistance. A manual for developing nations action plans. Version 1. http://apps.who.int/iris/bitstream/handle/10665/204470/9789241549530_eng.pdf;jsessionid=3B5EA17EC2A21038D896E2937CA7FAA9?sequence=1.
- 7.Platell JL, Johnson JR, Cobbold RN, Trott DJ. 2011. Multidrug-resistant extraintestinal pathogenic Escherichia coli of sequence type ST131 in animals and foods. Vet Microbiol 153:99–108. doi: 10.1016/j.vetmic.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Marcadé G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, Denamur E, Arlet G. 2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum beta-lactamases. J Antimicrob Chemother 63:67–71. doi: 10.1093/jac/dkn428. [DOI] [PubMed] [Google Scholar]
- 9.Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, Song JH. 2011. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents 38:160–163. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 10.de Been M, Lanza VF, de Toro M, Scharringa J, Dohmen W, Du Y, Hu J, Lei Y, Li N, Tooming-Klunderud A, Heederik DJ, Fluit AC, Bonten MJ, Willems RJ, de la Cruz F, van Schaik W. 2014. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet 10:e1004776. doi: 10.1371/journal.pgen.1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazarus B, Paterson DL, Mollinger JL, Rogers BA. 2015. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis 60:439–452. doi: 10.1093/cid/ciu785. [DOI] [PubMed] [Google Scholar]
- 12.Cottell JL, Webber MA, Coldham NG, Taylor DL, Cerdeño-Tárraga AM, Hauser H, Thomson NR, Woodward MJ, Piddock LJ. 2011. Complete sequence and molecular epidemiology of IncK epidemic plasmid encoding blaCTX-M-14. Emerg Infect Dis 17:645–652. doi: 10.3201/eid1704.101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson TP, Wertheim HF, Kakkar M, Kariuki S, Bu D, Price LB. 2016. Animal production and antimicrobial resistance in the clinic. Lancet 387:e1–e3. doi: 10.1016/S0140-6736(15)00730-8. [DOI] [PubMed] [Google Scholar]
- 14.Ludden C, Raven KE, Jamrozy D, Gouliouris T, Blane B, Coll F, de Goffau M, Naydenova P, Horner C, Hernandez-Garcia J, Wood P, Hadjirin N, Radakovic M, Brown NM, Holmes M, Parkhill J, Peacock SJ. 2019. One Health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 10:e02693-18. doi: 10.1128/mBio.02693-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higham LE, Deakin A, Tivey E, Porteus V, Ridgway S, Rayner AC. 2018. A survey of dairy cow farmers in the United Kingdom: knowledge, attitudes and practices surrounding antimicrobial use and resistance. Vet Rec 183:746. doi: 10.1136/vr.104986. [DOI] [PubMed] [Google Scholar]
- 16.Brunton LA, Duncan D, Coldham NG, Snow LC, Jones JR. 2012. A survey of antimicrobial usage on dairy farms and waste milk feeding practices in England and Wales. Vet Rec 171:296. doi: 10.1136/vr.100924. [DOI] [PubMed] [Google Scholar]
- 17.UK-VARSS. 2018. UK veterinary antibiotic resistance and sales surveillance report. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/750811/_1473963-v1-UK-VARSS_2017_Report_FINAL.pdf.
- 18.Schubert H, Morley K, Puddy EF, Arbon R, Findlay J, Mounsey O, Gould VC, Vass L, Evans M, Rees GM, Barrett DC, Turner KM, Cogan TA, Avison MB, Reyher KK. 2020. Reduced antibacterial drug resistance and blaCTX-M β-lactamase gene carriage in cattle-associated Escherichia coli at low temperatures, at sites dominated by older animals and on pastureland: implications for surveillance. bioRxiv doi: 10.1101/778407. [DOI] [PMC free article] [PubMed]
- 19.Alzayn M, Findlay J, Schubert H, Mounsey O, Gould VC, Heesom KJ, Turner KM, Barrett DC, Reyher KK, Avison MB. 2020. Characterization of AmpC hyper-producing Escherichia coli from humans and dairy farms collected in parallel in the same geographical region. J Antimicrob Chemother 75:2471–2479. doi: 10.1093/jac/dkaa207. [DOI] [PubMed] [Google Scholar]
- 20.Findlay J, Gould VC, North P, Bowker KE, Williams MO, MacGowan AP, Avison MB. 2020. Characterization of cefotaxime-resistant urinary Escherichia coli from primary care in South-West England 2017–18. J Antimicrob Chemother 75:65–71. doi: 10.1093/jac/dkz397. [DOI] [PubMed] [Google Scholar]
- 21.Cartelle M, del Mar Tomas M, Molina F, Moure R, Villanueva R, Bou G. 2004. High-level resistance to ceftazidime conferred by a novel enzyme, CTX-M-32, derived from CTX-M-1 through a single Asp240-Gly substitution. Antimicrob Agents Chemother 48:2308–2313. doi: 10.1128/AAC.48.6.2308-2313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson TJ, Elnekave E, Miller EA, Munoz-Aguayo J, Flores Figueroa C, Johnston B, Nielson DW, Logue CM, Johnson JR. 2018. Phylogenomic analysis of extraintestinal pathogenic Escherichia coli sequence type 1193, an emerging multidrug-resistant clonal group. Antimicrob Agents Chemother 63:e01913-18. doi: 10.1128/AAC.01913-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Day MJ, Rodríguez I, van Essen-Zandbergen A, Dierikx C, Kadlec K, Schink AK, Wu G, Chattaway MA, DoNascimento V, Wain J, Helmuth R, Guerra B, Schwarz S, Threlfall J, Woodward MJ, Coldham N, Mevius D, Woodford N. 2016. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J Antimicrob Chemother 71:1178–1182. doi: 10.1093/jac/dkv485. [DOI] [PubMed] [Google Scholar]
- 24.Wagner S, Lupolova N, Gally DL, Argyle SA. 2017. Convergence of plasmid architectures drives emergence of multi-drug resistance in a clonally diverse Escherichia coli population from a veterinary clinical care setting. Vet Microbiol 211:6–14. doi: 10.1016/j.vetmic.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin YT, Pan YJ, Lin TL, Fung CP, Wang JT. 2015. Transfer of CMY-2 cephalosporinase from Escherichia coli to virulent Klebsiella pneumoniae causing a recurrent liver abscess. Antimicrob Agents Chemother 59:5000–5002. doi: 10.1128/AAC.00492-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietsch M, Irrgang A, Roschanski N, Brenner Michael G, Hamprecht A, Rieber H, Käsbohrer A, Schwarz S, Rösler U, Kreienbrock L, Pfeifer Y, Fuchs S, Werner G, RESET Study Group. 2018. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genomics 19:601. doi: 10.1186/s12864-018-4976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valcek A, Roer L, Overballe-Petersen S, Hansen F, Bortolaia V, Leekitcharoenphon P, Korsgaard HB, Seyfarth AM, Hendriksen RS, Hasman H, Hammerum AM. 2019. IncI1 ST3 and IncI1 ST7 plasmids from CTX-M-1-producing Escherichia coli obtained from patients with bloodstream infections are closely related to plasmids from E. coli of animal origin. J Antimicrob Chemother 74:2171–2175. doi: 10.1093/jac/dkz199. [DOI] [PubMed] [Google Scholar]
- 28.EUCAST. 2018. Breakpoint tables for interpretation of MICs and zone diameters. Version 8.1. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf.
- 29.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 36.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grüning B, Dale R, Sjödin A, Chapman BA, Rowe J, Tomkins-Tinch CH, Valieris R, Köster J, Bioconda Team. 2018. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat Methods 15:475–476. doi: 10.1038/s41592-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connor TR, Loman NJ, Thompson S, Smith A, Southgate J, Poplawski R, Bull MJ, Richardson E, Ismail M, Thompson SE, Kitchen C, Guest M, Bakke M, Sheppard SK, Pallen MJ. 2016. CLIMB (the Cloud Infrastructure for Microbial Bioinformatics): an online resource for the medical microbiology community. Microb Genom 2:e000086. doi: 10.1099/mgen.0.000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 43.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 44.Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H, Spratt BG, Aanensen DM. 2016. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Newly determined sequence data have been deposited in GenBank under accession numbers MH121688 (CTX-M-214) and MK169211 (pMOO-32).


