Carbohydrate metabolism is central to the development of dental caries. A variety of sugars available to dental microorganisms influence the development of caries by affecting the physiology, ecology, and pathogenic potential of tooth biofilms. Using two well-characterized oral bacteria, one pathogen (Streptococcus mutans) and one commensal (Streptococcus gordonii), in an RNA deep-sequencing analysis, we studied the impact of two abundant amino sugars on bacterial gene expression and interspecies interactions. The results indicated large-scale remodeling of gene expression induced by GlcN in particular, affecting bacterial energy generation, acid production, protein synthesis, and release of antimicrobial molecules. Our study provides novel insights into how amino sugars modify bacterial behavior, information that will be valuable in the design of new technologies to detect and prevent oral infectious diseases.
KEYWORDS: amino sugar, commensal bacteria, transcriptomic analysis, pyruvate metabolism, dental caries
ABSTRACT
Amino sugars, particularly glucosamine (GlcN) and N-acetylglucosamine (GlcNAc), are abundant carbon and nitrogen sources supplied in host secretions and in the diet to the biofilms colonizing the human oral cavity. Evidence is emerging that these amino sugars provide ecological advantages to beneficial commensals over oral pathogens and pathobionts. Here, we performed transcriptome analysis on Streptococcus mutans and Streptococcus gordonii growing in single-species or dual-species cultures with glucose, GlcN, or GlcNAc as the primary carbohydrate source. Compared to glucose, GlcN caused drastic transcriptomic shifts in each species of bacteria when it was cultured alone. Likewise, cocultivation in the presence of GlcN yielded transcriptomic profiles that were dramatically different from the single-species results from GlcN-grown cells. In contrast, GlcNAc elicited only minor changes in the transcriptome of either organism in single- and dual-species cultures. Interestingly, genes involved in pyruvate metabolism were among the most significantly affected by GlcN in both species, and these changes were consistent with measurements of pyruvate in culture supernatants. Differing from what was found in a previous report, growth of S. mutans alone with GlcN inhibited the expression of multiple operons required for mutacin production. Cocultivation with S. gordonii consistently increased the expression of two manganese transporter operons (slo and mntH) and decreased expression of mutacin genes in S. mutans. Conversely, S. gordonii appeared to be less affected by the presence of S. mutans but did show increases in genes for biosynthetic processes in the cocultures. In conclusion, amino sugars profoundly alter the interactions between pathogenic and commensal streptococci by reprogramming central metabolism.
IMPORTANCE Carbohydrate metabolism is central to the development of dental caries. A variety of sugars available to dental microorganisms influence the development of caries by affecting the physiology, ecology, and pathogenic potential of tooth biofilms. Using two well-characterized oral bacteria, one pathogen (Streptococcus mutans) and one commensal (Streptococcus gordonii), in an RNA deep-sequencing analysis, we studied the impact of two abundant amino sugars on bacterial gene expression and interspecies interactions. The results indicated large-scale remodeling of gene expression induced by GlcN in particular, affecting bacterial energy generation, acid production, protein synthesis, and release of antimicrobial molecules. Our study provides novel insights into how amino sugars modify bacterial behavior, information that will be valuable in the design of new technologies to detect and prevent oral infectious diseases.
INTRODUCTION
Amino sugars are particularly abundant in the human oral cavity. In particular, N-acetylglucosamine (GlcNAc) and glucosamine (GlcN) decorate host proteins, such as mucins and basic proline-rich glycoproteins (PRG), and are the fundamental building blocks of the cell envelopes of bacteria and fungi (1, 2). These carbohydrates are also common in foodstuffs consumed by humans. GlcN and GlcNAc can be released from polymers through the collective activities of microbial glycohydrolases, providing a steady supply of carbon and nitrogen sources to the oral microflora, especially while the host is fasting. Most known oral bacteria have the capacity, or carry the necessary genes, for metabolizing exogenous sources of amino sugars. GlcN and GlcNAc catabolism generally requires a specialized membrane transporter, often a permease of the sugar-phosphotransferase system (PTS); a GlcNAc-6-P deacetylase (NagA) that converts GlcNAc-6-P into GlcN-6-P; and a GlcN-6-P deaminase (NagB) that converts GlcN-6-P into fructose-6-P (F-6-P). F-6-P can enter the Embden–Meyerhof–Parnas (EMP) pathway, while the molecule of ammonia released from GlcN or GlcNAc can complex with a proton inside or outside the cell to raise the pH or can be funneled into amino acid biosynthesis. Metabolism of GlcNAc also releases a molecule of acetate, which can enter catabolic or anabolic processes, depending on the organism and its bioenergetic and nutritional needs (3).
With only a partial tricarboxylic acid (TCA) cycle, the dental caries pathogen Streptococcus mutans depends almost exclusively on the EMP pathway for production of ATP (4, 5). Under carbohydrate excess and low-oxygen conditions, the bacterium converts the product of glycolysis, pyruvate, primarily into lactate via a lactate dehydrogenase (LDH) complex, with concomitant conversion of NADH to NAD+ (6). As a fairly strong organic acid, lactate drives down the environmental pH, accelerating the initiation and progression of human dental caries. LDH in S. mutans is allosterically activated by the glycolytic intermediate fructose-1,6-bisphosphate (F-1,6,-bP) and functions optimally in a pH range of 5.5 to 6.3 for pyruvate reduction (6, 7). Under carbohydrate-limiting conditions, the metabolism of pyruvate proceeds mainly by pyruvate dehydrogenase (PDH) or by an oxygen-sensitive pyruvate formate lyase (PFL), yielding acetyl-coenzyme A (CoA) and converting NAD+ into NADH (8, 9). Many commensal oral streptococci, e.g., Streptococcus gordonii and Streptococcus sanguinis, express a pyruvate oxidase enzyme (SpxB) that converts pyruvate into acetyl-phosphate (AcP) with the concomitant production of hydrogen peroxide (H2O2) (10). S. mutans is known to be sensitive to H2O2, with inhibition of certain enzymes, cellular processes, and growth being evident at the levels of H2O2 that can be produced and well tolerated by many commensal streptococci (11–13). Lacking SpxB, S. mutans converts acetyl-CoA into AcP using the enzyme phosphotransacetylase (Pta) (14). AcP can be acted on by an acetate kinase (AckA) in the presence of ADP to produce acetate and ATP. Notably, many bacteria allow the reverse reaction, converting acetate into AcP and eventually into acetyl-CoA. In addition to its involvement with pyruvate and acetyl-CoA metabolism, AcP appears to have important roles in gene regulation and can serve as a substrate for certain two-component signal transduction systems (15).
GlcN and GlcNAc can significantly impact bacterial fitness and interbacterial interactions in streptococci that are abundant in dental biofilms (16, 17). For example, amino sugars enhanced the ability of two health-associated commensal bacteria, S. gordonii and S. sanguinis, to outcompete S. mutans in vitro in planktonic cultures and in biofilms (16, 17). These observations were generally attributed to (i) more efficient utilization of amino sugars, GlcNAc in particular, by the commensals; (ii) increased release of H2O2 by the commensals growing on amino sugars; and (iii) moderation of acidification of the environment by release of ammonia from the amino sugars and from increased expression of the arginine deiminase (AD) system of the commensals growing on amino sugars (18). Conversely, growth on amino sugars was also reported to result in enhanced acid tolerance and increased bacteriocin (mutacin) production by S. mutans (16, 19). Furthermore, in a human saliva-derived ex vivo microcosm model, GlcNAc, but not GlcN, was shown to inhibit the persistence of S. mutans (16). In light of the profound impact of amino sugars on both beneficial properties of commensals and virulence-related attributes of S. mutans, a more comprehensive understanding of the effects of amino sugars on these oral bacteria is needed, particularly in the context of the ecological dynamics that influence the balance between oral health and diseases. By applying RNA deep sequencing (RNA-Seq) to S. mutans UA159 and S. gordonii DL1 in a batch culture model, we uncovered molecular mechanisms underlying physiological changes and behaviors elicited by growth on amino sugars and with cocultivation when amino sugars are present. The study reveals significant physiological reprogramming in both bacteria that can guide future work to manipulate bacterial ecology to control oral infectious diseases.
RESULTS AND DISCUSSION
RNA-Seq sample clustering.
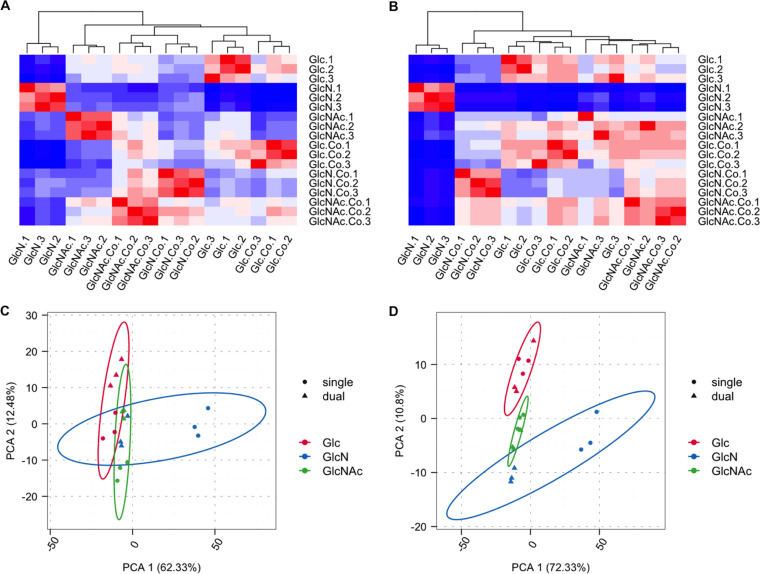
The two main objectives of this study were to catalogue the impacts of GlcN and GlcNAc, compared to glucose, on the transcriptomes of S. mutans UA159 and S. gordonii DL1 and to gain insights into the mechanisms by which these carbohydrates affect competition between these bacteria. To achieve these goals, each organism was grown in tryptone-vitamin (TV) base medium formulated with one of the three carbohydrates, either in single- or dual-species cultures. For each condition, there were three biological replicates. TV medium was chosen over synthetic FMC (20) and TY (3% tryptone, 0.5% yeast extract) medium for specific reasons: S. mutans experiences significant delays in growth on FMC-GlcNAc when transferred from a glucose-based medium (17), and TV lacks the yeast extract in TY that adds contaminating carbohydrates that could influence the interpretation of the results. In toto, 27 mRNA-enriched samples and cDNA libraries were prepared, 18 from single-species cultures and 9 from cocultures. After cDNA sequencing and data analysis, it was determined that there was a sufficient quantity of high-quality reads and sufficient coverage of the genomes of both organisms under all conditions tested, with each individual library yielding an average of about 9 million reads. After mapping all qualified reads to annotated genes in both genomes, 18 RNA-Seq samples of each bacterium were subjected to correlation, clustering, and principal-component analyses (PCA) to assess their overall sample quality and relationships (Fig. 1; see also Fig. S1 in the supplemental material). As visualized in Fig. 1, the replicates from each group showed good consistency in gene expression patterns. Interestingly, the single-species cultures grown on GlcN showed the greatest divergence from the groups grown on glucose or GlcNAc. Cocultivation of S. mutans and S. gordonii with GlcN also showed the greatest impact on both transcriptomes relative to the single-species cultures on GlcN. In contrast, each organism expressed more similar transcriptomes when growing on glucose or GlcNAc as a single species or in cocultures. The tendency for GlcN cultures to diverge from all other samples was even better illustrated by the results of the PCA in Fig. 1C and D.
FIG 1.
Clustering (A and B) and principal-component analysis (C and D) of 18 RNA-Seq samples for S. mutans (A and C) and S. gordonii (B and D). (A and B) Heatmap of the cluster analysis shows the Euclidean distances between the samples, as calculated from the variance-stabilizing transformation of the count data. (C and D) The circles in PCA diagrams represent single-species samples, and the triangles represent mixed-species samples. The red, blue, and green symbols represent samples grown on glucose (Glc), GlcN, and GlcNAc, respectively.
Differential expression analysis.
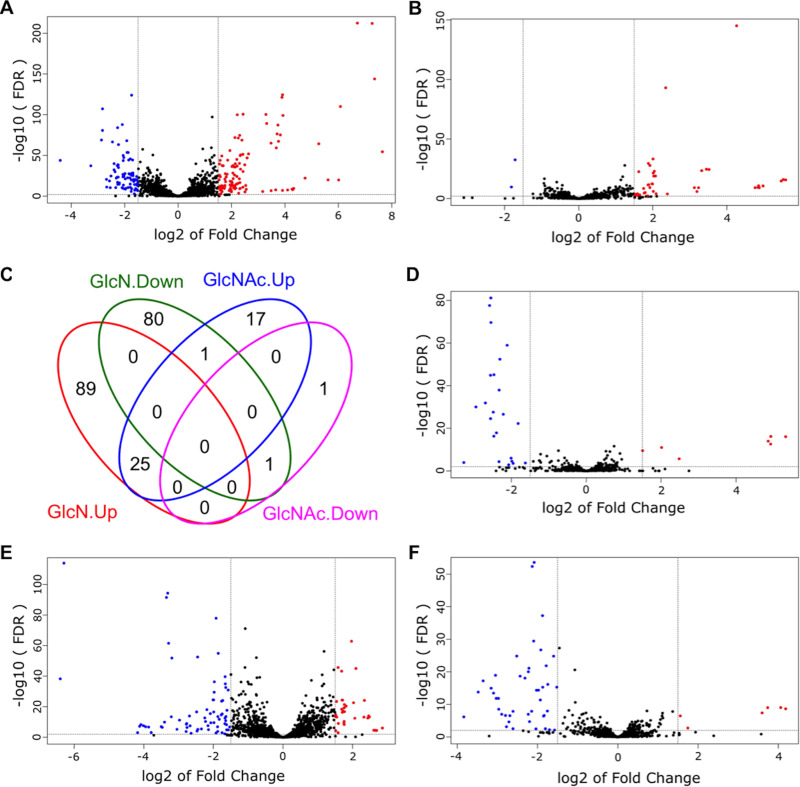
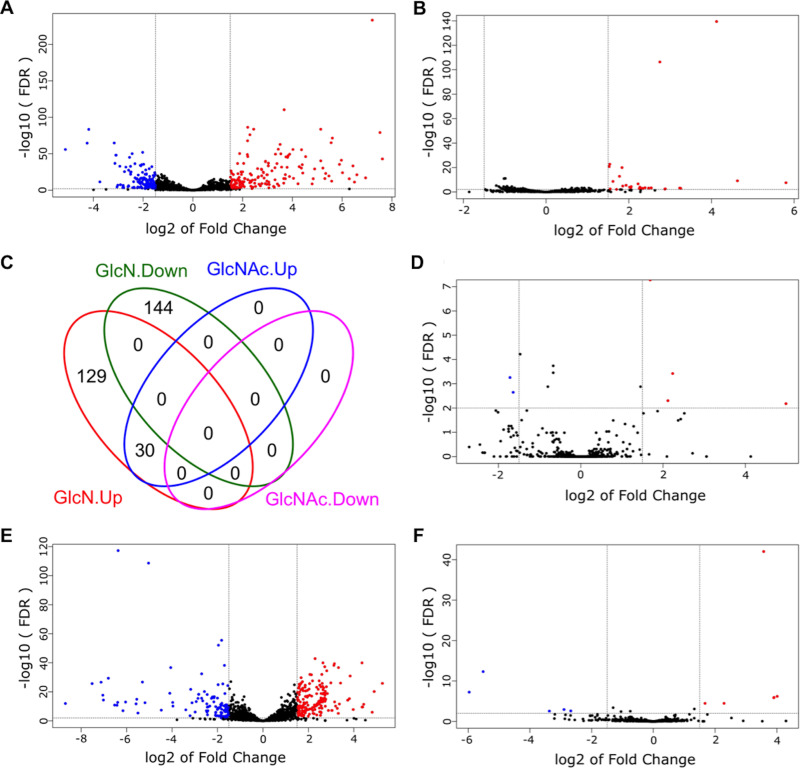
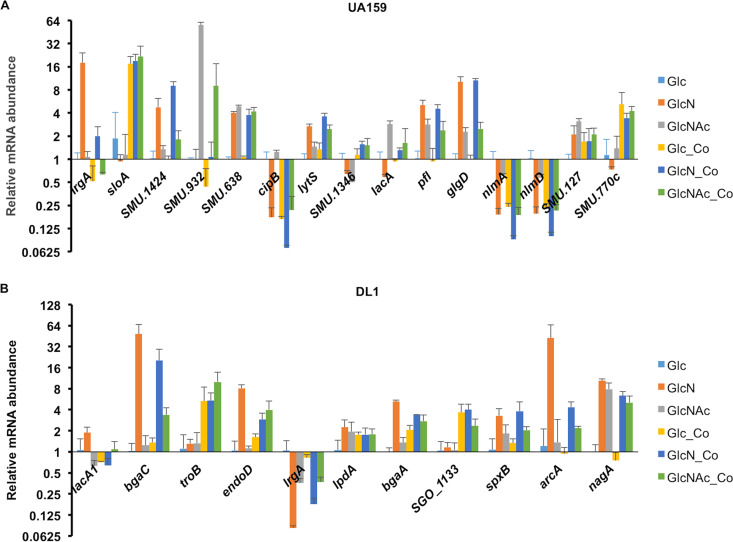
With carbohydrates and cultivation conditions as the two independent variables, we carried out 5 pairwise comparisons for each bacterium. For single-species samples, GlcN and GlcNAc conditions were each contrasted against the glucose (Glc) samples; for cocultures, results from each carbohydrate were compared with results from single-species cultures grown on the same carbohydrate. The cutoff for identifying differentially expressed genes (DEG) was a false discovery rate (FDR) of <0.01 and a |log2(fold change)| of ≥1.5 (or fold change of ≥2.8) in the number of reads for a given mRNA. These results are presented in volcano plots and Venn diagrams (Fig. 2 and 3), with the lists of DEG included in the supplemental material (Table S1 to S7). To better understand the biological processes associated with the DEG, functional enrichment analysis of gene ontology (GO) was performed using DEG for identification of specific pathways, which are shown as Kyoto Encyclopedia of Genes and Genomes (KEGG) maps (see the supplemental material). Subsequently, a subset of genes from both bacteria was selected for validation using reverse transcription quantitative PCR (RT-qPCR) (Fig. 4).
FIG 2.
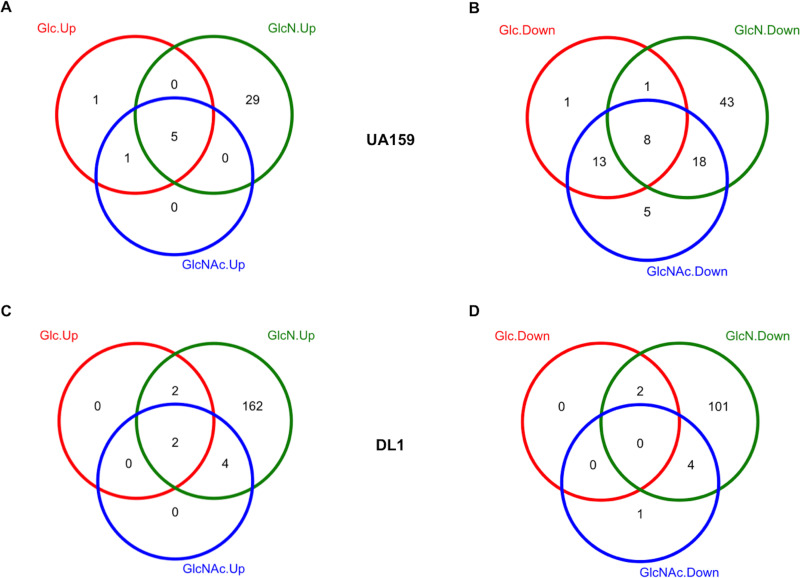
Differentially expressed genes (DEG) from S. mutans UA159 identified by RNA-Seq. Qualified reads were mapped to the genome of UA159, counted for each annotated gene, and normalized across the entire transcriptome. The DEG were identified by comparison of single-species cultures between GlcN and Glc (A) or GlcNAc and Glc (B) and by comparison of mixed-species cultures against single-species cultures, each grown on Glc (D), GlcN (E), or GlcNAc (F). Red circles depict DEG with increased expression, and blue circles depict those with decreased expression. (C) All DEG from panels A and B are presented as one Venn diagram.
FIG 3.
DEG from S. gordonii DL1 identified by RNA-Seq. The DEG were identified by comparison of single-species cultures between GlcN and Glc (A) or GlcNAc and Glc (B) and by comparison of mixed-species cultures against single-species cultures, each grown on Glc (D), GlcN (E), or GlcNAc (F). Red circles depict DEG with increased expression, and blue circles depict those with decreased expression. (C) All DEG from single-species cultures are presented as one Venn diagram.
FIG 4.
RT-qPCR of differentially expressed genes from S. mutans (A) and S. gordonii (B). The results of each gene, including single species and cocultures (-Co), are presented as relative mRNA abundance by comparison to levels measured in single-species cultures supported by glucose (Glc). Each bar represents the average from three biological repeats, with the error bar denoting the standard deviation. Culture conditions for RNA isolation were the same as that for the RNA-Seq analysis.
(i) S. mutans in single-species cultures.
Growth of S. mutans on GlcN and GlcNAc can elicit some similar behaviors, as we previously reported. In particular, both sugars enhance acid tolerance of S. mutans, presumably due to cytoplasmic alkalization by release of ammonia (19), along with increased mutacin production, as assessed in a deferred antagonism assay on TY agar formulated with the different sugars (16). Conversely, there are GlcNAc-specific effects that include an extended lag phase when S. mutans is transferred from glucose-containing medium to a medium with GlcNAc as the primary carbohydrate source. (17). There is also a significant reduction in persistence of the bacterium in a human-derived ex vivo consortium biofilm model provided with GlcNAc as the main carbohydrate (16).
(a) UA159 on GlcN alone. When S. mutans UA159 was cultivated alone in TV-GlcN rather than TV-glucose, 114 annotated genes showed significantly increased expression, while 82 others had decreased expression (Fig. 2A and 4A and Table S1). The differentially expressed genes mainly included pathways for carbohydrate metabolism: PTS, glycolytic enzymes, pyruvate metabolism, and gene products for use of cellobiose, galactose, lactose, sucrose, starch, and glycogen. Additionally, amino acid transport, biosynthesis, and metabolism of lysine, serine, threonine, and methionine, along with nucleotide metabolism and bacteriocin production, were substantially affected by growth in GlcN compared with that of cells growing on glucose (Fig. S2 depicts KEGG visualization). Notably, two operons in S. mutans UA159 showed the greatest increase in mRNA levels when growing on GlcN: the pyruvate dehydrogenase operon (PDH; SMU.1421 to SMU.1425 [SMU.1421-SMU.1425]; increased by 68- to ∼147-fold) and a pyruvate transporter operon (lrgAB; SMU.574c-SMU.575c; increased by 137- to ∼156-fold) (21) together with the genes encoding the two-component signal transduction system that activates lrgAB (lytTS; SMU.576-SMU.577; increased by 5-fold) (22). It was recently discovered that S. mutans excretes pyruvate as an overflow metabolite under glucose-rich conditions, only to reinternalize the pyruvate once glucose is depleted in the environment (21). Another apparent orthologue of the PDH complex, SMU.127-SMU.130, also showed about a 2-fold increase in mRNA levels (Fig. 4A). Genes homologous to this particular PDH operon are widely conserved in multiple streptococcal species, including S. gordonii, Streptococcus pneumoniae, and group A streptococci. Two other genes encoding enzymes for pyruvate metabolism, pyruvate formate lyase (PFL; SMU.402) and PFL-activating enzyme (pflC; SMU.490) (9), were upregulated about 4-fold in cells grown on GlcN compared to cells grown on glucose. Under carbohydrate-rich and low-oxygen tension conditions, S. mutans generally depends on glycolysis and lactate generation to meet its demand for energy production and growth. These alternate pyruvate metabolic pathways are often activated when energy sources are limiting, as the use of pyruvate by these routes yields greater amounts of ATP per molecule of carbohydrate catabolized.
Also of significance, a number of genes encoding mutacins and related proteins (23), including nlmA, nlmB, SMU.152-SMU.153, cipB (also known as nlmC), nlmD, nlmE, and nlmT, showed decreased mRNA levels in GlcN-grown cultures (Fig. 4A and Table S1). This was a surprising finding, as we reported previously that S. mutans UA159 showed enhanced killing of S. sanguinis SK150 in a deferred antagonism assay using TY agar, not the TV medium used here, when supplemented with amino sugars (16). Insights into why these differences exist are provided below.
(b) UA159 on GlcNAc alone. When S. mutans was cultured on TV-GlcNAc, 43 genes showed higher mRNA levels, and only 2 genes showed lower mRNA levels, relative to cultures growing on glucose (Fig. 2B and 4A and Table S2). Among the DEG with the highest increase in expression were an ABC transporter operon (SMU.651c-SMU.653c; >40-fold increase) with unknown substrate(s), along with a putative amino acid transporter operon (SMU.932-SMU.936; 23- to 28-fold increase) and its nearby genes (SMU.930c; 8-fold increase; SMU.937-SMU.939; ∼2-fold increase). A significant portion of these 43 genes was involved in energy metabolism, followed by genes annotated for nucleotide metabolism; most notably, a 20-gene cluster, ranging from SMU.29 to SMU.59 and annotated for purine biosynthesis, showed 2- to 6-fold increases in mRNA levels.
Twenty-five genes were more highly expressed in both GlcN and GlcNAc (Fig. 2C and Table S3) than glucose. Most notable were the nagA and nagB genes, encoding GlcNAc-6-P deacetylase and GlcN-6-P deaminase, respectively. The expression of these genes is known to be induced by growth on these amino sugars (19). Some genes linked to nagAB were also differentially expressed. Other genes showing increased expression under both conditions were the tagatose pathway (SMU.1492-SMU.1496) and the aforementioned two ABC transporter operons (SMU.932-SMU.936 and SMU.651c-SMU.653c). As reported previously, growth on amino sugars only downregulated the glmS gene, encoding the GlcN-6-P synthase (19). In light of the results showing the reduced expression of mutacin genes on GlcN, we further examined the expression of these genes in GlcNAc-grown cultures. As shown in Fig. 4A, compared to glucose, GlcNAc reduced the message levels of nlmAB and nlmD, but the reduction was not as significant as that seen with growth on GlcN. No effect of GlcNAc was noted for cipB.
Metabolism of GlcN differs from that of glucose primarily in the deamination of GlcN-6-P by NagB, thereby releasing an ammonia molecule intracellularly (19). In addition to the generation of ammonia, the degradation of GlcNAc by S. mutans also requires deacetylation of GlcNAc-6-P, yielding a molecule of acetate. This was confirmed by measurements of acetate in bacterial cultures of both S. mutans and S. gordonii, prepared using different carbohydrates (Fig. S3). Since the expression of both nagA and nagB is coregulated by the GntR-type regulator NagR in association with the allosteric effector GlcN-6-P, produced by catabolism of both amino sugars (24), the drastically disparate gene expression patterns among these three carbohydrate conditions most likely are the outcome of cellular responses to distinct physiological conditions. Under these batch culture conditions, where carbohydrate is not limiting, LDH is likely the main pathway for metabolizing pyruvate generated by glycolysis, concurrently regenerating NAD+ and yielding lactate as the end product. The enzymatic activity of LDH in S. mutans functions optimally between pH 5.5 and 6.3, and the cells also have more LDH protein when growing at pH 5.5 than 7.0 (7). We reason that the release of ammonia by the degradation of GlcN and GlcNAc increases intracellular pH (19) and reduces LDH activities, which in turn raises intracellular pyruvate levels, redirecting carbon flow from the generation of lactate to the PDH and/or PFL pathway for the generation of acetate or acetate and formate, respectively. Since the metabolism of GlcNAc also releases a molecule of acetate, acetate accumulation could create negative feedback pressure upstream of PDH and/or PFL (14); thus, the forward reactions of these alternative pyruvate dissimilation pathways may be inhibited, possibly allowing the accumulation of pyruvate and even acetyl phosphate. It is also worth noting that S. mutans organisms growing on GlcN generated more ammonia and reached higher final pH values than those growing on GlcNAc (19). This difference between two amino sugars could be due to the faster catabolism of GlcN or more ammonium ions being directed into anabolic pathways in cells growing on GlcNAc.
Another, perhaps equally important, outcome of reduced LDH activity could arise from changes in the NAD+/NADH ratios. Balancing NAD+/NADH is critical in S. mutans, and imbalances could disrupt glycolysis and intracellular redox balance, either of which could have a profound impact on global gene expression (25). One example of this likely compensatory response is the increased expression of the glucose-PTS (EIIMan; manLMN) that was shown to be mediated by the redox sensor Rex (26). Further examination of the RNA-Seq data indicated that the ldh gene in S. mutans showed a modest reduction (∼40%) in expression in cells growing on GlcN compared to glucose, whereas the pfl gene, pflC, and the second PDH operon (SMU.127-SMU.130) showed significantly increased expression in cells growing on GlcNAc, 3-fold, 3-fold, and 2-fold, respectively (Fig. 4A and RNA-Seq data not shown). Expression of the manLMN operon also was increased by about 2-fold in both GlcN and GlcNAc cultures. However, transcripts of the PDH operon (SMU.1421-SMU.1425) showed no change in GlcNAc.
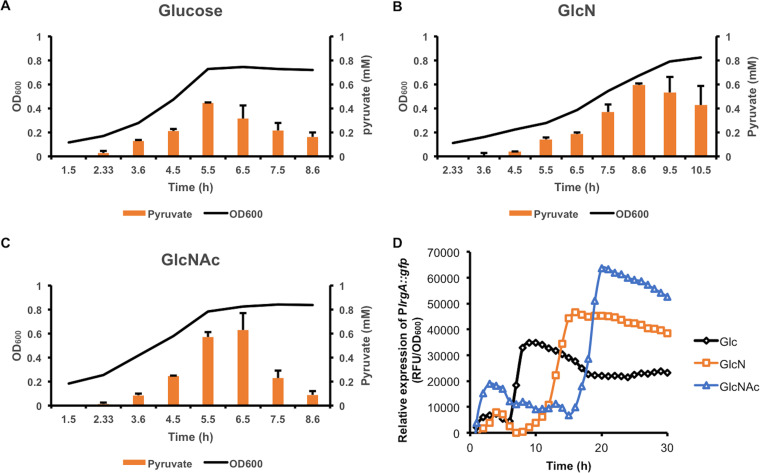
To further test the hypothesis that amino sugars affected pyruvate metabolism, we measured extracellular pyruvate levels in S. mutans UA159 cultured on glucose, GlcN, or GlcNAc in different growth phases. As pyruvate is known to be excreted into the environment by S. mutans, we expected that a reduction in LDH activity could increase the pyruvate concentrations in the cells and culture supernatants. The results (Fig. 5) showed that as the optical density of the cultures increased, so did the levels of extracellular pyruvate, regardless of the carbohydrate source. As the glucose cultures entered stationary phase, the pyruvate levels declined rapidly and all but disappeared in a matter of hours, likely due to reinternalization by the LrgAB transporter (21). In comparison, the peak extracellular pyruvate levels in cultures growing on GlcN were about 33% higher than those in glucose cultures (P < 0.001). Furthermore, although it took slightly longer to reach the peak levels on GlcN, the rate at which the pyruvate levels declined afterwards was also significantly slower. In GlcNAc-supported cultures, the peak pyruvate levels again were higher (by 30 to ∼40%) than those in glucose cultures (P < 0.05); however, the decline followed a rapid trajectory, similar to that of cells growing on glucose.
FIG 5.
Measurements of extracellular pyruvate levels and PlrgA::gfp expression in S. mutans cultures. Strain UA159 was cultured in an FMC base medium supplemented with 10 mM glucose (A), GlcN (B), or GlcNAc (C), with pyruvate levels measured at regular intervals at various stages of growth. The optical density (OD600; black line) was plotted along with the corresponding pyruvate levels (orange bars) against time. Each bar represents the average from three biological repeats, with the error bar denoting the standard deviation. (D) UA159/PlrgA::gfp was similarly cultured on FMC and monitored for growth and GFP signals for a period of 30 h.
Transcription of the lrgAB genes is induced by the release of pyruvate (21). As such, we performed a promoter::reporter fusion analysis on a UA159 derivative carrying a PlrgA::gfp fusion (21) by monitoring green fluorescent protein (GFP) levels in cells growing on glucose, GlcN, or GlcNAc. Our results (Fig. 5D) showed increased lrgA promoter activities when cells were growing on GlcN and especially on GlcNAc compared to glucose, although the peak promoter activities appeared later in cells growing on GlcN and even later in those growing on GlcNAc. It is worth noting that the UA159 cultures used in RNA-Seq were harvested during mid-exponential phase, when lrgA expression likely remained low in cells growing on GlcNAc. This detail provides an explanation for the lack of increase in lrgA transcript under GlcNAc conditions (Fig. 4A). Together, these results are consistent with our hypothesis that the reprogramming of pyruvate metabolism occurred as a result of the differences in physiological conditions in cells catabolizing the three different carbohydrates.
(ii) S. gordonii cultured alone.
Previous research on amino sugar metabolism by S. gordonii and related commensals (16, 17) has shown enhanced competitiveness of the commensals against the oral pathogen S. mutans, primarily through increased production of H2O2 (with both amino sugars) and higher expression of the arginine deiminase pathway, mostly notably with GlcN. However, little is known regarding the underlying mechanisms responsible for these important phenotypes or the more global responses of commensals to these sugars.
(a) DL1 on GlcN alone. When single-species cultures of S. gordonii DL1 were growing on TV-GlcN, 159 genes showed increased expression and 144 showed decreased expression compared to cells growing in TV-glucose (Fig. 3A and 4B and Table S4). The majority of these DEG belonged to carbohydrate metabolic pathways, but significant proportions also involved nucleotide and amino acid metabolic networks (Fig. S4 depicts KEGG visualization). Similar to S. mutans growing on GlcN, most of these carbohydrate metabolic genes showed significantly increased expression in the presence of GlcN, including two extracellular β-galactosidases, bgaC-EII operon (SGO_0043-SGO_0048; increased by 29- to ∼210-fold) (27, 28) and bgaA (SGO_1486; increased by 9-fold) (28), two putative glucan-binding proteins (SGO_0477-SGO_0478; increased by ∼40-fold), two Leloir pathway genes (galKT; increased by 6- to ∼7-fold), an endo-β-N-acetylglucosaminidase D (endoD; 6-fold increase) (27), a 17-gene, two-cluster system encoding the tagatose pathway and PTS permeases required for metabolism of lactose and galactose (SGO_1512-SGO_1526; increase ranging from 5- to 100-fold) (28), a putative glycogen biosynthetic operon (SGO_ 1549-SGO_1554; increased by 3- to 5-fold), a trehalose metabolic operon (SGO_1652-SGO_1653; increased by 69- and 116-fold, respectively), a putative fructose/mannose PTS operon (SGO_1890-SGO_1893; increased by ∼3-fold), and a putative sugar-binding ABC transporter operon (SGO_ 1763-SGO_1765; increased by 22- to 28-fold). Also increased in expression were two enzymes likely involved in the production of H2O2: the pyruvate oxidase spxB cluster (10) (SGO_0289-SGO_0293; increased by 5- to ∼20-fold) and the NADH oxidase gene (nox; SGO_1167; increased by 5-fold), whose homologue in S. sanguinis is required for H2O2-mediated inhibition of S. mutans (29). This result was supported by direct measurements of H2O2 levels in liquid cultures of DL1 on different carbohydrates (Fig. S5) as well as previous research using clinical isolates of S. gordonii (16). The only pyruvate dehydrogenase operon in S. gordonii (SGO_1130-SGO_1133; SGO_1130 = lpdA; Fig. 4B) showed 4- to 7-fold increases in expression on TV-GlcN. However, the expression of two cidAB-lrgAB orthologues (SGO_1268-SGO_1269) decreased by ∼20-fold. Pyruvate overflow metabolism has been reported in S. gordonii (30), although it is not known if the cidAB-lrgAB gene products contribute to pyruvate metabolism in ways similar to that in S. mutans.
Among DEG related to amino acid metabolism was the arginine deiminase (AD) operon (arcABCDT), which showed a 3- to 6-fold increase in expression in GlcN compared to that in glucose. This result is consistent with the reported increase in AD activities of S. gordonii growing on amino sugars (17). It is also known that arc operon expression in DL-1 is induced by arginine, repressed by oxygen, and sensitive to catabolite repression (31). Conversely, many genes required for protein biosynthesis, most notably 17 ribosomal proteins, 18 tRNAs, and 5 amino acid-tRNA synthetases, showed significantly reduced expression (Table S4). A substantial number of genes from the nucleotide metabolism network also showed reduced expression in GlcN-grown cells. The simplest interpretation of these findings is that S. gordonii growing on GlcN scaled down the biosynthesis of proteins and nucleotides, which are required for growth, while directing more carbon to energy production. Similar to S. mutans, it is probably safe to assume that metabolism of GlcN, as well as GlcNAc, increases the intracellular pH of S. gordonii. Although the effects of pH on LDH from S. gordonii are not well characterized, LDH activity is likely lower when intracellular pH is elevated (6). This notion is supported by the increased expression of the genes for the pyruvate-oxidizing enzymes PDH and SpxB and increased H2O2 production in the presence of GlcN (Fig. 4B and Table S4 and Fig. S5) (16).
(b) DL1 on GlcNAc alone. When S. gordonii DL1 was cultured on TV-GlcNAc, only 30 genes showed increased expression compared to glucose cultures, and no genes were identified that had decreased expression (Fig. 3B and 4B and Table S4). Interestingly, all 30 genes also showed increased expression in GlcN cultures (Fig. 3C). The 30 genes are almost exclusively dedicated to carbohydrate metabolism, encompassing the nag pathway, the lactose/galactose cluster, the trehalose operon, the extracellular β-galactosidases BgaA/BgaC, and EndoD. For comparison, in S. mutans these two amino sugar regulons overlap by 25 genes, yet the GlcNAc regulon of S. mutans contained an additional 18 unique DEG that were not differentially expressed in cells growing on GlcN.
Current research supports roles for EndoD and BgaC in the ability of S. gordonii to grow on saliva by sequentially releasing monosaccharides from the basic proline-rich glycoproteins, in concert with the activity of the β-N-acetylglucosaminidase StrH (27). Since amino sugars and galactose are likely released during glycoprotein degradation but GlcN and GlcNAc were the only carbohydrates used in our cultures, it is possible that amino sugars or their metabolic derivatives are sufficient to activate the transcription of these secreted glycohydrolases, along with the tagatose pathway that is required for catabolizing galactose. Alternatively, some of these genes showing altered expression on both amino sugars may be due to shifts in pyruvate metabolism, as discussed above.
(iii) S. mutans and S. gordonii cultured together.
(a) Coculture on Glc. Cocultivation with S. gordonii on glucose presented a clear oxidative stress to S. mutans, apparently due to H2O2 production, as UA159 showed enhanced expression in the slo operon (SloABCR; SMU.182-SMU.186), and a two-gene cluster, SMU.768c-SMU.770c, that encodes a metal ion transporter, MntH (Fig. 4A and 6 and Table S5), compared to S. mutans growing in monoculture on glucose. SloR is a manganese-responsive metalloregulator that is required for S. mutans response to oxidative and acidic stressors (32, 33), and SloABC is a Mn2+/Fe2+ transporter that works in concert with MntH as the primary means of Mn2+ uptake in S. mutans (34, 35). The capacity of SloR to regulate both the slo operon and mntH, along with the known positive effects of Mn2+ on S. mutans aerotolerance, suggest that H2O2 production by DL1 triggered the induction of these genes. At the same time, cocultivation with S. gordonii reduced expression of various mutacin genes, namely, the nlmAB operon, nlmD, and the cluster SMU.1902c-SMU.1914c, which includes cipB. The decrease in mutacin gene expression is likely due, at least in part, to production by DL-1 of the protease challisin (Sgc). Sgc is secreted by DL1 and can degrade competence-stimulating peptide (CSP), which is required for mutacin expression (36). Notably, the pyruvate transporter gene lrgA was also identified as being downregulated in cocultured S. mutans.
FIG 6.
Venn diagrams comparing DEG identified in S. mutans UA159 (A and B) and S. gordonii DL1 (C and D) in mixed-species cultures compared to single-species cultures supported with the same carbohydrates. The data were grouped based on the direction of change: up (A and C) or down (B and D).
For S. gordonii DL1, cocultivation with S. mutans on glucose had very little impact on its transcriptome, with only 3 tRNA genes showing increased transcription and an alcohol-acetaldehyde dehydrogenase being expressed at lower levels (Table S5).
(b) Coculture on GlcN. Relative to cocultures on glucose, cocultivation on GlcN affected both bacterial transcriptomes in substantial ways, especially for S. gordonii. A total of 34 genes in S. mutans showed higher expression than S. mutans growing alone on GlcN, including 5 from the slo operon and 6 from the Tnsmu2 genomic island that encodes enzymes required for biosynthesis of mutanobactin, a pigment important for oxidative stress resistance and for cross-kingdom interactions with Candida albicans (37, 38) (Fig. 6 and Table S6). Downregulated genes (a total of 70) in S. mutans included cipB and a variety of pathways involved in carbohydrate metabolism, including two fructose-PTS operons (SMU.113-SMU.116 and SMU.1956c-SMU.1961c), the PDH operon (SMU.1421-SMU.1425), pflC, and the pyruvate transporter encoded by lrgAB. Also downregulated was a putative amino acid transport cluster, SMU.930c-SMU.936. The downshift in pyruvate metabolic enzymes is an indication that cocultivation with S. gordonii reduced the availability of extracellular pyruvate for S. mutans, as pyruvate is a known H2O2 scavenger (39, 40). The reaction of pyruvate and H2O2 at neutral pH yields carbon dioxide, acetate, and water. The effect of H2O2 also could have affected mutacin production, as a major sensor kinase, VicK, which is upregulated under oxidative stress (41), is required for optimal mutacin gene expression in S. mutans (42). Compared to cocultures grown on glucose, further reduction in all known mutacin genes was noted in cocultures grown on GlcN (Fig. 4A and Fig. S6). Again, we believe this was due in part to the effect of the challisin protease, as the higher expression of the sgc gene was detected in cocultures prepared with GlcN than with the other two sugars. Interestingly, such increased expression in sgc itself is likely independent of carbohydrates, as in single-culture format sgc showed reduced expression on GlcN compared to that of the other two carbohydrates (Fig. S6).
In S. gordonii, 170 genes were upregulated in cocultures with S. mutans growing on GlcN compared to that of S. gordonii grown on GlcN alone. The most prominent part of the transcriptome affected by the presence of S. mutans belonged to the protein biosynthetic network, including 34 ribosomal proteins, 16 tRNAs, 7 amino acid-tRNA synthetases, and a number of amino acid transporter clusters (Fig. 6 and Table S6). Conversely, among the 107 genes showing reduced expression, a significant portion were involved in carbohydrate metabolism, including 4 PTS operons, the lactose/galactose cluster encoding the tagatose pathway, the fructanase gene fruA, β-galactosidase gene bgaA, the glycogen operon, and others (Table S6). Contrasting this with the impact of GlcN on S. gordonii growing alone (Table S4), it can be inferred that in cocultures with S. mutans, S. gordonii switched central metabolism from energy production into biosynthesis and growth (Fig. S7 depicts the KEGG map). The basis for this behavior is not clear, but understanding the triggers for the activation of growth by the commensal in the presence of S. mutans could provide some novel insights into ways to favorably modify oral biofilm ecology.
(c) Coculture on GlcNAc. The cocultivation of these two bacteria in TV-GlcNAc resulted in a limited impact on gene expression, especially for S. gordonii, a result (Fig. 6 and Table S7) that was more similar to that seen with TV-glucose than TV-GlcN. For S. mutans, cocultivation elevated the expression of the slo operon and mntH gene, which we again attribute at least in part to the generation of H2O2 by DL1. Again, the nlmAB, nlmD, and cipB cluster showed significant reductions in mRNA levels. S. gordonii showed only 6 genes with increased expression, including a putative Mn transporter cluster. Only 4 genes showed reduced expression in S. gordonii, all of which were related to carbohydrate metabolism.
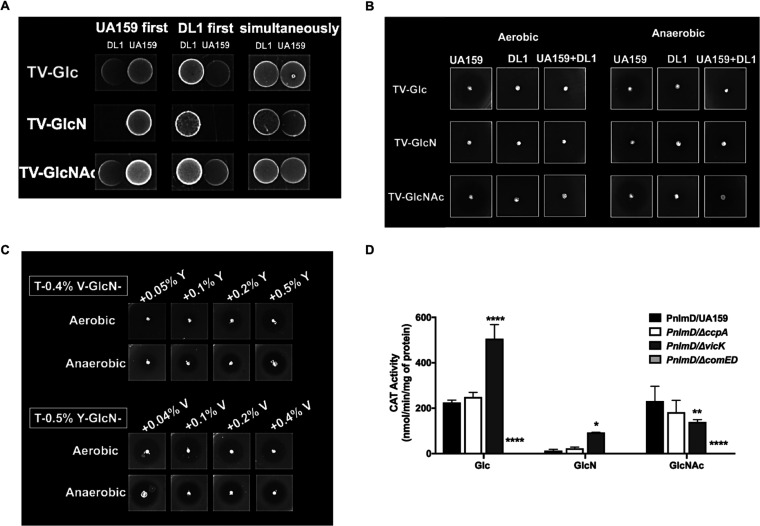
As a complement to the RNA-Seq analysis on mixed-species cultures, we carried out a plate-based competition assay by inoculating S. mutans and S. gordonii in close proximity, either sequentially or simultaneously, on TV base agar plates supplemented with glucose, GlcN, or GlcNAc. After incubation in the same aerobic environment as that used in the RNA-Seq study, it was clear that while these two bacteria generally coexisted on plates containing glucose or GlcNAc, they exhibited significant antagonism in the presence of GlcN (Fig. 7A). This was especially clear when one of the species was introduced 1 day earlier than the other, where the earlier colonizer consistently outcompeted the other species. These observations are consistent with the drastic shifts in transcriptomes shown by both bacteria when cocultivated on GlcN.
FIG 7.
Bacteriocin expression by S. mutans is regulated by multiple environmental factors. (A) S. mutans UA159 and S. gordonii DL1 were spotted on TV agar plates supported by glucose, GlcN, or GlcNAc, either simultaneously or 1 day apart, followed by incubation in an aerobic environment supplemented with 5% CO2. (B and C) UA159 was tested in a deferred antagonism assay under specified nutritional and atmospheric conditions in the presence (B) or absence (B and C) of DL1. (D) To study the regulation of a mutacin gene, nlmD, a PnlmD::cat fusion was examined in the wild-type UA159 and mutant derivatives deficient in ccpA, vicK, or comED, with carbohydrate being the other variable. Each bar represents the average CAT activity from three biological repeats, with the error bar denoting standard deviation. Asterisks indicate statistical significance compared to the wild type growing on the same carbohydrates according to two-way analysis of variance (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
The effects of amino sugars on bacteriocin production is impacted by nitrogen source and redox signal.
As noted earlier, growth on TV-GlcN resulted in reduced expression of mutacin gene clusters in S. mutans (Fig. 4A and Table S1), and this contradicted our previous findings on TY agar plates, where amino sugars appeared to increase the ability of S. mutans to kill in a deferred antagonism assay (16). To address this contradiction, we carried out a deferred antagonism assay on TV agar plates containing glucose, GlcN, or GlcNAc using S. sanguinis SK150 as an indicator strain (16). The results (Fig. 7B) showed substantially less killing of the indicator strain by S. mutans only on plates containing GlcN, in aerobic or anaerobic environments, confirming the results observed by RNA-Seq analysis. The coinoculation of S. gordonii together with S. mutans suppressed the production of mutacins under all conditions, which is also consistent with the results of RNA-Seq showing reduced expression of nlm genes in cocultures.
The main difference between TV- and TY-based media is the lack of yeast extract in the former and the lack of a vitamin mixture in the latter. To determine which factor was responsible for the differences in bacteriocin production, we created a modified TV agar by adding increasing amounts of yeast extract (0.05% to 0.5%) to the medium and a modified TY agar with increasing amounts of vitamin mix (0.04% to 0.4%), all with the same amounts of GlcN as the sole carbohydrate. When we repeated the deferred antagonism assay on these plates, S. mutans (Fig. 7C) showed a concentration-dependent response to yeast extract in the size of the zones of inhibition formed on TV media, producing greater inhibition zones with higher concentrations of yeast extract. Conversely, the addition of increasing amounts of the vitamin mix to TY agar had no impact on antagonism. Previous research (23, 43, 44) suggested that the production of mutacins and the expression of relevant genes require both nitrogen-rich ingredients, such as yeast extract, and specific carbohydrates, each provided at optimal levels. Here, we show that amino sugars can affect mutacin production by S. mutans in both directions, dependent on the overall nutritional state of the bacterium.
To further explore the transcriptional regulation of mutacins by carbohydrates, we utilized a PnlmD::cat reporter fusion (45) and studied the response of the nlmD promoter to amino sugars. Based on behaviors seen in the RNA-Seq experiments, we also included mutant strains lacking CcpA (46), VicK (42), or the CSP-responsive two-component system ComED, required for mutacin expression (47). The study was carried out using TV media in an aerobic atmosphere containing 5% CO2. In agreement with the transcriptomic analysis (Fig. 4A), the results of the chloramphenicol acetyltransferase (CAT) assays in wild-type UA159 (Fig. 7D) showed a clear reduction in nlmD expression in GlcN-grown cultures relative to that of glucose cultures; however, little difference was noted when using GlcNAc. The elimination of catabolite control protein CcpA had no impact on nlmD expression on any of the carbohydrates tested. However, the loss of the histidine kinase VicK altered the expression of nlmD under all three conditions: the promoter activity more than doubled with glucose or GlcN, yet it showed significant reductions with GlcNAc. As a control, loss of the ComED two-component system caused complete loss of nlmD promoter activity under all conditions. Mutacins are regulated by various environmental factors and by multiple genetic systems that often interact and overlap in functionality (23, 48). Previous research has shown the capacity for the VicRK system to negatively regulate mutacin genes by affecting the expression and function of the ComED system (42), as well as effects of VicRK on oxidative stress (41) and in trans-phosphorylating GcrR/CovR in a Mn-dependent manner (48); GcrR/CovR is an orphan response regulator involved in acid tolerance and biofilm development (49, 50). Together, these results and the RNA-Seq analysis suggest that the intersection of redox signaling, essential ions, and nutritional status drives the bacterium to balance the needs for growth, energy production, and defense.
Concluding remarks.
The release of pyruvate during carbohydrate metabolism has been reported in a few oral bacteria, including S. mutans and the commensal bacteria S. gordonii and S. sanguinis. Overflow metabolism allows the bacterium to achieve rapid growth and lactic acid production when carbon sources are plentiful, and subsequent uptake of extracellular pyruvate allows extended generation of ATP when resources become limited. Since S. gordonii also releases H2O2 via the action of SpxB on pyruvate and pyruvate is an H2O2 scavenger, the role of extracellular pyruvate in the physiology and ecology of commensal streptococci warrants further study. However, one reasonable explanation is that the release of H2O2 evolved as a weapon against bacteria such as S. mutans, while pyruvate serves to protect the commensal from self-harm (30). In the case of S. mutans, which lacks SpxB and does not produce significant quantities of H2O2, pyruvate should also provide some protection from oxidative damage from H2O2. Utilizing RNA-Seq instead of a reductive approach, we were able to identify the impact of two amino sugars on pyruvate metabolism on S. mutans and S. gordonii individually and when growing together. Through the release of metabolic by-products, namely, ammonia and acetate, amino sugars could initiate significant perturbations in pyruvate metabolism, a homeostasis that is subject to influence by environmental factors, including carbohydrate and nitrogen sources, oxidative stressors (O2 and H2O2), and essential metal ions. These adaptations in turn result in carbon flux redistribution among energy and acid production, growth, and biosynthetic pathways. We also recognize that the current study is limited by the amount of biochemical evidence and knowledge available in literature and assays performed here, including enzymatic activities and levels of key metabolites being produced by oral bacteria growing on amino sugars, in particular for commensal streptococci. While we continue improving this working model through other comprehensive approaches, these findings allow us to expand our insights into amino sugar metabolism by other species of the dental biofilm and explore the potential of utilizing metabolic modulation toward caries management and intervention.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Three bacterial species were used in this study: S. mutans UA159 and derivatives therefrom, S. gordonii DL1, and S. sanguinis (strain SK150). Brain heart infusion (BHI; Difco Laboratories, Detroit, MI) medium was used in both liquid form and agar for maintenance of bacterial strains. For experiments carried out with carbohydrates as the sole variable, a semidefined tryptone-vitamin (TV) base medium (51), a TY base medium (3% tryptone, 0.5% yeast extract), or a modified chemically defined FMC medium (20) was used with addition of the specified amounts of carbohydrates. TV and TY base media could be prepared in advance, with carbohydrates added immediately before use and the pH of the medium adjusted to 7.0 after addition of the carbohydrate. All bacterial cultures were maintained at 37°C in an aerobic atmosphere supplemented with 5% CO2 or an anaerobic environment established using an AnaeroPack (Thermo Fisher Scientific, Waltham, MA). Growth analyses were performed on UA159 and DL1 under three carbohydrate conditions, and the results showed that while UA159 grew at a slightly lower rate on two amino sugars than glucose, DL1 grew equally well on all three sugars (see Fig. S8 in the supplemental material).
For RNA extraction, and for most assays performed in this report, bacterial strains UA159 and DL1 were first cultivated overnight at 37°C in an aerobic atmosphere supplemented with 5% CO2, each with at least 3 individual cultures, on TV medium supplemented with 20 mM glucose, GlcN, or GlcNAc (pH 7.0) and then diluted 1:20 ratio into fresh TV medium containing the same carbohydrate that was used to grow the inocula. When the optical density at 600 nm (OD600) of an individual culture reached 0.5, a portion of the culture was placed on ice temporarily before use. The rest of the culture was harvested by centrifugation, treated with the RNAprotect bacterial reagent (Qiagen, Germantown, MD), and stored at −80°C. Once all cultures were ready on ice, UA159 and DL1 that had been grown on the same carbohydrates were mixed in a 20:1 ratio based on their OD600 values, and then the mixture was diluted 1:50 ratio into the same medium used to prepare the inocula. To ensure that approximately equal numbers of bacteria from each species were present at the end of the cultivation, the ratio at which these two strains were being mixed was determined empirically by enumerating their final CFU on agar plates with and without bacitracin. After the dual-species culture reached an OD600 of 0.5, cells were harvested as before and stored at −80°C.
RNA extraction, RNA-Seq, data analysis, and RT-qPCR.
Total RNA samples from single- and dual-species cultures were extracted using the RNeasy minikit (Qiagen) by following an established protocol detailed elsewhere (19). To enrich mRNA by removing 16S and 23S rRNAs from each RNA sample, 10 μg of total RNA was treated, twice, using the MICROBExpress bacterial mRNA enrichment kit (Ambion Life Technologies, Grand Island, NY), as directed by the suppliers. The quality of enriched mRNA samples was assessed using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). cDNA libraries were constructed from 100 ng of enriched mRNA samples by using a NEBNext Ultra directional RNA library preparation kit for Illumina and a NEBNext multiplex oligonucleotides for Illumina (New England BioLabs, Ipswich, MA) for barcoding. Parallel deep sequencing was carried out by the NextGen DNA Sequencing Core Laboratory of ICBR at the University of Florida (Gainesville, FL).
The analysis of the RNA-Seq expression data was conducted on R (52), versions 3.3.2 “Sincere Pumpkin” and 3.3.3 “Another Canoe.” The compositional matrix of the expression data was normalized with the “voom” (53) function from the R package “limma” (54). These normalized data sets were used to generate heatmaps, correlation plots, and PCA plots. Correlation and heatmaps of the compositional matrix were plotted with the “corrplot” package (55) and the enhanced “heatmap.2” function from the “gplots” (56) package, respectively. The data for PCA plots were extracted from the “prcomp” base function output and used to generate plots with the package “ggplot2” (57) in combination with the “ellipse” (58) function to produce the ellipse confidence regions. The overlapping set of genes under different conditions were plotted as Venn diagrams with the function “vennPlot” from the package “systemPipeR” (59). The statistical analysis of the expression data was conducted by using the DESeq2 pipeline (4) from the homonymous R package. To monitor pairwise comparisons required by the experiment, the contrasts were parsed to the pipeline by the results function. The pipeline output data included both a P value and a multiple-comparison-adjusted P value. A false discovery rate (FDR) of 0.01 was used as a cutoff value to call the significant genes in the results and for the volcano plots as well. Subsequently, KEGG functional pathways and gene ontology (GO) terms were generated individually for all data sets using DAVID bioinformatic resources (60). Pathway maps were generated using the KEGG mapping tools from KEGG resources (61).
RT-qPCR was carried out for confirmation of selected genes after RNA-Seq analysis. RT was carried out using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) on 0.5 μg of total RNA in a 10-μl reaction mixture, with each gene-specific, antisense primer added at 200 nM (Table S8). qPCR was performed on a CFX96 Touch real-time PCR detection system, using the SsoAdvanced Universal SYBR green Supermix (Bio-Rad) and a 1:10 dilution of the original RT reaction as the template for amplification. cDNA levels were calculated using the ΔΔCq method, with a housekeeping gene, gyrA, from each bacterium as the internal control. Considering the relatively close phylogenetic relationship of S. mutans and S. gordonii, additional control experiments were performed to assess the stringency of RT-qPCRs when using RNA samples from mixed-species cocultures. Briefly, antisense primers for a select number of S. mutans genes were used in RT reactions together with RNA extracted from 3 S. gordonii cultures (TV-Glc), while in 3 separate reactions, primers for S. gordonii genes were used to prime RNA from S. mutans cultures (TV-Glc). qPCR results derived from these RT reactions confirmed the absence of any significant cross-species priming in reactions that used RNA samples from mixed-species cultures, with the majority of the Cq values being >30 under the current experimental setup (data not shown).
Deferred antagonism assay.
Overnight cultures of S. mutans UA159 and S. gordonii DL1 grown in TV containing 20 mM Glc, GlcN, or GlcNAc were each diluted into fresh TV medium containing the same carbohydrate and incubated until the OD600 reached 0.5. UA159 alone, DL1 alone, or UA159 together with DL1 at a ratio of 1:1 was inoculated by stabbing onto TV agar plates prepared with 20 mM Glc, GlcN, or GlcNAc, in accordance with carbohydrates used in cultures. After 24 h of incubation at 37°C, each plate was overlaid with 8 ml of warm soft agar (0.75% TV agar, prepared with the same carbohydrate as that in the plate) containing 107 CFU of S. sanguinis SK150 as an indicator strain. Plates were incubated for an additional 24 h before measuring zones of inhibition.
To understand the different impacts of TV and TY media on the production of mutacins by S. mutans, various amounts of yeast extract, ranging from 0.05% to 0.5%, were added into TV base medium containing 20 mM GlcN. For comparison, increasing amounts (0.04% to 0.4%) of the stock TV vitamin mix (10 μg/ml p-aminobenzoic acid, 50 μg/ml thiamine-HCl, 250 μg/ml nicotinamide, and 50 μg/ml riboflavin) were added into TY base medium supplemented with 20 mM GlcN. Using agar plates prepared with these modified TV and TY media, a UA159 culture from the exponential phase was used as described above in a deferred antagonism assay with S. sanguinis SK150.
Plate-based competition assay.
TV-based cultures of UA159 and DL1 from the exponential phase were prepared as described above, using 3 different carbohydrates. Six microliters of each of the cell suspensions of UA159 and DL1, prepared with the same carbohydrate, was spotted in close proximity on TV agar plates formulated with 20 mM Glc, GlcN, or GlcNAc, in accordance with the carbohydrates used in the cultures. Depending on the experimental design, UA159 or DL1 was inoculated alone first, incubated for 24 h, and inoculated by the other strain, or they were spotted simultaneously. Plates were incubated under aerobic conditions with 5% CO2 for another 24 h at 37°C before being photographed.
Promoter::reporter fusion and enzymatic assays.
Chloramphenicol acetyltransferase (CAT) assays were carried out to quantify promoter activities according to an established protocol (62). S. mutans derivatives containing an nlmD promoter::cat fusion (45) were cultivated to mid-exponential phase (OD600 of 0.5) in TV prepared with 20 mM Glc, GlcN, or GlcNAc before being harvested for assays. To assess the expression of a PlrgA::gfp (green fluorescent protein) reporter fusion, the S. mutans derivative hosting the fusion plasmid (21) was inoculated into a synthetic FMC medium supported by 10 mM Glc, GlcN, or GlcNAc and monitored for both optical density and green fluorescence signals for 30 h in a fluorescence-capable plate reader (63).
To measure pyruvate levels in supernatant fluids, overnight BHI cultures of S. mutans were washed and resuspended in FMC base medium, followed by 1:20 dilution into an FMC constituted with 10 mM Glc or GlcN. To avoid the long lag often associated with transferring UA159 from glucose-based media onto FMC-GlcNAc (17), we prepared a set of TV-GlcNAc cultures as starters and diluted them similarly into FMC containing 10 mM GlcNAc. At various time points (∼1-h interval), aliquots of the bacterial culture were removed for measurement of OD600 or spun down to harvest the supernatants. Pyruvate levels in these culture supernatants were measured using an EnzyChrom pyruvate assay kit (BioAssay Systems, Hayward, CA) by following the supplier’s instructions. The results were plotted together with their corresponding OD600 values. FMC was chosen over TV in pyruvate-related experiments to minimize medium components interfering with the fluorescence signal and the pyruvate assay. Acetate levels were similarly measured in the culture supernatants using an acetate colorimetric assay kit (Sigma), in this case using TV-based medium supplemented with 20 mM Glc, GlcN, or GlcNAc. TV was chosen over FMC for the acetate assays because substantial amounts of acetate (44 mM) are present in FMC.
Data availability.
The high-throughput sequencing data from this study have been deposited in the Gene Expression Omnibus (GEO) and assigned accession number GSE152021.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by DE12236 and DE25832 from NIDCR.
We thank Sang-Joon Ahn for useful discussions and providing the PlrgA::gfp reporter strain.
L.Z. and R.A.B. designed the studies; L.C. and L.Z. performed the experiments; A.R.W., L.C., and L.Z. analyzed the data; L.Z. and R.A.B. wrote the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Battistone GC, Burnett GW. 1961. The free amino acid composition of human saliva. Arch Oral Biol 3:161–170. doi: 10.1016/0003-9969(61)90133-9. [DOI] [PubMed] [Google Scholar]
- 2.Schaffer C, Messner P. 2005. The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together. Microbiology 151:643–651. doi: 10.1099/mic.0.27749-0. [DOI] [PubMed] [Google Scholar]
- 3.Rogers HJ. 1949. The metabolism of the amino sugars. 1. The breakdown of N-acetylglucosamine by strains of Streptococcus haemolyticus and other streptococci. Biochem J 45:87–93. doi: 10.1042/bj0450087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moye ZD, Zeng L, Burne RA. 2014. Fueling the caries process: carbohydrate metabolism and gene regulation by Streptococcus mutans. J Oral Microbiol 6:24878–24892. doi: 10.3402/jom.v6.24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banas JA. 2004. Virulence properties of Streptococcus mutans. Front Biosci 9:1267–1277. doi: 10.2741/1305. [DOI] [PubMed] [Google Scholar]
- 6.Garvie EI. 1980. Bacterial lactate dehydrogenases. Microbiol Rev 44:106–139. doi: 10.1128/MMBR.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwami Y, Abbe K, Takahashi-Abbe S, Yamada T. 1992. Acid production by streptococci growing at low pH in a chemostat under anaerobic conditions. Oral Microbiol Immunol 7:304–308. doi: 10.1111/j.1399-302x.1992.tb00593.x. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson J, Kujala U, Edlund MB. 1985. Pyruvate dehydrogenase activity in Streptococcus mutans. Infect Immun 49:674–678. doi: 10.1128/IAI.49.3.674-678.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi-Abbe S, Abe K, Takahashi N. 2003. Biochemical and functional properties of a pyruvate formate-lyase (PFL)-activating system in Streptococcus mutans. Oral Microbiol Immunol 18:293–297. doi: 10.1034/j.1399-302x.2003.00081.x. [DOI] [PubMed] [Google Scholar]
- 10.Zheng L, Itzek A, Chen Z, Kreth J. 2011. Environmental influences on competitive hydrogen peroxide production in Streptococcus gordonii. Appl Environ Microbiol 77:4318–4328. doi: 10.1128/AEM.00309-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas EL, Milligan TW, Joyner RE, Jefferson MM. 1994. Antibacterial activity of hydrogen peroxide and the lactoperoxidase-hydrogen peroxide-thiocyanate system against oral streptococci. Infect Immun 62:529–535. doi: 10.1128/IAI.62.2.529-535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Furio M, Ahn SJ, Burne RA, Hagen SJ. 2017. Oxidative stressors modify the response of Streptococcus mutans to its competence signal peptides. Appl Environ Microbiol 83:e01345-17. doi: 10.1128/AEM.01345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JN, Ahn SJ, Burne RA. 2015. Genetics and physiology of acetate metabolism by the Pta-Ack pathway of Streptococcus mutans. Appl Environ Microbiol 81:5015–5025. doi: 10.1128/AEM.01160-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCleary WR, Stock JB, Ninfa AJ. 1993. Is acetyl phosphate a global signal in Escherichia coli? J Bacteriol 175:2793–2798. doi: 10.1128/jb.175.10.2793-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Chakraborty B, Zou J, Burne RA, Zeng L. 2019. Amino sugars modify antagonistic interactions between commensal oral streptococci and Streptococcus mutans. Appl Environ Microbiol 85:e00370-19. doi: 10.1128/AEM.00370-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng L, Farivar T, Burne RA. 2016. Amino sugars enhance the competitiveness of beneficial commensals with Streptococcus mutans through multiple mechanisms. Appl Environ Microbiol 82:3671–3682. doi: 10.1128/AEM.00637-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Y, Chen YY, Snyder JA, Burne RA. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl Environ Microbiol 68:5549–5553. doi: 10.1128/aem.68.11.5549-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moye ZD, Burne RA, Zeng L. 2014. Uptake and metabolism of N-acetylglucosamine and glucosamine by Streptococcus mutans. Appl Environ Microbiol 80:5053–5067. doi: 10.1128/AEM.00820-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun 11:649–655. doi: 10.1128/IAI.11.4.649-655.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn SJ, Deep K, Turner ME, Ishkov I, Waters A, Hagen SJ, Rice KC. 2019. Characterization of LrgAB as a stationary phase-specific pyruvate uptake system in Streptococcus mutans. BMC Microbiol 19:223. doi: 10.1186/s12866-019-1600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn SJ, Qu MD, Roberts E, Burne RA, Rice KC. 2012. Identification of the Streptococcus mutans LytST two-component regulon reveals its contribution to oxidative stress tolerance. BMC Microbiol 12:187. doi: 10.1186/1471-2180-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merritt J, Qi F. 2012. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol 27:57–69. doi: 10.1111/j.2041-1014.2011.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng L, Burne RA. 2015. NagR differentially regulates the expression of the glmS and nagAB genes required for amino sugar metabolism by Streptococcus mutans. J Bacteriol 197:3533–3544. doi: 10.1128/JB.00606-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker JL, Derr AM, Karuppaiah K, MacGilvray ME, Kajfasz JK, Faustoferri RC, Rivera-Ramos I, Bitoun JP, Lemos JA, Wen ZT, Quivey RG Jr. 2014. Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J Bacteriol 196:2166–2177. doi: 10.1128/JB.01542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker JL, Derr AM, Faustoferri RC, Quivey RG Jr. 2015. Loss of NADH oxidase activity in Streptococcus mutans leads to Rex-mediated overcompensation in NAD+ regeneration by lactate dehydrogenase. J Bacteriol 197:3645–3657. doi: 10.1128/JB.00383-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Zhou Y, Zhang L, Shah N, Jin C, Palmer RJ Jr, Cisar JO. 2016. Cell surface glycoside hydrolases of Streptococcus gordonii promote growth in saliva. Appl Environ Microbiol 82:5278–5286. doi: 10.1128/AEM.01291-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng L, Martino NC, Burne RA. 2012. Two gene clusters coordinate galactose and lactose metabolism in Streptococcus gordonii. Appl Environ Microbiol 78:5597–5605. doi: 10.1128/AEM.01393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge X, Yu Y, Zhang M, Chen L, Chen W, Elrami F, Kong F, Kitten T, Xu P. 2016. Involvement of NADH oxidase in competition and endocarditis virulence in Streptococcus sanguinis. Infect Immun 84:1470–1477. doi: 10.1128/IAI.01203-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redanz S, Treerat P, Mu R, Redanz U, Zou Z, Koley D, Merritt J, Kreth J. 2020. Pyruvate secretion by oral streptococci modulates hydrogen peroxide dependent antagonism. ISME J 14:1074–1088. doi: 10.1038/s41396-020-0592-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Y, Chen YY, Burne RA. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J Bacteriol 186:2511–2514. doi: 10.1128/jb.186.8.2511-2514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolerson E, Swick A, Newlon L, Palmer C, Pan Y, Keeshan B, Spatafora G. 2006. The SloR/Dlg metalloregulator modulates Streptococcus mutans virulence gene expression. J Bacteriol 188:5033–5044. doi: 10.1128/JB.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crepps SC, Fields EE, Galan D, Corbett JP, Von Hasseln ER, Spatafora GA. 2016. The SloR metalloregulator is involved in the Streptococcus mutans oxidative stress response. Mol Oral Microbiol 31:526–539. doi: 10.1111/omi.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paik S, Brown A, Munro CL, Cornelissen CN, Kitten T. 2003. The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J Bacteriol 185:5967–5975. doi: 10.1128/jb.185.20.5967-5975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajfasz JK, Katrak C, Ganguly T, Vargas J, Wright L, Peters ZT, Spatafora GA, Abranches J, Lemos JA. 2020. Manganese uptake, mediated by SloABC and MntH, is essential for the fitness of Streptococcus mutans. mSphere 5:e00764-19. doi: 10.1128/mSphere.00764-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang BY, Kuramitsu HK. 2005. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol 71:354–362. doi: 10.1128/AEM.71.1.354-362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu C, Cichewicz R, Li Y, Liu J, Roe B, Ferretti J, Merritt J, Qi F. 2010. Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl Environ Microbiol 76:5815–5826. doi: 10.1128/AEM.03079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joyner PM, Liu J, Zhang Z, Merritt J, Qi F, Cichewicz RH. 2010. Mutanobactin A from the human oral pathogen Streptococcus mutans is a cross-kingdom regulator of the yeast-mycelium transition. Org Biomol Chem 8:5486–5489. doi: 10.1039/c0ob00579g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long LH, Halliwell B. 2009. Artefacts in cell culture: pyruvate as a scavenger of hydrogen peroxide generated by ascorbate or epigallocatechin gallate in cell culture media. Biochem Biophys Res Commun 388:700–704. doi: 10.1016/j.bbrc.2009.08.069. [DOI] [PubMed] [Google Scholar]
- 40.Giandomenico AR, Cerniglia GE, Biaglow JE, Stevens CW, Koch CJ. 1997. The importance of sodium pyruvate in assessing damage produced by hydrogen peroxide. Free Radic Biol Med 23:426–434. doi: 10.1016/s0891-5849(97)00113-5. [DOI] [PubMed] [Google Scholar]
- 41.Deng DM, Liu MJ, ten Cate JM, Crielaard W. 2007. The VicRK system of Streptococcus mutans responds to oxidative stress. J Dent Res 86:606–610. doi: 10.1177/154405910708600705. [DOI] [PubMed] [Google Scholar]
- 42.Senadheera DB, Cordova M, Ayala EA, Chavez de Paz LE, Singh K, Downey JS, Svensater G, Goodman SD, Cvitkovitch DG. 2012. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J Bacteriol 194:1307–1316. doi: 10.1128/JB.06071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicolas G, Auger I, Beaudoin M, Halle F, Morency H, LaPointe G, Lavoie MC. 2004. Improved methods for mutacin detection and production. J Microbiol Methods 59:351–361. doi: 10.1016/j.mimet.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Qi F, Chen P, Caufield PW. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl Environ Microbiol 65:652–658. doi: 10.1128/AEM.65.2.652-658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watts MO. 2014. The non-lantibiotic mutacin gene nlmD and the pyruvate dehydrogenase complex (pdh) operon: models for the regulation of gene expression in Streptococcus mutans in response to aeration. University of Florida, Gainesville, FL. [Google Scholar]
- 46.Zeng L, Choi SC, Danko CG, Siepel A, Stanhope MJ, Burne RA. 2013. Gene regulation by CcpA and catabolite repression explored by RNA-Seq in Streptococcus mutans. PLoS One 8:e60465. doi: 10.1371/journal.pone.0060465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kreth J, Merritt J, Shi W, Qi F. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol Microbiol 57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downey JS, Mashburn-Warren L, Ayala EA, Senadheera DB, Hendrickson WK, McCall LW, Sweet JG, Cvitkovitch DG, Spatafora GA, Goodman SD. 2014. In vitro manganese-dependent cross-talk between Streptococcus mutans VicK and GcrR: implications for overlapping stress response pathways. PLoS One 9:e115975. doi: 10.1371/journal.pone.0115975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dmitriev A, Mohapatra SS, Chong P, Neely M, Biswas S, Biswas I. 2011. CovR-controlled global regulation of gene expression in Streptococcus mutans. PLoS One 6:e20127. doi: 10.1371/journal.pone.0020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stipp RN, Boisvert H, Smith DJ, Hofling JF, Duncan MJ, Mattos-Graner RO. 2013. CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans. PLoS One 8:e58271. doi: 10.1371/journal.pone.0058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burne RA, Wen ZT, Chen YY, Penders JE. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J Bacteriol 181:2863–2871. doi: 10.1128/JB.181.9.2863-2871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Core Development Team. 2010. R: a language and environment for statistical computing. R foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 53.Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei T, Simko V. 2017. Package “corrplot: visualization of a correlation matrix” (v.0.84). https://CRAN.R-project.org/package=corrplot.
- 56.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T. 2016. Package “gplots”: various R programming tools for plotting data. https://github.com/talgalili/gplots.
- 57.Wickham H. 2017. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 58.Murdoch D, Chow ED. 2007. ellipse: functions for drawing ellipses and ellipse-like confidence regions. https://cran.r-project.org/web/packages/ellipse/index.html.
- 59.H Backman TW, Girke T. 2016. systemPipeR: NGS workflow and report generation environment. BMC Bioinformatics 17:388. doi: 10.1186/s12859-016-1241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 61.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng L, Wen ZT, Burne RA. 2006. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol Microbiol 62:187–200. doi: 10.1111/j.1365-2958.2006.05359.x. [DOI] [PubMed] [Google Scholar]
- 63.Zeng L, Chen L, Burne RA. 2018. Preferred hexoses influence long-term memory in and induction of lactose catabolism by Streptococcus mutans. Appl Environ Microbiol 84:e00864-18. doi: 10.1128/AEM.00864-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The high-throughput sequencing data from this study have been deposited in the Gene Expression Omnibus (GEO) and assigned accession number GSE152021.