Dietary strategies (e.g., phytase supplementation and lactic acid [LA] treatment of cereals) used to improve the availability of phytate-phosphorus (P) from pig feed reduce the amount of P flowing into the large intestine, whereas LA treatment-induced changes in nutrient fractions alter the substrate being available to the microbiota. In ruminants, lower intestinal P availability compromises the fibrolytic activity of the microbiome. Here, we report that the functional capacities were less dramatically affected than the taxonomic composition by phytase-supplemented and LA-treated cereals. The bacterial community appeared to be partly capable of functionally compensating for the altered flow of P by replacing taxa with higher P needs by those with lower P needs. Therefore, by acting as mucosal immune stimulants, alterations in microbiota-associated molecular patterns (MAMPs) due to the taxonomic shifts may play a greater role for host physiology and health than functional differences caused by differing intestinal P availabilities, which merits further research.
KEYWORDS: cecum, metagenome, lactic acid treatment of cereal grains, pig, phytase
ABSTRACT
Phosphorus (P) is an essential nutrient for the gut bacteria and the host. Nevertheless, little information exists that indicates to what extent an improved level of P availability in the small intestine leads to functional adaptations in bacterial metabolic pathways in the large intestine. Therefore, we investigated the changes in the taxonomic and functional bacterial metagenome in cecal digesta of growing pigs fed diets containing phytase and/or cereals treated with 2.5% lactic acid (LA) for 19 days (n = 8/diet) using shotgun metagenome sequencing. The phytase supplementation resulted in strikingly distinct bacterial communities, affecting almost all major bacterial families, whereas functional changes were less dramatic among the feeding groups. While phytase treatment decreased predominant Prevotellaceae levels, it seemed that Clostridiaceae, Ruminococcaceae, and Lachnospiraceae filled the opening metabolic niches (P < 0.05). The LA-treated cereals mediated reduced levels of Bacteroidaceae and increased levels of Veillonellaceae, but those results were mainly seen when the cereals were fed as a single treatment (P < 0.05). In association with the taxonomic alterations, phytase caused changes within the major functional pathways corresponding to amino acid metabolism; translation; membrane transport; folding, sorting, and degradation; and energy metabolism, whereas the LA treatment of cereals resulted in decreased enzymatic capacities within the carbohydrate metabolism and energy metabolism pathways (P < 0.05). Metabolic dependencies corresponding to the starch and sucrose metabolism, glycolysis/gluconeogenesis, and citrate cycle pathways were indicated by diet-associated changes in enzymatic capacities related to short-chain fatty acid, methane, vitamin, and bacterial antigen synthesis. Accordingly, the present results support the idea of the importance of the availability of intestinal P for bacterial metabolism. However, the functional profiles were less different than the taxonomic profiles among the dietary treatment results, indicating a certain degree of metabolic plasticity within the cecal metagenome.
IMPORTANCE Dietary strategies (e.g., phytase supplementation and lactic acid [LA] treatment of cereals) used to improve the availability of phytate-phosphorus (P) from pig feed reduce the amount of P flowing into the large intestine, whereas LA treatment-induced changes in nutrient fractions alter the substrate being available to the microbiota. In ruminants, lower intestinal P availability compromises the fibrolytic activity of the microbiome. Here, we report that the functional capacities were less dramatically affected than the taxonomic composition by phytase-supplemented and LA-treated cereals. The bacterial community appeared to be partly capable of functionally compensating for the altered flow of P by replacing taxa with higher P needs by those with lower P needs. Therefore, by acting as mucosal immune stimulants, alterations in microbiota-associated molecular patterns (MAMPs) due to the taxonomic shifts may play a greater role for host physiology and health than functional differences caused by differing intestinal P availabilities, which merits further research.
INTRODUCTION
Diet is the most important factor influencing the composition and function of the porcine bacterial microbiome along the gastrointestinal tract (1, 2). An appropriate nutrient balance is therefore essential not only to support the health and well-being of the pig but also for the maintenance of a stable gut microbiota. Changes in the dietary composition cause compositional and metabolic adaptations in the gut bacterial community that depend on the bacterial substrate preferences (3). Bacteria may compete with the pig host for easily digestible dietary components in the jejunum and ileum, whereas they provide the host with absorbable nutrients, such as short-chain fatty acids (SCFA). By fermenting dietary fiber, bacteria make energy in an otherwise unusable dietary component available to the host (4). Simultaneously, alterations in bacterial abundances are accompanied by changes in microbiota-associated molecular patterns (MAMPs), which are structural motifs that are highly conserved in microbes, e.g., bacterial flagellin, peptidoglycan, and lipopolysaccharides (LPS) (5, 6). In particular, LPS, which is abundant in the outer cell membrane of Gram-negative bacteria, acts as an immune stimulant, upregulating a variety of innate immune responses while weakening the mucosal barrier function (7). The immune reactivity of LPS, however, depends on the fatty acid composition of the lipid A component, which is responsible for the activation of the Toll-like receptor-4 (TLR-4) response and differs among the various Gram-negative bacteria (8). The LPS of Proteobacteria commonly provokes a strong immune response, whereas that of other Gram-negative genera, such as Prevotella and Bacteroides, is a less strong immune stimulant (9). Revealing diet-induced changes in the functional capacity of the gut microbiota may therefore help in understanding their impact on host physiology (2). Despite this awareness, our knowledge about dietary effects on functional capabilities of the gut microbiome and concomitant MAMPs is still limited.
Phosphorus (P) is an essential macroelement for the gut microbiota due to its roles as an important component of nucleic acids and structural membranes (phospholipids) as well as a cofactor of many enzymes (10). Therefore, various metabolic features such as osmotic and acid-base balance, energy metabolism, amino acid metabolism, and protein synthesis in bacterial cells are compromised by insufficient P supply (11, 12). In pig production, dietary supplementation with inorganic P has been reduced due to environmental and economic reasons (13). Cereals and protein-rich by-products, as the main components of pig’s diet, contain per se sufficient amounts of P; however, it is present mostly in the form of phytate-P, which is not digestible for the host due to missing endogenous enzymes (11). In contrast to the host animal, gut bacteria comprise the necessary enzymatic capacities to utilize phytate-P, but they express phytase only when the P concentration in their environment becomes very low (14). In order to render the phytate-P more available to the pig, different dietary strategies are applied, including the most common strategy of exogenous microbial phytase supplementation and the more traditional soaking of dietary cereals in mild organic acids such as lactic acid (LA) (15, 16). These treatments mainly increase the P availability and absorption in the upper digestive tract, subsequently lowering the amount of P available to the microbes in the large intestine (17). In accordance with previous observations in ruminants (18), it can be hypothesized that fibrolytic enzymatic pathways of bacteria in digesta of pigs are especially affected by the lower P flow into the large intestine. In addition, the soaking of cereals in LA may change the availability of other nutrients in the cereal grains by altering nutrient fractions, such as protein, sugar, (resistant) starch, and hemicellulose fractions (e.g., arabinoxylans and β-glucans) (19–24). For instance, Harder et al. (23) showed that treatment of barley grains with 5% LA at room temperature for 24 h caused changes in the structure of the starch molecules. These molecular changes resulted in a higher concentration of resistant starch in the soaked grains, while the neutral detergent fiber (NDF) and crude protein content was reduced. This LA-soaking-related modification of the dietary carbohydrates and proteins may lead to changes in the ileal flow of fermentable substrate, altering not only the bacterial taxonomy but also the enzymatic capabilities needed to ferment the available substrate. In combination with the lower P availability, the LA treatment of cereals may specifically promote the presence of bacterial taxa that possess the required enzymatic capabilities while coping better with the lower P availability. We were recently able to show that phytase supplementation of pig’s diet greatly affected the fecal metabolically active community, especially impacting the fecal abundances of saccharolytic Lactobacillaceae and hemicellulolytic Ruminococcaceae (25). It is feasible that LA treatment of cereals released phenolic compounds as observed during sourdough fermentation (21), which may have exerted antimicrobial activity. In line with that reasoning, we found lower abundances of metabolically active pathobionts within Proteobacteria (i.e., Pasteurellaceae, Helicobacteraceae, and Campylobacteraceae, all comprising highly immune-stimulating LPS) in ileal digesta and at the mucosa of pigs fed LA-treated cereals, which also influenced the bacterial translocation into the adjacent lymph nodes (26). If this scenario were also true for the cecum, inclusion of both phytase-supplemented and LA-treated cereal in pig’s diet may alter the abundances and availability of immune-stimulating MAMPs. Therefore, this study aimed to investigate the effect of phytase supplementation and of soaking of cereals in 2.5% lactic acid on the cecal metagenome at the functional and taxonomic levels. We hypothesized that the alterations in taxonomy and functional abilities caused by the dietary treatments would reveal functional pathways that were specifically susceptible to the decreased P flow into the cecum, thereby impacting bacterial cellulolytic and hemicellulolytic capacities.
RESULTS
Animals and diets.
Four diets consisting of wheat, corn, and soybean meal were formulated and fed to the pigs in a 2 (0 versus 500 phytase units/kg complete feed) × 2 (soaking of cereals in 2.5% LA for 48 h or not) factorial design. All pigs were clinically healthy throughout the experiment, and the inner organs did not show any sign of disease during necropsy. Pigs across dietary treatment groups ate the same amount of feed (784 ± 20.4 g/days). With regard to the dietary composition, the LA treatment of cereals increased dry matter content but decreased ash, protein, neutral detergent fiber (NDF), and nonresistant starch in the diet (see Table S1 in the supplemental material).
Annotating cecal metagenomes.
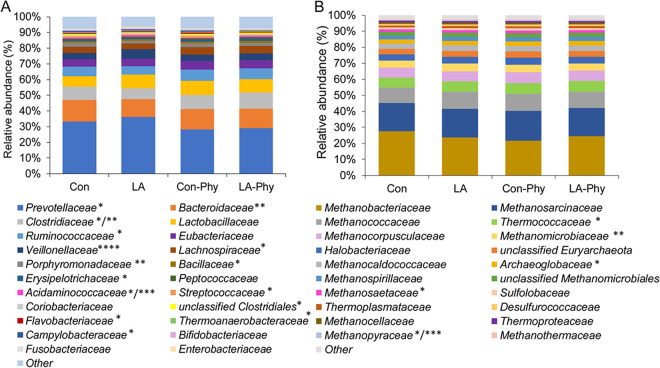
Shotgun metagenomic sequencing generated a total of 295,927,943 reads in the 32 cecal samples from pigs fed diets with or without phytase and LA treatment of cereals, with an average of 9,247,748 reads per sample and an average read length of 149 bp. On average, 2,923,226 proteins were functionally assigned, which amounted to 31.6% of the total reads in each cecal sample. Identified sequence proteins clustered into 133 and 325 bacterial families and genera at >0.05% relative abundances, respectively. Overall, the archaeal 16S rRNA abundances represented proportionally 0.60% of the total eubacterial 16S rRNA abundances (Table 1). At the family level, the cecal communities were dominated by Prevotellaceae and Bacteroidaceae, with mean relative abundances of 30.81% and 12.39%, respectively, followed by Clostridiaceae (8.36%), Lactobacillaceae (7.99%), Ruminococcaceae (6.15%), Eubacteriaceae (5.02%), Veillonellaceae (4.39%), and Lachnospiraceae (4.21%) (Fig. 1A; see also Table 2). The 29 archaeal families were dominated by Methanobacteriaceae (24.38%), Methanosarcinaceae (17.99%), and Methanococcaceae (10.30%), which were followed by Thermococcaceae (6.50%), Methanocorpusculaceae (6.46%), Methanomicrobiaceae (4.66%), and Halobacteriaceae (4.15%) (Fig. 1B; see also Table 3).
TABLE 1.
Total number of 16S rRNA gene copies of Eubacteria and Archaea in cecal digesta of pigs fed diets with or without phytase and lactic acid treatment of cerealsa
| Domain | Value under indicated grain treatment conditions |
SEM |
P value |
|||||
|---|---|---|---|---|---|---|---|---|
| No phytase |
Phytase |
|||||||
| Con | LA | Con | LA | Phytase | LA | Phytase × LA | ||
| Eubacteria (log10 gene copies/g) | 12.3 | 12.5 | 12.4 | 12.4 | 0.042 | 0.792 | 0.058 | 0.225 |
| Archaea (log10 gene copies/g) | 9.0 a | 8.9 a | 7.9 b | 8.9 a | 0.260 | 0.044 | 0.080 | 0.038 |
| Archaea (% of total bacteria) | 1.18 a | 0.43 ab | 0.22 b | 0.55 ab | 0.314 | 0.195 | 0.506 | 0.097 |
Values are presented as least-square means ± SEM (n = 8 pigs per dietary treatment). Different lowercase letters (a and b) in cell entries within a row indicate a significant difference (P ≤ 0.05). Con, control; LA, lactic acid-treated cereals.
FIG 1.
Relative abundances of families of eubacteria (A) and archaea (B) in cecal digesta of pigs (>0.05% relative abundance). Values are presented as least-square means (n = 32). Different sets of asterisks indicate a significant difference by the dietary treatment (*, phytase [Phy] effect; **, LA effect; ***, phytase × LA effect; ****, all treatment effects) at P ≤ 0.05. Sequence data were analyzed before April 2020, when the family Lactobacillaceae was reclassified (52). Due to this reclassification, Leuconostocaceae were united with Lactobacillaceae, becoming the novel Lactobacillaceae family. Accordingly, data from Lactobacillaceae and Leuconostocaceae were merged.
TABLE 2.
Selected bacterial families in cecal digesta of pigs fed diets with or without phytase and lactic acid treatment of cerealsa
| Family | Mean % relative abundance under indicated grain treatment conditions |
SEM |
P value |
|||||
|---|---|---|---|---|---|---|---|---|
| No phytase |
Phytase |
|||||||
| Con | LA | Con | LA | Phytase | LA | Phytase × LA | ||
| Prevotellaceae | 32.40 | 35.23 | 27.42 | 28.19 | 2.035 | 0.007 | 0.385 | 0.617 |
| Bacteroidaceae | 13.41 | 11.32 | 12.74 | 12.10 | 0.419 | 0.898 | 0.003 | 0.096 |
| Clostridiaceae | 8.16 bc | 6.49 c | 8.60 ab | 10.20 a | 0.729 | 0.009 | 0.960 | 0.035 |
| Ruminococcaceae | 5.80 | 5.21 | 6.73 | 6.84 | 0.457 | 0.010 | 0.609 | 0.457 |
| Veillonellaceae | 3.93 b | 5.97 a | 3.64 b | 4.02 b | 0.322 | 0.002 | 0.001 | 0.016 |
| Lachnospiraceae | 3.96 | 3.49 | 4.83 | 4.58 | 0.395 | 0.020 | 0.373 | 0.778 |
| Porphyromonadaceae | 2.15 | 1.61 | 2.14 | 1.99 | 0.105 | 0.088 | 0.003 | 0.074 |
| Bacillaceae | 1.06 | 1.03 | 1.19 | 1.28 | 0.057 | 0.003 | 0.668 | 0.277 |
| Erysipelotrichaceae | 0.88 b | 0.86 b | 1.00 b | 1.32 a | 0.076 | 0.001 | 0.055 | 0.037 |
| Acidaminococcaceae | 0.77 | 0.92 | 0.71 | 0.71 | 0.044 | 0.005 | 0.086 | 0.119 |
| Streptococcaceae | 0.76 | 0.51 | 0.87 | 0.60 | 0.108 | 0.381 | 0.026 | 0.935 |
| Unclassified Clostridiales | 0.67 | 0.52 | 0.80 | 0.75 | 0.073 | 0.020 | 0.167 | 0.494 |
| Flavobacteriaceae | 0.74 | 0.60 | 0.72 | 0.66 | 0.026 | 0.388 | 0.001 | 0.112 |
| Thermoanaerobacteraceae | 0.53 | 0.53 | 0.60 | 0.61 | 0.026 | 0.014 | 0.798 | 0.807 |
| Campylobacteraceae | 0.68 | 0.84 | 0.56 | 0.24 | 0.162 | 0.037 | 0.623 | 0.155 |
| Fusobacteriaceae | 0.441 | 0.403 | 0.476 | 0.491 | 0.022 | 0.010 | 0.600 | 0.237 |
| Enterobacteriaceae | 0.467 | 0.473 | 0.333 | 0.313 | 0.037 | 0.001 | 0.863 | 0.728 |
| Thermoanaerobacterales III | 0.350 | 0.327 | 0.397 | 0.404 | 0.020 | 0.005 | 0.710 | 0.473 |
| Cytophagaceae | 0.311 | 0.258 | 0.303 | 0.276 | 0.011 | 0.627 | 0.001 | 0.248 |
| Paenibacillaceae | 0.252 | 0.236 | 0.283 | 0.292 | 0.016 | 0.013 | 0.828 | 0.434 |
| Pasteurellaceae | 0.292 | 0.234 | 0.212 | 0.206 | 0.025 | 0.042 | 0.217 | 0.302 |
| Clostridiales XI | 0.224 | 0.196 | 0.250 | 0.255 | 0.012 | 0.002 | 0.360 | 0.197 |
| Enterococcaceae | 0.203 | 0.192 | 0.229 | 0.242 | 0.012 | 0.003 | 0.948 | 0.293 |
| Fibrobacteraceae | 0.213 | 0.173 | 0.208 | 0.195 | 0.011 | 0.435 | 0.025 | 0.254 |
| Chlorobiaceae | 0.193 | 0.172 | 0.194 | 0.186 | 0.004 | 0.124 | 0.004 | 0.187 |
| Helicobacteraceae | 0.210 | 0.238 | 0.170 | 0.110 | 0.031 | 0.011 | 0.607 | 0.165 |
| Vibrionaceae | 0.217 | 0.172 | 0.156 | 0.146 | 0.017 | 0.020 | 0.124 | 0.332 |
| Sphingobacteriaceae | 0.182 | 0.142 | 0.182 | 0.163 | 0.008 | 0.192 | 0.001 | 0.194 |
| Thermotogaceae | 0.151 | 0.135 | 0.162 | 0.166 | 0.008 | 0.016 | 0.509 | 0.226 |
| Syntrophomonadaceae | 0.145 | 0.141 | 0.165 | 0.160 | 0.008 | 0.021 | 0.591 | 0.961 |
| Burkholderiaceae | 0.177 | 0.129 | 0.139 | 0.131 | 0.013 | 0.172 | 0.035 | 0.134 |
| Aerococcaceae | 0.136 | 0.113 | 0.159 | 0.153 | 0.013 | 0.022 | 0.282 | 0.528 |
| Staphylococcaceae | 0.119 | 0.117 | 0.138 | 0.150 | 0.007 | 0.001 | 0.497 | 0.328 |
| Rikenellaceae | 0.144 | 0.101 | 0.142 | 0.120 | 0.008 | 0.317 | 0.001 | 0.228 |
| Shewanellaceae | 0.148 | 0.123 | 0.116 | 0.110 | 0.009 | 0.020 | 0.096 | 0.305 |
| Pseudomonadaceae | 0.144 | 0.119 | 0.119 | 0.113 | 0.006 | 0.019 | 0.022 | 0.164 |
| Listeriaceae | 0.110 | 0.109 | 0.124 | 0.132 | 0.005 | 0.001 | 0.524 | 0.403 |
| Neisseriaceae | 0.124 a | 0.076 b | 0.084 b | 0.073 b | 0.008 | 0.014 | 0.001 | 0.032 |
| Rhodobacteraceae | 0.092 | 0.077 | 0.089 | 0.084 | 0.004 | 0.706 | 0.027 | 0.228 |
| Halanaerobiaceae | 0.080 | 0.081 | 0.089 | 0.090 | 0.004 | 0.031 | 0.800 | 0.987 |
| Comamonadaceae | 0.098 | 0.073 | 0.081 | 0.076 | 0.006 | 0.242 | 0.021 | 0.113 |
| Unclassified Sphingobacteriales | 0.087 | 0.073 | 0.084 | 0.078 | 0.003 | 0.704 | 0.004 | 0.189 |
| Aeromonadaceae | 0.104 | 0.083 | 0.054 | 0.048 | 0.014 | 0.005 | 0.348 | 0.591 |
| Mycoplasmataceae | 0.064 | 0.058 | 0.071 | 0.079 | 0.003 | 0.000 | 0.865 | 0.067 |
| Unclassified Bacillales | 0.062 | 0.059 | 0.071 | 0.075 | 0.004 | 0.002 | 0.781 | 0.330 |
| Bradyrhizobiaceae | 0.069 | 0.061 | 0.068 | 0.064 | 0.003 | 0.860 | 0.041 | 0.502 |
| Peptostreptococcaceae | 0.062 ab | 0.051 b | 0.065 a | 0.074 a | 0.005 | 0.013 | 0.765 | 0.049 |
| Moraxellaceae | 0.074 | 0.058 | 0.059 | 0.057 | 0.004 | 0.034 | 0.026 | 0.079 |
| Unclassified Bacteroidetes | 0.069 | 0.046 | 0.070 | 0.062 | 0.005 | 0.082 | 0.002 | 0.114 |
| Alteromonadaceae | 0.068 | 0.053 | 0.052 | 0.051 | 0.004 | 0.022 | 0.032 | 0.054 |
| Rhodospirillaceae | 0.064 | 0.052 | 0.056 | 0.047 | 0.005 | 0.181 | 0.031 | 0.763 |
| Unclassified Burkholderiales | 0.062 | 0.043 | 0.055 | 0.048 | 0.004 | 0.871 | 0.006 | 0.145 |
Values are presented as least-square means ± SEM (n = 8 pigs per dietary treatment). Different lowercase letters (a, b, and c) in cell entries within a row indicate a significant difference (P ≤ 0.05). Con, control; LA, lactic acid-treated cereals. Only families that showed relative abundances of >0.05% of all reads and that were significantly different or tended to be different among treatments are presented. Sequence data were analyzed before April 2020, when the family Lactobacillaceae was reclassified (52). Due to this reclassification, Leuconostocaceae were united with Lactobacillaceae, becoming the novel Lactobacillaceae family. Accordingly, data from Lactobacillaceae and Leuconostocaceae were merged.
TABLE 3.
Selected archaeal families in cecal digesta of pigs fed diets with or without phytase and lactic acid treatment of cerealsa
| Family | Mean % relative abundance under indicated grain treatment conditions |
SEM |
P value |
|||||
|---|---|---|---|---|---|---|---|---|
| No phytase |
Phytase |
|||||||
| Con | LA | Con | LA | Phytase | LA | Phytase × LA | ||
| Thermococcaceae | 6.10 | 6.43 | 6.84 | 6.61 | 0.182 | 0.019 | 0.775 | 0.133 |
| Methanomicrobiaceae | 4.36 b | 4.94 a | 4.77 ab | 4.56 ab | 0.176 | 0.923 | 0.304 | 0.031 |
| Archaeoglobaceae | 2.46 | 2.66 | 2.80 | 2.72 | 0.087 | 0.030 | 0.504 | 0.113 |
| Methanosaetaceae | 1.48 | 1.45 | 1.68 | 1.57 | 0.072 | 0.036 | 0.300 | 0.602 |
| Methanopyraceae | 0.493 b | 0.569 a | 0.509 b | 0.463 b | 0.022 | 0.050 | 0.501 | 0.009 |
| Thermofilaceae | 0.394 | 0.401 | 0.470 | 0.462 | 0.023 | 0.007 | 0.980 | 0.753 |
| Picrophilaceae | 0.331 | 0.312 | 0.417 | 0.399 | 0.032 | 0.012 | 0.568 | 0.974 |
| Unclassified Korarchaeota | 0.326 | 0.302 | 0.372 | 0.368 | 0.017 | 0.003 | 0.405 | 0.557 |
| Acidilobaceae | 0.074 c | 0.106 a | 0.103 a | 0.073 b | 0.009 | 0.894 | 0.904 | 0.002 |
| Unclassified Nanoarchaeota | 0.033 | 0.027 | 0.041 | 0.039 | 0.004 | 0.043 | 0.382 | 0.665 |
Values are presented as least-square means ± SEM (n = 8 pigs per dietary treatment). Different lowercase letters (a, b, and c) in cell entries within a row indicate a significant difference (P ≤ 0.05). Con, control; LA, lactic acid-treated cereals. Only archaeal families that showed relative abundances of >0.05% of all reads and whose results were significantly different (P ≤ 0.05) or tended to be different (P ≤ 0.10) among treatments are presented.
Diet-related shift in bacterial community in cecum.
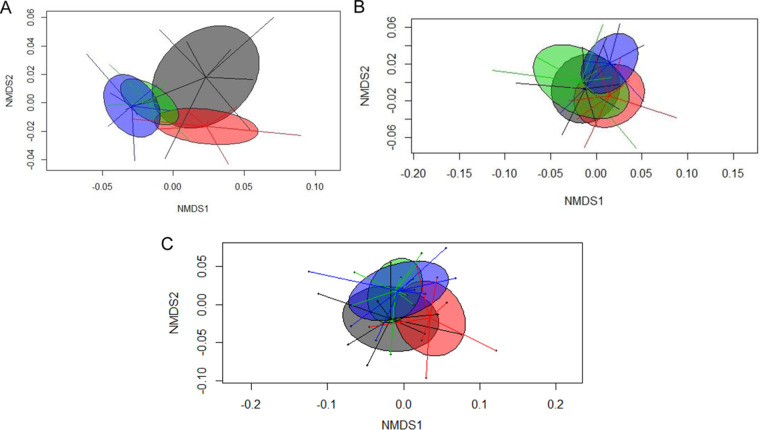
While the total eubacterial 16S rRNA gene copy numbers were more or less similar among treatment groups, total archaeal 16S rRNA gene copy numbers were reduced by the presence of phytase by 1 log unit (Table 1). However, the phytase × LA-treated cereal interaction indicated that this was the case only when the phytase was added to the control diet and not when it was added to the LA diet (P = 0.038). The permutational multivariate analysis of variance (PERMANOVA) results showed that bacterial communities clustered according to dietary treatments (Table S2; see also Fig. 2A); in particular, the dietary supplementation with phytase caused drastic changes in the composition of the bacterial and archaeal species (P = 0.028) (Table S2; see also Fig. 2A). In contrast, the LA treatment of dietary cereals was less decisive for the clustering of the cecal communities at the taxonomic level (Fig. 2A). This conclusion was supported by α-diversity indices showing that both Shannon and Simpson values were increased by the phytase supplementation (P < 0.05) but were unaffected by the LA treatment of cereals (Table 4).
FIG 2.
Nonmetric multidimensional scaling (NMDS) plot of pairwise Bray-Curtis dissimilarities among (A) bacterial communities, (B) KEGG orthology functions, and (C) KEGG orthology functions for carbohydrate metabolism pathways in cecal digesta of pigs fed diets with or without phytase and lactic acid treatment of cereals. Diet effect is indicated as follows: gray, control diet; red, diet containing lactic acid-treated cereals; green, diet with phytase supplementation; blue, diet with phytase supplementation and lactic acid-treated cereals.
TABLE 4.
Alpha-diversity indices in cecal digesta of pigs fed diets with or without phytase and lactic acid treatment of cerealsa
| Parameter | Value under indicated grain treatment conditions |
SEM |
P value |
|||||
|---|---|---|---|---|---|---|---|---|
| No phytase |
Phytase |
|||||||
| Con | LA | Con | LA | Phytase | LA | Phytase × LA | ||
| No. of species (species richness) | 1,535 | 1,536 | 1,535 | 1,533 | 1.67 | 0.341 | 0.658 | 0.341 |
| Simpson index | 0.977 | 0.967 | 0.982 | 0.981 | 0.003 | 0.006 | 0.120 | 0.183 |
| Shannon index | 5.11 | 4.90 | 5.20 | 5.16 | 0.065 | 0.013 | 0.062 | 0.205 |
Values are presented as least-square means ± SEM (n = 8 pigs per dietary treatment). Con, control diet; LA, diet containing lactic acid-treated cereals.
Likewise, taxonomy results showed that the presence of phytase affected almost all major eubacterial and archaeal families (Table 1; see also Table 3), whereas fewer taxa were influenced by the LA-treated cereals. With respect to the 9 most abundant families (relative abundance > 1% of all reads), phytase supplementation decreased the levels of the dominating Prevotellaceae and Veillonellaceae families by 17.8% and 22.6%, respectively, while increasing Clostridiaceae, Ruminococcaceae, and Lachnospiraceae levels by 28.3%, 23.3%, and 26.33%, respectively (P < 0.05). In contrast, LA treatment of cereals decreased the relative abundance of Bacteroidaceae by 22.63% but increased that of Veillonellaceae by 31.97%. As indicated by the phytase × LA-treated cereal interaction, the effect on Veillonellaceae was stronger when the LA treatment of cereals was applied as a single treatment. Moreover, cereals treated with phytase and LA showed interacting effects with respect to Clostridiaceae and Erysipelatrichaceae abundances, with the effects depending on whether the treatments were provided as single or combined treatments. Similar dietary effects were observed at the genus and species levels, with, for instance, the levels of Prevotella and Mitsuokella being decreased and those of Roseburia being increased with the phytase supplementation (Table S3 and S4). The dietary inclusion of the LA-treated cereals, in turn, increased the cecal numbers of Mitsuokella but decreased the abundance of Bacteroides (Table S3). Likewise, the phytase supplementation mainly affected the abundance of archaeal families, whereby the interaction of phytase with the LA-treated cereals showed that the LA-treated cereals influenced the abundance of Methanomicrobiaceae and Methanopyraceae only when the cereals were fed as a single treatment (P < 0.05; Table 3).
Diet-related functional composition of cecal metagenome.
To identify potential effects of the treatment of cereals with phytase and LA on the functional composition, we linked the genes in the present metagenomes to Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO) classes and Clusters of Orthologous Groups (COG) in order to obtain the functional profiles of the present metagenomes (27). While the PERMANOVA for KO functions indicated similar functional profiles across dietary treatment groups (Table S5; see also Fig. 2B), comparison of the single functional annotations indicated certain effects on the relative abundances of KO pathways at level 2 (Table 5) and level 3 (Table 6) and functions due to the phytase addition (Table 5 and 6) (see Fig. S1 in the supplemental material; see also Table S6). Coinciding with the alterations in taxonomic abundances, phytase affected almost all major pathways within the KO level 2, including amino acid metabolism; translation; membrane transport; folding, sorting, and degradation; and energy metabolism pathways, whereas the LA treatment of cereals mainly decreased the abundance of genes coding for the carbohydrate metabolism and energy metabolism pathways (P < 0.05; Table 5). As indicated by the interactions seen with the phytase × LA-treated cereals, the LA treatment of cereals increased gene abundances for the pathway corresponding to metabolism of cofactors and vitamins but only when applied as a single treatment. Among the 120 KO level 3 pathways, 10 were enriched and 13 were less frequent due to the phytase (Table 6). The LA treatment of dietary cereals, in turn, enriched 4 and decreased 4 pathways at KO level 3 (Table 6). The majority of the pathways altered by the dietary treatments could be found for reads related to the amino acid metabolism and carbohydrate metabolism pathways.
TABLE 5.
Selected KEGG orthology level 2 pathways in cecal digesta of pigs fed diets with or without phytase and lactic acid treatment of cerealsa
| KEGG pathway | % relative abundance under indicated grain treatment conditions |
SEM |
P value |
|||||
|---|---|---|---|---|---|---|---|---|
| No phytase |
Phytase |
|||||||
| CON | LA | CON | LA | Phytase | LA | Phytase × LA | ||
| Amino acid metabolism | 32.71 | 35.49 | 27.73 | 28.47 | 2.036 | 0.007 | 0.396 | 0.621 |
| Carbohydrate metabolism | 13.76 a | 11.47 b | 13.11 ab | 12.63 b | 0.435 | 0.557 | 0.004 | 0.048 |
| Translation | 8.16 bc | 6.48 c | 8.60 b | 10.20 a | 0.729 | 0.009 | 0.960 | 0.035 |
| Membrane transport | 5.80 | 5.21 | 6.73 | 6.84 | 0.457 | 0.010 | 0.609 | 0.457 |
| Metabolism of cofactors and vitamins | 4.39 b | 6.44 a | 3.97 b | 4.33 b | 0.319 | < 0.01 | < 0.01 | 0.014 |
| Folding, sorting, and degradation | 3.96 | 3.49 | 4.83 | 4.58 | 0.395 | 0.020 | 0.373 | 0.778 |
| Energy metabolism | 2.37 | 1.81 | 2.39 | 2.25 | 0.109 | 0.046 | 0.003 | 0.063 |
| Signal transduction | 1.28 | 1.22 | 1.40 | 1.48 | 0.068 | 0.012 | 0.921 | 0.318 |
| Glycan biosynthesis and metabolism | 0.88 b | 0.86 b | 1.00 b | 1.32 a | 0.076 | < 0.01 | 0.055 | 0.037 |
| Lipid metabolism | 0.98 | 1.10 | 0.91 | 0.91 | 0.041 | 0.005 | 0.220 | 0.172 |
| Cell growth and death | 0.76 | 0.51 | 0.87 | 0.60 | 0.108 | 0.381 | 0.026 | 0.935 |
| Metabolism of terpenoids and polyketides | 0.67 | 0.52 | 0.80 | 0.75 | 0.073 | 0.020 | 0.167 | 0.494 |
| Cell motility | 0.74 | 0.60 | 0.72 | 0.66 | 0.026 | 0.388 | 0.001 | 0.112 |
| Membrane transport | 6.33 | 5.75 | 7.33 | 7.45 | 0.472 | 0.009 | 0.630 | 0.463 |
| Transport and catabolism | 0.68 | 0.84 | 0.56 | 0.24 | 0.162 | 0.037 | 0.623 | 0.155 |
| Xenobiotics biodegradation and metabolism | 0.64 | 0.59 | 0.70 | 0.73 | 0.032 | 0.005 | 0.737 | 0.236 |
| Cell communication | 0.35 | 0.33 | 0.40 | 0.40 | 0.020 | 0.005 | 0.710 | 0.473 |
Values are presented as least-square means ± SEM (n = 8 pigs per dietary treatment). Different lowercase letters (a, b, and c) in cell entries within a row indicate a significant difference (P ≤ 0.05). KEGG, Kyoto Encyclopedia of Genes and Genomes; Con, control; LA, lactic acid-treated cereals. Only KO level 2 pathways whose results were significantly different (P ≤ 0.05) or tended to be different (P ≤ 0.10) among treatments are presented.
TABLE 6.
Selected KEGG orthology level 3 pathways in cecal digesta of pigs fed diets with or without phytase and lactic acid treatment of cerealsa
| KEGG pathway no. and category | % relative abundance under indicated grain treatment conditions |
SEM |
P value |
|||||
|---|---|---|---|---|---|---|---|---|
| No phytase |
Phytase |
|||||||
| CON | LA | CON | LA | Phytase | LA | Phytase × LA | ||
| Amino acid metabolism | ||||||||
| 00260 Glycine, serine, and threonine metabolism [PATH:ko00260] | 5.66 | 5.49 | 5.62 | 5.52 | 0.036 | 0.858 | 0.001 | 0.293 |
| 00280 Valine, leucine, and isoleucine degradation [PATH:ko00280] | 0.93 | 0.83 | 1.00 | 0.94 | 0.035 | 0.015 | 0.034 | 0.595 |
| 00290 Valine, leucine, and isoleucine biosynthesis [PATH:ko00290] | 2.71 | 2.67 | 2.60 | 2.56 | 0.051 | 0.047 | 0.413 | 0.972 |
| 00300 Lysine biosynthesis [PATH:ko00300] | 3.31 | 3.46 | 3.37 | 3.42 | 0.040 | 0.788 | 0.028 | 0.238 |
| 00330 Arginine and proline metabolism [PATH:ko00330] | 3.56 | 3.59 | 3.50 | 3.51 | 0.037 | 0.052 | 0.617 | 0.760 |
| 00350 Tyrosine metabolism [PATH:ko00350] | 0.38 | 0.42 | 0.44 | 0.47 | 0.017 | 0.007 | 0.072 | 0.908 |
| 00400 Phenylalanine, tyrosine, and tryptophan biosynthesis [PATH:ko00400] | 2.29 | 2.29 | 2.20 | 2.18 | 0.035 | 0.007 | 0.816 | 0.798 |
| Carbohydrate metabolism | ||||||||
| 00010 Glycolysis/gluconeogenesis [PATH:ko00010] | 3.45 | 3.41 | 3.58 | 3.58 | 0.052 | 0.006 | 0.667 | 0.666 |
| 00020 Citrate cycle (TCA cycle) [PATH:ko00020] | 2.07 | 2.03 | 1.92 | 1.89 | 0.044 | 0.003 | 0.405 | 0.937 |
| 00030 Pentose phosphate pathway [PATH:ko00030] | 2.15 | 2.19 | 2.29 | 2.28 | 0.051 | 0.041 | 0.766 | 0.608 |
| 00052 Galactose metabolism [PATH:ko00052] | 3.45 | 3.54 | 3.63 | 3.74 | 0.059 | 0.003 | 0.103 | 0.835 |
| 00500 Starch and sucrose metabolism [PATH:ko00500] | 2.96 | 2.91 | 3.11 | 3.07 | 0.068 | 0.031 | 0.516 | 0.948 |
| 00620 Pyruvate metabolism [PATH:ko00620] | 2.81 | 2.80 | 2.89 | 2.95 | 0.048 | 0.026 | 0.591 | 0.514 |
| 00650 Butanoate metabolism [PATH:ko00650] | 0.29 a | 0.26 b | 0.27 ab | 0.30 a | 0.012 | 0.294 | 0.553 | 0.024 |
| Energy metabolism | ||||||||
| 00190 Oxidative phosphorylation [PATH:ko00190] | 3.35 | 3.23 | 3.37 | 3.38 | 0.027 | 0.007 | 0.055 | 0.022 |
| 00680 Methane metabolism [PATH:ko00680] | 0.60 ab | 0.57 b | 0.58 b | 0.64 a | 0.019 | 0.188 | 0.423 | 0.032 |
| 00720 Carbon fixation pathways in prokaryotes [PATH:ko00720] | 0.607 | 0.636 | 0.647 | 0.665 | 0.011 | 0.006 | 0.051 | 0.645 |
| Glycan biosynthesis and metabolism | ||||||||
| 00540 Lipopolysaccharide biosynthesis [PATH:ko00540] | 1.31 | 1.39 | 1.18 | 1.09 | 0.057 | 0.001 | 0.915 | 0.156 |
| Lipid metabolism | ||||||||
| 00062 Fatty acid elongation [PATH:ko00062] | 0.001 | 0.001 | 0.002 | 0.003 | 0.001 | 0.040 | 0.777 | 0.262 |
| 00071 Fatty acid metabolism [PATH:ko00071] | 0.014 | 0.010 | 0.010 | 0.007 | 0.002 | 0.043 | 0.050 | 0.642 |
| Metabolism of cofactors and vitamins | ||||||||
| 00130 Ubiquinone and other terpenoid-quinone biosynthesis [PATH:ko00130] | 0.54 | 0.58 | 0.48 | 0.49 | 0.024 | 0.004 | 0.315 | 0.569 |
| 00740 Riboflavin metabolism [PATH:ko00740] | 0.77 | 0.82 | 0.74 | 0.73 | 0.018 | 0.002 | 0.315 | 0.078 |
| 00750 Vitamin B6 metabolism [PATH:ko00750] | 0.54 | 0.55 | 0.51 | 0.50 | 0.014 | 0.012 | 0.816 | 0.629 |
| 00780 Biotin metabolism [PATH:ko00780] | 0.48 | 0.52 | 0.47 | 0.47 | 0.014 | 0.037 | 0.126 | 0.151 |
| 00785 Lipoic acid metabolism [PATH:ko00785] | 0.041 a | 0.034 b | 0.038 ab | 0.040 a | 0.002 | 0.397 | 0.213 | 0.039 |
| 00790 Folate biosynthesis [PATH:ko00790] | 0.498 | 0.530 | 0.502 | 0.510 | 0.006 | 0.255 | 0.005 | 0.071 |
| Metabolism of terpenoids and polyketides | ||||||||
| 00281 Geraniol degradation [PATH:ko00281] | 0.115 | 0.092 | 0.113 | 0.102 | 0.006 | 0.570 | 0.014 | 0.321 |
| Xenobiotics biodegradation and metabolism | ||||||||
| 00633 Nitrotoluene degradation [PATH:ko00633] | 0.027 | 0.041 | 0.017 | 0.025 | 0.004 | 0.009 | 0.018 | 0.525 |
| 00791 Atrazine degradation [PATH:ko00791] | 0.016 a | 0.011 b | 0.011 b | 0.016 a | 0.002 | 0.740 | 0.832 | 0.003 |
| 00983 Drug metabolism—other enzymes [PATH:ko00983] | 0.113 | 0.131 | 0.096 | 0.103 | 0.009 | 0.018 | 0.183 | 0.507 |
Values are presented as least-square means ± SEM (n = 8 pigs per dietary treatment). Different lowercase letters (a and b) in cell entries within a row indicate a significant difference (P ≤ 0.05). KEGG, Kyoto Encyclopedia of Genes and Genomes; Con, control; LA, lactic acid-treated cereals. Only KO level 3 pathways that were significantly different (P ≤ 0.05) or tended to be different (P ≤ 0.10) among treatments are presented. TCA, tricarboxylic acid.
Strong evidence exists for rumen metabolism indicating that changes in the rumen P availability affect the microbial enzymatic capacities to degrade the various carbohydrates, including cellulose, hemicelluloses, and starches (18). Moreover, due to the importance of fermentation in the large intestine for the energy supply of the host and knowing that metabolites from saccharolytic fermentation, such as acetate, propionate, and butyrate, exert anti-inflammatory properties, we screened the functional enzymes mapping for the carbohydrate metabolism pathway at the function level using both KO (Fig. S1; see also Table S6) and COG (Table S7) of proteins. Although the PERMANOVA data indicated very similar functional profiles of genes coding for the carbohydrate metabolism pathway across dietary treatment groups (Fig. 2C; see also Table S8), comparison of the abundances associated with individual genes showed altered enzymatic capacities for starch and fiber degradation. At KO level 3, read abundances that were functionally annotated for key catabolic steps in the degradation of glycans were altered mostly by phytase (Table 6). Accordingly, cecal samples from pigs fed the phytase diets were enriched in genes mapping for the glycolysis/gluconeogenesis, galactose metabolism, pentose phosphate, starch and mannose metabolism, and pyruvate metabolism pathways, while function genes coding for the citrate cycle pathway were less abundant in these pigs (P < 0.05; Table 6). However, in the screening of the functional enzymes, both the phytase-treated and LA-treated cereals showed a great impact on gene abundances within the starch and sucrose metabolism, glycolysis/gluconeogenesis, and citrate cycle pathways (Table S6). For instance, while genes coding for alpha-amylases and beta-xylosidases were not affected by diet, it seemed that some genes coding for enzymes needed for the degradation of cellulose were less abundant with the LA-treated cereals (P < 0.05; Table S6). Moreover, phytase supplementation and LA treatment of cereals oppositely affected abundances of genes mapping for key enzymes within the citrate cycle pathway, e.g., pyruvate carboxylase and succinyl coenzyme A (succinyl-CoA) synthetases (P < 0.05; Table S6). Also, altered primary fermentation metabolite production was indicated by abundances of genes coding for enzymes within the acetate, propionate, butyrate, lactate, and succinate synthesis pathways (P < 0.05; Table S6). For instance, the presence of phytase altered the respective gene abundances mapping for enzymes within acetate (e.g., increased acetate kinase) and propionate metabolism (i.e., decreased 2-methylcitrate dehydratase, methylisocitrate lyase, and 1-aminocyclopropane-1-carboxylate deaminase). In contrast, the LA-treated cereals tended (P < 0.10) to enrich gene abundance for the propionate-CoA transferase (pct).
Both dietary treatments led to a lower capacity of the cecal metagenome to generate butyrate, as indicated by the trend for a reduced abundance of butyrate kinase, a key enzyme in the last step of butyrate synthesis, but only when fed as single treatments as indicated by the interaction seen with the phytase × LA-treated cereals (P < 0.05; Table S6). Of note, both the phytase-treated and LA-treated cereals were associated with increased enzymatic capacities for galactose formation within the galactose metabolism pathway (P < 0.05; Table S6). Also of note, reads coding for LPS biosynthesis were less abundant with the phytase supplementation (Table S6). Moreover, interactive effects of phytase and LA treatment of cereals on methane metabolism indicated that the combination of the two treatments led to an enrichment in these genes compared to the single treatments (P < 0.05). Linking gene abundances to COG resulted in similar function profiles, showing that of the 149 COG functions, LA treatment of cereals affected 28 COG functions by enriching the genomes by 11 COG functions and depleting them by 17 COG functions (P < 0.05; Table S7). In contrast, the phytase supplementation altered 21 COG functions, depleting 9 COG functions and increasing 12 COG functions (P < 0.05; Table S7).
DISCUSSION
In the present study, we used two different dietary strategies, i.e., phytase supplementation and LA treatment of dietary cereals, to increase intestinal phytate P availability for pigs. The implementation of these strategies greatly modified the taxonomic and functional composition of the cecal bacterial metagenome, supporting the idea of the importance of the intestinal P availability for bacterial metabolism and growth. LA soaking of cereals modifies nutrient fractions other than P, including the protein, starch, and hemicellulose fractions (e.g., arabinoxylans), thereby confirming findings from our group and others (20, 22–24). These changes were likely behind the taxonomic and functional alterations in the cecal metagenome seen when only the LA-treated cereals affected taxa and enzymatic capacities. Supported by PERMANOVA results, the effect of the presence of phytase and hence the cecal P availability on alpha-diversity and on taxonomic and functional gene abundances appeared to be stronger than that of the LA treatment of cereals. Although effects on functional gene abundances were detected, the effect of the dietary treatments on the taxonomic composition was generally stronger than that on the functional composition, indicating a certain degree of compensation in the functional gene pool. All major bacterial taxa, including Prevotellaceae, Clostridiaceae, Ruminococcaceae, and Lachnospiraceae, as well as the majority of the detected functional genes with the most abundant mapping for the amino acid metabolism and carbohydrate metabolism pathways were modified by phytase supplementation. In contrast, the LA treatment of cereals largely affected the Bacteroidacaeae and Veillonellaceae communities as well as enzymatic capacities related to the carbohydrate metabolism and energy metabolism pathways, potentially reflecting a lower level of cecal provision associated with hemicelluloses (e.g., arabinoxylans), resistant starch, and protein. Unfortunately, we did not distinguish the various hemicellulose fractions and host and microbial protein degradability in the present study. Nevertheless, we can assume that the composition of carbohydrates flowing into the cecum among treatment groups was different due to the LA treatment of cereals as indicated by the differences in the gene abundances for carbohydrate metabolism pathways. This probably had consequences for the abundances of enzymatic capacities of downstream pathways for the production of primary and secondary microbial metabolites, including short- and medium-chain fatty acids and vitamin synthesis. Moreover, it should be kept in mind when interpreting the present data that, although the present shotgun metagenomics approach provided a deeper insight into the functional composition than would have been possible using 16S rRNA amplicon sequencing, it is still a prediction of gene abundances and enzymatic capacities based on short, 150-bp fragments. Moreover, the prediction of the functional capabilities was based on the KO database and the corresponding alignment of genes into “metabolic clusters,” which may not always reflect the physiologic situation in the porcine cecum (28).
Phytase supplementation generally increases phytate-P availability, especially in the upper digestive tract, leading to increased absorption of P in the small intestine (17). As reported previously (19–24), the LA treatment of cereals also improved the intestinal P availability; however, the treatment was less efficient than phytase supplementation. The greatest improvement in P retention was achieved by the combination of the two treatments (29). The latter may explain some interactive effects of the two treatments on bacterial taxonomy and metabolism. Accordingly, less P would flow into the cecum with the phytase-treated than with the LA-treated cereal diets, resulting in a reduced amount of P being available for bacterial metabolism and causing the distinct bacterial community clusters visualized in the NMDS plots. Nevertheless, species richness was not compromised by the potentially lower P availability in cecal digesta with the phytase supplementation, suggesting that taxa with higher P requirements were replaced by taxa with lower P demands. Following this reasoning, members of Clostridiaceae, Ruminococcaceae, and Lachnospiraceae may have filled the metabolic niches that opened due to the phytase-related decline in the populations of the most abundant families, Prevotellaceae and Veillonellaceae. In accordance with that, Shannon and Simpson indices indicated an increase in the diversification of the bacterial communities due to the phytase. Since greater diversity has been often linked with higher levels of gut health (30), this finding may point toward greater stability of the microbial community with the phytase supplementation. A stable bacterial community may have been also indicated by the overall functional profiles among treatment groups, which diverged less than the taxonomic profiles, demonstrating the metabolic plasticity of the cecal metagenome across dietary treatments. However, a closer look at the actual changes in species abundances is warranted as, for instance, the predominating family Clostridiaceae comprises certain pathobionts. For instance, the combination of phytase supplementation with LA treatment of cereals increased the abundances of Clostridium difficile and C. perfringens. Their abundances may not necessarily compromise the health of the pig, but they may have zoonotic potential if shed in feces or as contaminants of pork during the slaughtering process (31). Lower abundances of pathobionts may be favorable for cecal health in stressful periods of pig’s life. Following this reasoning, the phytase diet decreased the abundances of groups representing lower abundance within Proteobacteria, such as Enterobacteriaceae and Campylobacteraceae. These families comprise mostly commensals but also contain some swine-relevant pathobionts. Moreover, many of these bacteria carry strongly immune-reactive MAMPs on their cell surface, thereby contributing to the activation of proinflammatory pathways in the mucosa (6). Functional profiles indicated that fewer genes mapping for enzymes within the LPS biosynthesis pathway were present with the phytase diet, which potentially led to a lower Toll-like receptor-4 (TLR-4)-activated immune response in the cecum of these pigs. Albeit with each representing less than 1% of the cecal community, it can be speculated whether the phytase-related decline in Campylobacteraceae and Enterobacteriaceae abundances might have had a stronger effect (reduced immune activation) than the declines in the abundances of the other major Gram-negative taxa reduced by the phytase diets, the members of the Prevotellaceae family. In fact, the fatty acid composition of the lipid A component of LPS, which is responsible for TLR-4 activation, in Proteobacteria provokes a stronger immune response than LPS in the cell wall of most Bacteroidetes (9). Moreover, the present data appear to support the idea of a positive relationship between intestinal P availability and the abundance of alphaproteobacteria such as Enterobacteriaceae (25, 32).
In contrast, compromised fiber fermentation due to insufficient intestinal P flow, especially in ruminants, due to potentially higher P requirements of fibrolytic than amylolytic bacteria has been previously reported (10). Since fiber fermentation contributes to the overall energy supply for the pig (33) and since the primary fermentation metabolites, such as SCFA, have anti-inflammatory properties (34), modulation of the dietary P supply also needs to be regarded from this angle. Following this line of reasoning, cellulolytic and hemicellulolytic taxa within the very versatile and diverse members of the Prevotellaceae family may have been reduced in abundance with the phytase diet, including the most dominant amplicon sequence variants (ASV) being assigned as Prevotella copri, confirming the negative link between fibrolytic bacteria and luminal P availability. In fact, P. copri utilizes various complex polysaccharides from plant material (35). Against the generalization that intestinal fibrolytic bacteria are negatively affected by lower levels of P in their environment were the present findings indicating that certain fibrolytic taxa within the families Clostridiaceae, Ruminococcaceae, and Lachnospiraceae thrived on or were unaffected by the phytase diet. These included predominant cellulolytic and hemicellulolytic species, such as C. phytofermentans, C. thermocellum, C. cellulolyticum, Ruminococcus albus, and R. flavefaciens, potentially indicating lower bacterial P requirements of these fibrolytic bacteria. Apparently, the P content in the cecal digesta was sufficient to support their growth, replacing the Prevotella ASVs. This assumption might be supported by the fact that the abundances of functional genes coding for hemicellulases (e.g., xylanases) and beta-glucanases were similar across the dietary treatment groups. Likewise, the abundances of amylolytic taxa, including C. beijerinckii, C. acetobutylicum, C. saccharolyticum, and Roseburia spp., increased in cecal digesta of pigs receiving the phytase-containing diets, coinciding with the enrichment of functional genes mapping for the starch and sucrose metabolism pathway seen with the phytase supplementation, supporting greater bacterial degradation of residual dietary starch in cecal digesta. In particular, capabilities identified as associated with starch phosphorylase and cyclomaltodextrinase increased, whereas alpha-amylases were not differently abundant. Nevertheless, starch-degrading Eubacterium rectale, representing the fourth most abundant ASV, decreased in abundance in cecal digesta of pigs fed the phytase diets, challenging the assumption that amylolytic bacteria may have lower P requirements per se than fibrolytic bacteria.
The level of the effect of the LA treatment on phytate-P release was lower than that seen with the phytase supplementation (24); therefore, more P was flowing into the cecum of pigs fed the diet with the LA-treated cereals than into that of pigs fed the phytase diet. In accordance with that result, the decline in the abundances of Bacteroides, Parabacteroides, and Streptococcus seen with the LA-treated cereals may rather have been related to altered availability of complex carbohydrates, such as resistant and nonresistant starch and hemicelluloses, and of protein in the LA-soaked cereals. It may be assumed that the taxa capable of degrading arabinoxylans were mainly affected as wheat and corn comprise higher levels of insoluble and soluble arabinoxylans than β-glucans (36). However, changes in the ileal flow of starch residuals and degradation products may have led to differences in cross-feeding relationships, altering the abundances of bacteria (e.g., Bacteroides, Parabacteroides, and Streptococcus) as well as their nutritive dependencies. This assumption might be supported by the reduced gene abundances mapping for endoglucanase or the presumed increased trehalose-6-phosphate hydrolase activity, to give two examples. Nevertheless, it needs to be considered that these gene abundances were predicted from KO functions, which hardly described the bacterial relationships and consortia needed for cellulose (e.g., cellulosomes) and hemicellulose degradation. The lower protein content and assumingly altered protein degradability of the LA diet may have specifically altered metabolic pathways related to the glycine, serine, and threonine metabolism pathway and the lysine biosynthesis pathway.
Alterations in metabolite release from the starch and sucrose metabolism, galactose metabolism, and amino sugar and nucleotide metabolism pathways associated with the phytase-supplemented and LA-treated cereals were likely the reason for the functional changes observed in the downstream glycolysis/gluconeogenesis, citrate cycle, and pyruvate metabolism pathways, thereby altering the supply of metabolites for the LPS biosynthesis, propanoate metabolism, and butanoate metabolism pathways. It is noteworthy that enzymatic capacities within the citrate cycle appeared to be greatly affected by the cecal P availability, being one of the key metabolic pathways for generation of NADH and FADH and, eventually, ATP (37). The phytase-associated decrease in pathways related to the vitamin metabolism pathway (i.e., the riboflavin, pyridoxine, and biotin pathways) should have had no consequences for the host pig as sufficient dietary supplementation was offered and the vitamin absorption from the hindgut is generally low (38, 39). Nevertheless, the increase in genes mapping for the folate biosynthesis pathway seen with the LA-treated cereals may have provided the pigs with additional methyl-group donors, being beneficial for systemic carbohydrate and amino acid metabolism in fast-growing animals (37).
Changes in taxonomic and functional abundances also indicated alterations in bacterial cross-feeding relationships (4). For instance, phytase supplementation decreased while LA treatment of cereals increased the abundances of lactate-utilizing and acetate- and succinate-producing Mitsuokella species, including M. multiacida (40). Of note, although changes in the functional features, e.g., SCFA synthesis pathways, represent a conglomerate of genes from various taxa, the enzyme profiles for butyrate synthesis (phytase × LA-treated cereal interaction) pointed toward Clostridium (41) as the main contributor to this gene pool.
Results further indicated that it was mainly the phytase supplementation that influenced the abundance and metabolism of the archaeal community (e.g., methane metabolism) as the three major archaeal families, i.e., Methanobacteriaceae, Methanosarcinaceae, and Methanococcaceae, representing together more than 50% of the archaeal reads, were unaffected by diet. Phytase effects on archaeal taxon abundances diverged from the dietary effect on methane metabolism, suggesting that metabolic changes were independent of taxon alterations. Since the archaeal metabolism relies on the supply of nutrients, such as monosaccharides and hydrogen, from other bacteria (42), the increased abundance of genes mapping for enzymes associated with methane metabolism may point toward there having been greater substrate availability in cecal digesta when the two dietary treatments were combined. There were similar interactive effects of phytase-supplemented and LA-treated cereals on the butanoate metabolism pathway, potentially indicating that the methane metabolism and butyrate production pathways had been linked. Supporting this assumption, parallel hydrogen production up to a certain level of butyrate generation has been reported to occur, for instance, in marine organisms (i.e., up to 25 mmol butyrate/liter) (43). As the cecal butyrate concentrations were lower than 25 mmol/liter, the occurrence of enhanced levels of hydrogen supply with both treatments may be a reasonable explanation for the increased enzymatic capacities related to methane metabolism.
In conclusion, the present results demonstrated the dramatic impact of dietary P availability on the taxonomic and functional composition of the cecal metagenome in growing pigs. Due to the stronger effect on phytase-P release, phytase supplementation affected the abundances of almost all predominant eubacterial and many archaeal taxa, whereas the LA-treated cereals mainly reduced Bacteroidaceae abundance. Results further indicated that taxonomic alterations were more drastic than functional changes. Notably, a greater number of Gram-negative taxa, such as the dominant Prevotellaceae, were reduced in abundance by the phytase, whereas Gram-positive taxa (e.g., Clostridiaceae, Ruminococcaceae, and Lachnospiraceae) seemed to fill the opening metabolic niches, with consequences for the MAMP profiles stimulating the cecal mucosal immune response, which should be pursued in further research. The LA-treated cereals mainly modified enzymatic capacities corresponding to the amino acid metabolism and carbohydrate metabolism pathways, being, aside from the changes in P availability, linked to LA-soaking-associated changes of the protein, starch, and fiber fraction in the cereals.
MATERIALS AND METHODS
Experimental design and diets.
All procedures involving animal handling and treatment were approved by the institutional ethics committee of the University of Veterinary Medicine and the national authority according to paragraph 8 of the Law for Animal Experiments, Tierversuchsgesetz (TVG) (BMWFW-68.205/0158-W F/V/3b/2016). A total of 32 castrated male pigs (Large White, 13.1 ± 2.3 kg; age, 6 to 8 weeks) were obtained from the research farm of the University of Veterinary Medicine, Vienna. Only healthy pigs were selected for the experiments. Housing and preexperimental conditions were as reported previously by Klinsoda et al. (25, 26). Pigs were randomly allocated to one of four dietary treatments in a 2 (phytase supplementation, 0 versus 500 phytase units [FTU])/kg complete feed) × 2 (LA-treated versus nontreated cereals) factorial design with four replicate batches (n = 8 per replicate batch) as follows: control diet, diet containing LA-treated cereals, control diet with phytase, and diet with phytase-supplemented and LA-treated cereals. Two pigs were fed one diet per replicate batch. Each replicate batch lasted 19 days, with collection of intestinal digesta on days 18 and 19.
The basal diet consisted of wheat, corn, and soybean meal and was formulated to meet or exceed current recommendations for nutrient requirements for growing pigs (44, 45; see also Table S1 in the supplemental material). Two mineral-vitamin premixes, one without phytase and one with phytase (500 FTU/kg complete feed), were formulated according to the standard inclusion level of phytase (14). The diets were mixed with either the premix without phytase or the premix with phytase (500 FTU/kg complete feed). Cereals (i.e., wheat and corn) were left untreated or soaked in a 2.5% LA solution for 48 h (24). The LA concentration and incubation time were selected based on the results of our previous in vitro experiment (24). After soaking, the cereals were dried at 70°C for 1 h followed by 60°C for 23 h. The dried grains were ground to pass through a 5-mm-pore-size sieve followed by preparation of the diets. Diets were analyzed for dry matter, Ca, P, neutral detergent fiber, acid detergent fiber, resistant starch, and nonresistant starch as previously described (25, 26). Pigs were fed three meals per day, at 8 a.m., 12 a.m., and 4 p.m., as mash, with feed allowances amounting to 3 times the maintenance requirement {[(body weight0.6 × 197)/238.68] × 3} (44).
Sample collection.
Two hours after their last feeding on experimental days 18 and 19, pigs were anesthetized (ketamine hydrochloride [Narketan; Vétoquinol GmbH, Germany] [100 mg/ml, 1 ml/10 kg body weight] and azaperone [Stresnil; Elanco Deutschland GmbH, Germany] [40 mg/ml, 0.5 ml/10 kg body weight]) and euthanized (intracardiac injection of 10 ml/kg embutramide [MSD Animal Health, Vienna, Austria]). After the abdominal cavity was opened, the entire gastrointestinal tract was removed, the small and large intestines were carefully dissected from the mesentery, and clamps were used to prevent mixing of digesta between intestinal segments. After separation of the cecum, the total cecal digesta was aseptically collected. Homogenized aliquots of cecal digesta were snap-frozen in liquid nitrogen and stored at –80°C. All samples were collected within 20 to 30 min after the death of the animal.
DNA isolation and shotgun metagenomic sequencing.
Total DNA was isolated from approximately 250 mg of cecal digesta using a PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA) with a few modifications. There was an additional heating step at 70°C for 10 min performed between the steps during which the digesta samples were mixed with buffer C1. The samples were beaten (6.5 m/s) 3 times for 1 min each time on a FastPrep-24 instrument (MP Biomedicals, Heidelberg, Germany) with cooling on ice performed between the individual bead-beating steps. The DNA concentration of each sample was quantified by a Qubit 4.0 fluorometer (Life Technologies, Carlsbad, CA) using a Qubit double-stranded DNA HS (high sensitivity) assay kit (Life Technologies, Carlsbad, CA) following the manufacturer’s instructions. The DNA isolates were sent for whole-genome shotgun metagenome sequencing using an Illumina NextSeq 500 v2 sequencing platform (Illumina Inc., San Diego, CA) as described before (2). For this, the total genomic DNA was prepared with Illumina TruSeq reagent chemistry and the Illumina TruSeq nano protocol for whole-genome shotgun sequencing of multiplexed 150-bp libraries by Microsynth AG (Balgach, Switzerland) using a high-output, single-end protocol. The barcoded sample libraries were sequenced. The obtained FASTQ files were demultiplexed, quality filtered, and trimmed of Illumina adaptor residuals to 150 bp by Microsynth AG.
Bioinformatic processing of sequences.
The quality-filtered sequences were processed using the MG-RAST pipeline (Metagenomics Rapid Annotation using Subsystem Technology, v4.0; Argonne National Laboratories; https://www.mg-rast.org) (46–48) in order to build the taxonomic and functional profiles. Detailed information on the bioinformatics workflow used by the MG RAST pipeline can be found at https://www.mg-rast.org. In brief, after removal of host genomic DNA, prediction of duplicate reads, 16S rRNA reads, and putative protein coding features was performed using the GragGeneScan method and the reads were clustered at a high identity level (90%). Sequence protein similarity searches against the M5NR protein database were computed in sBLAT (49). The REfSeq database, KEGG orthology (KO), and Clusters of Orthologous Groups (COG) annotation sources of MG-RAST were used for taxonomic and functional analysis of cecal samples (50, 51). The criteria applied for inclusion were a maximum E value of 1e−05, a minimum identity threshold value of 60%, and a minimum alignment length of 15 (2). MG RAST processing of the sequence data took place before April 2020, when the family Lactobacillaceae was reclassified (52). Due to this reclassification, Leuconostocaceae were united with Lactobacillaceae representing the novel Lactobacillaceae family (52). Accordingly, results at the family level for Lactobacillaceae and Leuconostococcaceae were merged after the bioinformatic processing in the present work was performed.
Differences in bacterial community were assessed using R studio (version 1.0.136) as previously described by Klinsoda et al. (25). Statistical assessment of dissimilarity matrices (Bray-Curtis) using the ‘adonis2’ function (PERMANOVA) in the R package ‘vegan’ (version 2.5.2) was used to analyze β-diversity and was visualized in two-dimensional nonmetric multidimensional scaling (NMDS) ordination plots obtained with the ‘metaMDS’ function in the vegan R package (53). PERMANOVA was thereby applied to the Bray-Curtis distance matrices between dietary factors (phytase supplementation and LA treatment of cereals and their interaction). Statistical significance was calculated after 999 random permutations.
Statistical analysis.
A power test analysis was performed to identify the number of observations needed for the present pig experiment in order to reject the null hypothesis (H0), if H0 was false (P = 1 − β), using SAS software (version 9.4; SAS Institute Inc., Cary, NC, USA) as described by Metzler-Zebeli et al. (2, 54). Data representing the relative abundance at each taxonomic and functional level were ranked and analyzed in SAS. Data were analyzed for normal distribution using the Shapiro-Wilk test with the UNIVARIATE procedure in SAS. Taxonomic data at the phylum, family, genus, and ASV levels as well as predicted KEGG pathways and COG functions between dietary treatments were subjected to analysis of variance (ANOVA) using the MIXED procedure in SAS. The fixed effects included in the model were phytase supplementation and LA treatment of grains and their two-way interaction. The random effect was “replicate,” and the experimental unit was “pig.” The approximate degrees of freedom were estimated by the Kenward-Rogers method (ddfm = kr). Relative abundances of taxa and predicted KEGG pathways were reported as least-square means ± standard errors of the means (SEM). If two-way interactions occurred, pairwise comparisons between least-squares means were performed using the pdiff function in SAS. For hypothesis testing, the level of significance was declared at a P value of <0.05 and trends at 0.05 < P ≤ 0.10.
Data availability.
Raw sequence data can be found at MG RAST (project identifier [ID] mjg89507).
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Enzinger, A. Sener, M. Hollmann, A. Dockner, M. Wild, and S. Sharma of the Institute of Animal Nutrition and Functional Plant Compounds for assistance in the animal trial, sample collection, and laboratory analysis.
J.K. is grateful for funding provided under the project of the ASEAN-European Academic University Network (ASEA UNINET) from the Austrian Agency for International Cooperation (Österreichischer Austauschdienst, OEAD) for the financial support in the Ph.D. program in Austria.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Liu B, Wang W, Zhu X, Sun X, Xiao J, Li D, Cui Y, Wang C, Shi Y. 2018. Response of gut microbiota to dietary fiber and metabolic interaction with SCFAs in piglets. Front Microbiol 9:2344. doi: 10.3389/fmicb.2018.02344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzler-Zebeli BU, Newman MA, Grüll D, Zebeli Q. 2019. Functional adaptations in the cecal and colonic metagenomes associated with the consumption of transglycosylated starch in a pig model. BMC Microbiol 19:87. doi: 10.1186/s12866-019-1462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zarrinpar A, Chaix A, Yooseph S, Panda S. 2014. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flint HJ, Duncan SH, Scott KP, Louis P. 2015. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc 74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- 5.Jones RM, Neish S. 2011. Recognition of bacterial pathogens and mucosal immunity. Cell Microbiol 13:670–676. doi: 10.1111/j.1462-5822.2011.01579.x. [DOI] [PubMed] [Google Scholar]
- 6.Broom L, Kogut M. 2018. Gut immunity: its development and reasons and opportunities for modulation in monogastric production animals. Anim Health Res Rev 19:46–52. doi: 10.1017/S1466252318000026. [DOI] [PubMed] [Google Scholar]
- 7.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander C, Rietschel E. 2001. Invited review: bacterial lipopolysaccharides and innate immunity. J Endotoxin Res 7:167–202. doi: 10.1177/09680519010070030101. [DOI] [PubMed] [Google Scholar]
- 9.Wyns H, Plessers E, De Backer P, Meyer E, Croubels S. 2015. In vivo porcine lipopolysaccharide inflammation models to study immunomodulation of drugs. Vet Immunol Immunopathol 166:58–69. doi: 10.1016/j.vetimm.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Durand M, Komisarczuk S. 1988. Influence of major minerals on rumen microbiota. J Nutr 118:249–260. doi: 10.1093/jn/118.2.249. [DOI] [PubMed] [Google Scholar]
- 11.Heyer C, Weiss E, Schmucker S, Rodehutscord M, Hoelzle L, Mosenthin R, Stefanski V. 2015. The impact of phosphorus on the immune system and the intestinal microbiota with special focus on the pig. Nutr Res Rev 28:67–82. doi: 10.1017/S0954422415000049. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Zhang D, Yang T, Bryden W. 2016. Phosphorus bioavailability: a key aspect for conserving this critical animal feed resource with reference to broiler nutrition. Agriculture 6:25. doi: 10.3390/agriculture6020025. [DOI] [Google Scholar]
- 13.Oster M, Gerlinger C, Heide K, Just F, Borgelt L, Wolf P, Polley C, Vollmar B, Muráni E, Ponsuksili S, Wimmers K. 2018. Lower dietary phosphorus supply in pigs match both animal welfare aspects and resource efficiency. Ambio 47:20–29. doi: 10.1007/s13280-017-0969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dersjant-Li Y, Awati A, Schulze H, Partridge G. 2015. Phytase in non-ruminant animal nutrition: a critical review on phytase activities in the gastrointestinal tract and influencing factors. J Sci Food Agric 95:878–896. doi: 10.1002/jsfa.6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredlund K, Asp NG, Larsson M, Marklinder I, Sandberg AS. 1997. Phytate reduction in whole grains of wheat, rye, barley and oats after hydrothermal treatment. J Cereal Sci 25:83–91. doi: 10.1006/jcrs.1996.0070. [DOI] [Google Scholar]
- 16.Lyberg K, Lundh T, Pedersen C, Lindberg JE. 2006. Influence of soaking, fermentation and phytase supplementation on nutrient digestibility in pigs offered a grower diet based on wheat and barley. Anim Sci 82:853–858. doi: 10.1017/ASC2006109. [DOI] [Google Scholar]
- 17.Metzler BU, Mosenthin R, Baumgärtel T, Rodehutscord M. 2008. The effect of dietary phosphorus and calcium level, phytase supplementation, and ileal infusion of pectin on the chemical composition and carbohydrase activity of fecal bacteria and the level of microbial metabolites in the gastrointestinal tract of pigs. J Anim Sci 86:1544–1555. doi: 10.2527/jas.2007-0267. [DOI] [PubMed] [Google Scholar]
- 18.Metzler-Zebeli BU, Haselmann A, Klevenhusen F, Knaus W, Zebeli Q. 2018. Lactic acid treatment of by-products and phosphorus level in the diet modulate bacterial microbiome and the predicted metagenome functions using the rumen simulation technique. J Dairy Sci 101:9800–9814. doi: 10.3168/jds.2018-14821. [DOI] [PubMed] [Google Scholar]
- 19.Rimsten L, Haraldsson A, Andersson R, Alminger M, Sandberg A, Åman P. 2002. Effects of malting on β-glucanase and phytase activity in barley grain. J Sci Food Agric 82:904–912. doi: 10.1002/jsfa.1135. [DOI] [Google Scholar]
- 20.Haraldsson A, Rimsten L, Alminger M, Andersson R, Andlid T, Åman P, Sandberg A. 2004. Phytate content is reduced and β-glucanase activity suppressed in malted barley steeped with lactic acid at high temperature. J Sci Food Agric 84:653–662. doi: 10.1002/jsfa.1724. [DOI] [Google Scholar]
- 21.Gänzle MG. 2014. Enzymatic and bacterial conversions during sourdough fermentation. Food Microbiol 37:2–10. doi: 10.1016/j.fm.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Metzler-Zebeli BU, Deckardt K, Schollenberger M, Rodehutscord M, Zebeli Q. 2014. Lactic acid and thermal treatments trigger the hydrolysis of myo-inositol hexakisphosphate and modify the abundance of lower myo-inositol phosphates in barley (Hordeum vulgare L.). PLoS One 9:e101166. doi: 10.1371/journal.pone.0101166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harder H, Khol-Parisini A, Metzler-Zebeli BU, Klevenhusen F, Zebeli Q. 2015. Treatment of grain with organic acids at 2 different dietary phosphorus levels modulates ruminal microbial community structure and fermentation patterns in vitro. J Dairy Sci 98:8107–8120. doi: 10.3168/jds.2015-9913. [DOI] [PubMed] [Google Scholar]
- 24.Vötterl JC, Zebeli Q, Hennig-Pauka I, Metzler-Zebeli BU. 2019. Soaking in lactic acid lowers the phytate-phosphorus content and increases the resistant starch in wheat and corn grains. Anim Feed Sci Tech 252:115–125. doi: 10.1016/j.anifeedsci.2019.04.013. [DOI] [Google Scholar]
- 25.Klinsoda J, Vötterl J, Zebeli Q, Metzler-Zebeli BU. 2019. Lactic acid treatment of cereals and dietary phytase modified fecal microbiome composition without affecting expression of virulence factor genes in growing pigs. Front Microbiol 10:2345. doi: 10.3389/fmicb.2019.02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klinsoda J, Vötterl J, Zebeli Q, Metzler-Zebeli BU. 2019. Alterations of the viable ileal microbiota of gut-mucosa-lymph node axis in pigs fed phytase and lactic acid-treated cereals. Appl Environ Microbiol 86:e02128-19. doi: 10.1128/AEM.02128-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jovel J, Patterson J, Wang W, Hotte N, O'Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, Wong GK. 2016. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol 7:459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Hu H, Zijlstra R, Zheng J, Gänzle M. 2019. Metagenomic reconstructions of gut microbial metabolism in weanling pigs. Microbiome 7:48. doi: 10.1186/s40168-019-0662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vötterl J, Klinsoda J, Zebeli Q, Hennig-Pauka I, Metzler-Zebeli BU. 2019. Effects of lactic acid treatment of cereals and dietary phytase on calcium and phosphorus balance and serum parameters in growing pigs, p 25 Proceedings of the 73rd Conference. Society of Nutrition Physiology, Göttingen, Germany. [Google Scholar]
- 30.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baer A, Miller M, Dilger A. 2013. Pathogens of interest to the pork industry: a review of research on interventions to assure food safety. Compr Rev Food Sci Food Saf 12:183–217. doi: 10.1111/1541-4337.12001. [DOI] [Google Scholar]
- 32.Metzler-Zebeli BU, Mann E, Schmitz-Esser S, Wagner M, Ritzmann M, Zebeli Q. 2013. Changing dietary calcium-phosphorus level and cereal source selectively alters abundance of bacteria and metabolites in the upper gastrointestinal tracts of weaned pigs. Appl Environ Microbiol 79:7264–7272. doi: 10.1128/AEM.02691-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, Bai Y, Zhang G, Liu L, Lai C. 2020. Relationship between dietary fiber fermentation and volatile fatty acids’ concentration in growing pigs. Animals 10:263. doi: 10.3390/ani10020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenzie C, Tan J, Macia L, Mackay CR. 2017. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol Rev 278:277–295. doi: 10.1111/imr.12556. [DOI] [PubMed] [Google Scholar]
- 35.Fehlner-Peach H, Magnabosco C, Raghavan V, Scher J, Tett A, Cox L, Gottsegen C, Watters A, Wiltshire-Gordon J, Segata N, Bonneau R, Littman D. 2019. Distinct polysaccharide utilization profiles of human intestinal Prevotella copri isolates. Cell Host Microbe 26:680–690.e5. doi: 10.1016/j.chom.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamphues J, Hartung C, Wilke V, Grone R. 2019. Rye: renaissance of a traditional grain type in animal feeding? Übers Tierernährg 43:107–163. [Google Scholar]
- 37.Michal G. 1999. Biochemical pathways: an atlas of biochemistry and molecular biology. Spektrum Akademischer Verlag, Heidelberg, Germany. [Google Scholar]
- 38.Kopinski JS, Leibholz J, Love RJ. 1989. Biotin studies in pigs: the post-ileal absorption of biotin. Br J Nutr 62:781–789. doi: 10.1079/bjn19890079. [DOI] [PubMed] [Google Scholar]
- 39.Asrar FM, O'Connor DL. 2005. Bacterially synthesized folate and supplemental folic acid are absorbed across the large intestine of piglets. J Nutr Biochem 16:587–593. doi: 10.1016/j.jnutbio.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Tsukahara T, Koyama H, Okada M, Ushida K. 2002. Stimulation of butyrate production by gluconic acid in batch culture of pig cecal digesta and identification of butyrate-producing bacteria. J Nutr 132:2229–2234. doi: 10.1093/jn/132.8.2229. [DOI] [PubMed] [Google Scholar]
- 41.Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol 186:2099–2106. doi: 10.1128/jb.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nkamga VD, Henrissat B, Drancourt M. 2017. Archaea: essential inhabitants of the human digestive microbiota. Hum Microb J 3:1–8. doi: 10.1016/j.humic.2016.11.005. [DOI] [Google Scholar]
- 43.Lee J, Klaus D, Maness P, Spear J. 2007. The effect of butyrate concentration on hydrogen production via photofermentation for use in a Martian habitat resource recovery process. Int J Hydrogen Energy 32:3301–3307. doi: 10.1016/j.ijhydene.2007.05.029. [DOI] [Google Scholar]
- 44.Gesellschaft fur Ernahrungsphysiologie. 2006. Empfehlungen zur Energie- und Nährstoffversorgung von Schweinen. DLG, Frankfurt am Main, Germany. [Google Scholar]
- 45.NRC. 2012. Nutrient requirements of swine, 11th revised ed National Academy Press, Washington, DC. [Google Scholar]
- 46.Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. 2010. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc 2010:pdb.prot5368. doi: 10.1101/pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- 47.Keegan KP, Glass EM, Meyer F. 2016. MG-RAST, a metagenomics service for analysis of microbial community structure and function. Methods Mol Biol 1399:207–233. doi: 10.1007/978-1-4939-3369-3_13. [DOI] [PubMed] [Google Scholar]
- 48.Wilke A, Bischof J, Gerlach W, Glass E, Harrison T, Keegan KP, Paczian T, Trimble WL, Bagchi S, Grama A, Chaterji S, Meyer F. 2016. The MG-RAST metagenomics database and portal in 2015. Nucleic Acids Res 44:D590–D594. doi: 10.1093/nar/gkv1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kent WJ. 2002. BLAT - the BLAST-like alignment tool. Genome Res 12:656–654. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng J, Wittouck S, Salvetti E, Franz CMAP, Harris HMB, Mattarelli P, O'Toole PW, Pot B, Vandamme P, Walter J, Watanabe K, Wuyts S, Felis GE, Gänzle MG, Lebeer S. 2020. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70:2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 53.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Eduard Szoecs E, Wagner H. 2018. vegan: Community Ecology Package R package version 2.5-2. https://CRAN.R-project.org/package=vegan.
- 54.Metzler-Zebeli BU, Schmitz-Esser S, Mann E, Grüll D, Molnar T, Zebeli Q. 2015. Adaptation of the cecal bacterial microbiome of growing pigs in response to resistant starch type 4. Appl Environ Microbiol 81:8489–8499. doi: 10.1128/AEM.02756-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data can be found at MG RAST (project identifier [ID] mjg89507).