Abstract
Background
Single-gene tests and hotspot panels targeting specific subsets of biomarkers constitute the Canadian genomic testing landscape for non-small-cell lung cancer (nsclc). However, newer testing options such as comprehensive genomic profiling (cgp) offer improved detection rates and identification of multiple classes of genomic alterations in a single assay, minimizing tissue requirements and turnaround time. The objective of the present analysis was to assess the health and budget impacts of adopting cgp testing for nsclc in Canada.
Methods
This study assessed the impact of funding the cgp tests FoundationOne CDx and FoundationOne Liquid (Foundation Medicine, Cambridge, MA, U.S.A.) over a 3-year time horizon using a Canadian societal perspective for Ontario. Conventional testing strategies were summarized into two reference scenarios: a series of single-gene tests only, and reflex single-gene testing followed by a hotspot panel for negative results. Four adoption scenarios for cgp testing were considered: replacing all single-gene and hotspot panel testing, replacing hotspot panel testing only, use after negative single-gene and hotspot testing, and use of FoundationOne Liquid in individuals with insufficient tissue for conventional testing.
Results
When cgp testing was assumed to replace all conventional testing with 50% uptake, the budget impact per person per year ranged from $0.71 to $0.87, depending on the reference scenario, with a 3-year gain of 680.9 life–years and 3831 working days over the full cohort.
Conclusions
Given the present testing landscape for patients with nsclc in Canada, listing cgp testing could optimize the selection of appropriately targeted treatments, and thus add life–years and productivity for this population, with a minimal budget impact.
Keywords: Foundation Medicine, health and budget impact analyses, genomic profiling, next-generation sequencing, non-small-cell lung cancer, single-gene tests, hotspot panels, Canadian private payers
INTRODUCTION
Lung cancer is the most common cause of cancer-related death in Canada, and more than 80% of lung cancer cases are non-small-cell lung cancer (nsclc)1. Therapies targeting specific genomic mutations have shown promising results for treating this type of cancer. In Canada, approved treatments include crizotinib for ALK-positive tumours; pembrolizumab and nivolumab for PD-L1–positive tumours; erlotinib, gefitinib, and afatinib for EGFR-positive tumours; and crizotinib for ROS1-positive tumours2,3. Critical in the selection of nsclc therapies is early genomic testing to ascertain which mutations a patient might be harbouring and thus the feasibility of receiving therapies that target such mutations. There is currently no national oversight in Canada with respect to standard-of-care genetic testing4, which has led to provincial variation in the proportion of patients with cancer who are tested for genomic alterations5 and in the timing and modalities of tests received6.
Current genomic testing in Canada uses a combination of single-gene testing methods such as polymerase chain reaction, Sanger sequencing, fluorescence in situ hybridization, and immunohistochemistry, and multi-gene testing, which typically uses next-generation sequencing hotspot panels. Although accessible, those conventional testing methods have documented limitations. In one study, 70% of patients with actionable ALK rearrangements missed by single-gene testing methods responded to treatment7. In other studies, 65% of tumours deemed by single-gene testing methods to be negative for a driver alteration were subsequently found to have clinically-relevant and actionable alterations8, and 58% of patients who tested negative for EGFR, KRAS, and ALK had clinically relevant alterations suggesting potential for benefit from targeted therapy9. Furthermore, sequential testing can potentially exhaust available tissue before all relevant biomarkers are measured10–13.
Given variation across provinces (and regional variation within provinces), there is no standard approach to testing in Canada. Some treatment facilities have in-house laboratories able to conduct testing, including hotspot panels, resulting in testing strategy flexibility. However, smaller facilities lack access to the same infrastructure and must ship samples to reference laboratories. Those differences can lead to discrepancies in testing accessibility and subsequent treatment options for patients. In addition, the increased need for testing can be a burden to the testing centres.
Comprehensive genomic profiling (cgp) refers to a method by which multiple classes of genomic alterations can be identified in a single assay, thus addressing high tissue requirements and minimizing turnaround time, while providing comprehensive results. The process is more patient-centric, requiring less-frequent invasive tissue collection and offering an option for patients for whom tissue collection is not possible. The FoundationOne CDx and FoundationOne Liquid tests from Foundation Medicine (Cambridge, MA, U.S.A.) represent one such cgp approach. The tests detect all known classes of alterations within a single assay. For nsclc in particular, FoundationOne CDx can simultaneously test for EGFR, ALK, BRAF, ROS1, and various other alterations of interest, potentially reducing the amount of adequate tissue required and the turnaround time relative to strategies requiring multiple or sequential testing. The FoundationOne Liquid test provides analogous results by liquid biopsy of a blood sample, making cgp testing feasible in individuals with insufficient tissue for traditional testing methods. In nsclc, available tumour tissue is a practical concern for some patients, and in addition, a liquid test is desirable at the time of disease progression because it can help to avoid a repeat biopsy.
There is increasing evidence that, although still relatively new, personalized medicine—including optimized diagnostic testing—has the potential to improve health outcomes14,15. However, the costs of the tests themselves can be an important consideration in uptake, and test costs can be challenging to incorporate into cost-effectiveness assessments of treatments5. Economic analyses specific to testing can help to provide insights.
The objective of the present study was to estimate the health and budget impacts of listing FoundationOne CDx and FoundationOne Liquid for patients with advanced or metastatic nsclc as a companion to current testing strategies. For the estimate, a health and budget impact model was developed that characterized differences in costs and overall survival (os) associated with varying use of the Foundation Medicine tests as an alternative to conventional testing methods for patients with nsclc in Canada.
METHODS
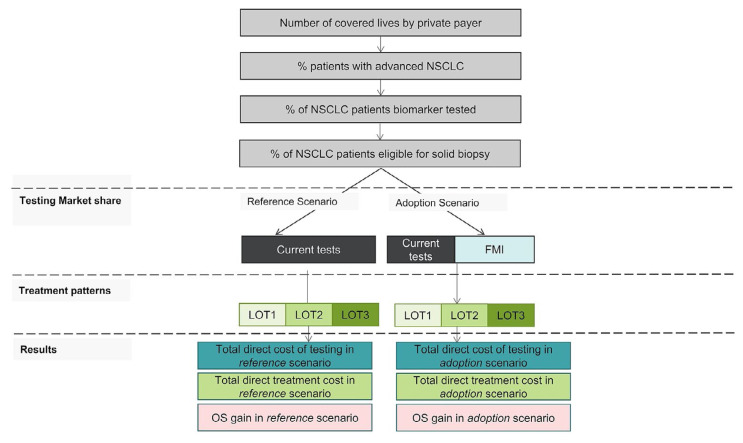
To quantify the impact of funding Foundation Medicine testing, a health and budget impact model was developed, describing testing strategy and flow, resulting mutation detection and treatment decisions, and costs and life–years associated with various testing strategies in a cohort of individuals (Figure 1). The starting population included individuals with advanced nsclc in Ontario eligible for testing. Within that population, testing practices were associated with treatments received and resulting clinical outcomes.
FIGURE 1.
Budget impact model overview. BIA = budget impact analysis; NSCLC = non-small-cell lung cancer; FMI = Foundation Medicine Incorporated; LOT = line of therapy; OS = overall survival
A Canadian societal perspective was taken, with Ontario as a sample province, in which workplace productivity was considered in addition to testing costs and clinical outcomes. Patients considered eligible for Foundation Medicine testing included individuals who would normally receive first-line reflex testing, those who would undergo subsequent panel testing, and those with tissue insufficient or unavailable for testing either initially or at progression of disease.
Model Structure
The model is related to another that has previously been described16 and adapted to the Canadian setting. The model is defined by a reference scenario, in which only conventional tests are used, and by an adoption scenario, in which some testing (for EGFR, ALK, ROS1, BRAF, MET, ERBB2, RET, and KRAS alterations) is replaced by Foundation Medicine testing.
Epidemiologic Data
The incidence of lung cancer was based on observed age- and sex-specific Canadian incidence rates17. From that overall lung cancer population, it was estimated that 88% would have nsclc17, and that 52.5% would have advanced disease (5.4% stage iiib and 47.1% stage iv).
Testing Efficacy
Certain assumptions were made in the development of the model. Table I lists the detection rates for individual alterations in conventional tests compared with the Foundation Medicine tests (prevalence rates for conventional testing are given in Appendix 1, Table I, of the supplemental material). Values for both were obtained from the published literature7,18–22,24. The rates reflect the improved sensitivity of the Foundation Medicine tests, resulting in higher detection rates across all genes, excluding PD-L1. In addition, all genomic alteration values for nonsquamous nsclc were multiplied by 0.7 to account for the fact that about 30% of all patients with nsclc have squamous cell carcinoma and are unlikely to carry those alterations25. The detection rate for MET alteration consisted of a combination of squamous and nonsquamous rates, using the same 70:30 split (conventional testing detection rates, Appendix 1, Table 2, and Foundation Medicine testing, Appendix 1, Table 4, in the supplemental material). The same nonsquamous:squamous proportion was applied for detection rates with Foundation Medicine testing (prevalence rates for Foundation Medicine testing, Appendix 1, Table 3, in the supplemental material)22,24.
TABLE I.
Detection rates for individual alterations and biomarkers in conventional tests compared with Foundation Medicine Incorporated (FMI) tests
| Gene or biomarker | Detection rate (%) by test type | |
|---|---|---|
| Conventional | FMI | |
| ALK18,19 | 2.1 | 2.9 |
| PD-L1a,20 | 30.2 | |
| EGFR7,21,22 | 11.9 | 14.0 |
| KRAS7,22 | 17.5 | 22.4 |
| BRAF7,22,23 | 1.4 | 4.0 |
| ROS17,22 | 0.7 | 1.0 |
| ERBB27,22 | 2.1 | 4.2 |
| MET7,22 | 2.6 | 5.8 |
| RET7,22 | 0.7 | 1.7 |
Tested using immunohistochemical assays and used in conjunction with FoundationOne testing.
Treatment Efficacy
Survival outcomes for individuals treated with targeted therapy were derived from a French national database study reporting survival outcomes in nsclc for patients with and without a matched gene target26. Table II shows the distribution of os and progression-free survival duration by line of therapy. Because all treatments (excluding palliative care) are associated with at least 12 months of os time, the benefits of targeted and immunotherapies are not fully recognized until the 2nd and 3rd years after initiation. Overall survival figures in the database study were reported for patients with an identified gene alteration. Survival for the subset of patients receiving targeted therapy (approximately 30% of those with an identified gene modification) was derived based on the assumption that 70% of the gene alteration group would experience survival outcomes comparable to the group without a matched gene alteration. The remaining survival outcomes for immunotherapy were taken from various keynote trials comparing pembrolizumab with chemotherapy, or pembrolizumab–chemotherapy with chemotherapy alone20,27–29.
TABLE II.
Progression-free (PFS) and overall survival (OS) by first-line treatment received
| First-line therapy received | PFS (months on treatment) | OS (months from line-of-therapy initiation to death) | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Line 1 | Line 2 | Line 3 | Line 1 | Line 2 | Line 3 | |
| Matched targeted therapy26 | 16 | 4 | 2 | 22.75a/26 | 10 | 6 |
|
| ||||||
| Matched immunotherapy27 | 10 | 5 | 3 | 20 | 10 | 5 |
|
| ||||||
| Chemotherapy plus immunotherapy28 | 6 | 5 | 3 | 16 | 10 | 5 |
|
| ||||||
| Chemotherapy27 | 6 | 4 | 2 | 12 | 6 | 2 |
|
| ||||||
| No treatment or palliative | 3 | 2 | 1 | 5 | 3 | 2 |
Conventional tests. First-line targeted therapy adjusted for delays in turnaround time.
For conventional testing methods, an adjustment was made based on potential delays in turnaround time associated with sending tests to external labs (that is, a longer turnaround relative to the required turnaround for Foundation Medicine testing), leading to a delay in results and the potential effect on treatment decisions and outcomes. For urban and academically-affiliated centres, turnaround time is anticipated to be less of a concern than it is for centres in rural communities that lack in-house options and are affected by physical distance to nearest testing centres. In the latter case, turnaround might take up to 4 weeks, potentially delaying detection and treatment initiation past the window of greatest efficacy. In a Canadian population, it was found that, for 79% of patients with nonsquamous nsclc undergoing biomarker testing, test results were not available to the oncologist at the time of initial consultation, leading to delays in treatment decisions and initiation relative to patients for whom such information was available through rapid reflex testing30. In the budget impact model, based on expert opinion, the conservative assumption was made that delay would affect 25% of patients receiving conventional tests, and that treatment and survival time for targeted therapies in that subset of patients would be reduced by 50% (similar to untargeted therapies). As a result, the 26-month estimated survival time is reduced, overall, to 22.75 months for patients tested conventionally.
Use of Foundation Medicine Testing in Canadian Practice
For the eligible population, a variety of scenarios were considered with respect to current testing practices (reference scenarios) and potential uptake of Foundation Medicine testing (adoption scenarios), based on the range of practices observed and anticipated in the Canadian setting. To inform the assumptions about current testing practice, structured interviews were conducted with experts from a variety of Canadian oncology centres. The experts provided algorithms based on their current practice.
These reference scenarios were considered:
-
■ Reference scenario 1: a series of single-gene tests with no use of hotspot panels
The following assumptions about testing frequency were made: 95% ALK and PD-L1, 75% EGFR, 60% ROS1, and 5% KRAS and BRAF.
-
■ Reference scenario 2: single-gene tests followed by hotspot panels in patients with negative results
The following assumptions were made about testing frequency: 95% ALK and PD-L1, 60% ROS1, and 75% hotspot panel testing including EGFR, KRAS, and BRAF.
These adoption scenarios were considered:
■ Adoption scenario 1: FoundationOne CDx upfront, replacing all reflex testing (that is, all single-gene and panel testing with the exception of PD-L1)
■ Adoption scenario 2: FoundationOne CDx used after single-gene testing for patients who have tested negative in the initial tests (replacing hotspot panel)
■ Adoption scenario 3: FoundationOne CDx used after all standard testing in patients who have tested negative for all alterations tested so far (that is, after both single-gene testing and hotspot panel testing)
■ Adoption scenario 4: Use of FoundationOne Liquid testing in the 5% of patients who initially present for testing and treatment, but who do not receive any testing in the reference scenario, based on the assumption that insufficient tissue availability (irrelevant to the Liquid test) is a key driver of the decision
Note that reference scenario 1 (single-gene testing only) is presented only in combination with adoption scenario 1 (FoundationOne CDx used upfront) because adoption scenarios 2 and 3 (FoundationOne CDx replacing panel testing or conducted after panel testing respectively) result in no first-line FoundationOne CDx use and are thus not applicable. For all scenarios, PD-L1 expression testing and subsequent immunotherapy treatment decisions are assumed to be independent of Foundation Medicine testing, because Foundation Medicine testing will not replace that test.
Testing Uptake
For the assumptions about FoundationOne testing uptake, given large variation in testing practice from centre to centre, a mid-range figure of 50% for uptake was chosen: that is, across adoption scenarios, it was assumed that, of patients meeting the criteria for Foundation Medicine testing, 50% would receive it, and the remaining 50% would receive testing consistent with current routine practice, reflecting the fact that some facilities (for example, those with existing in-house laboratory facilities) might choose to continue with the status quo. Testing frequency rates were based on clinical experience and communications with Canadian specialists.
Although the 50% uptake assumption was made in the first 3 adoption scenarios, because adoption scenario 4 involves a population with no other testing options, it was assumed that 95% of those individuals would receive the FoundationOne Liquid test.
Given that uptake was based largely on assumption, further sensitivity analyses were conducted to understand the potential impacts of incorporating Foundation Medicine testing into Canadian practice: a lower proportion of 25% and an upper proportion of 100% were selected as the range for potential uptake.
Second-Line Testing
Although most testing occurs in a first-line setting, second-line testing is also useful in the subset of patients with an EGFR mutation identified in the first line who are treated with a tyrosine kinase inhibitor, to test for second-line T790M resistance mutations and to identify any missed alterations. Conventional testing in such patients could require a repeat biopsy, which might be infeasible depending on the availability of accessible tissue, patient frailty, and disease progression. Conversely, because the FoundationOne Liquid biopsy test can be conducted using a blood sample only, that test was considered to be an alternative mechanism in this patient subgroup. This second-line testing was included in all Foundation Medicine adoption scenarios; uptake of Foundation Medicine testing was assumed to be 50%.
Unit Costs
The costs of FoundationOne CDx and FoundationOne Liquid are based on current Canadian list prices ($6,193.90). The costs of conventional testing vary with the specific test and method used (for example, in-house compared with external laboratory), and structured interviews were conducted with laboratory directors to comprehensively estimate the cost of such tests, including allocation of capital and staffing costs, adjusted for the volume of laboratory work associated with nsclc specifically (inputs and calculations described in Appendix 2 of the supplemental material). Based on that assessment, the assumed cost was $652 per single-gene test and $1,919 per hotspot panel test. Testing costs by year are based on the assumption that first-line testing occurs in the first year of treatment flow, and that second-line testing occurs in the second year.
Workplace productivity was also incorporated into the model, based on the assumption that more individuals remaining in the progression-free survival health state would be able to remain in the workforce. Based on clinical experience, the following assumptions were made to incorporate productivity into the analysis: 24% of patients with progression-free disease would be able to work half-time per week, compared with 0% of patients with progressive disease. The subsequent adjustment was based on a Canadian general population workforce working 1804.4 hours per year31, with a workforce participation rate of 73.2%32, and assuming an hourly wage of $25.06 and a retirement age of 6533.
RESULTS
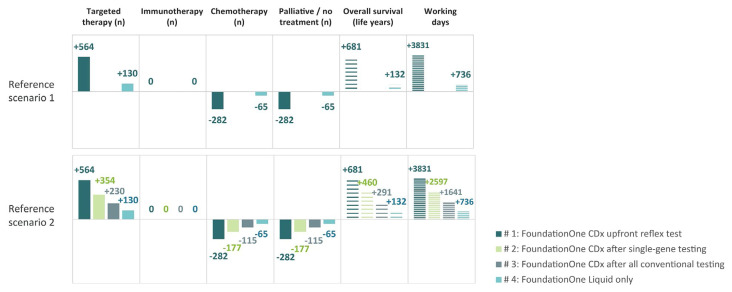
Based on the provincial population and the age and sex distribution in Ontario, and Canadian nsclc incidence rates, the total number of patients eligible for testing each year was estimated to be 5109. Adoption scenarios were all associated with fewer total tests per person (Appendix 1, Table 5, in the supplemental material), particularly for reference scenario 1, in which the initial series of single-gene tests is replaced by a single FoundationOne CDx test. In addition, the FoundationOne Liquid test provides the opportunity for testing where it was not possible before. Given the increased detection rate of Foundation Medicine tests compared with conventional tests (Table I and Appendix 1, Table 4, in the supplemental material), adoption scenarios are all also associated with increased mutation detection and subsequent use of targeted therapies (Table III, Figure 2). As a result, cost efficiencies in testing and improved outcomes could be expected from the decrease in the use of inappropriate therapies.
TABLE III.
Genes and PD-L1 biomarker detected in the reference and adoption scenariosa
| Gene or biomarker | Detections (n) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Adoption scenario | |||||||||||||
|
| |||||||||||||
| Reference scenario (single-gene testing only) | 1b | 2c | 3d | 4e | |||||||||
|
|
|
|
|
||||||||||
| Conv. | FO | Total | Conv. | FO | Total | Conv. | FO | Total | Conv. | FO | Total | ||
| Reference scenario 1: Single-gene testing only | |||||||||||||
|
| |||||||||||||
| ALK | 102 | 51 | 77 | 128 | Not applicable | Not applicable | 102 | 7 | 109 | ||||
|
| |||||||||||||
| PD-L1 | 1466 | 1466 | 0 | 1466 | 1466 | 0 | 1466 | ||||||
|
| |||||||||||||
| EGFR | 456 | 228 | 374 | 602 | 456 | 34 | 490 | ||||||
|
| |||||||||||||
| KRAS | 45 | 22 | 598 | 620 | 45 | 54 | 99 | ||||||
|
| |||||||||||||
| BRAF | 4 | 2 | 107 | 109 | 4 | 10 | 13 | ||||||
|
| |||||||||||||
| ROS1 | 21 | 11 | 27 | 37 | 21 | 2 | 24 | ||||||
|
| |||||||||||||
| ERBB2 | 0 | 0 | 112 | 112 | 0 | 10 | 10 | ||||||
|
| |||||||||||||
| MET | 0 | 0 | 155 | 155 | 0 | 14 | 14 | ||||||
|
| |||||||||||||
| RET | 0 | 0 | 45 | 45 | 0 | 4 | 4 | ||||||
|
| |||||||||||||
| Unmatched | 3016 | 1834 | 2880 | ||||||||||
| Reference scenario 2: Single-gene testing followed by hotspot panel | |||||||||||||
|
| |||||||||||||
| ALK | 102 | 51 | 77 | 128 | 62 | 63 | 124 | 80 | 37 | 117 | 102 | 7 | 109 |
|
| |||||||||||||
| PD-L1 | 1466 | 1466 | 0 | 1466 | 1466 | 0 | 1466 | 1466 | 0 | 1466 | 1466 | 0 | 1466 |
|
| |||||||||||||
| EGFR | 456 | 228 | 374 | 602 | 228 | 302 | 530 | 332 | 180 | 512 | 456 | 34 | 490 |
|
| |||||||||||||
| KRAS | 671 | 335 | 598 | 933 | 335 | 484 | 819 | 488 | 288 | 776 | 671 | 54 | 725 |
|
| |||||||||||||
| BRAF | 54 | 27 | 107 | 134 | 27 | 86 | 113 | 39 | 51 | 90 | 54 | 10 | 63 |
|
| |||||||||||||
| ROS1 | 21 | 11 | 27 | 37 | 21 | 22 | 43 | 14 | 13 | 27 | 21 | 2 | 24 |
|
| |||||||||||||
| ERBB2 | 0 | 0 | 112 | 112 | 0 | 91 | 91 | 0 | 54 | 54 | 0 | 10 | 10 |
|
| |||||||||||||
| MET | 0 | 0 | 155 | 155 | 0 | 125 | 125 | 0 | 75 | 75 | 0 | 14 | 14 |
|
| |||||||||||||
| RET | 0 | 0 | 45 | 45 | 0 | 37 | 37 | 0 | 22 | 22 | 0 | 4 | 4 |
|
| |||||||||||||
| Unmatched | 2340 | 1497 | 1761 | 1970 | 2204 | ||||||||
Scenarios 1–3 assume 50% uptake for FoundationOne CDx (Foundation Medicine, Cambridge, MA, U.S.A.). PD-L1 expression is immunohistochemical assays and used in conjunction with FoundationOne testing.
FoundationOne CDx replaces all reflex testing.
FoundationOne CDx replaces hotspot panel.
FoundationOne CDx used after negative reflex and hotspot panel.
FoundationOne Liquid testing only.
Conv. = conventional testing; FO = FoundationOne testing.
FIGURE 2.
Overview of treatment and impact associated with the FoundationOne CDx and FoundationOne Liquid tests from Foundation Medicine (Cambridge, MA, U.S.A.) in the various use scenarios. (FoundationOne Liquid is used in all scenarios.) Table III presents a detailed breakdown by gene.
Across all reference and adoption scenarios, Foundation Medicine tests were associated with an increase in budget impact and with increased life–years and workplace productivity (Table IV). Adoption scenarios 1–4 are associated with decreasing use of Foundation Medicine testing, from an upfront replacement of all conventional testing in adoption scenario 1, to additional testing only for patients ineligible for conventional testing in adoption scenario 4. The budget and health impacts therefore decline over the scenarios. The impact of the adoption scenarios is similar for the two reference scenarios (excluding adoption scenarios 2 and 3, which are relevant only for reference scenario 2). The total number of tests administered (reflecting increased use of sequential testing in the reference scenarios) is reported in Appendix 1, Table 5, in the supplemental material.
TABLE IV.
Health and budget impacts by scenarioa
| Scenario and variable | Reference scenario 1: Single-gene testing only | Reference scenario 2: Single-gene testing plus hotspot panel | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Overall | Year 1 | Year 2 | Year 3 | Overall | Year 1 | Year 2 | Year 3 | |
| Reference scenario costs ($M) | 343.3 | 61.0 | 114.5 | 167.8 | 356.9 | 65.5 | 119.0 | 172.3 |
| Adoption scenario 1: FoundationOne CDx replaces all reflex tests | ||||||||
|
| ||||||||
| Incremental budget impact ($M) | 37.1 | 12.4 | 12.3 | 12.3 | 30.3 | 10.1 | 10.1 | 10.1 |
|
| ||||||||
| Incremental budget impact PMPY ($) | 0.87 | 0.87 | 0.87 | 0.87 | 0.71 | 0.71 | 0.71 | 0.71 |
|
| ||||||||
| Incremental overall survival (years) | 680.9 | 54.9 | 273.2 | 352.8 | 680.9 | 54.9 | 273.2 | 352.8 |
|
| ||||||||
| Incremental working days | 3831 | 784 | 1524 | 1524 | 3831 | 784 | 1524 | 1524 |
|
| ||||||||
| NNT to gain 1 life–year | 174 Patients | 174 Patients | ||||||
|
| ||||||||
| Adoption scenario 2: FoundationOne CDx replaces hotspot panel | ||||||||
|
| ||||||||
| Incremental budget impact ($ million) | Not applicable | 28.7 | 9.6 | 9.5 | 9.5 | |||
|
| ||||||||
| Incremental budget impact PMPY ($) | 0.67 | 0.68 | 0.67 | 0.67 | ||||
|
| ||||||||
| Incremental overall survival (years) | 460.0 | 34.5 | 180.6 | 245.0 | ||||
|
| ||||||||
| Incremental working days | 2597 | 492 | 1053 | 1053 | ||||
|
| ||||||||
| NNT to gain 1 life–year | 250 Patients | |||||||
|
| ||||||||
| Adoption scenario 3: FoundationOne CDx after negative reflex testing and hotspot panel | ||||||||
|
| ||||||||
| Incremental budget impact ($ million) | Not applicable | 23.6 | 7.9 | 7.9 | 7.9 | |||
|
| ||||||||
| Incremental budget impact PMPY ($) | 0.56 | 0.56 | 0.55 | 0.55 | ||||
|
| ||||||||
| Incremental overall survival (years) | 291.0 | 22.4 | 115.1 | 153.5 | ||||
|
| ||||||||
| Incremental working days | 1641 | 320 | 661 | 661 | ||||
|
| ||||||||
| NNT to gain 1 life–year | 399 Patients | |||||||
|
| ||||||||
| Adoption scenario 4: FoundationOne Liquid only | ||||||||
|
| ||||||||
| Incremental budget impact ($ million) | 4.4 | 1.5 | 1.5 | 1.5 | 4.4 | 1.5 | 1.5 | 1.5 |
|
| ||||||||
| Incremental budget impact PMPY ($) | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 |
|
| ||||||||
| Incremental overall survival (years) | 132.1 | 12.7 | 56.1 | 63.3 | 132.1 | 12.7 | 56.1 | 63.3 |
|
| ||||||||
| Incremental working days | 736 | 181 | 277 | 277 | 736 | 181 | 277 | 277 |
|
| ||||||||
| NNT to gain 1 life–year | 968 Patients | 968 Patients | ||||||
Scenarios 1–3 assume 50% uptake for FoundationOne CDx (Foundation Medicine, Cambridge, MA, U.S.A.).
$M = millions of dollars; PMPY = per member per year; NNT = number needed to treat.
Over the full 3-year time horizon, the budget impact of replacing all upfront testing with Foundation Medicine testing (based on an uptake of 50%), is $37.1 million compared with reference scenario 1 of single-gene testing only, and $30.3 million compared with reference scenario 2 of single-gene plus hotspot panel testing. That translates to a cost per person in Ontario per year of $0.87 for reference scenario 1 and $0.71 for reference scenario 2. For both reference scenarios, a gain of 680.9 additional life–years and 3831 working days is estimated for the cohort, which is associated with a number-needed-to-test (nnt) of 174 to gain 1 life–year. Note that the nnt is based on the number of people included in the full scenario, including the 50% still receiving conventional testing in the adoption scenario. That number can therefore be considered to be the total number of people receiving the combination of tests associated with each scenario, rather than the number of individuals actually receiving a Foundation Medicine test.
Adoption scenarios 2 and 3 are characterized by lesser use of Foundation Medicine testing, specifically to replace the panel or to be used only after a negative panel. Correspondingly, they are associated with a lower budget impact ($28.7 million and $23.6 million respectively over 3 years) and with lesser gains in os and working days when compared with adoption scenario 1 of reflex testing with Foundation Medicine. In current practice patterns, only a subset of patients who receive single-gene reflex tests would go on to receive a multi-gene panel (regardless of results); thus, if Foundation Medicine testing is replacing only testing in the subgroup currently receiving multi-gene testing, it would not be used for as many patients as if it had been given in an upfront reflex scenario. The nnt for those scenarios is 250 and 399 patients respectively.
Finally, adoption scenario 4 is associated with substantially lower use of Foundation Medicine testing. Because only individuals not otherwise receiving conventional testing are tested, the results are the same for both reference scenarios. The budget impact over 3 years is $4.4 million, with a nnt of 968 patients.
Sensitivity analyses varying the uptake of Foundation Medicine testing (Appendix 1, Table 5, in the supplemental material) largely affected the overall budget impact across all scenarios, ranging from −46% to −19% of the base-case budget impact for the option of 25% uptake, and ranging from 38% to 94% of the base-case budget impact for the option of 100% uptake.
DISCUSSION
A health and budget impact analysis from a societal perspective was undertaken to quantify the potential effects of providing Foundation Medicine testing as a companion to conventional testing in Canadian clinical practice for nsclc. Recent real-world evidence studies in Canada34 and the United States35 have reported on the survival benefits and cost-effectiveness of conventional multigene testing as an alternative to single-gene testing in lung cancer. Given the increased detection rates and potential for shorter turnaround time associated with Foundation Medicine testing relative to all available conventional tests, the adoption scenarios were associated with greater use of targeted therapies and resulting improvements in os and work productivity. A similar modelling approach has been used in a U.S. setting16; differences in outcomes result primarily from different assumptions about the details of current reference and future adoption scenarios.
Challenges in the economic evaluation of molecular testing have been noted previously36,37. One advantage of the current approach is that the primary focus of the analysis is on testing specifically, rather than on incorporating testing into a treatment-specific model. The model was therefore specifically designed to incorporate the nuances of testing strategies, taking a population-based approach to advanced nsclc, rather than evaluating individual therapies.
In the absence of empirical data, a number of assumptions were necessary, particularly in the area of testing uptake for FoundationOne. To support the assumption of 50% uptake, scenarios ranging from 25% to 100% uptake were considered to provide a range of potential budget impacts. Over time, as data continue to accrue about the efficacy of various testing modalities, those estimates could be refined. Large observational datasets have been suggested as a future direction in assessing outcomes in molecular testing36. In addition to detection rates, the anticipated uptake of Foundation Medicine testing and the relationships between test modality, turnaround time, and clinical outcomes were based on assumptions derived from feedback obtained from experts familiar with Canadian clinical practice. Comprehensive assessment of laboratory practice and nsclc-related expenses in Canada was conducted to derive robust and empirical estimates of conventional testing costs.
When outcomes and adoption scenario results are considered in aggregate, the benefits of Foundation Medicine testing include a reduction in the absolute number of tests conducted, increased use of targeted therapies, and optimization of the timing of those therapies for individuals who currently face an extended turnaround time for test results. The potential to conduct testing in patients for whom it would otherwise be infeasible because of lack of tissue availability is a further advantage. As a result, Foundation Medicine testing is ultimately associated with increased life–years and work productivity.
Although quality of life was not incorporated into the present analysis, the reduced toxicity associated with targeted therapies relative to traditional chemotherapy makes it likely that Foundation Medicine testing would be associated with improved quality and not just length of life. The budget impact was estimated to be relatively modest, ranging from $0.10 to $0.87 per person in Ontario per year, with the scenarios associated with the greatest uptake of Foundation Medicine testing associated with both the highest budget impact and the greatest benefits.
CONCLUSIONS
The present study demonstrates that, as a result of improved testing efficacy, Foundation Medicine testing offers clinical benefits to the population for a relatively modest budget impact. Given that Foundation Medicine testing does not require access to an in-house laboratory, provision of this type of testing modality could also potentially promote equability in the Canadian health care system.
Supplementary Information
ACKNOWLEDGMENTS
This research was funded by Hoffmann–La Roche Ltd. Editorial support for this manuscript was provided by Elizabeth Badillo, an employee of Broadstreet Health Economics and Outcomes Research, which received payment from Hoffmann–La Roche Limited during the conduct of this study.
An abstract and poster containing some of the results reported herein was presented at the International Society for Pharmacoeconomics and Outcomes Research’s Europe 2019 meeting; Copenhagen, Denmark; 2–6 November 2019.
Footnotes
Supplemental material available at http://www.current-oncology.com
CONFLICT OF INTEREST DISCLOSURES
We have read and understood Current Oncology’s policy on disclosing conflicts of interest, and we declare the following interests: JN is an employee of Hoffmann–La Roche Ltd., a pharmaceutical company whose products include FoundationOne CDx and FoundationOne Liquid. KMJ, PL, and CQ are or were employed by Broadstreet Health Economics and Outcomes Research, which was commissioned by Roche to conduct this analysis. BSS and SY received fees as consultants in the conduct of this study.
REFERENCES
- 1.Canadian Cancer Statistics Advisory Committee. Canadian Cancer Statistics 2018. Toronto, ON: Canadian Cancer Society; 2018. [Google Scholar]
- 2.Ellis PM, Vella ET, Ung YC and the Lung Cancer Disease Site Group. Systemic Treatment for Patients with Advanced Non–Small Cell Lung Cancer. Toronto, ON: Ontario Health (Cancer Care Ontario); 2016. [Google Scholar]
- 3.Alberta Health Services (ahs) Non–small Cell Lung Cancer Stage IV. Edmonton, AB: AHS; 2013. [Google Scholar]
- 4.Enewold L, Thomas A. Real-world patterns of EGFR testing and treatment with erlotinib for non–small cell lung cancer in the United States. PLoS One. 2016;11:e0156728. doi: 10.1371/journal.pone.0156728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weymann D, Dragojlovic N, Pollard S, Regier DA. Allocating healthcare resources to genomic testing in Canada: latest evidence and current challenges. J Community Genet. 2019 doi: 10.1007/s12687-019-00428-5. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirsh V, Melosky B, Goss G, Morris D, Morzycki W. A personalized approach to treatment: use of egfr tyrosine kinase inhibitors for the treatment of non-small-cell lung cancer in Canada. Curr Oncol. 2012;19:78–90. doi: 10.3747/co.19.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali SM, Hensing T, Schrock AB, et al. Comprehensive genomic profiling identifies a subset of crizotinib responsive ALK rearranged non–small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist. 2016;21:762–70. doi: 10.1634/theoncologist.2015-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture–based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res. 2015;21:3631–9. doi: 10.1158/1078-0432.CCR-14-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel dna sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen SJ, Liu H, Liao CT, et al. Ultra-deep targeted sequencing of advanced oral squamous cell carcinoma identifies a mutation-based prognostic gene signature. Oncotarget. 2015;6:18066–80. doi: 10.18632/oncotarget.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteva FJ, Sahin AA, Cristofanilli M, et al. Prognostic role of a multigene reverse transcriptase–pcr assay in patients with node-negative breast cancer not receiving adjuvant systemic therapy. Clin Cancer Res. 2005;11:3315–19. doi: 10.1158/1078-0432.CCR-04-1707. [DOI] [PubMed] [Google Scholar]
- 13.Du XL, Lin CC, Johnson NJ, Altekruse S. Effects of individual-level socioeconomic factors on racial disparities in cancer treatment and survival: findings from the National Longitudinal Mortality Study, 1979–2003. Cancer. 2011;117:3242–51. doi: 10.1002/cncr.25854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt KT, Chau CH, Price DK, Figg WD. Precision oncology medicine: the clinical relevance of patient-specific biomarkers used to optimize cancer treatment. J Clin Pharmacol. 2016;56:1484–99. doi: 10.1002/jcph.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol. 2013;31:1039–49. doi: 10.1200/JCO.2012.45.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Signorovitch J, Zhou Z, Ryan J, Anhorn R, Chawla A. Budget impact analysis of comprehensive genomic profiling in patients with advanced non–small cell lung cancer. J Med Econ. 2019;22:140–50. doi: 10.1080/13696998.2018.1549056. [DOI] [PubMed] [Google Scholar]
- 17.Canadian Cancer Statistics Advisory Committee. Canadian Cancer Statistics: A 2018 Special Report on Cancer Incidence by Stage. Toronto, ON: Canadian Cancer Society; 2018. [Google Scholar]
- 18.pCODR Expert Review Committee (pERC): Final Recommendation. [re. trifluridine–tipiracil (Lonsurf) for gastric cancer]. Ottawa, ON: pCODR; 2019. pan-Canadian Oncology Drug Review (pcodr) [Available online at: https://www.cadth.ca/sites/default/files/pcodr/Reviews2020/10197TrifluridineTipiracilGC_fnRec_EarlyConv_ApprovedbypERCChair_Post024Mar2020_final.pdf; cited 16 November 2020] [Google Scholar]
- 19.Hirshfield KM, Tolkunov D, Zhong H, et al. Clinical actionability of comprehensive genomic profiling for management of rare or refractory cancers. Oncologist. 2016;21:1315–25. doi: 10.1634/theoncologist.2016-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reck M, Rodríguez-Abreu D, Robinson AG, et al. on behalf of the keynote-024 investigators. Pembrolizumab versus chemotherapy for PD-L1–positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–33. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 21.pCODR Expert Review Committee (pERC): Final Recommendation. [re. osimertinib (Tagrisso)]. Ottawa, ON: pCODR; 2017. pan-Canadian Oncology Drug Review (pcodr) [Available online at: https://www.cadth.ca/sites/default/files/pcodr/pcodr_osimertinib_tagrisso_nsclc_fn_rec.pdf; cited 16 November 2020] [Google Scholar]
- 22.Heist RS, Sequist LV, Engelman JA. Genetic changes in squamous cell lung cancer: a review. J Thorac Oncol. 2012;7:924–33. doi: 10.1097/JTO.0b013e31824cc334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.pan-Canadian Oncology Drug Review (pcodr) pCODR Expert Review Committee (pERC): Final Recommendation. [re. combination dabrafenib (Tafinlar) and trametinib (Mekinist) for metastatic melanoma]. Ottawa, ON: pCODR; 2019. [Available online at: https://www.cadth.ca/sites/default/files/pcodr/Reviews2019/10152DabrafenibTrametinibMAT_Final%20Recc_Approved%20by%20Chair_2May2019_REDACT_Post_03May2019_final.pdf; cited 16 November 2020] [Google Scholar]
- 24.Suh JH, Johnson A, Albacker L, et al. Comprehensive genomic profiling facilitates implementation of the national comprehensive cancer network guidelines for lung cancer biomarker testing and identifies patients who may benefit from enrollment in mechanism-driven clinical trials. Oncologist. 2016;21:684–91. doi: 10.1634/theoncologist.2016-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.American Cancer Society(acs) Key Statistics for Lung Cancer 2018. Atlanta, GA: ACS; 2018. [Google Scholar]
- 26.Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (ifct) Lancet. 2016;387:1415–26. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 27.De Lima Lopes G, Wu YL, Sadowski S, et al. Pembrolizumab vs platinum-based chemotherapy for PD-L1+ nsclc: phase 3, randomized, open-label keynote-042 ( NCT02220894) [abstract P2.43] J Thorac Oncol. 2016;11(suppl):S244–5. doi: 10.1016/j.jtho.2016.08.114. [DOI] [Google Scholar]
- 28.Paz-Ares L, Luft A, Vicente D, et al. on behalf of the keynote-407 investigators. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med. 2018;379:2040–51. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 29.Somasundaram A, Burns TF. Pembrolizumab in the treatment of metastatic non-small-cell lung cancer: patient selection and perspectives. Lung Cancer (Auckl) 2017;8:1–11. doi: 10.2147/LCTT.S105678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol. 2015;26:1415–21. doi: 10.1093/annonc/mdv208. [DOI] [PubMed] [Google Scholar]
- 31.Institut de la statistique Québec. Heures hebdomadaires habituelles moyennes pour l’ensemble des emplois, résultats selon le sexe pour diverses caractéristiques de la main-d’oeuvre et de l’emploi, Québec, Ontario et Canada. Québec, QC: Institut de la statistique Québec; 2019. [Google Scholar]
- 32.Statistics Canada. Labour Force Characteristics by Province, Monthly, Unadjusted for Seasonality. Ottawa, ON: Statistics Canada; 2019. [Google Scholar]
- 33.Statistics Canada. Average Usual Hours and Wages by Selected Characteristics, Monthly, Unadjusted for Seasonality (×1,000) Ottawa, ON: Statistics Canada; 2019. [Google Scholar]
- 34.Perdrizet K, Stockley T, Law JH, et al. Non–small cell lung cancer (nsclc) next generation sequencing (ngs) using the Oncomine Comprehensive Assay (oca) v3: integrating expanded genomic sequencing into the Canadian publicly funded health care model [abstract 2620] J Clin Oncol. 2019;37 doi: 10.1200/JCO.2019.37.15_suppl.2620. [Available online at: https://ascopubs.org/doi/abs/10.1200/JCO.2019.37.15_suppl.2620; cited 9 November 2020] [DOI] [Google Scholar]
- 35.Steuten L, Goulart B, Meropol NJ, Pritchard D, Ramsey SD. Cost effectiveness of multigene panel sequencing for patients with advanced non-small-cell lung cancer. JCO Clin Cancer Inform. 2019;3:1–10. doi: 10.1200/CCI.19.00002. [DOI] [PubMed] [Google Scholar]
- 36.Phillips KA, Deverka PA, Marshall DA, et al. Methodological issues in assessing the economic value of next-generation sequencing tests: many challenges and not enough solutions. Value Health. 2018;21:1033–42. doi: 10.1016/j.jval.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payne K, Gavan SP, Wright SJ, Thompson AJ. Cost-effectiveness analyses of genetic and genomic diagnostic tests. Nat Rev Genet. 2018;19:235–46. doi: 10.1038/nrg.2017.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.