Abstract
We evaluated the rapid immunochromatographic test for severe acute respiratory coronavirus 2 (SARS-CoV-2) antigen detection using 16 saliva specimens collected from 6 COVID-19 hospitalized patients, and detected N-antigen in 4 of 7 RT-PCR positive specimens. This POCT detected SARS-CoV-2 antigen in saliva and would be useful for COVID-19 diagnosis.
Keywords: COVID-19, SARS-CoV-2, POCT, Antigen test, Immunochromatography
1. Introduction
Respiratory disease (COVID-19) caused by 2019 novel coronavirus, severe acute respiratory coronavirus 2 (SARS-CoV-2), is diagnosed by nucleic acid amplification test, as reverse transcription polymerase chain reaction (RT-PCR), for viral genome RNA in nasopharyngeal swab specimens. The collection procedure of nasopharyngeal specimens poses a risk of secondary infection to health-care workers. The risk could be reduced if saliva specimens could be used for diagnosis of COVID-19. Recently, several reports indicated that SARS-CoV-2 was detected in saliva specimens of equally or higher sensitivity than the nasopharyngeal specimens [[1], [2], [3], [4]].
A rapid antigen detection kit, ESPLINE® SARS-CoV-2 reagent (Fujirebio Inc., Tokyo), for detection of the viral N antigens in nasopharyngeal swabs was recently developed [5]. The ESPLINE test is an enzyme-immunoassay based on immunochromatographic technology using monoclonal antibodies specific to SARS-CoV and CoV-2 N antigens.
In this report, we evaluated the saliva specimens for diagnosis of COVID-19 by RT-PCR and the point of care testing (POCT) antigen test.
1.1. Case report
Sets of saliva and nasopharyngeal specimens were collected within several days from 6 COVID-19 patients hospitalized in Omori Hospital, who had been diagnosed by RT-PCR test. The study protocol was approved by the Ethics Committee of Faculty of Medicine, Toho University (No. A20028_A20020_A20014_A19099). Informed consent was obtained from all participants in the study. Saliva was collected by passive drool method according to previous report [6,7]. Nasopharyngeal swab specimens were collected directly following the ESPLINE kit manufacturer's instructions except for 4 cases, in which the swab specimens were resuspended in universal virus transport medium (UVT). The specimen-treatment solution supplied with the kits was used for assay of both kit's swab and saliva specimens and the assays were performed following the kit manufacturer's instructions. Ct values were obtained by RT-PCR test using N2 probe according to the manual provided by National Institute of Infectious Diseases. SARS-CoV-2 antigen was assessed using ESPLINE kit according to the manufacturer's instruction. All assays were carried out on the day of specimen collection.
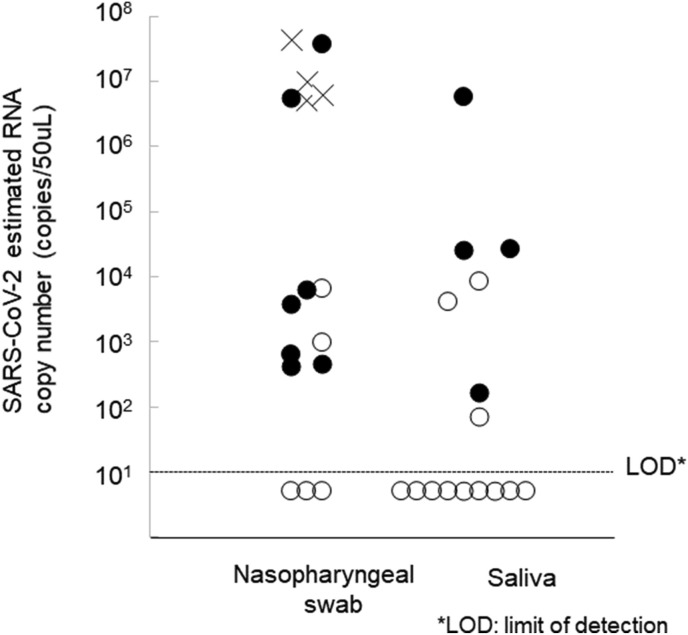
SARS-CoV-2 RNA was detected in 13 of 16 (81.3%) of nasopharyngeal specimens. On the other hand, RT-PCR detected RNA in only 7 of 16 saliva specimens (43.8%). Following exclusion of the 4 nasopharyngeal specimens that had been suspended in UVT, 7 of the remaining 9 (77.8% sensitivity) were positive by the ESPLINE test. The antigen was detected in 4 of 7 RT-PCR positive saliva specimens (47.8%) (Fig. 1 ). The sensitivity of the antigen test seems to depend on the RNA-copy concentrations in the specimens; however, no significant difference was observed (Fig. 1).
Fig. 1.
Detection of SARS-CoV-2 N antigen in nasopharyngeal swab and saliva specimens. N antigen-positive specimens are shown as solid circles; negative specimens are shown as open circles. Crosses indicate specimens that were not assayed by the antigen test. There were 4.525 in antigen-positive and 3.444 and log10 mean RNA copies/mL antigen-negative specimens were, and the difference was not significant (p = 0.1088, t-test).
2. Discussion
Sensitivity of SARS-CoV-2 RT-PCR in saliva specimens was lower than those reported previously [1,2],[4],[[8], [9], [10]]. Among several procedures of saliva collection which have been applied, we chose the passive drool method to reduce the burden of the patients and the secondary risk to healthcare workers. However, collection procedure should be examined for further studies. In addition, symptoms, periods after the onset of symptoms and infection could affect the sensitivity of the RNA detection in saliva. These factors would also affect the sensitivity of antigen detection using the ESPLINE kit.
This study was carried out using specimens collected from hospitalized patients. The mechanism and hazard of secondary infection caused by asymptomatic carriers and super spreaders through biological materials from them have been discussed [11]. To assess the secondary infection hazards, it would be important to measure infectious virus particles in saliva and nasal fluid. Low prevalence of RNA in saliva specimens from the hospitalized patients might provide clues to assessment of the secondary infection hazards. The important study limitations include the limited number of specimens and bias associated with specimen collection from hospitalized COVID-19 patients. An extensive evaluation of rapid antigen testing using saliva samples would require additional studies using a variety of specimens collected from a larger number of patients.
The combination of saliva specimen and rapid antigen detection test could reduce burden of medical settings by decreasing not only the risk of secondary infection, but also time of diagnosis and cost of the specialized expensive equipment. A saliva-antigen test would be useful in ordinal clinics to identify persons with high-viral loads.
Ethical approval
The study protocol was approved by the Ethics Committee of Faculty of Medicine, Toho University (No. A20028_A20020_A20014_A19099).
Funding
A part of this research was supported by the Japan Agency for Medical Research and Development under Grant Number JP19fk0108113.
Author contributions
YI, SY, KaA and KT contributed to design this study. KK, TMa, TM, and SaY contributed to collect specimens, KoA and YI contributed to conduct and perform RT-PCR and antigen tests, YI, KoA, SY and KaA contributed to analyze data, and SY, YI, KoA and KT contributed to prepare this manuscript.
Declaration of competing interest
SY is an employee and KaA is the director of Fujirebio Inc.
Acknowledgments
We would like to express our thanks to Ms. Tang Cheuk Gee for the proofreading of the manuscript.
References
- 1.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020 doi: 10.1016/s1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., et al. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. Medrxiv. 2020 doi: 10.1101/2020.04.16.20067835. 2020. [DOI] [Google Scholar]
- 3.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease-2019 (COVID-19): a cross-sectional study. Medrxiv. 2020 doi: 10.1101/2020.04.17.20070045. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020 doi: 10.1128/jcm.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamakawa K., Fujimoto A., Miyamoto K., Ohshima T., Suzuki T., Nagata N., et al. Development of rapid lmmunochromatographic enzyme lmmunoassay for SARS-CoV-2 nucleocapsid protein. Jpn J Med Pharm Sci. 2020;77:937–944. (In Japanese) [Google Scholar]
- 6.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., et al. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis. 2020;73 doi: 10.7883/yoken.jjid.2020.061. JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 7.Manual for the Detection of Pathogen 2019-nCoV Ver.2.6. https://www.niid.go.jp/niid/images/epi/corona/2019-nCoVmanual20200217-en.pdf
- 8.To K.K.-W., Tsang O.T., Yip C.C.-Y., Chan K.-H., Wu T.-C., Chan J.M.C., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong F., Liang Y., Xu J., Chu M., Tang G., Hu F., et al. Continuously high detection sensitivity of saliva, viral shedding in salivary glands and high viral load in patients with COVID-19. Ssrn Electron J. 2020 doi: 10.2139/ssrn.3576869. [DOI] [Google Scholar]
- 10.Chen L., Zhao J., Peng J., Li X., Deng X., Geng Z., et al. Detection of 2019-nCoV in saliva and characterization of oral symptoms in COVID-19 patients. Ssrn Electron J. 2020 doi: 10.2139/ssrn.3556665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu R., Cui B., Duan X., Zhang P., Zhou X., Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int J Oral Sci. 2020;12:11. doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]