Abstract
Shewanella shows good application potentials in the decolorization and detoxification of azo dye wastewater. However, the molecular mechanism of decolorization is still lacking. In this study, it was found that Shewanella putrefaciens CN32 exhibited good decolorization ability to various azo dyes, and a global regulatory protein cAMP receptor protein (Crp) was identified to be required for the decolorization of acid yellow 36 (AY) by constructing a transposon mutant library. Then, the molecular mechanism of AY decolorization regulated by Crp was further investigated. RT-qPCR and electrophoretic mobility shift assay (EMSA) results showed that Crp was able to directly bind to the promoter region of the cymA gene and promote its expression. Riboflavin acting as an electron shuttle could accelerate the AY decolorization efficiency of S. putrefaciens CN32 wild-type (WT) but did not show a promoting effect to Δcrp mutant and ΔcymA mutant, further confirming that Crp promotes the decolorization through regulating electron transport chains. Moreover, the mutant with cymA overexpression could slightly enhance the AY decolorization efficiency compared with the WT strain. In addition, it was found that MtrA, MtrB, and MtrC partially contribute to the electron transfer from CymA to dye molecules, and other main electron transport chains need to be identified in future experiments. This study revealed the molecular mechanism of a global regulator Crp regulating the decolorization of azo dye, which is helpful in understanding the relationship between the decolorization and other metabolic processes in S. putrefaciens CN32.
Keywords: Shewanella putrefaciens CN32, decolorization, azo dye, acid yellow, Crp
Introduction
With the rapid development of the textile, printing, and dyeing industries, more than 700,000 tons of commercial synthetic dyes are produced worldwide each year (Guo et al., 2020b), of which azo dyes account for 60–70% (Hameed and Ismail, 2018; Kong et al., 2018), mainly due to their properties of low cost, easy synthesis, high coloring efficiency, and good stability to various oxidizing agents. The dyeing industries consume huge amounts of azo dyes, and about 10–15% of dye molecules that are not effectively bound to clothing is discharged into the surrounding environment in the form of dyeing wastewater (Zahir et al., 2014; Zhang et al., 2019), causing serious environmental pollution (Yesilada et al., 2003). Dyeing wastewater not only affects the transparency of the surrounding water but also poses a serious threat to the ecological environment and even human health because azo dye molecules and their degradation products have strong mutagenic, teratogenic, and carcinogenic effects (Guo et al., 2020a; Samir et al., 2020). The most remarkable characteristic of azo dyes is that their molecules contain one or more azo groups (Yan et al., 2012). Because of their stubborn structural properties, azo dyes are very stable in nature and difficult to degrade (Liu W. et al., 2017). Therefore, decolorization and detoxification are very necessary before the discharge of azo dye-containing wastewater (Liu et al., 2017b).
Physical, chemical, and biological strategies are generally applied to treat azo dye-containing wastewater (Pandey et al., 2007). Compared with physicochemical methods, which are limited by high energy consumption and secondary pollution, biological methods attracted more and more attention due to their advantages of high decolorization efficiency, low operation cost, and environmental friendliness (Saratale et al., 2011; Samir et al., 2020; Wang et al., 2020). The mechanism of biological decolorization of azo dyes mainly includes bioflocculation (Liu et al., 2009), biological adsorption (Saratale et al., 2011), electron reduction (Cai et al., 2012), and enzymatic degradation (Dawkar et al., 2010; Baweja et al., 2016; Yang et al., 2018, 2020). Moreover, the decolorization efficiency of azo dyes can also be enhanced by the combined use of different biological methods at the same time. For example, the degradation efficiency of insoluble Sudan red can be accelerated by the synergistic effect of enzyme-catalyzed biodegradation and non-specific reductive decolorization (Liu et al., 2018). In addition, much effort has been made to improve the decolorization efficiency of azo dyes (Imran et al., 2016). For example, some electron shuttles, such as riboflavin and methylene blue, have been found to be able to promote electron transfer from the cell surface to the dye molecules, thereby accelerating the biodegradation of azo dyes (Liu et al., 2016).
Many strains have been reported to be able to decolorize azo dyes; among them, Shewanella strains are the most concerned species due to their excellent decolorization performance (Cai et al., 2012; Liu et al., 2016, 2018). Shewanella, as a species of facultative anaerobic bacteria with remarkable respiratory pathways, is able to utilize various terminal electron acceptors under anaerobic conditions, including various pollutants, such as azo dyes and heavy metal ions (Fredrickson et al., 2008; Ding et al., 2014). Therefore, Shewanella strains show good potential in the field of environmental remediation. In Shewanella, many components in the electron transfer pathway are necessary for its decolorization ability (Cai et al., 2012; Xiao et al., 2012; Liu et al., 2016). In the cytomembrane, the tetraheme c-type cytochrome CymA receives electrons from the quinone pool and then transfers them to multiple respiratory pathways, such as Mtr, Dms, NarfA/B, and NrfA pathways (Schwalb et al., 2003; Gao et al., 2009). In Shewanella oneidensis MR-1, the Mtr respiratory pathway, including MtrA, MtrB, MtrC, and OmcA, has been found to be involved in decolorization processes of various azo dyes (Cai et al., 2012; Xiao et al., 2012). However, studies on the molecular mechanism of the decolorization of azo dyes by Shewanella are still lacking, especially on the global regulatory factors related to the decolorization of azo dyes.
In this study, cAMP receptor protein (Crp), a global transcription regulator (Gao et al., 2012), was found to be essential for the decolorization of azo dyes in Shewanella putrefaciens CN32 through the construction of a transposon mutant library and the selection of mutants with different decolorization abilities and transposon locations. And then, the molecular mechanism of the decolorization of azo dyes regulated by Crp was also investigated. We found that, with the assistance of cAMP, Crp was able to directly bind to the promoter region of the cymA gene and promote its expression, thereby promoting decolorization through regulating electron transport chains. Thus, this study has a certain theoretical significance for revealing the molecular mechanism of the decolorization of azo dyes by Shewanella strains.
Materials and Methods
Bacterial Strains, Plasmids, Primers, and Culture Conditions
The strains and plasmids used or constructed in this study are shown in Table 1, and the primers used in this study are listed in Table 2. Escherichia coli and S. putrefaciens CN32 strains were grown aerobically at 37°C and 30°C, respectively, in Luria–Bertani (LB) medium which contains tryptone 10 g/L, yeast extract 5 g/L, and NaCl 10 g/L, for genetic manipulation. If necessary, kanamycin of 50 μg/ml was added into the medium. The decolorization medium for S. putrefaciens CN32 consisted of yeast extract 2 g/L, NH4Cl 1 g/L, NaCl 0.5 g/L, Na2HPO4⋅12H2O 7.52 g/L, NaH2PO4⋅2H2O 7.13 g/L, and filter-sterilized sodium lactate 20 mM. The transformants were screened on the LKT medium, which is based on the LB medium added with 50 μg/ml of kanamycin and 20 μg/ml of potassium tellurite.
TABLE 1.
The strains and plasmids used in this study.
| Strains/plasmids | Descriptions | Sources |
| E. coli strains | ||
| DH5α | Host for cloning | Lab stock |
| BL21(DE3) | Expression host for pET-28a(+) | Lab stock |
| UQ3021 | DH5a/λpir | Larsen et al., 2002 |
| UQ3022 | UQ3021/pRL27, Kmr | Larsen et al., 2002 |
| S. putrefaciens strains | ||
| CN32 | Wild type | Lab stock |
| Δcrp | In-frame deletion mutant of crp (sputcn32_0652) in CN32 | This study |
| C-crp | Δcrp mutant carrying complement pBBR1MCS-2-PaacC1-crp | This study |
| ΔcymA | In-frame deletion mutant of cymA (sputcn32_0286) in CN32 | This study |
| ΔcrpΔcymA | Mutant with in-frame deletion of crp and cymA in CN32 | This study |
| C-cymA | ΔcymA mutant carrying complement pBBR1MCS-2-PaacC1-cymA | This study |
| O-cymA | CN32 with overexpression plasmid pBBR1MCS-2-PaacC1-cymA | This study |
| ΔmtrA | In-frame deletion mutant of mtrA (sputcn32_1477) in CN32 | This study |
| ΔmtrB | In-frame deletion mutant of mtrB (sputcn32_1476) in CN32 | This study |
| ΔmtrC | In-frame deletion mutant of mtrC (sputcn32_1478) in CN32 | This study |
| ΔmtrCAB | Mutant with in-frame deletion of mtrC, mtrA, and mtrB in CN32 | This study |
| ΔundA | In-frame deletion mutant of undA (sputcn32_1479) in CN32 | This study |
| ΔundAΔmtrCAB | Mutant with in-frame deletion of undA, mtrC, mtrA, and mtrB | This study |
| Plasmids | ||
| pRL27 | Contains mini-Tn5 transposon (oriR6K) delivery vector, Kmr | Larsen et al., 2002 |
| pK19mobsacB | Suicide plasmid for strain CN32, Kmr | Schäfer et al., 1994 |
| pRK2013 | Helper plasmid in triparental conjugation | Figurski and Helinski, 1979 |
| pBBR1MCS-2-PaacC1 | Broad-host-range plasmid with the promoter of aacC1; Kmr | Liu et al., 2017a |
| pET-28a(+) | Vector for protein overexpression in BL21(DE3), Kmr | Lab stock |
TABLE 2.
The primer sequences used in this study.
| Primers | Sequences (5′–3′) | Functions or target genes |
| Tn5-seqF | CAGCAACACC TTCTTCACGA | For Tn5 Sequence |
| Tn5-seqR | AACAAGCCAGG GATGTAACG | For Tn5 Sequence |
| Crp-D5F | CTCAAAGAATTCTAAG ATGAGTCCAATCACTGTGCCC | crp deletion |
| Crp-D5R | GTATACTCTAGAGGTAC CCGTTAAGTTAGTCTTCAGC | crp deletion |
| Crp-D3F | TTTTACTCTAGAGATGTAA TAAAGGGTATCTGAATCT | crp deletion |
| Crp-D3R | AGGCAGAAGCTTCAGCGA GGTTATCTAAATTAGTGGG | crp deletion |
| Crp-UF | GTTGGATACACCAGTG CGAACAGAC | crp deletion |
| Crp-DR | TCTAAACTAAGACT TCTATCAAGTT | crp deletion |
| Crp-OF | CCAGCATGATATG TTCAAGATCTT | crp deletion |
| Crp-OR | GCAGCACTAAAAT CACCAATTTCT | crp deletion |
| Crp-InF | CCAATCTCTTGA CGAGTGATCTTG | crp deletion |
| Crp-InR | AAGGTTCTGTTG CCGTATTGATTAA | crp deletion |
| Crp-CF | CAAAGGGATCCCGA CCACACCATAAAGTTAGCCTG | crp complementation |
| Crp-CR | TTTAATGAATTCGAA ACAGGCTTAAATCAAGCTGAAG | crp complementation |
| Crp-EF | ATCAACGGATCCATGGC TCTGATTGGTAAGCCAAAACC | express His6-Crp protein |
| Crp-ER | TGGCCTAAGCTTAGAAT TTATGCTAGGCCACTTTAATG | express His6-Crp protein |
| CymA-D5F | CCGGAATTCCCATTG CAGTATCGCTTATG | cymA deletion |
| CymA-D5R | GATACAGAACT GATCCGTACTTG | cymA deletion |
| CymA-D3F | GCTCACCCATA TCCAAAAG | cymA deletion |
| CymA-D3R | AAAACTGCAGC TACCTATCCAAGATCTCGAAG | cymA deletion |
| CymA-UF | TATTGTCCTGAT AGTTAGAGCT | cymA deletion |
| CymA-DR | CCTGTTAGTTTA TCGTCAGC | cymA deletion |
| CymA-OF | GCCGAAGACAAA GAGATAG | cymA deletion |
| CymA-OR | AAACCGCCAAAA ATAAAC | cymA deletion |
| CymA-InF | CCTGTCACAGC AACCATT | cymA deletion |
| CymA-InR | CTGCGGAAATA CTTAAGTGC | cymA deletion |
| CymA-CF | TTCTAAGGATCCTAAGTGAAAT AGCATAAACTAGACTT | cymA complementation |
| CymA-CR | GTTCAAGAATTCGAATGAATCG CTAAAACCTATTATCC | cymA complementation |
| CymA-PF | GACTAAGAGTTTG ATGCATAAGTATT | cymA EMSA probe |
| CymA-PR | AATCATCAAACA ATCGCAAGTTAT | cymA EMSA probe |
| CymA-QF | GAACTGGCGT GCACTATT | cymA RT-qPCR |
| CymA-QR | ATACAGAACT GATCCGTACTTG | cymA RT-qPCR |
| 16S QF | GCAGGCGGTTT GTTAAGCGAGATG | internal standard for RT-qPCR |
| 16S QR | CTTCGCCACCGG TATTCCTCCAGA | internal standard for RT-qPCR |
| CymA-FootF | TGTAAAACGACGGCCA GTAATAATGAACGGCTCGAT | cymA DNase I footprinting |
| CymA-FootR | CAGGAAACAGCTATGACCAAT AGTGCACGCCAGTTC | cymA DNase I footprinting |
| MtrC-D5F | AAAACTGCAGCTGTGTTAG CTGTCATAATGA | mtrC deletion |
| MtrC-D5R | AAACTTGAAAT ATTGAAGCC | mtrC deletion |
| MtrC-D3F | GCAGTGCAG TCAGAAACC | mtrC deletion |
| MtrC-D3R | CCGGAATTCT GCTCACCTCCATGACAT | mtrC deletion |
| MtrC-UF | GCATTAACTTA AGTCGCCTC | mtrC deletion |
| MtrC-DR | TGTTCTTCATA ATAGGCTTCC | mtrC deletion |
| MtrC-OF | GTGGTTGTAGCA GTTGTCATAC | mtrC deletion |
| MtrC-OR | ATAATGCCCCTT ACTACTGG | mtrC deletion |
| MtrC-InF | GGTACCTACAG CTATGACTTCG | mtrC deletion |
| MtrC-InR | TGGTGTAATGT TTGGCGT | mtrC deletion |
| MtrA-D5F | CCGGAATTCAA GTATTGTTGACGGTAAGCT | mtrA deletion |
| MtrA-D5R | ATAACTCCCTT CAGCGAAC | mtrA deletion |
| MtrA-D3F | AATTGCCATA GTCAGGTTCA | mtrA deletion |
| MtrA-D3R | AAAACTGCAG CCAGATATCACATTGGTATTGTC | mtrA deletion |
| MtrA-UF | GCAGTGCAGTC AGAAACCT | mtrA deletion |
| MtrA-DR | AGCCCTTACAG CTCCATG | mtrA deletion |
| MtrA-OF | TTGAAATTATT ACTAACGTTGGCC | mtrA deletion |
| MtrA-OR | CTTGGCTCATT TGTCCCG | mtrA deletion |
| MtrA-InF | GCAATGAACC GATGATCAC | mtrA deletion |
| MtrA-InR | CGTGACAGGC ATAACAGGT | mtrA deletion |
| MtrB-D5F | CCGGAATTCG CCACCTTAGATAAAAAGTTCG | mtrB deletion |
| MtrB-D5R | AACGATGCA GCCCTTACAG | mtrB deletion |
| MtrB-D3F | GACGCCGCGA ATGATATC | mtrB deletion |
| MtrB-D3R | AAAACTGCAG TGCTAATAAAGATGTCATGGATGC | mtrB deletion |
| MtrB-UF | GCTGCTTAAAT TGCCATAGT | mtrB deletion |
| MtrB-DR | CTTGTCGTAGC GCTTAAAC | mtrB deletion |
| MtrB-OF | TTGCTACGAGT GCTCATG | mtrB deletion |
| MtrB-OR | GGCGACTTGCT TGTAATAT | mtrB deletion |
| MtrB-InF | GGCAAATTT GACGCTGAC | mtrB deletion |
| MtrB-InR | TACTATCCA GTTATCAGGCAATGT | mtrB deletion |
| UndA-D5F | AAAACTGCAGA CTGCGCCTATTGTAGCT | undA deletion |
| UndA-D5R | GGTCAGTACTA TATCAACGCTG | undA deletion |
| UndA-D3F | AACGGTGGTGT GTACAATG | undA deletion |
| UndA-D3R | TTGGATCCGCG AACATAGTTATTCAGTACAAT | undA deletion |
| UndA-UF | TGATGATGATTA CAACTATTGC | undA deletion |
| UndA-DR | TCTGTCAACTAT TGCTGCTT | undA deletion |
| UndA-OF | CCGATTTCAGA AATAATGC | undA deletion |
| UndA-OR | AGTAAAGACA GGTAGCGTGG | undA deletion |
| UndA-InF | TGGATCAGC TATATCAACTCAGT | undA deletion |
| UndA-InR | GGTGTATCAA GGTCGGGT | undA deletion |
| MtrC-D5F1 | CCCAAGCTTC TGTGTTAGCTGTCATAATGA | mtrCAB deletion |
| MtrC-D5R1 | TTGGATCCAAACT TGAAATATTGAAGCC | mtrCAB deletion |
| UndA-D5F1 | CCCAAGCTTACTGCGCC TATTGTAGCT | mtrCAB/undA deletion |
| UndA-D5R1 | TTGGATCCGGTCAGTA CTATATCAACGCTG | mtrCAB/undA deletion |
| MtrB-D3F1 | TTGGATCCGACGCCGC GAATGATATC | mtrCAB/undA deletion |
| MtrB-D3R1 | AAAACTGCAGTGCTAA TAAAGATGTCATGGATGC | mtrCAB/undA deletion |
Decolorization Assay of Azo Dyes
S. putrefaciens CN32 was cultured in LB medium for 12 h to obtain the seed culture and then inoculated into 60 ml serum vials containing 50 ml of the decolorization medium with azo dyes to an initial OD600 of 0.1. To ensure anaerobic condition, decolorization systems were purged with nitrogen gas for 2 min. Subsequently, serum vials were sealed with rubber stoppers immediately and then cultured in an incubator at 30°C without shaking for decolorization. The decolorization samples of different times were centrifuged at 10,000 rpm, 25°C for 1 min. The absorbance of the supernatant was determined at 497 nm for Congo red, 520 nm for amaranth, 414 nm for acid yellow 36 (AY), 465 nm for methyl orange, 618 nm for amino black, and 714 nm for naphthol green to determine the concentration of residual azo dyes. The decolorization rate (%) was calculated based on the following equation: (ODx − ODy)/ODx × 100%, where ODx and ODy refer to the absorbance of the initial and decolorized samples, respectively. Experiments were repeated independently at least three times.
Transposon Mutagenesis and Location of Transposon Insertion Sites
S. putrefaciens CN32 (recipient strain) was mixed with E. coli UQ3022 (donor strain), which carries a plasmid pRL27 containing a mini-Tn5 transposon and a kanamycin resistance gene (Larsen et al., 2002). Both recipient strain and donor strain were cultured overnight in LB medium. The donor strain was washed with LB medium twice and mixed with the recipient strain in a 1:1 ratio. The mixture of these two strains was spotted on a solid LB agar plate and incubated at 30°C for 8 h. Then the cells were scraped off from the agar plate and plated onto an LB agar plate with 50 μg/ml of kanamycin and 20 μg/ml of potassium tellurite (to inhibit the cell growth of the donor strain) and incubated at 30°C for 36 h. Single black colonies presented were purified, and their decolorization abilities were determined in the decolorization medium with 200 mg/L AY. The mutants with significantly altered decolorization capacity were selected and preserved. Then, the genomic DNA of these mutants was extracted for localization of the transposon insertion site. The extracted genomic DNA was self-linked after digestion by BamHI or SpeI and then transformed into E. coli UQ3021 (Larsen et al., 2002) and selected on the LB agar plate added with 50 μg/ml of kanamycin. Plasmids extracted from the transformant were sequenced using primer Tn5-seqF and Tn5-seqR and BLAST in NCBI to map the location of the mini-Tn5 transposon. The target genes inserted by the mini-Tn5 transposon were deleted to confirm its function in regulating AY decolorization in S. putrefaciens CN32.
Construction of Deletion Mutants and Complementation Strains
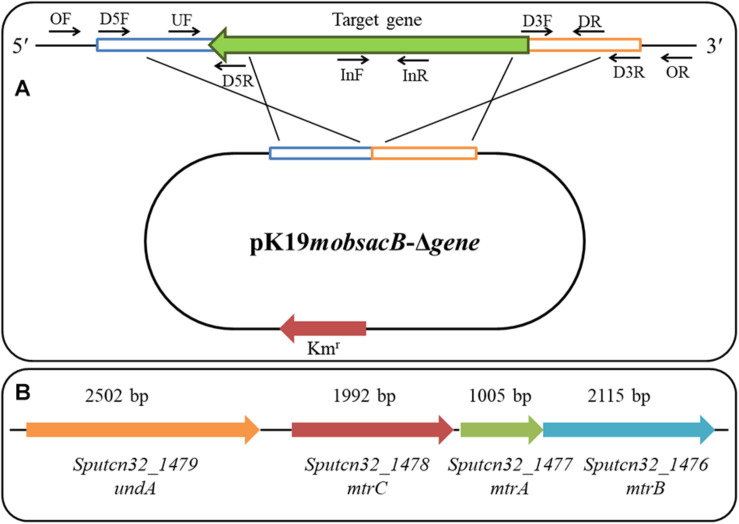
The in-frame deletion of mutants for crp, mtrA, mtrB, mtrC, undA, and cymA was performed based on the principle of homologous recombination. The primer positions for the target gene in-frame deletion are shown in Figure 1A, and the genomic arrangement of mtrA, mtrB, mtrC, undA, and cymA in S. putrefaciens CN32 is shown in Figure 1B. To delete the target gene, approximately 1,000-bp fragments upstream and downstream the targeted gene were amplified with respective primers D5F/D5R and D3F/D3R, using the S. putrefaciens CN32 genome DNA as a PCR template. These two fragments were ligated into the suicide vector pK19mobsacB after being digested by the corresponding restriction endonuclease (Schäfer et al., 1994). Then, the constructed plasmid was transformed into E. coli DH5α and introduced from E. coli DH5α into S. putrefaciens CN32 wild type (WT) using a helper plasmid, pRK2013, by triparental conjugation (Figurski and Helinski, 1979). The conjugation experiment was performed according to a previous study (Liu et al., 2017a). Briefly, the donor and recipient strains were conjugated in a 1:1 ratio and spotted on an LB agar plate. LKT medium plates were used to screen for transconjugants after conjugation for 8 h at 30°C. The single-crossover recombinant strain was selected using the primer UF/DR. After overnight culture in the LB medium added with 50 μg/ml of kanamycin, a single-crossover recombinant strain was transferred to a NaCl-free LB medium in a ratio of 0.1%, and the double-crossover disruptants were screened according to the sucrose sensitivity and finally checked by four pairs of primers: OF/OR, UF/OR, OF/DR, and InF/InR. All the mutants were confirmed by sequencing analysis. For the construction of the ΔmtrCAB mutant, upstream and downstream fragments were amplified with primers MtrC-D5F1/MtrC-D5R1 and MtrB-D3F1/MtrB-D3R1, respectively, and for the construction of ΔundAΔmtrCAB, upstream and downstream fragments were amplified with primers UndA1-D5F1/UndA1-D5R1 and MtrB-D3F1/MtrB-D3R1, respectively.
FIGURE 1.
The primer positions for target gene in-frame deletion (A) and genomic arrangement of mtrA, mtrB, mtrC, and undA in S. putrefaciens CN32 (B).
To construct a complementation plasmid, a DNA fragment carrying the ribosome binding site (RBS) and open reading frame (ORF) of the targeted gene was amplified by primers CF/CR using the S. putrefaciens CN32 genome as a PCR template and then ligated with the plasmid pBBR1MCS-2-PaacC1 with the corresponding restriction site (Liu et al., 2017a). And then the complementation plasmid was introduced into corresponding mutants for complementation or into S. putrefaciens CN32 WT for overexpression.
RNA Extraction and Real-Time RT-PCR Analysis
In order to analyze the regulatory effect of Crp on the cymA gene, the seed samples of WT and Δcrp mutant were inoculated into the decolorization medium with 200 mg/L AY and cultured for 4 h. The cells were collected by centrifugation at 10,000 rpm, 4°C for 5 min, and RNA was extracted by the TRIzol reagent (Tiangen, China) according to the protocol provided by the manufacturer. Real-time RT-PCR was performed using primers CymA-QF/CymA-QR, with SYBR Green Master Mix (Biosharp, China) according to the specification from the manufacturer. Signal intensities of PCR products were standardized to those of the 16S rRNA gene amplified with primers 16S QF/16S QR. The experiments were performed with at least three replicates.
Purification of His6-Crp Protein
To prepare the His6-Crp protein, the coding region of crp was amplified by PCR using primers Crp-EF/Crp-ER. The PCR product was digested with BamHI/HindIII and cloned into pET-28a (+) to generate pET28-crp. The resulting plasmid was confirmed by sequencing and then transformed into E. coli BL21(DE3) for overexpression of His6-Crp (six-histidine tag on the N-terminus of the Crp protein). After induction with 0.4 mM isopropyl β-D-thiogalactoside (IPTG) at 16°C for 20 h, the soluble recombinant His6-Crp protein was purified by Ni-agarose resin chromatography (CoWin Biosciences, China).
Electrophoretic Mobility Shift Assay (EMSA)
DNA probe-carrying promoter regions of cymA were PCR-amplified by using primers CymA-PF and CymA-PR. The PCR products purified from agarose gel were labeled with digoxigenin (DIG) using terminal transferase. The 3′-terminal DIG-labeled probe of 0.15 nM was incubated with various quantities of His6-Crp and 1 μM cAMP in a binding reaction. EMSA experiments were operated as described previously (Sun et al., 2016). To confirm specificity of protein–DNA interaction, a 300-fold excess of unlabeled specific probe or non-specific DNA was added into the binding mixture before incubation.
DNase I Footprinting
DNase I footprinting assay was carried out to determine the binding site of Crp in the promoter region of cymA. PCR was conducted using 5′-terminal FAM-labeled forward primer CymA-FootF and regular reverse primer CymA-FootR, and the PCR products were purified to obtain FAM-labeled footprinting probes. The mixtures of 20-μl volume containing various concentrations of purified His6-Crp, 300-ng probes, and 2 μM of cAMP were incubated at 25°C for 40 min to achieve the binding reaction of the Crp protein with the probe. Then 1 U of DNase I (NEB, United States) was added to the mixture and incubated at 37°C for 10 s. The DNase I treatment was terminated by adding 10 μl of 0.5 M EDTA solution and heated at 80°C for 10 min. The DNA fragments were purified and capillary sequenced in a 3730XL DNA Genetic Analyzer (ABI, United States). The data were processed and analyzed with the GeneMarker program, v2.2.0.
Effect of Exogenous Riboflavin on Decolorization
In order to analyze the effect of exogenous riboflavin on AY decolorization of S. putrefaciens CN32 WT, Δcrp, and ΔcymA, the strains were cultured in LB medium to an OD600 of around 1.0 and used as seed culture. Then, the cells in seed culture were collected by centrifugation at 25°C, 8,000 rpm for 5 min, and were washed once using the decolorization base medium (decolorization medium without adding yeast extract). The washed cells were inoculated into 60 ml serum vials containing 50 ml of the decolorization base medium with 200 mg/L of AY to an initial OD600 of 0.1. After purging with nitrogen gas for 2 min, the decolorization ability of WT, Δcrp, and ΔcymA with and without adding 2 μM riboflavin was determined at different time intervals.
Results and Discussion
Decolorization of Various Azo Dyes by S. putrefaciens CN32
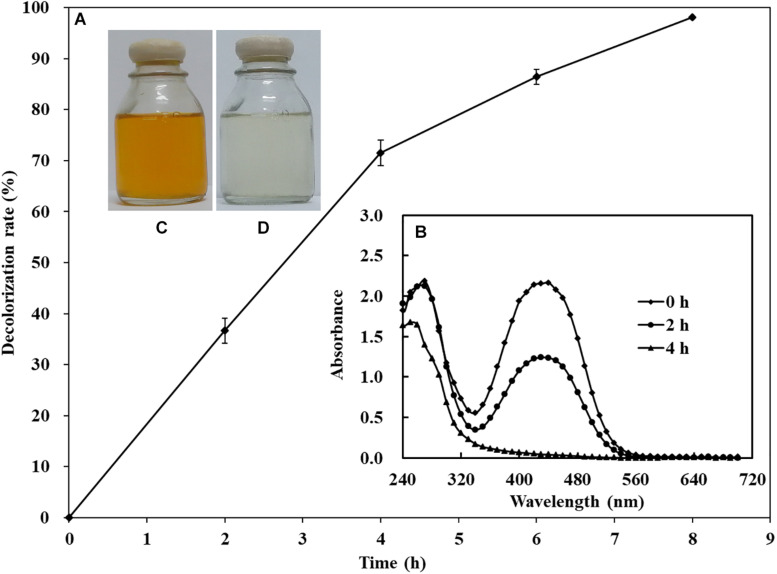
The decolorization ability of S. putrefaciens CN32 to various azo dyes was analyzed under anaerobic condition (Table 3). The results showed that S. putrefaciens CN32 was able to decolorize all the tested azo dyes within 48 h. Especially to AY (Figure 2) and methyl orange, more than 90% of decolorization rates were achieved within 8 h. As shown in Figure 2, the scanning spectrum of AY and its degradation products in a range from 240 to 720 nm was investigated. It can be seen that the characteristic absorption peak at around 414 nm disappeared after decolorization of 4 h, suggesting that S. putrefaciens CN32 shows good potential in the treatment of azo dye-containing wastewater. In previous studies, several Shewanella strains have been used to decolorize azo dyes, such as S. oneidensis MR-1 (Liu et al., 2016; Li et al., 2018), Shewanella decolorationis S12 (Xu et al., 2007), Shewanella sp. RQs-106 (Zhou et al., 2018), Shewanella aquimarina (Meng et al., 2012), and Shewanella algae (Meng et al., 2014). In S. oneidensis MR-1, a transmembrane electron transport chain Mtr respiratory pathway, which includes cytochromes MtrC and OmcA and related proteins MtrA and MtrB, has been proven to be responsible for the anaerobic decolorization of azo dyes (Xiao et al., 2012). However, the further regulatory mechanism of the decolorization of azo dyes in Shewanella species is largely unknown. In this study, AY was selected for investigating the molecular mechanism of azo dye decolorization by S. putrefaciens CN32 due to its very high decolorization efficiency.
TABLE 3.
Decolorization ability of S. putrefaciens CN32 to tested azo dyes.
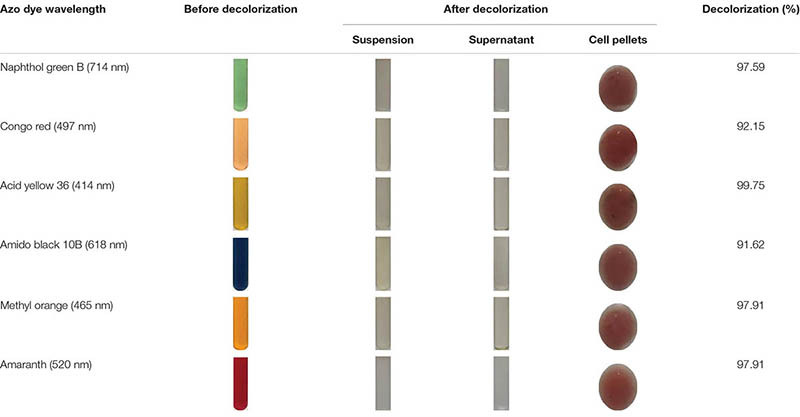 |
The initial concentration of azo dyes is 50 mg/L. The images of azo dye solution obtained before decolorization and after 24 h of decolorization. The cell pellets were collected from decolorization solution using a 2 ml tube with 10,000 rpm.
FIGURE 2.
Decolorization of 200 mg/L of AY by S. putrefaciens CN32 (A). Scanning spectrum analysis of 50 mg/L of AY decolorized by S. putrefaciens CN32 at different time intervals (B). The image of 50 mg/L AY before (C) and after (D) decolorization.
Crp Promotes the Decolorization Ability of S. putrefaciens CN32 Under Anaerobic Conditions
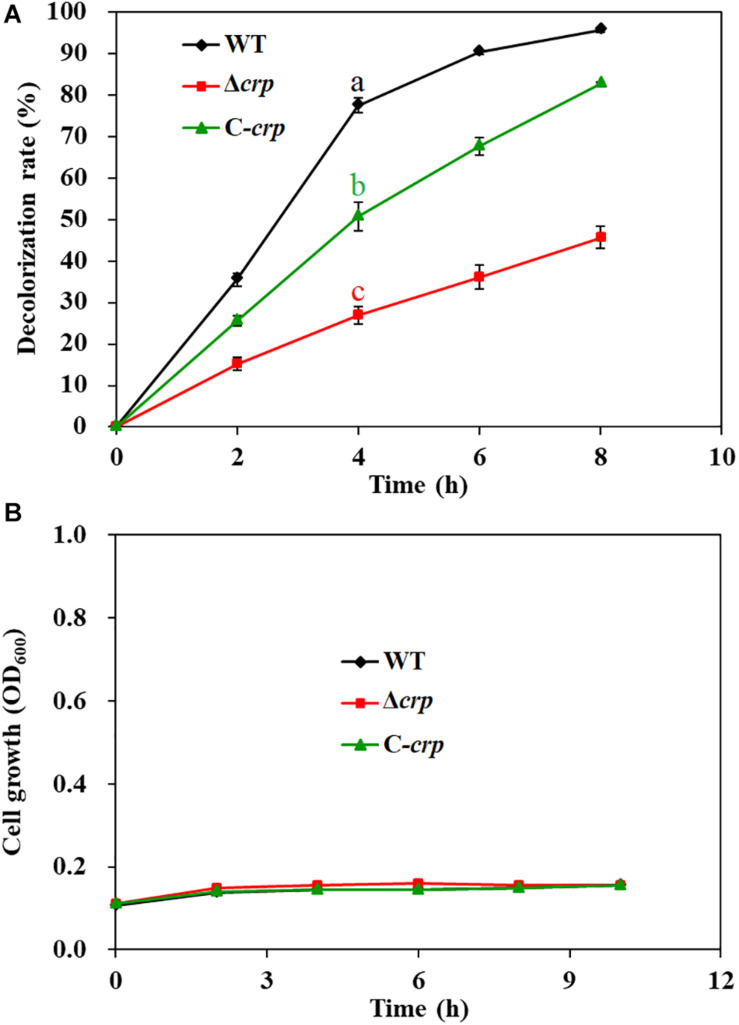
To identify the underlying decolorization mechanisms of AY by S. putrefaciens CN32 in anaerobic respiration, approximately 1,000 mini-Tn5 transposon-inserted mutants screened by a kanamycin agar plate were selected for decolorization ability analysis using 60 ml serum vials. Compared with the CN32 WT strain, the decolorization efficiencies of 10 transposon-inserted mutants were significantly altered (more than 50% change), in which three mutants that exhibited poor decolorization ability to AY were inserted into different sites of the crp gene (sputcn32_0652), suggesting that Crp may be involved in biodecolorization. Thus, an in-frame deletion mutant of the crp gene (Δcrp) and a complementation strain (C-crp) were constructed. As shown in Figure 3A, the anaerobic decolorization efficiency of the Δcrp mutant to AY was seriously decreased compared with the WT, and the C-crp mutant was obviously restored to the WT level. At the same time, the growth of WT, Δcrp mutant, and C-crp mutant was determined under the anaerobic decolorization condition. The results (Figure 3B) showed that the WT, Δcrp mutant, and C-crp mutant did not show obvious cell growth during the anaerobic AY decolorization process. Therefore, these results indicated that Crp promotes AY decolorization in S. putrefaciens CN32. Crp is a global regulatory factor which regulates different metabolic processes in many bacteria by forming complexes with cAMP; for example, Crp is involved in carbon catabolite repression in many bacteria (Deutscher et al., 2006); in E. coli, Crp regulates many stress responses to protect cells from harmful environments including starvation and osmotic shock (Battesti et al., 2011; Kalia et al., 2013); in Pseudomonas aeruginosa, Crp (Vfr) regulates biofilm formation through controlling type IV pili (Beatson et al., 2002; Persat et al., 2015). In this study, it was found that Crp is necessary for biological decolorization in S. putrefaciens CN32.
FIGURE 3.
Decolorization of AY (A) and cell growth (B) of the WT, Δcrp mutant, and C-crp mutant. The initial concentration of AY is 200 mg/L. The cell growth was determined in the decolorization medium under anaerobic conditions. Values for decolorization and cell growth are means ± SD (n = 3). Significance analysis of decolorization rates of different strains at 4 h was performed using one-way analysis of variance (ANOVA) with the Tukey–Kramer comparison test (p < 0.05).
CymA Is Necessary for the Anaerobic Decolorization of AY in S. putrefaciens CN32
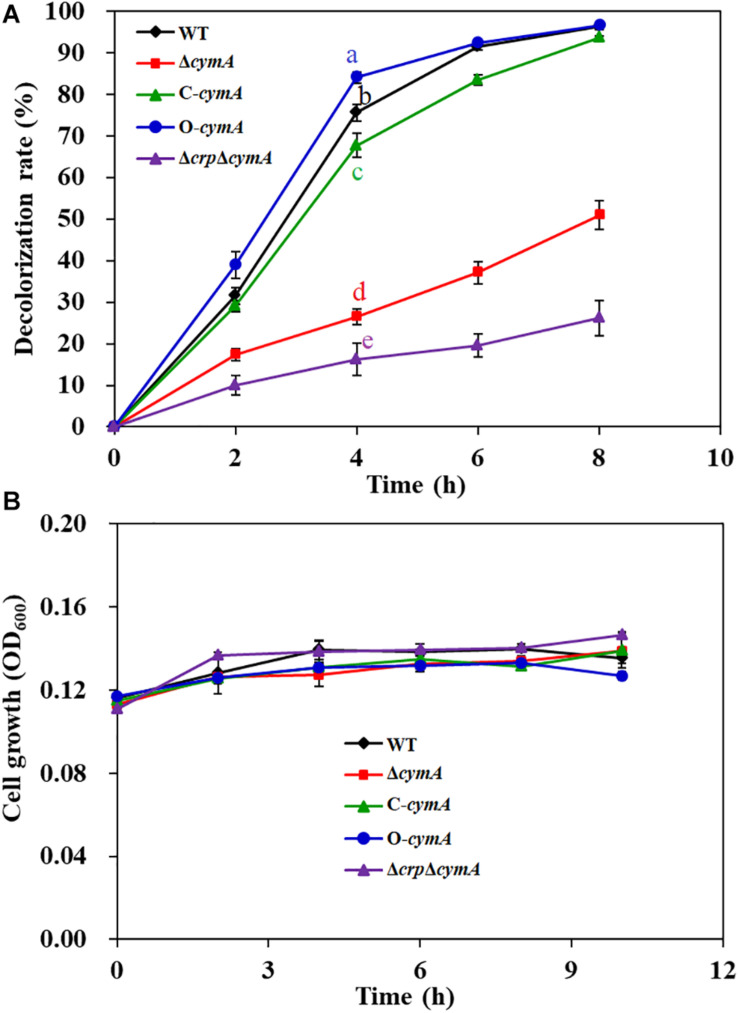
Except for the crp gene, two transposon-inserted mutants with a significant decrease in decolorization efficiency were identified with different insertion sites in the cymA gene (sputcn32_0286). CymA, a c-type cytochrome, is a component of the Mtr respiratory pathway, which is a critical transmembrane electron transfer channel in dissimilatory metal-reducing strains (Xiao et al., 2012). A previous study showed that the ΔcymA mutant of S. oneidensis MR-1 almost lost complete decolorization capability (Xiao et al., 2012). Genome analysis found that the amino acid sequence of CymA in S. putrefaciens CN32 exhibits 95.7% similarity to that in S. oneidensis MR-1. To confirm the function of CymA in S. putrefaciens CN32 during AY decolorization, an in-frame gene deletion mutant ΔcymA and a complementation strain C-cymA were constructed. The anaerobic decolorization ability of the ΔcymA mutant was seriously decreased compared with that of the WT strain (Figure 4A); the C-cymA mutant was obviously restored to the WT level. And the WT, ΔcymA mutant, and C-cymA mutant exhibited similar cell growth (Figure 4B), suggesting that CymA is necessary for the anaerobic decolorization of AY in S. putrefaciens CN32. As the ΔcymA mutant showed a similar phenotype to the Δcrp mutant, a double mutant ΔcrpΔcymA was constructed to analyze their relationship. The results showed that the ΔcrpΔcymA mutant exhibited a similar decolorization efficiency to the Δcrp and ΔcymA mutants, which is obviously lower than that of the WT strain, indicating that Crp and CymA may be involved in the same decolorization pathway.
FIGURE 4.
Decolorization of 200 mg/L of AY (A) and cell growth (B) of the WT, ΔcymA mutant, C-cymA mutant, and ΔcrpΔcymA mutant. Values for decolorization and cell growth are means ± SD (n = 3). The cell growth was determined in the decolorization medium under anaerobic conditions. The decolorization rates of different strains at 4 h were analyzed by ANOVA with the Tukey–Kramer comparison test (p < 0.05).
Crp Directly Activates the Transcription of cymA
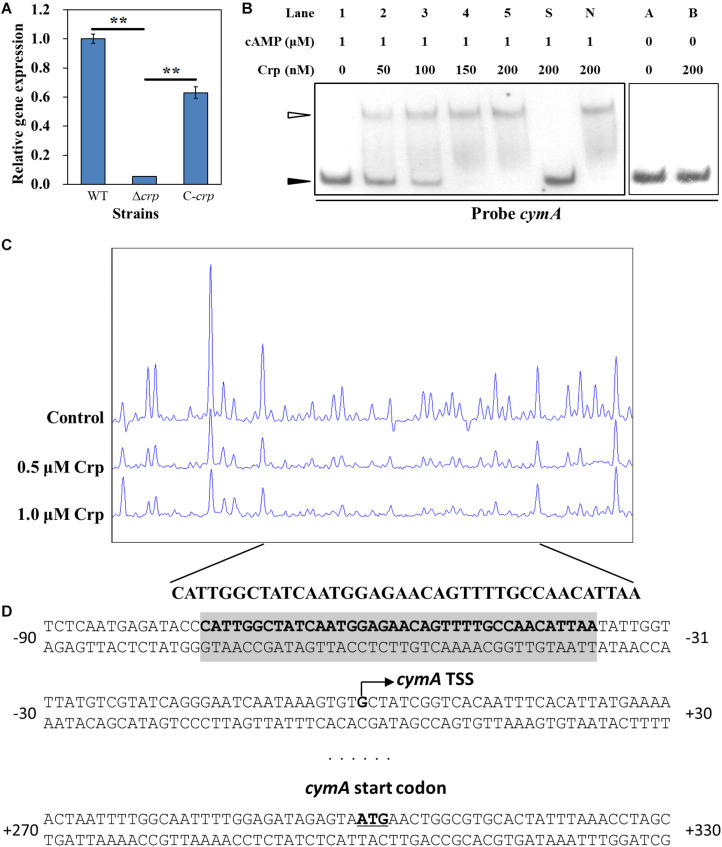
Crp is a critical global transcriptional regulator in prokaryotes through forming a complex with cAMP. In S. oneidensis MR-1, the complex of Crp and cAMP can regulate the transcription of multiple cytochrome c genes including omcA and mtrC (Kasai et al., 2015). However, the relationship between Crp and the cymA gene is still unclear. To determine whether Crp regulates the transcription of the cymA gene in S. putrefaciens CN32, real-time reverse-transcription PCR (RT-qPCR) was carried out (Figure 5A). Compared with that in the WT strain, the cymA expression level in the Δcrp mutant was significantly decreased, and almost no cymA expression was detected in the Δcrp mutant, indicating that Crp plays a critical role in activating the expression of the cymA gene. To identify the regulation that Crp exerts on the transcription of the cymA gene, EMSA was performed to investigate whether Crp directly binds to the upstream regions of the cymA gene. When a labeled DNA probe containing the region from −183 to −477 bp upstream of cymA was incubated with the Crp–cAMP complex, shifted bands were observed (Figure 5B). When there is no cAMP in the mixture, no shifted band was observed (Figure 5B). These findings suggested that the Crp–cAMP complex can bind to the promoter region of the cymA gene. Taken together, Crp directly activates the transcription of the cymA gene through forming a complex with cAMP. To further investigate the mechanism by which the Crp–cAMP complex activates cymA, a DNase I footprinting assay was carried out to identify the precise binding site of the Crp–cAMP complex at the upstream regions of the cymA gene, and a protected region from −375 to −338 nt upstream of the cymA start codon was revealed (Figure 5C). Subsequently, the transcription start site (TSS) of cymA was predicted using an online BDGM promoter prediction tool1. A possible TSS was found to be a “G” located at 300 nt upstream of the cymA start codon (Figure 5D). Based on the above results, it is possible that the Crp–cAMP complex activates cymA expression by recruiting the RNA polymerase to the promoter region of cymA.
FIGURE 5.
Regulatory effect of Crp on cymA gene. (A) The relative expression level of the cymA gene in the WT, Δcrp mutant, and C-crp mutant. cymA expression level in the WT strain was normalized as 1. Error bars indicate standard deviation of three samples. Significance analysis was performed using Student’s t-test; **p < 0.01. (B) EMSAs of the interaction of different concentrations of His6-Crp with DNA probes; 0.15 nM labeled probe and 1.0 μM cAMP were added to reaction mixtures with different concentrations of His6-Crp (Lanes 1, 2, 3, 4, and 5). A 300-fold-excess unlabeled specific competitor (Probe cymA, Lane S) and non-specific competitor (Lane N) were added to the mixture to perform specific or non-specific competition assays, respectively. Lanes A and B: Reaction mixtures without addition of cAMP used as negative controls. Filled triangles: Free probes. Empty triangle: Protein–DNA complex. (C) DNase I footprinting assay of different concentrations (0.5 and 1.0 μM) of His6-Crp and cymA in the upstream region. Each reaction mixture contains 300-ng FAM-labeled DNA probe and 2 μM cAMP. Control: No His6-Crp protein in the reaction mixture. The nucleotide sequences of the protected region were shown below the graph. (D) Nucleotide sequences of cymA promoter region and His6-Crp binding site. Bent arrow: Predicted cymA TSS. Shaded region: Region protected by Crp in the DNase I footprinting assay. Underline: cymA start codon. Numbers indicate the distance (nt) from TSS.
Effect of Exogenous Riboflavin on Decolorization
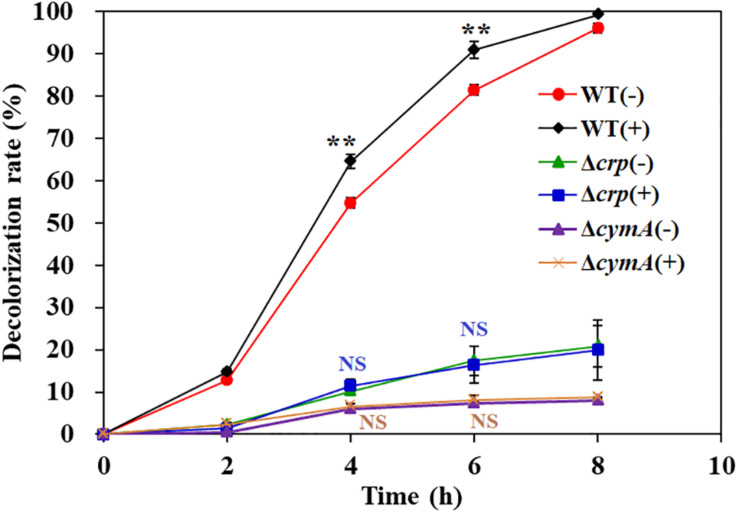
Previous studies have reported that flavins produced from Shewanella genus (Marsili et al., 2008) and other chemical substances, such as methylene blue (Liu et al., 2016) and humic acids (Liu et al., 2011), were able to act as shuttles to accelerate the electron transfer from the cell surface to pollutant molecules, such as azo dyes and heavy metal ions. In order to analyze the effect of electron shuttle on AY decolorization by S. putrefaciens CN32, 2 μM riboflavin was added into the decolorization systems of WT, Δcrp, and ΔcymA. The results showed that riboflavin could significantly improve the decolorization efficiency of the WT strain, but no significant promotion effect was observed for the Δcrp mutant and ΔcymA mutant (Figure 6). These results further demonstrated that the regulation of Crp to the AY decolorization efficiency is achieved by regulating the members of electron transport chains including CymA.
FIGURE 6.
The effect of exogenous riboflavin on AY decolorization. Values are means ± SD (n = 3). The cells were cultured in decolorization medium with (+) or without (–) adding 2 μM riboflavin. The initial AY concentration of is 200 mg/L. Significance analysis of decolorization rates of the same strain with and without riboflavin was performed using Student’s t-test; asterisks **p < 0.01, and NS indicates not significant.
Analysis of the Electron Transport Pathway From CymA to AY
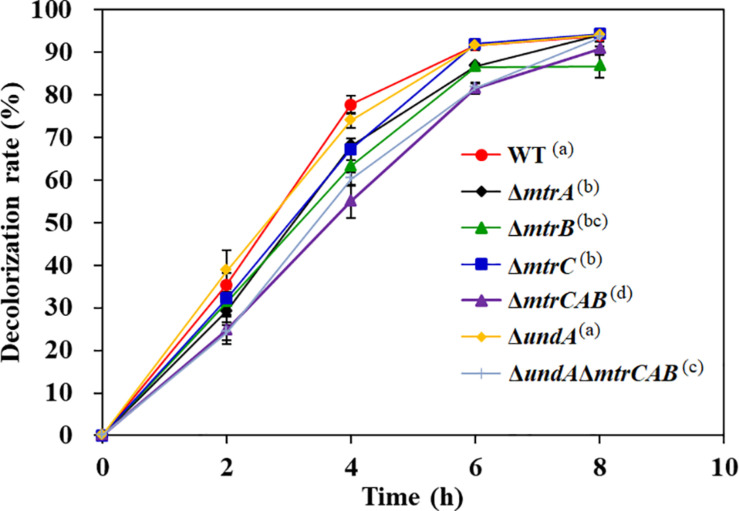
CymA is an electron transfer hub supporting multi-branched respiratory chains (Marritt et al., 2012). Recent studies showed that the Mtr respiratory pathway of S. oneidensis MR-1 not only participates in the dissimilatory reduction of multiple metal ions (Toporek et al., 2019) but also plays a critical role in the decolorization process of a variety of textile dyes (Cai et al., 2012). Genome analysis found that an mtr-like gene cluster exists in the genome of S. putrefaciens CN32, including MtrC, MtrA, MtrB, and UndA, and their amino acid sequences exhibit 53.5, 88.6, 83.4, and 26.6% similarity to MtrC, MtrA, MtrB, and OmcA, respectively, in S. oneidensis MR-1. To determine whether the Mtr respiratory pathway is involved in AY decolorization in S. putrefaciens CN32, the in-frame deletion mutants ΔmtrC, ΔmtrA, ΔmtrB, and ΔundA and the triple mutants ΔmtrCAB and ΔundAΔmtrCAB were constructed. As shown in Figure 7, ΔmtrC, ΔmtrA, ΔmtrB, ΔundA, ΔmtrCAB, and ΔundAΔmtrCAB showed 10.6, 9.8, 14.5, 3.8, 22.8, and 17.6% decreases in the decolorization efficiency of AY at 4 h, respectively, indicating that the Mtr respiratory pathway only partially contributes to the transfer of electron required for the AY decolorization process in S. putrefaciens CN32. This is consistent with the results previously reported (Xiao et al., 2012). At the same time, we also noticed that the blocking of the Mtr pathway could not completely inhibit the AY decolorization efficiency and that the influence of the Mtr pathway on the decolorization of MR-1 was more significant than that of CN32. In S. putrefaciens CN32, the decolorization efficiency of Mtr mutants (except ΔcymA mutant) decreased only about 20% compared with the WT strain; however, in S. oneidensis MR-1, a decrease of more than 60% in decolorization efficiency was reported (Xiao et al., 2012), indicating that MtrA, MtrB, and MtrC only partially contribute to the AY decolorization in S. putrefaciens CN32. This is similar to the latest published result, which showed that MtrA, MtrB, and MtrC did not play a major role in the decolorization of methyl orange by S. putrefaciens CN32 under microaerobic conditions in 96-well plates (Min et al., 2020). Thus, S. putrefaciens CN32 exhibits a more complex electron transfer process than did S. oneidensis MR-1 in the decolorization of azo dyes. Therefore, it is necessary to identify other main electron transfer pathways from the CymA electron transfer hub to azo dye acceptors in future studies.
FIGURE 7.
The decolorization of 200 mg/L AY by the WT and Mtr pathway mutants. Values are means ± SD (n = 3). The decolorization rates of different strains at 4 h were analyzed by ANOVA with the Tukey–Kramer comparison test (p < 0.05).
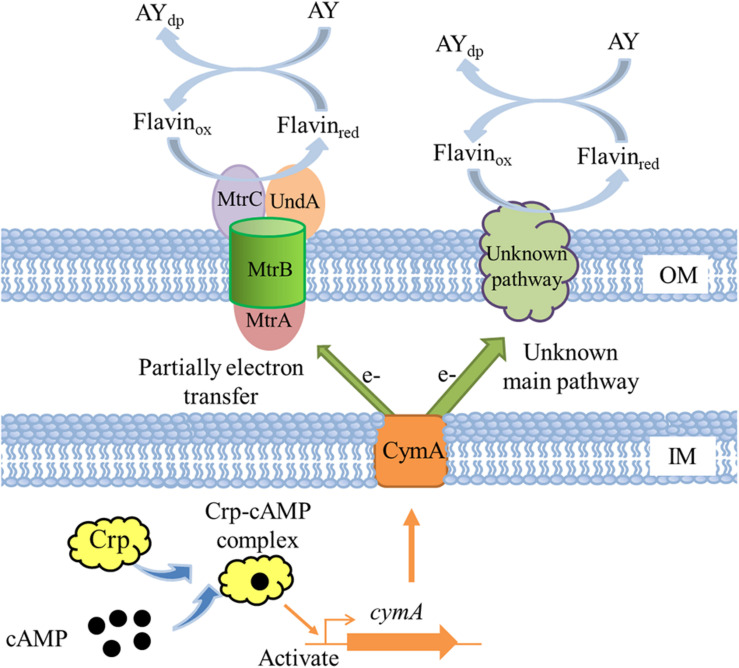
Mechanism of Crp Regulating the AY Decolorization in S. putrefaciens CN32
A molecular mechanism model of Crp regulating AY biodecolorization was proposed in Figure 8. Crp and cAMP form a complex and then directly activate the transcription of cymA; MtrA, MtrB, and MtrC partially contribute to electron transfer from cells to AY acceptors, and other major electron transfer pathways from CymA to dye molecules need to be identified in future studies; flavin acts as a shuttle to accelerate the electron transfer from cells to dye molecules through the switch between the oxidation state and reduction state. AY dye molecules are decomposed into colorless degradation products under the action of electron reduction. Based on the above molecular regulation mechanism, we tried to improve the decolorization ability of S. putrefaciens CN32 by overexpression of cymA in the WT strain (O-cymA mutant). The result showed that the decolorization ability of the O-cymA mutant was slightly improved compared to that of the WT strain. The above results indicate that Crp is necessary to activate the expression of cymA, thereby promoting AY decolorization through accelerating electron transfer from cells to dye molecules.
FIGURE 8.
A proposed molecular model of Crp regulating AY decolorization in S. putrefaciens CN32. OM indicates outer membrane; IM indicates inner membrane; AYdp indicates degradation products of acid yellow.
Conclusion
In this study, AY was selected as an electron acceptor to reveal the molecular mechanism of S. putrefaciens CN32 decolorization of azo dyes. By constructing a transposon mutant library, the cAMP receptor protein Crp was identified as a necessary regulator for AY decolorization in S. putrefaciens CN32. Crp can directly bind to the promoter region of the cymA gene and activate the expression of the cymA gene, thereby supporting AY decolorization. MtrA, MtrB, and MtrC partially contribute to the electron transfer from CymA to AY molecules, and other major electron transfer pathways need to be identified in future studies. Furthermore, the overexpression of cymA could slightly enhance the decolorization efficiency of AY in S. putrefaciens CN32. This study will help us understand the molecular mechanism of azo dye decolorization in other Shewanella strains.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
WL and CL designed the research. WL and YC performed decolorization experiments. YC and XZ operated gene deletion and complementation experiments. JL and JZ carried out protein expression and purification experiments. YC, SW, and DS performed RNA extraction, RT-qPCR and DNase I footprinting assays. WL, CL, and DS wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study is supported by the National Natural Science Foundation of China (31970036, 31900401, and 31800020), Natural Science Foundation of Jiangsu Province (BK20171163 and BK20181009), Natural Science Foundation of Xuzhou City (KC19196), Six Talent Peaks Project of Jiangsu Province (JNHB-103), Qing Lan Project of Jiangsu Province, Postgraduate Research & Practice Innovation Program of Jiangsu Province (2019XKT404), and Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- Battesti A., Majdalani N., Gottesman S. (2011). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65 189–213. 10.1146/annurev-micro-090110-102946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baweja M., Nain L., Kawarabayasi Y., Shukla P. (2016). Current technological improvements in enzymes toward their biotechnological applications. Front. Microbiol. 7:965. 10.3389/fmicb.2016.00965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatson S. A., Whitchurch C. B., Sargent J. L., Levesque R. C., Mattick J. S. (2002). Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacterial 184 3605–3613. 10.1128/jb.184.13.3605-3613.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P. J., Xiao X., He Y. R., Li W. W., Chu J., Wu C., et al. (2012). Anaerobic biodecolorization mechanism of methyl orange by Shewanella oneidensis MR-1. Appl. Microbiol. Biotechnol. 93 1769–1776. 10.1007/s00253-011-3508-8 [DOI] [PubMed] [Google Scholar]
- Dawkar V. V., Jadhav U. U., Tamboli D. P., Govindwar S. P. (2010). Efficient industrial dye decolorization by Bacillus sp. VUS with its enzyme system. Ecotoxicol. Environ. Saf. 73 1696–1703. 10.1016/j.ecoenv.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Deutscher J., Francke C., Postma P. W. (2006). How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70 939–1031. 10.1128/MMBR.00024-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Peng N., Du Y., Ji L., Cao B. (2014). Disruption of putrescine biosynthesis in Shewanella oneidensis enhances biofilm cohesiveness and performance in Cr(VI) immobilization. Appl. Environ. Microbiol. 80 1498–1506. 10.1128/AEM.03461-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76 1648–1652. 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson J. K., Romine M. F., Beliaev A. S., Auchtung J. M., Driscoll M. E., Gardner T. S., et al. (2008). Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6 592–603. 10.1038/nrmicro1947 [DOI] [PubMed] [Google Scholar]
- Gao C., Mulder D., Yin C., Elliot M. A. (2012). Crp is a global regulator of antibiotic production in Streptomyces. mBio 3:e00407-12. 10.1128/mBio.00407-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Yang Z. K., Barua S., Reed S. B., Zhou J. (2009). Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J. 3:966. 10.1038/ismej.2009.40 [DOI] [PubMed] [Google Scholar]
- Guo G., Hao J., Tian F., Liu C., Ding K., Xu J., et al. (2020a). Decolorization and detoxification of azo dye by halo-alkaliphilic bacterial consortium: systematic investigations of performance, pathway and metagenome. Ecotoxicol. Environ. Saf. 204:111073. 10.1016/j.ecoenv.2020.111073 [DOI] [PubMed] [Google Scholar]
- Guo G., Li X., Tian F., Liu T., Yang F., Ding K., et al. (2020b). Azo dye decolorization by a halotolerant consortium under microaerophilic conditions. Chemosphere 244:125510. 10.1016/j.chemosphere.2019.125510 [DOI] [PubMed] [Google Scholar]
- Hameed B. B., Ismail Z. Z. (2018). Decolorization, biodegradation and detoxification of reactive red azo dye using non-adapted immobilized mixed cells. Biochem. Eng. J. 137 71–77. 10.1016/j.bej.2018.05.018 [DOI] [Google Scholar]
- Imran M., Arshad M., Negm F., Khalid A., Shaharoona B., Hussain S., et al. (2016). Yeast extract promotes decolorization of azo dyes by stimulating azoreductase activity in Shewanella sp. strain IFN4. Ecotoxicol. Environ. Saf. 124 42–49. 10.1016/j.ecoenv.2015.09.041 [DOI] [PubMed] [Google Scholar]
- Kalia D., Merey G., Nakayama S., Zheng Y., Zhou J., Luo Y., et al. (2013). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP,(p) ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42 305–341. 10.1039/c2cs35206k [DOI] [PubMed] [Google Scholar]
- Kasai T., Kouzuma A., Nojiri H., Watanabe K. (2015). Transcriptional mechanisms for differential expression of outer membrane cytochrome genes omcA and mtrC in Shewanella oneidensis MR-1. BMC Microbiol. 15:68. 10.1186/s12866-015-0406-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F., Ren H.-Y., Pavlostathis S. G., Wang A., Nan J., Ren N.-Q. (2018). Enhanced azo dye decolorization and microbial community analysis in a stacked bioelectrochemical system. Chem. Eng. J. 354 351–362. 10.1016/j.cej.2018.08.027 [DOI] [Google Scholar]
- Larsen R. A., Wilson M. M., Guss A. M., Metcalf W. W. (2002). Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178 193–201. 10.1007/s00203-002-0442-2 [DOI] [PubMed] [Google Scholar]
- Li Q., Feng X., Lu X., Li T., Han X., Xiao X., et al. (2018). Combined intra-and extracellular reduction involved in the anaerobic biodecolorization of cationic azo dye by Shewanella oneidensis MR-1. Chemosphere 211 701–708. 10.1016/j.chemosphere.2018.08.006 [DOI] [PubMed] [Google Scholar]
- Liu C., Yang J., Liu L., Li B., Liu W. (2017a). Sodium lactate negatively regulates biofilm formation of Shewanella putrefaciens CN32 via a three-component regulatory system (LrbS-LrBA-LrbR). Appl. Environ. Microbiol. 83:AEM.00712-17. 10.1128/AEM.00712-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., You Y., Zhao R., Sun D., Zhang P., Jiang J., et al. (2017b). Biosurfactant production from Pseudomonas taiwanensis L1011 and its application in accelerating the chemical and biological decolorization of azo dyes. Ecotoxicol. Environ. Saf. 145 8–15. 10.1016/j.ecoenv.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Liu G., Zhou J., Wang J., Wang X., Jin R., Lv H. (2011). Decolorization of azo dyes by Shewanella oneidensis MR-1 in the presence of humic acids. Appl. Microbiol. Biotechnol. 91 417–424. 10.1007/s00253-011-3273-8 [DOI] [PubMed] [Google Scholar]
- Liu W., Liu C., Liu L., You Y., Jiang J., Zhou Z., et al. (2017). Simultaneous decolorization of sulfonated azo dyes and reduction of hexavalent chromium under high salt condition by a newly isolated salt-tolerant strain Bacillus circulans BWL1061. Ecotoxicol. Environ. Saf. 141 9–16. 10.1016/j.ecoenv.2017.03.005 [DOI] [PubMed] [Google Scholar]
- Liu W., Liu L., Liu C., Hao Y., Yang H., Yuan B., et al. (2016). Methylene blue enhances the anaerobic decolorization and detoxication of azo dye by Shewanella onediensis MR-1. Biochem. Eng. J. 110 115–124. 10.1016/j.bej.2016.02.012 [DOI] [Google Scholar]
- Liu W., You Y., Sun D., Wang S., Zhu J., Liu C. (2018). Decolorization and detoxification of water-insoluble Sudan dye by Shewanella putrefaciens CN32 co-cultured with Bacillus circulans BWL1061. Ecotoxicol. Environ. Saf. 166 11–17. 10.1016/j.ecoenv.2018.09.055 [DOI] [PubMed] [Google Scholar]
- Liu W. J., Yuan H. L., Yang J. S., Li B. Z. (2009). Characterization of bioflocculants from biologically aerated filter backwashed sludge and its application in dying wastewater treatment. Bioresour. Technol. 100 2629–2632. 10.1016/j.biortech.2008.12.017 [DOI] [PubMed] [Google Scholar]
- Marritt S. J., Lowe T. G., Bye J., McMillan D. G., Shi L., Fredrickson J., et al. (2012). A functional description of CymA, an electron-transfer hub supporting anaerobic respiratory flexibility in Shewanella. Biochem. J. 444 465–474. 10.1042/BJ20120197 [DOI] [PubMed] [Google Scholar]
- Marsili E., Baron D. B., Shikhare I. D., Coursolle D., Gralnick J. A., Bond D. R. (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105 3968–3973. 10.1073/pnas.0710525105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Liu G., Zhou J., Fu Q. S. (2014). Effects of redox mediators on azo dye decolorization by Shewanella algae under saline conditions. Bioresour. Technol. 151 63–68. 10.1016/j.biortech.2013.09.131 [DOI] [PubMed] [Google Scholar]
- Meng X., Liu G., Zhou J., Fu Q. S., Wang G. (2012). Azo dye decolorization by Shewanella aquimarina under saline conditions. Bioresour. Technol. 114 95–101. 10.1016/j.biortech.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Min D., Cheng L., Liu D.-F., Li W.-W., Yu H.-Q. (2020). Electron transfer via the non-Mtr respiratory pathway from Shewanella putrefaciens CN-32 for methyl orange bioreduction. Process Biochem. 95 108–114. 10.1016/j.procbio.2020.05.015 [DOI] [Google Scholar]
- Pandey A., Singh P., Iyengar L. (2007). Bacterial decolorization and degradation of azo dyes. Int. Biodeter. Biodegr. 59 73–84. 10.1016/j.ibiod.2006.08.006 [DOI] [Google Scholar]
- Persat A., Inclan Y. F., Engel J. N., Stone H. A., Gitai Z. (2015). Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112 7563–7568. 10.1073/pnas.1502025112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samir S., Al-Tohamy R., Kenawy E. R., Sun J. (2020). Performance of a newly isolated salt-tolerant yeast strain Sterigmatomyces halophilus SSA-1575 for azo dye decolorization and detoxification. Front. Microbiol. 11:1163. 10.3389/fmicb.2020.01163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saratale R. G., Saratale G. D., Chang J.-S., Govindwar S. P. (2011). Bacterial decolorization and degradation of azo dyes: a review. J. Taiwan Inst. Chem. E. 42 138–157. 10.1016/j.jtice.2010.06.006 [DOI] [Google Scholar]
- Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145 69–73. 10.1016/0378-1119(94)90324-7 [DOI] [PubMed] [Google Scholar]
- Schwalb C., Chapman S. K., Reid G. A. (2003). The tetraheme cytochrome CymA is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis. Biochemistry 42 9491–9497. 10.1021/bi034456f [DOI] [PubMed] [Google Scholar]
- Sun D., Zhu J., Chen Z., Li J., Wen Y. (2016). SAV742, a novel AraC-family regulator from Streptomyces avermitilis, controls avermectin biosynthesis, cell growth and development. Sci. Rep. 6:36915. 10.1038/srep36915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toporek Y. J., Mok J. K., Shin H. D., Lee B. D., Lee M. H., DiChristina T. J. (2019). Metal reduction and protein secretion genes required for iodate reduction by Shewanella oneidensis. Appl. Environ. Microbiol. 85:e02115-18. 10.1128/AEM.02115-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang Y., Ning S., Shi S., Tan L. (2020). Improving azo dye decolorization performance and halotolerance of Pichia occidentalis A2 by static magnetic field and possible mechanisms through comparative transcriptome analysis. Front. Microbiol. 11:712 10.3389/fmicb.2020.00712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Xu C.-C., Wu Y.-M., Cai P.-J., Li W.-W., Du D.-L., et al. (2012). Biodecolorization of Naphthol Green B dye by Shewanella oneidensis MR-1 under anaerobic conditions. Bioresour. Technol. 110 86–90. 10.1016/j.biortech.2012.01.099 [DOI] [PubMed] [Google Scholar]
- Xu M., Guo J., Sun G. (2007). Biodegradation of textile azo dye by Shewanella decolorationis S12 under microaerophilic conditions. Appl. Microbiol. Biotechnol. 76 719–726. 10.1007/s00253-007-1032-7 [DOI] [PubMed] [Google Scholar]
- Yan B., Du C., Xu M., Liao W. (2012). Decolorization of azo dyes by a salt-tolerant Staphylococcus cohnii strain isolated from textile wastewater. Front. Env. Sci. Eng. 6:806–814. [Google Scholar]
- Yang Q., Zhang M., Zhang M., Wang C., Liu Y., Fan X., et al. (2018). Characterization of a novel, cold-adapted, and thermostable laccase-like enzyme with high tolerance for organic solvents and salt and potent dye decolorization ability, derived from a marine metagenomic library. Front. Microbiol. 9:2998. 10.3389/fmicb.2018.02998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wu Y., Zhang Y., Yang E., Yan J. (2020). A thermo-active laccase isoenzyme from Trametes trogii and its potential for dye decolorization at high temperature. Front. Microbiol. 11:241. 10.3389/fmicb.2020.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesilada O., Asma D., Cing S. (2003). Decolorization of textile dyes by fungal pellets. Process Biochem. 38 933–938. 10.1016/S0032-9592(02)00197-8 [DOI] [Google Scholar]
- Zahir H., Naidoo M., Kostadinova R.-M., Ortiz K. A., Sun-Kou R., Navarro A. E. (2014). Decolorization of hair dye by lignocellulosic waste materials from contaminated waters. Front. Environ. Sci. 2:28 10.3389/fenvs.2014.00028 [DOI] [Google Scholar]
- Zhang F., Guo X., Qian D.-K., Sun T., Zhang W., Dai K., et al. (2019). Decolorization of Acid Orange 7 by extreme-thermophilic mixed culture. Bioresour. Technol. 291:121875. 10.1016/j.biortech.2019.121875 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Lu H., Wang J., Zhou J., Leng X., Liu G. (2018). Catalytic performance of quinone and graphene-modified polyurethane foam on the decolorization of azo dye Acid Red 18 by Shewanella sp. RQs-106. J. Hazard. Mater. 356 82–90. 10.1016/j.jhazmat.2018.05.043 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.