Abstract
Dendritic cells (DCs) are pivotal stimulators of T cell responses. They provide essential signals (epitope presentation, proinflammatory cytokines, co-stimulation) to T cells and prime adaptive immunity. Therefore, they are paramount to immunization strategies geared to generate T cell immunity. The inflammatory signals DCs respond to, classically occur in the context of acute virus infection. Yet, enlisting viruses for engaging DCs is hampered by their penchant for targeting DCs with sophisticated immune evasive and suppressive ploys. In this review, we discuss our work on devising vectors based on a recombinant polio:rhinovirus chimera for effectively targeting and engaging DCs. We are juxtaposing this approach with commonly used, recently studied dsDNA virus vector platforms.
Introduction
DCs were initially identified after isolation from mouse spleen [1,2]. A role as professional antigen presenting cells emerged after DCs were found to express MHC II and to act as potent stimulators of lymphocytes [3]. While monocytes, macrophages and B cells also are capable of presenting antigens, DCs are estimated to be at least 100-times more efficient at stimulating leukocytes [3]. Therefore, DCs have since been established as the ‘sentinel of the immune system’.
DCs detect invading pathogens through sensors known as Pathogen Recognition Receptors (PRR) and they capture, process and present antigens (both endogenous and exogenous) to T cells. Antigen presentation in combination with cytokine release and co-stimulatory signals from DCs educate the generation of T cell responses. Depending on the cytokine release pattern and the co-stimulatory range, DCs can prime Th1-, Th2-, Th17-, cytotoxic CD8 T cell-, or tolerogenic responses such as regulatory T cells (Tregs) [4]. Antigen presentation in the absence of appropriate co-stimulation leads to tolerance and immune escape [5]. Because of the central role of DCs in priming of immune responses, a wide range of vaccination strategies have sought to target antigens and proinflammatory stimulation to DCs.
Viral vectors are particularly pertinent for this, because many viruses naturally infect DCs, enabling antigen delivery to DCs, and because viruses naturally provide the range of PRR engagement and proinflammatory stimuli DCs have evolved to respond to. In this review, we discuss fundamental obstacles of effectively targeting DCs with viral vectors and our approach to overcome them.
Viruses target DCs for infection and immune suppression
Given their central role in innate and adaptive antiviral immunity, many human pathogenic viruses have evolved to target host DCs for infection. Ostensibly, infecting DCs offers two advantages to viruses: 1. evade and suppress host antiviral immunity (see below); 2. exploit DC trafficking as a dissemination vehicle [6]. Natural DC targeting is prominent with all viruses commonly harnessed for therapeutic purposes, either as vectors or immunostimulatory agents in cancer immunotherapy. DCs sense viruses via a range of PRRs that detect pathogen signatures revealed during endocytosis or upon appearance of viral genomic material in the host cell interior, unleashing the innate antiviral response. This comprises PRR signaling to innate kinase assemblies TBK1 and IKKα:β; activation of IRF3 and NFκB transcriptional programs; proinflammatory cytokine induction; autocrine and paracrine cytokine signaling; induction of costimulatory molecules.
The innate antiviral response, e.g. in infected DCs, can provide the appropriate costimulatory context for generating T cell immunity. However, human pathogenic viruses have been shown to intercept this process at every conceivable step, ranging from mild immunomodulatory diversion to outright DC killing [7].
Viruses evade or antagonize innate and adaptive immunity
For most members of the human herpesvirus family elaborate immune evasive and suppressive relationships with DCs have been reported, exemplified by cytomegalovirus (CMV) [8] and herpes simplex virus 1 (HSV1) [9]. They evade detection by the host innate antiviral system, e.g. the cytoplasmic vDNA sensor cGAS [10-17] amongst other PRRs [16], and actively intercept signaling through the TBK1/IKKα:β innate kinase assemblies and downstream IRF3/NFκB transcriptional networks [18-21]. Human orthopoxviruses, exemplified by vaccinia virus (VACV), also target DCs [22] and evade host innate sensing [23,24]. This is accompanied by multiple viral strategies to intercept downstream innate antiviral signaling, e.g. blocking of TBK1 [25] and NFκB [26] through actions of the C6 and K1 viral gene products, respectively [26]. Also, the VACV proteins N1 and E3 were shown to inhibit type-I IFN production upon DC infection [27]. Orthopoxviruses express a vast array of viral proteins implicated in host immune evasion and suppression [28]; thus, multiple other viral gene products (A46, A52, B15, K7) are known to interfere with innate immunity (reviewed in [29]). Adenoviruses evade innate sensing through the E1A oncogene product that blocks the cGAS-STING pathway [30], and inhibit IFN signaling by blocking histone ubiquitination [31], and via Virus-Associated (VA) RNA that inhibits the dsRNA-dependent protein kinase (PKR) [32].
Complimenting their elaborate strategies of evading and suppressing innate antiviral responses in infected DCs, herpesviruses (e.g. CMV [33] and HSV1 [34]), orthopoxviruses, e.g. VACV [35], and adenoviruses [32] encode arrays of immunomodulatory proteins that interfere with virtually every step of antigen processing and presentation [36], including: (i) inhibition of the immunoproteasome/antigen processing [37]; (ii) interference with the functions of the transporter associated with antigen processing (TAP) [38-40] or tapasin [41] in antigen loading; (iii) disruption of MHC I intracellular transport and interference with MHC I surface presentation [42]; (iv) downregulation of MHC I/II expression [43] or sequestration of β2 microglobulin [44]; (v) active MHC I degradation [45]. Moreover, these viruses also suppress the upregulation of costimulatory molecules and production of pro-inflammatory cytokines in DCs, e.g. blocked CD40, CD80, CD86 induction and active suppression of T/B cell activation by CMV [46-48], or absent CD40, CD86, CD83 and CCR7/CXCR4 induction [49] combined with a paucity of pro-inflammatory cytokines (IL-12, TNF-α, IL-6) resulting in poor T cell stimulation by DC infection with HSV1 [50]. Similarly, VACV actively suppresses the induction of maturation markers [22] and proinflammatory cytokine responses, e.g. type-I IFN [51], type-II IFN [52], TNF-α [53] and IL-1β [54] in infected DCs.
In contrast to human pathogenic dsDNA viruses with their sizeable genomes encoding for arrays of complimentary immunomodulatory factors, the extreme genetic austerity of enteroviruses (EV) [55] does not permit elaborate immune evasion and suppression strategies. Yet, even EVs have the capacity to interfere with host innate immune activation and DC function. They accomplish this through simple but effective means of overwhelming their host cells without a need for specialized immunomodulatory proteins.
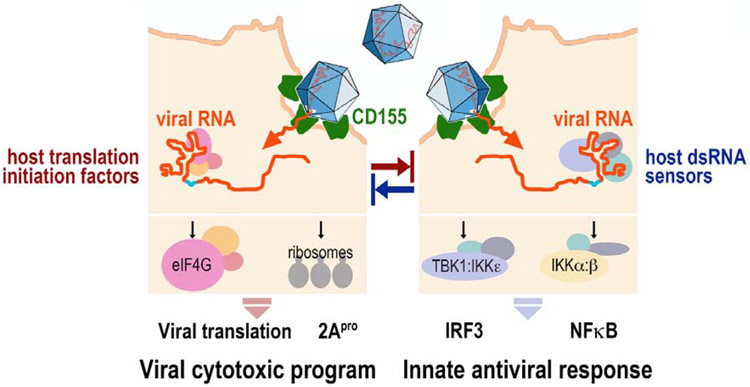
The flagship EV, poliovirus (PV), infects DCs/antigen-presenting cells in vivo [56] and destroys them effectively [57]. EVs target viral translation and replication to a privileged site at the endoplasmic reticulum [58,59], providing relative shelter from PKR-mediated eIF2α (S51) phosphorylation and, possibly, other innate antiviral events [59]. Viral m7G (‘cap’)-independent translation via internal ribosomal entry sites (IRESs) provides a highly efficient means for immediate early translation of incoming viral genomic RNA [60,61]. The first viral protein released, via co-translational autocatalytic processing [62], is the viral 2Apro protease (Fig. 1). Autocatalytic release of 2Apro not only provides the essential cleavage of the P1 and P2 precursors for processing of the viral polyprotein [63], but also is the virus’ main agent for subverting the host cell innate antiviral program [64]. A proteolytic program directed by 2Apro degrade the central scaffold of the translation initiation apparatus, eukaryotic initiation factor (eIF) 4G [65], and the nuclear pore complex [66], thereby broadly suppressing gene expression in infected host cells. Simultaneously, the ‘viral cytotoxic program’, dependent on immediate early viral translation of 2Apro, is antagonized by detection of vRNA signatures in the host cell interior triggering an early innate host response (Fig. 1).
Figure 1.
Enterovirus 5’ m7G-cap independent translation via its IRES drives viral translation and a cytotoxic program unleashed by immediate early release of 2Apro. Simultaneously, vRNA signatures are detected by host cell dsRNA sensors that initiate the host innate antiviral response. The outcome of the infection is determined by the balance of viral, IRES-mediated translation vs. host sensing of incoming vRNA templates.
Viral vectors and the problem of intrinsic immune evasion and suppression
Given the prevalence of viral immune evasion and suppression strategies targeting DCs, what is the status of devising viral immunization vectors—based on diverse families of viruses—to overcome them? Vectors delivering simian immunodeficiency virus (SIV) signatures based on Rhesus macaque CMV (RhCMV), attenuated through fibroblast adaptation analogous to the TOWNE and AD169 live attenuated CMV strains [67], yielded atypical antigen presentation with unconventional, Class II/MHC-E restricted CD8 T cell responses against non-canonical epitopes [68]. RhCMV vector vaccination achieved outstanding results in Rhesus SIV challenge models [69]. However, clinical studies with human fibroblast-adapted (hybrid TOWNE) CMV strains revealed conventional CD8 T cell responses in humans, suggesting that Rhesus species-specific features are the likely cause for the unconventional T cell response to RhCMV vector immunization [70].
Poxvirus vector platforms include VACV [71], MVA (Modified Vaccinia Ankara) or avipox-based agents such as ALVAC (Canarypox) [72], which are replication-incompetent in mammalian cells. MVA, a result of serial passage of VACV (Ankara) in avian cells [73], carries deletions with some immunomodulatory gene loss [74]. Yet, recent studies with inactivated MVA as an intratumor immune stimulatory agent in mouse tumor models yielded better antitumor responses and pro-inflammatory cytokine production than live MVA [75], suggesting that MVA immune evasion/suppression limits the effectiveness of viral adjuvancy. In fact, MVA retains expression of the soluble IL-1β decoy receptor [54] and the viral C6 inhibitor of IRF3/7 phosphorylation [76]. Deletion of these genes from MVA led to enhanced activity as a T cell adjuvant in mice [76,77]. Immunization with an ALVAC-based vector plus recombinant HIV gp120 subunit boost yielded promising early results [78], but failed in a subsequent, pivotal trial (https://niaid.nih.gov/newsevents/experimental-hiv-vaccine-regimen-ineffective-preventing-hiv).
New prototype Adenovirus vectors based on chimpanzee Ad (ChAd) strains, to circumvent the problem of human preexisting anti-Ad neutralizing immunity, demonstrated promising results with the generation of humoral and T cell responses against Ebola [79,80]. Registration trials of this approach are ongoing [81]. ChAd3 is replication-defective with viral E1/E3 gene deletions; E1 and E3 Ad gene products have immune modulatory properties, but even E1/E3-deleted Ad vectors inhibit T cell proliferation due to vector targeting of DCs [82].
Thus, balancing viral replication-dependent epitope expression with virus-mediated immune evasion is likely critical towards developing efficacious viral vector platforms.
We recently designed a recombinant PV viral vector prototype with a peculiar, unexpected potential to engage DCs [83]. This approach has the disadvantage of coding capacity restrictions due to the small (~7.4kB) size of the PV genome, and the inherent genetic instability of (+)ssRNA viruses [84]. However, it has the advantage of robust DC co-stimulation, in the absence of apparent viral immune suppressive or evasive strategies [83] (Fig. 2). Our strategy is based on the highly attenuated rhino:poliovirus chimera PVSRIPO, the type 1 live attenuated poliovirus (SABIN) vaccine replicating under control of a heterologous human rhinovirus type 2 (HRV2) IRES [85]. PVSRIPO, in non-vector form [86], is being employed as an effective cancer immunotherapy intervention that has safely been used by high-dose intracerebral inoculation in >150 patients with recurrent glioblastoma [87], including children. PVSRIPO has lower basal virulence compared to its type 1 SABIN vaccine precursor due to a—naturally evolved—lower propensity of the HRV2 IRES to recruit host 40S ribosomal subunits [88]. This phenotype is engrained in the structure of the HRV2 IRES stem loop domains V and VI, which harbor the footprint of the eIF4G:4A:4B translation initiation helicase on all EV IRESs [85,89-96].
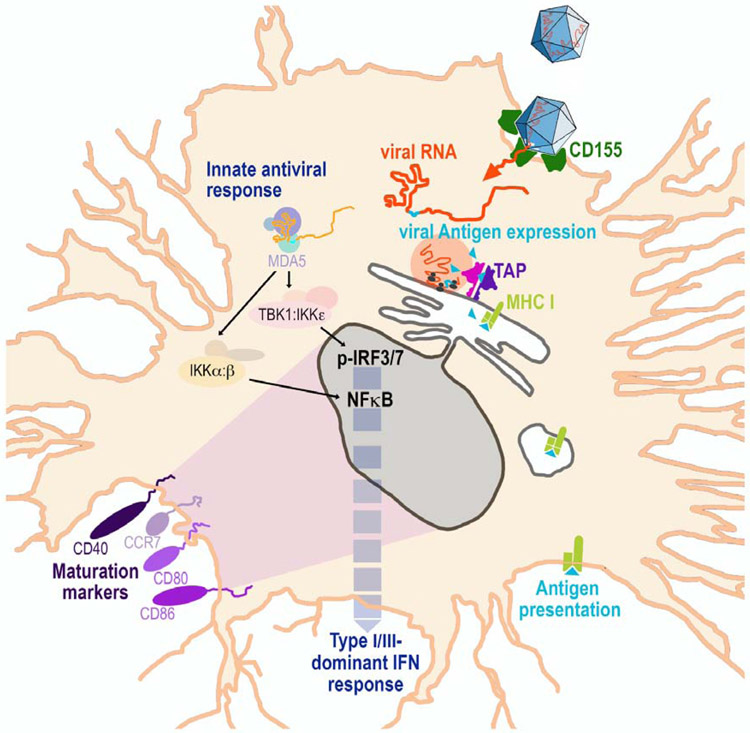
Figure 2.
Illustration of PVSRIPO’s peculiar host relationship with DCs (see [83] for context). Polioviruses naturally target DCs/macrophages via CD155. PVSRIPO-based vectors express encoded antigens internally, for presentation via the classical pathway. Marginal, ongoing viral propagation provides a steady supply of antigen, but subdued viral translation is insufficient to damage the host. Potent, sustained type-I/III IFN-dominant inflammation induces maturation markers and profuse cytokine release.
There are 3 known sites of productive poliovirus propagation in humans: an unknown population of cells within the enteric tract epithelium [97], spinal cord anterior horn motor neurons [98,99], and CD11c+ DCs/macrophages [56]. PVSRIPO’s phenotype in the enteric tract is unknown. The finding that PVSRIPO’s growth potential in primary explant human renal epithelial cells is lower than type 1 poliovirus (SABIN) [88] suggests a similar scenario for enteric epithelial cells. PVSRIPO has profound neuron-specific incompetence [88], due to a ribonucleoprotein complex forming at the foreign HRV2 IRES in neuronal cells that intercepts eIF4G binding/40S subunit recruitment [100,101]. Similarly, in contrast to wild type PV or the SABIN vaccines [57], PVSRIPO infection is non-cytopathogenic in DCs [102]. Rather, viral translation is sluggish and transitory, and 2Apro mediated cleavages of host proteins, e.g. eIF4G, do not occur [83,102]. Yet, low-level viral propagation in infected DCs is surprisingly durable, with lingering replication ongoing for several days [83]. Sublethal PVSRIPO vector infection of DCs produces a constant source of vector-encoded antigen expressed in DCs that is presented via the classical pathway [83].
We hypothesize that PVSRIPO’s intrinsically lowered capacity for recruiting host cell translation machinery tips the balance in favor of the host innate antiviral response (Fig. 1). This is the reason for a potent, sustained innate inflammatory response with type-I/III IFN-dominant inflammation in PVSRIPO-infected DCs, accompanied by upregulation of co-stimulatory molecules CD40, CD80, CD86 and CCR7 [83,102] (Fig. 2). Due to HRV2 IRES recombination, delaying immediate early translation and autocatalytic release of 2Apro, PVSRIPO lacks any capacity to intercept or suppress innate antiviral immunity in infected DCs, [103]. We have devised a sophisticated strategy to derive genetically stable vector prototypes through recombination with foreign inserts in the viral 5’ untranslated region [83].
Conclusions
The main attraction of viral vectors devised for immunization purposes is their capacity to provide the natural, evolved costimulatory signals needed to generate effective T cell immunity. This valid objective has to contend with the elaborate, pernicious strategies of viruses to evade and suppress host innate and adaptive immunity, in large part achieved by targeting DCs directly. In this review, we discuss our strategy of harnessing recombinant PV for vector use, based on a peculiar host relationship in human DCs providing for a proinflammatory pattern with ideal costimulatory properties (Fig. 2).
Highlights.
Dendritic cells are central protagonists in viral vector immunization approaches
Many viruses naturally target dendritic cells for immune suppression
Viral vector-mediated immune suppression intercepts innate and adaptive immunity
Recombinant poliovirus unfolds peculiar non-cytopathogenic relations with dendritic cells
These result in profound dendritic cell engagement for co-stimulation of T cell immunity
Acknowledgments
This research was supported by Public Health Service Grant R01 NS108773 (M.G.); a Defeat DIPG Research Grant, a Research Grant from the V Foundation, a Kirschstein National Research Service Award F32CA224593 (M.C.B.) and the National Cancer Center Breast Cancer Fellowship (M.C.B).
Footnotes
Summary Declaration of Interest Statement. M.G. and M.C.B. are inventors of intellectual property related to the discussed research that was licensed to Istari Oncology, Inc. M.G. holds equity in-, and is an advisor and compensated consultant of Istari Oncology, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steinman RM, Cohn ZA: Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med 1973, 137:1142–1162.** Identification of dendritic cells (also see 2.).
- 2.Steinman RM, Lustig DS, Cohn ZA: Identification of a novel cell type in peripheral lymphoid organs of mice. 3. Functional properties in vivo. J Exp Med 1974, 139:1431–1445.** Identification of dendritic cells (also see 1.).
- 3.Steinman RM, Witmer MD: Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. Proc Natl Acad Sci U S A 1978, 75:5132–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh KP, Mills KH: Dendritic cells and other innate determinants of T helper cell polarisation. Trends Immunol 2013, 34:521–530. [DOI] [PubMed] [Google Scholar]
- 5.Bonifaz L, Bonnyay D, Mahnke K, Rivera M, Nussenzweig MC, Steinman RM: Efficient targeting of protein antigen to the dendritic cell receptor DEC-205 in the steady state leads to antigen presentation on major histocompatibility complex class I products and peripheral CD8+ T cell tolerance. J Exp Med 2002, 196:1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu L, KewalRamani VN: Dendritic-cell interactions with HIV: infection and viral dissemination. Nat Rev Immunol 2006, 6:859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen TH, Bouvier M: MHC class I antigen presentation: learning from viral evasion strategies. Nat Rev Immunol 2009, 9:503–513. [DOI] [PubMed] [Google Scholar]
- 8.Sinclair J, Reeves M: The intimate relationship between human cytomegalovirus and the dendritic cell lineage. Front Microbiol 2014, 5:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobelt D, Lechmann M, Steinkasserer A: The interaction between dendritic cells and herpes simplex virus-1. Curr Top Microbiol Immunol 2003, 276:145–161. [DOI] [PubMed] [Google Scholar]
- 10.Choi HJ, Park A, Kang S, Lee E, Lee TA, Ra EA, Lee J, Lee S, Park B: Human cytomegalovirus-encoded US9 targets MAVS and STING signaling to evade type I interferon immune responses. Nat Commun 2018, 9:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen MH, Jensen SB, Miettinen JJ, Luecke S, Prabakaran T, Reinert LS, Mettenleiter T, Chen ZJ, Knipe DM, Sandri-Goldin RM, et al. : HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J 2016, 35:1385–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu YZ, Su S, Gao YQ, Wang PP, Huang ZF, Hu MM, Luo WW, Li S, Luo MH, Wang YY, et al. : Human Cytomegalovirus Tegument Protein UL82 Inhibits STING-Mediated Signaling to Evade Antiviral Immunity. Cell Host Microbe 2017, 21:231–243. [DOI] [PubMed] [Google Scholar]
- 13.Huang ZF, Zou HM, Liao BW, Zhang HY, Yang Y, Fu YZ, Wang SY, Luo MH, Wang YY: Human Cytomegalovirus Protein UL31 Inhibits DNA Sensing of cGAS to Mediate Immune Evasion. Cell Host Microbe 2018, 24:69–80 e64. [DOI] [PubMed] [Google Scholar]
- 14.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B: Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A 2015, 112:E4306–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su C, Zheng C: Herpes Simplex Virus 1 Abrogates the cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway via Its Virion Host Shutoff Protein, UL41. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tognarelli EI, Palomino TF, Corrales N, Bueno SM, Kalergis AM, Gonzalez PA: Herpes Simplex Virus Evasion of Early Host Antiviral Responses. Front Cell Infect Microbiol 2019, 9:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Zhao J, Xu S, Li J, He S, Zeng Y, Xie L, Xie N, Liu T, Lee K, et al. : Species-Specific Deamidation of cGAS by Herpes Simplex Virus UL37 Protein Facilitates Viral Replication. Cell Host Microbe 2018, 24:234–248 e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu YZ, Su S, Zou HM, Guo Y, Wang SY, Li S, Luo MH, Wang YY: Human Cytomegalovirus DNA Polymerase Subunit UL44 Antagonizes Antiviral Immune Responses by Suppressing IRF3- and NF-kappaB-Mediated Transcription. J Virol 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL: The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J Virol 2004, 78:1675–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Wang K, Lin R, Zheng C: Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol 2013, 87:12814–12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Liu Y, Wang P, Guan X, He S, Luo S, Li C, Hu K, Jin W, Du T, et al. : HSV-2 immediate-early protein US1 inhibits IFN-beta production by suppressing association of IRF-3 with IFN-beta promoter. J Immunol 2015, 194:3102–3115. [DOI] [PubMed] [Google Scholar]
- 22.Engelmayer J, Larsson M, Subklewe M, Chahroudi A, Cox WI, Steinman RM, Bhardwaj N: Vaccinia virus inhibits the maturation of human dendritic cells: a novel mechanism of immune evasion. J Immunol 1999, 163:6762–6768. [PubMed] [Google Scholar]
- 23.Georgana I, Sumner RP, Towers GJ, Maluquer de Motes C: Virulent Poxviruses Inhibit DNA Sensing by Preventing STING Activation. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phelan T, Little MA, Brady G: Targeting of the cGAS-STING system by DNA viruses. Biochem Pharmacol 2020, 174:113831. [DOI] [PubMed] [Google Scholar]
- 25.Unterholzner L, Sumner RP, Baran M, Ren H, Mansur DS, Bourke NM, Randow F, Smith GL, Bowie AG: Vaccinia virus protein C6 is a virulence factor that binds TBK-1 adaptor proteins and inhibits activation of IRF3 and IRF7. PLoS Pathog 2011, 7:e1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shisler JL, Jin XL: The vaccinia virus K1L gene product inhibits host NF-kappaB activation by preventing IkappaBalpha degradation. J Virol 2004, 78:3553–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, Joyce JA, Li XD, Chen Z, Merghoub T, et al. : Modified vaccinia virus Ankara triggers type I IFN production in murine conventional dendritic cells via a cGAS/STING-mediated cytosolic DNA-sensing pathway. PLoS Pathog 2014, 10:e1003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seet BT, Johnston JB, Brunetti CR, Barrett JW, Everett H, Cameron C, Sypula J, Nazarian SH, Lucas A, McFadden G: Poxviruses and immune evasion. Annu Rev Immunol 2003, 21:377–423. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez JM, Esteban M: A poxvirus Bcl-2-like gene family involved in regulation of host immune response: sequence similarity and evolutionary history. Virol J 2010, 7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau L, Gray EE, Brunette RL, Stetson DB: DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015, 350:568–571. [DOI] [PubMed] [Google Scholar]
- 31.Fonseca GJ, Thillainadesan G, Yousef AF, Ablack JN, Mossman KL, Torchia J, Mymryk JS: Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe 2012, 11:597–606. [DOI] [PubMed] [Google Scholar]
- 32.Mahr JA, Gooding LR: Immune evasion by adenoviruses. Immunol Rev 1999, 168:121–130. [DOI] [PubMed] [Google Scholar]
- 33.Powers C, DeFilippis V, Malouli D, Fruh K: Cytomegalovirus immune evasion. Curr Top Microbiol Immunol 2008, 325:333–359. [DOI] [PubMed] [Google Scholar]
- 34.Wiertz EJ, Devlin R, Collins HL, Ressing ME: Herpesvirus interference with major histocompatibility complex class II-restricted T-cell activation. J Virol 2007, 81:4389–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li P, Wang N, Zhou D, Yee CS, Chang CH, Brutkiewicz RR, Blum JS: Disruption of MHC class II-restricted antigen presentation by vaccinia virus. J Immunol 2005, 175:6481–6488. [DOI] [PubMed] [Google Scholar]
- 36.Alcami A, Koszinowski UH: Viral mechanisms of immune evasion. Trends Microbiol 2000, 8:410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG: Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 1995, 375:685–688. [DOI] [PubMed] [Google Scholar]
- 38.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D: Herpes simplex virus turns off the TAP to evade host immunity. Nature 1995, 375:411–415. [DOI] [PubMed] [Google Scholar]
- 39.Roder G, Geironson L, Bressendorff I, Paulsson K: Viral proteins interfering with antigen presentation target the major histocompatibility complex class I peptide-loading complex. J Virol 2008, 82:8246–8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verweij MC, Horst D, Griffin BD, Luteijn RD, Davison AJ, Ressing ME, Wiertz EJ: Viral inhibition of the transporter associated with antigen processing (TAP): a striking example of functional convergent evolution. PLoS Pathog 2015, 11:e1004743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park B, Kim Y, Shin J, Lee S, Cho K, Fruh K, Lee S, Ahn K: Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity 2004, 20:71–85. [DOI] [PubMed] [Google Scholar]
- 42.Petersen JL, Morris CR, Solheim JC: Virus evasion of MHC class I molecule presentation. J Immunol 2003, 171:4473–4478. [DOI] [PubMed] [Google Scholar]
- 43.York IA, Roop C, Andrews DW, Riddell SR, Graham FL, Johnson DC: A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell 1994, 77:525–535. [DOI] [PubMed] [Google Scholar]
- 44.Browne H, Smith G, Beck S, Minson T: A complex between the MHC class I homologue encoded by human cytomegalovirus and beta 2 microglobulin. Nature 1990, 347:770–772. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson PG, Boname JM, de Lima B, Efstathiou S: A battle for survival: immune control and immune evasion in murine gamma-herpesvirus-68 infection. Microbes Infect 2002, 4:1177–1182. [DOI] [PubMed] [Google Scholar]
- 46.Bitra A, Nemcovicova I, Picarda G, Doukov T, Wang J, Benedict CA, Zajonc DM: Structure of human cytomegalovirus UL144, an HVEM orthologue, bound to the B and T cell lymphocyte attenuator. J Biol Chem 2019, 294:10519–10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moutaftsi M, Mehl AM, Borysiewicz LK, Tabi Z: Human cytomegalovirus inhibits maturation and impairs function of monocyte-derived dendritic cells. Blood 2002, 99:2913–2921. [DOI] [PubMed] [Google Scholar]
- 48.Sedy JR, Bjordahl RL, Bekiaris V, Macauley MG, Ware BC, Norris PS, Lurain NS, Benedict CA, Ware CF: CD160 activation by herpesvirus entry mediator augments inflammatory cytokine production and cytolytic function by NK cells. J Immunol 2013, 191:828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prechtel AT, Turza NM, Kobelt DJ, Eisemann JI, Coffin RS, McGrath Y, Hacker C, Ju X, Zenke M, Steinkasserer A: Infection of mature dendritic cells with herpes simplex virus type 1 dramatically reduces lymphoid chemokine-mediated migration. J Gen Virol 2005, 86:1645–1657. [DOI] [PubMed] [Google Scholar]
- 50.Salio M, Cella M, Suter M, Lanzavecchia A: Inhibition of dendritic cell maturation by herpes simplex virus. Eur J Immunol 1999, 29:3245–3253. [DOI] [PubMed] [Google Scholar]
- 51.Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM: Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem 1995, 270:15974–15978. [DOI] [PubMed] [Google Scholar]
- 52.Alcami A, Smith GL: Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J Virol 1995, 69:4633–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alcami A, Khanna A, Paul NL, Smith GL: Vaccinia virus strains Lister, USSR and Evans express soluble and cell-surface tumour necrosis factor receptors. J Gen Virol 1999, 80 (Pt 4):949–959. [DOI] [PubMed] [Google Scholar]
- 54.Zimmerling S, Waibler Z, Resch T, Sutter G, Schwantes A: Interleukin-1beta receptor expressed by modified vaccinia virus Ankara interferes with interleukin-1beta activity produced in various virus-infected antigen-presenting cells. Virol J 2013, 10:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wimmer E, Hellen CU, Cao X: Genetics of poliovirus. Annu Rev Genet 1993, 27:353–436. [DOI] [PubMed] [Google Scholar]
- 56.Shen L, Chen CY, Huang D, Wang R, Zhang M, Qian L, Zhu Y, Zhang AZ, Yang E, Qaqish A, et al. : Pathogenic Events in a Nonhuman Primate Model of Oral Poliovirus Infection Leading to Paralytic Poliomyelitis. J Virol 2017, 91.**First study documenting primary host cell targets after oral poliovirus infection.
- 57.Wahid R, Cannon MJ, Chow M: Dendritic cells and macrophages are productively infected by poliovirus. J Virol 2005, 79:401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, et al. : Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 2010, 141:799–811.**Deciphering of the mechanisms of the enterovirus replication site.
- 59.Kastan JP, Dobrikova EY, Bryant JD, Gromeier M: CReP mediates selective translation initiation at the endoplasmic reticulum. Sci Adv 2020, 6:eaba0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jang SK, Pestova TV, Hellen CU, Witherell GW, Wimmer E: Cap-independent translation of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme 1990, 44:292–309.**Identification of picornavirus IRESes (also see 61.).
- 61.Pelletier J, Sonenberg N: Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334:320–325.**Identification of picornavirus IRESes (also see 60.).
- 62.Nicklin MJ, Krausslich HG, Toyoda H, Dunn JJ, Wimmer E: Poliovirus polypeptide precursors: expression in vitro and processing by exogenous 3C and 2A proteinases. Proc Natl Acad Sci U S A 1987, 84:4002–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toyoda H, Nicklin MJ, Murray MG, Anderson CW, Dunn JJ, Studier FW, Wimmer E: A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 1986, 45:761–770.**Identification of the poliovirus 2A protease.
- 64.Morrison JM, Racaniello VR: Proteinase 2Apro is essential for enterovirus replication in type I interferon-treated cells. J Virol 2009, 83:4412–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW: Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem 1982, 257:14806–14810.**Description of eIF4G cleavage by poliovirus.
- 66.Gustin KE, Sarnow P: Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J 2001, 20:240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilkinson GW, Davison AJ, Tomasec P, Fielding CA, Aicheler R, Murrell I, Seirafian S, Wang EC, Weekes M, Lehner PJ, et al. : Human cytomegalovirus: taking the strain. Med Microbiol Immunol 2015, 204:273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, Scholz I, Gilbride RM, Lewis MS, Gilliam AN, Ventura AB, et al. : Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 2013, 340:1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen SG, Piatak M Jr., Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, et al. : Immune clearance of highly pathogenic SIV infection. Nature 2013, 502:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murray SE, Nesterenko PA, Vanarsdall AL, Munks MW, Smart SM, Veziroglu EM, Sagario LC, Lee R, Claas FHJ, Doxiadis IIN, et al. : Fibroblast-adapted human CMV vaccines elicit predominantly conventional CD8 T cell responses in humans. J Exp Med 2017, 214:1889–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Panicali D, Paoletti E: Construction of poxviruses as cloning vectors: insertion of the thymidine kinase gene from herpes simplex virus into the DNA of infectious vaccinia virus. Proc Natl Acad Sci U S A 1982, 79:4927–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taylor J, Trimarchi C, Weinberg R, Languet B, Guillemin F, Desmettre P, Paoletti E: Efficacy studies on a canarypox-rabies recombinant virus. Vaccine 1991, 9:190–193. [DOI] [PubMed] [Google Scholar]
- 73.Mayr R, Hochstein-Mintzel V, Stickl H: Passage history, properties, and applicability of the attenuated vaccinia virus strain MVA. Infection 1975, 3:6–14. [Google Scholar]
- 74.Price PJ, Torres-Dominguez LE, Brandmuller C, Sutter G, Lehmann MH: Modified Vaccinia virus Ankara: innate immune activation and induction of cellular signalling. Vaccine 2013, 31:4231–4234. [DOI] [PubMed] [Google Scholar]
- 75.Dai P, Wang W, Yang N, Serna-Tamayo C, Ricca JM, Zamarin D, Shuman S, Merghoub T, Wolchok JD, Deng L: Intratumoral delivery of inactivated modified vaccinia virus Ankara (iMVA) induces systemic antitumor immunity via STING and Batf3-dependent dendritic cells. Sci Immunol 2017, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia-Arriaza J, Najera JL, Gomez CE, Tewabe N, Sorzano CO, Calandra T, Roger T, Esteban M: A candidate HIV/AIDS vaccine (MVA-B) lacking vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS One 2011, 6:e24244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Staib C, Kisling S, Erfle V, Sutter G: Inactivation of the viral interleukin 1beta receptor improves CD8+ T-cell memory responses elicited upon immunization with modified vaccinia virus Ankara. J Gen Virol 2005, 86:1997–2006. [DOI] [PubMed] [Google Scholar]
- 78.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. : Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 2009, 361:2209–2220. [DOI] [PubMed] [Google Scholar]
- 79.Ledgerwood JE, DeZure AD, Stanley DA, Coates EE, Novik L, Enama ME, Berkowitz NM, Hu Z, Joshi G, Ploquin A, et al. : Chimpanzee Adenovirus Vector Ebola Vaccine. N Engl J Med 2017, 376:928–938. [DOI] [PubMed] [Google Scholar]
- 80.Ewer K, Rampling T, Venkatraman N, Bowyer G, Wright D, Lambe T, Imoukhuede EB, Payne R, Fehling SK, Strecker T, et al. : A Monovalent Chimpanzee Adenovirus Ebola Vaccine Boosted with MVA. N Engl J Med 2016, 374:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kennedy SB, Bolay F, Kieh M, Grandits G, Badio M, Ballou R, Eckes R, Feinberg M, Follmann D, Grund B, et al. : Phase 2 Placebo-Controlled Trial of Two Vaccines to Prevent Ebola in Liberia. N Engl J Med 2017, 377:1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Newton KR, Sala-Soriano E, Varsani H, Stephenson JR, Goldblatt D, Wedderburn LR: Human dendritic cells infected with an adenoviral vector suppress proliferation of autologous and allogeneic T cells. Immunology 2008, 125:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mosaheb MM, Dobrikova EY, Brown MC, Yang Y, Cable J, Okada H, Nair SK, Bigner DD, Ashley DM, Gromeier M: Genetically stable poliovirus vectors activate dendritic cells and prime antitumor CD8 T cell immunity. Nat Commun 2020, 11:524.**First description of a feasible recombinant poliovirus vector strategy for engaging DCs.
- 84.Ward CD, Stokes MA, Flanegan JB: Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J Virol 1988, 62:558–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gromeier M, Alexander L, Wimmer E: Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc Natl Acad Sci U S A 1996, 93:2370–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gromeier M, Lachmann S, Rosenfeld MR, Gutin PH, Wimmer E: Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc Natl Acad Sci U S A 2000, 97:6803–6808.**Description of the concept of cancer immunotherapy with recombinant poliovirus.
- 87.Desjardins A, Gromeier M, Herndon JE, Beaubier N, Bolognesi DP, Friedman AH, Friedman HS, McSherry F, Muscat AM, Nair S, et al. : Recurrent Glioblastoma Treated with Recombinant Poliovirus. N Engl J Med 2018, 379:150–161.**First in-human study of recombinant poliovirus immunotherapy.
- 88.Dobrikova EY, Goetz C, Walters RW, Lawson SK, Peggins JO, Muszynski K, Ruppel S, Poole K, Giardina SL, Vela EM, et al. : Attenuation of neurovirulence, biodistribution, and shedding of a poliovirus:rhinovirus chimera after intrathalamic inoculation in Macaca fascicularis. J Virol 2012, 86:2750–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown MC, Bryant JD, Dobrikova EY, Shveygert M, Bradrick SS, Chandramohan V, Bigner DD, Gromeier M: Induction of viral, 7-methyl-guanosine cap-independent translation and oncolysis by mitogen-activated protein kinase-interacting kinase-mediated effects on the serine/arginine-rich protein kinase. J Virol 2014, 88:13135–13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown MC, Dobrikov MI, Gromeier M: Mitogen-activated protein kinase-interacting kinase regulates mTOR/AKT signaling and controls the serine/arginine-rich protein kinase-responsive type 1 internal ribosome entry site-mediated translation and viral oncolysis. J Virol 2014, 88:13149–13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brown MC, Gromeier M: MNK Controls mTORC1:Substrate Association through Regulation of TELO2 Binding with mTORC1. Cell Rep 2017, 18:1444–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Campbell SA, Lin J, Dobrikova EY, Gromeier M: Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J Virol 2005, 79:6281–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU: Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci U S A 2009, 106:9197–9202.**Identification of the mechanism of translation initiation at enterovirus IRESes.
- 94.Dobrikov MI, Dobrikova EY, Gromeier M: Dynamic Regulation of the Translation Initiation Helicase Complex by Mitogenic Signal Transduction to Eukaryotic Translation Initiation Factor 4G. Mol Cell Biol 2013, 33:937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gromeier M, Bossert B, Arita M, Nomoto A, Wimmer E: Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J Virol 1999, 73:958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sweeney TR, Abaeva IS, Pestova TV, Hellen CU: The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J 2014, 33:76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iwasaki A, Welker R, Mueller S, Linehan M, Nomoto A, Wimmer E: Immunofluorescence analysis of poliovirus receptor expression in Peyer's patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection. J Infect Dis 2002, 186:585–592. [DOI] [PubMed] [Google Scholar]
- 98.Bodian D: Emerging concept of poliomyelitis infection. Science 1955, 122:105–108. [DOI] [PubMed] [Google Scholar]
- 99.Sabin AB: Pathogenesis of poliomyelitis; reappraisal in the light of new data. Science 1956, 123:1151–1157. [DOI] [PubMed] [Google Scholar]
- 100.Merrill MK, Dobrikova EY, Gromeier M: Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J Virol 2006, 80:3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Merrill MK, Gromeier M: The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol 2006, 80:6936–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brown MC, Holl EK, Boczkowski D, Dobrikova E, Mosaheb M, Chandramohan V, Bigner DD, Gromeier M, Nair SK: Cancer immunotherapy with recombinant poliovirus induces IFN-dominant activation of dendritic cells and tumor antigen-specific CTLs. Sci Transl Med 2017, 9.**Elucidation of a peculiar virus:DC host relationship for recombinant poliovirus, PVSRIPO.
- 103.Walton RW, Brown M, Sacco MT, Gromeier M: Engineered oncolytic poliovirus PVSRIPO subverts MDA5-dependent innate immune responses in cancer cells. J Virol 2018, [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]