SUMMARY
Epithelial tissues form the boundaries of organs, where they perform a range of functions, including secretion, absorption, and protection. These tissues are commonly composed of discrete cell layers—sheets of cells that are one-cell thick. In multiple systems examined, epithelial cells round up and move in the apical direction before dividing, likely in response to neighbor-cell crowding [1–6]. Because of this movement, daughter cells may be born displaced from the tissue layer. Reintegration of these displaced cells supports tissue growth and maintains tissue architecture [4]. Two conserved IgCAMs (immunoglobulin superfamily cell adhesion molecules), neuroglian (Nrg) and fasciclin 2 (Fas2), participate in cell reintegration in the Drosophila follicular epithelium [4]. Like their vertebrate orthologs L1CAM and NCAM1/2, respectively, Nrg and Fas2 are cell adhesion molecules primarily studied in the context of nervous system development [7–10]. Consistent with this, we identify another neural IgCAM, Fasciclin 3 (Fas3), as a reintegration factor. Nrg, Fas2, and Fas3 are components of the insect septate junction, the functional equivalent of the vertebrate tight junction, but proliferating follicle cells do not have mature septate junctions, and we find that the septate junction protein neurexin IV does not participate in reintegration [11, 12]. Here, we show that epithelial reintegration works in the same way as IgCAM-mediated axon growth and pathfinding; it relies not only on extracellular adhesion but also mechanical coupling between IgCAMs and the lateral spectrin-based membrane skeleton. Our work indicates that reintegration is mediated by a distinct epithelial adhesion assembly that is compositionally and functionally equivalent to junctions made between axons.
Graphical Abstract
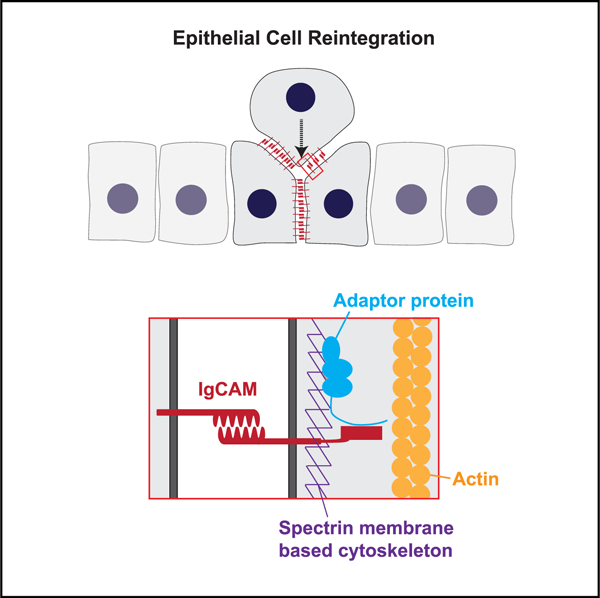
In Brief
Epithelial cells born protruding from a tissue monolayer will reintegrate into it. Cammarota, Finegan, et al. show that cell reintegration in the Drosophila follicular epithelium occurs via a mechanism previously implicated in axon pathfinding.
RESULTS AND DISCUSSION
Fas2 and Nrg Work in Parallel to Mediate Reintegration
When the adhesion molecules Nrg and Fas2 are disrupted, cells can fail to reintegrate and instead remain positioned above (apical to) the epithelial layer (Figure 1A). We refer to this phenotype as “popping out.” Whether Nrg and Fas2 act in parallel or inter-dependently has not been tested [4]. We disrupted them genetically using the null alleles Nrg14 or Fas2G0336 and measured the number of popped-out cells in stage 6–8 egg chambers. Whereas genetic loss of either Fas2 or Nrg causes ~5 popped-out cells per egg chamber (~1/150 cells), losing both causes ~50 popped-out cells per egg chamber (~1/15 cells; Figures 1C, 1D, and S1A; Video S1).
Figure 1. Nrg and Fas2 Act in Parallel to Drive Reintegration.
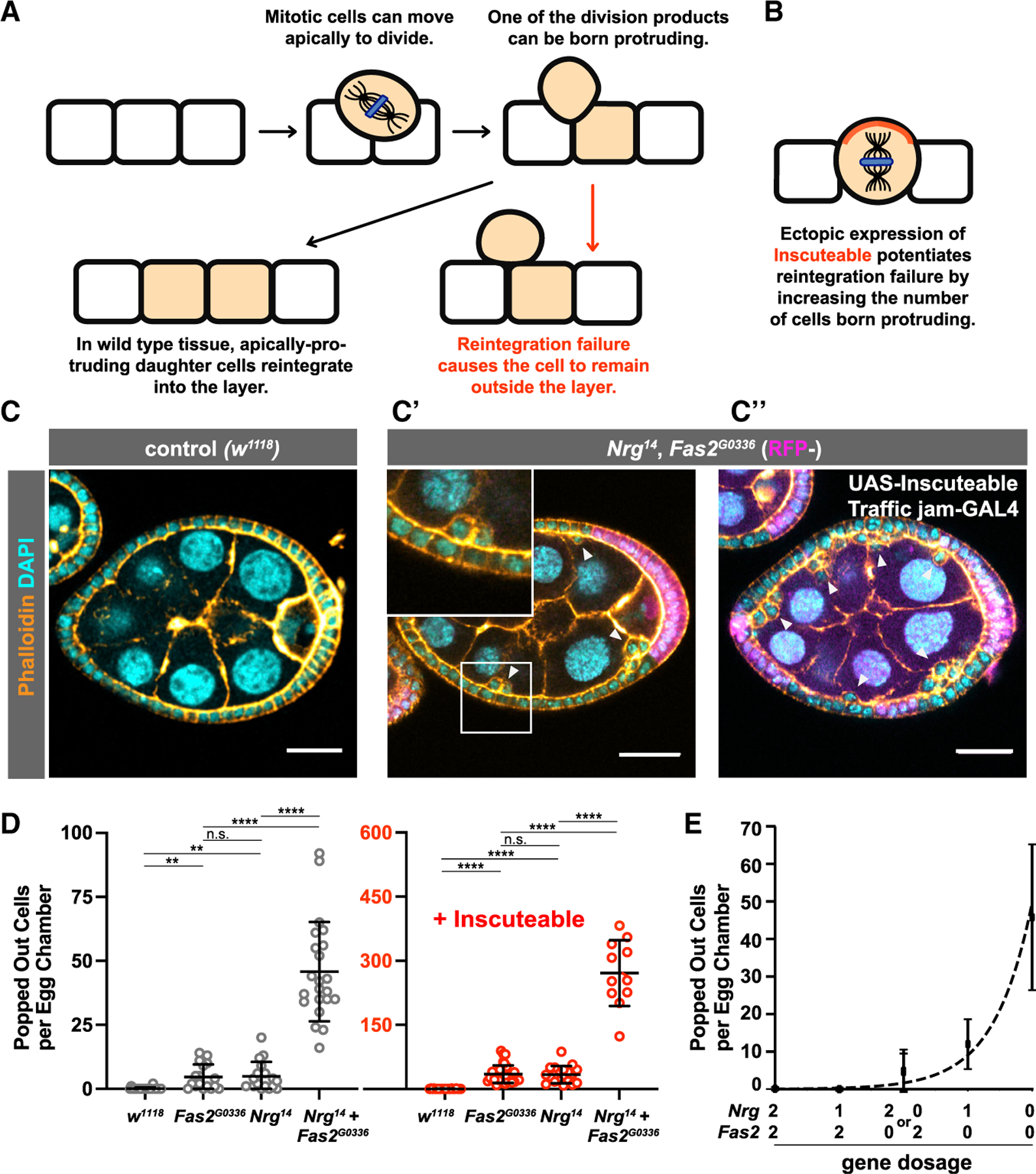
(A) Diagram showing apical cell movement during mitosis and subsequent cell reintegration.
(B) Inscuteable increases out-of-plane divisions and therefore the number of reintegration events.
(C and D) Genetic disruption of both Fas2 and Nrg causes a synergistic increase in reintegration failure over disruption of either gene alone. Ectopic expression of Inscuteable has a multiplicative effect on reintegration failure. Representative images are shown in (C) and quantification in (D). Each data point represents the number of extralayer (popped-out) cells in the entirety of one egg chamber. Each column represents three or more dissections of five or more flies. Significance was determined using an unpaired, two-tailed Student’s t test with Welch’s correction. p values left to right: p = 0.0018, p = 0.0031, p = 0.8723, p < 0.0001, p < 0.0001, p < 0.0001, p < 0.0001, p = 0.8201, p < 0.0001, p < 0.0001. Average and error bars correspond to mean and SD. Scale bars in (C) correspond to 20 µm.
(E) The relationship between reintegration failure and gene function is not linear. A plot of gene dosage versus misplaced cell number fits an exponential curve (R2 = 0.972), providing a speculative model for reintegration function. Average and error bars correspond to mean and SD.
We tested whether this effect reflected an increase in failed reintegration events. Ectopic expression of Inscuteable (Insc) reorients mitotic spindles in the follicular epithelium (FE), causing them to align along the apical-basal axis of the tissue rather than perpendicular to it (Figure 1B). This effect increases the frequency with which daughter cells are born apical to the tissue layer and therefore potentiates reintegration failure in Nrg- and Fas2-mutant tissue [4]. Inscuteable expression has a multiplicative effect on popping out, increasing the number of misplaced cells in tissue mutant for either Nrg14 or Fas2G0336 by 6-fold (Figure 1D). Because Inscuteable has the same effect for Nrg14, Fas2G0336 double mutants, we conclude that the increase in popped-out cells in double mutant tissue represents an increase in failed reintegrations and therefore that Nrg and Fas2 act in parallel (Figures 1C” and 1D; Video S1). In agreement, we found that Nrg localizes normally in Fas2G0336 mutant follicle tissue and Fas2 localizes normally in Nrg14 tissue (Figures S1B and S1C). These proteins are likewise functionally redundant in the control of axon guidance [13].
Although Nrg and Fas2 mutants have identical phenotypes, indicating equal contribution to reintegration, the loss of both factors causes a synergistic rather than additive increase in reintegration failure. To investigate the relationship between reintegration function and Fas2/Nrg activity, we built a speculative mathematical model by plotting popped-out cells versus gene dosage on a Cartesian plane (Figure 1E). The assumption that function correlates with gene dosage has not been tested directly but agrees with the correlation between Nrg dosage and Nrg expression (Figure S2). To expand our dataset, we used a genomic-rescue strategy to restore one copy of Nrg in Nrg14 single and Nrg14, Fas2G0336 double mutants. The resulting data points fit an exponential curve (Figures 1E and 2A). This curve suggests that reintegration is robust, as it is typically successful even at low IgCAM function.
Figure 2. Dissection of Nrg and Fas2 Function.
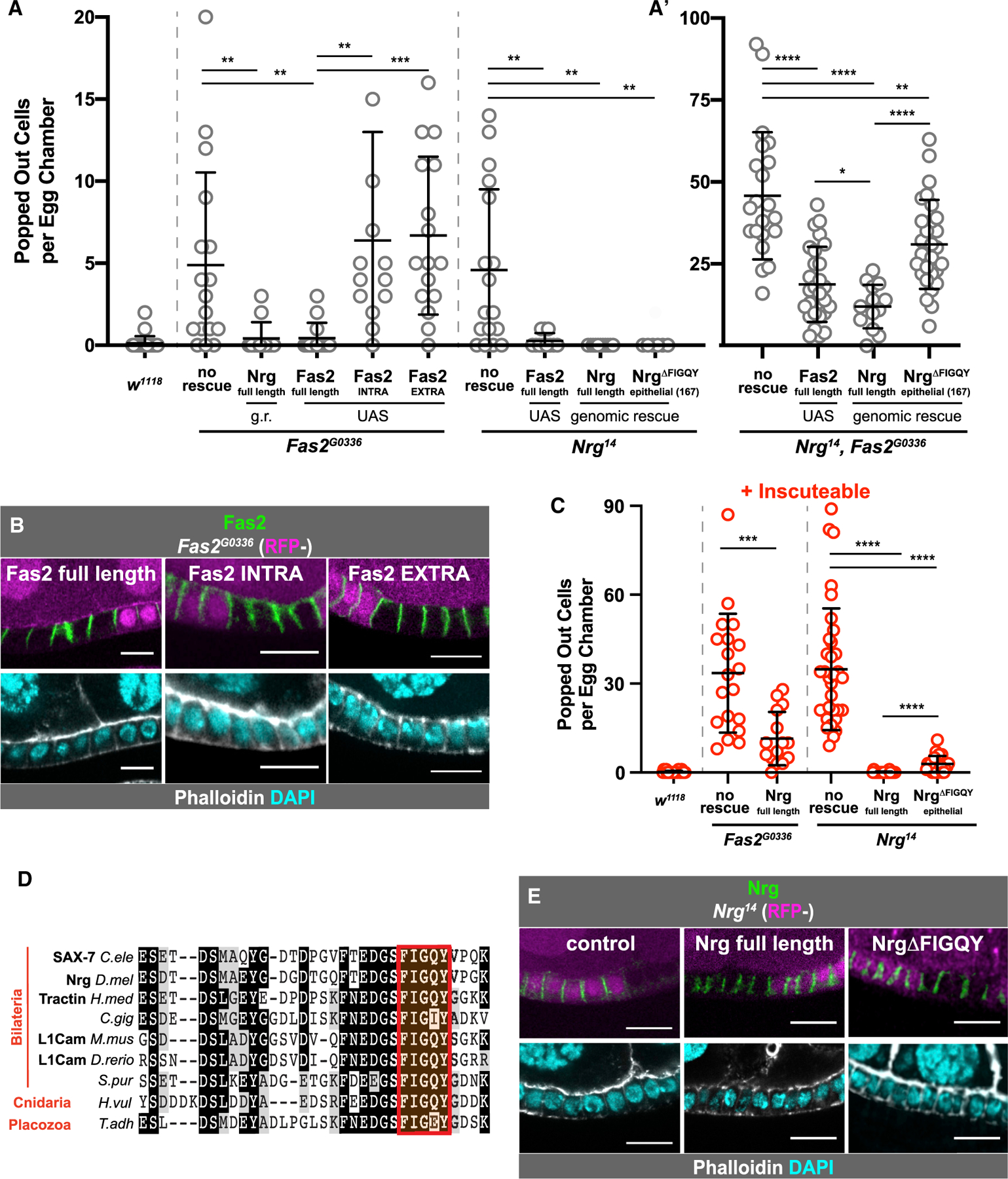
(A) Genetic rescue experiments reveal the relative roles of Nrg and Fas2 in reintegration, through assessment of the “popping out” phenotype. Average and error bars correspond to mean and SD. Expression of full-length Fas2 rescues reintegration failure in Nrg- and Fas2-null tissue, but both the extracellular and intracellular domains are required to rescue Fas2G0336. Neuroglian rescues reintegration failure in Nrg- and Fas2-null tissue. Removal of the FIGQY subsequence from the epithelial Nrg isoform allows for partial rescue in Nrg-null tissue. p values left to right: p = 0.0052, p = 0.0052, p = 0.0071, p = 0.0001, p = 0.0023, p = 0.0014, p = 0.0014. (A’) Neither expression of full-length Fas2 or Nrg fully rescues reintegration failure caused by Nrg14,Fas2G0336 double mutants. The Nrg variant with a disruption in the epithelial FIGQY subsequence rescues reintegration failure significantly less than the full Nrg rescue. UAS-Fas2-YFP and variants were expressed using the driver Traffic Jam-GAL4. Neuroglian and Nrg variants were expressed from a single genomic rescue construct. p values left to right: p < 0.0001, p < 0.0001, p = 0.0044, p = 0.0221, p < 0.0001.
(B) Fas2 localizes to cell-cell borders without either its extracellular or intracellular domain. UAS-Fas2-YFP and its truncated variants, expressed using Traffic Jam-Gal4, could all be detected at follicle cell-cell borders, whether in the presence (RFP+) or absence (RFP−) of endogenous Fas2.
(C) Ectopic expression of Insc reveals that both Nrg overexpression in Fas2-null and expression of Nrg missing the FIGQY subsequence in Nrg-null tissues incompletely rescue the reintegration failure phenotype. Average and error bars correspond to mean and SD. p values left to right: p = 0.0002, p < 0.0001, p < 0.0001, p < 0.0001.
(D) The ankyrin-binding domain of Nrg/L1CAM is evolutionarily conserved.
(E) The FIGQY subsequence is not required for Nrg localization to epithelial cell-cell borders. Representative images are shown in (E) and quantified in Figure S2C. Quantification and statistical tests in (A) and (C) were performed as in Figure 1. Average and error bars in (A) and (C) correspond to mean and SD. Scale bars in (B) and (E) correspond to 5 µm. See also Figure S2.
The Intracellular Region of Fas2 Participates in Reintegration
There are seven isoforms of Fas2 in Drosophila. Five of these have transmembrane domains and are considered to be neuronal [14]. The other two, which have been studied in non-neuronal cells in the trachea, Malpighian (renal) tubule, and in glial cells, are tethered to the membrane by glycosylphosphatidylinositol (GPI) anchors, indicating that they have only adhesive function [10, 14–16]. Because the FE is not neuronal, these observations suggest the possibility that Fas2 might mediate epithelial cell reintegration through adhesion alone. However, the FE expresses at least one “neuronal” isoform (i.e., including a transmembrane domain), as revealed by previous immunostaining with the 1D4 monoclonal antibody [4, 7, 14].
A previous study showed that upstream activating sequence (UAS)-Fas2 rescues photoreceptor axon pathfinding defects in Nrg mutants and vice versa [13]. In agreement, we found that UAS-Fas2-full-length, which includes a cytoplasmic region (isoform PD), rescues reintegration failure in Nrg14 and Fas2G0336 tissue (Figure 2A). Furthermore, UAS-Fas2-full-length partially rescues the popped-out cell phenotype in Nrg14, Fas2G0336 mutant tissue (Figure 2A’). According to our model, this restoration of reintegration function is similar, though slightly smaller, to that provided by one copy of Nrg (Figures 1E and 2A’). The difference might be attributed to diminished expression or function of UAS-Fas2-full-length relative to endogenous Fas2.
We further explored Fas2 function using variant forms of UAS-Fas2 [17]. UAS-Fas2-extracellular, which includes the extracellular and transmembrane regions but lacks an intracellular domain, rescues a non-neuronal function of Fas2, namely the regulation of brush border length in the Malpighian tubule epithelium [15]. However, we found that neither UAS-Fas2-extracellular nor UAS-Fas2-intracellular (the transmembrane and intracellular region of isoform PD) rescues reintegration failure in Fas2G0336 tissue, though both variants localize at lateral cell junctions (Figures 2A and 2B). These results show that the reintegration role played by Fas2 requires a combination of its adhesive and cytoplasmic domains.
The Cytoplasmic FIGQY Subsequence of Neuroglian Participates in Reintegration
We next investigated the functional contribution of Nrg to reintegration. Use of a genomic rescue strategy allowed us to consider gene dosage in these experiments. A single dose of Nrg rescued Nrg14 (Figures 1 and 2A). Expressing a third copy of Nrg rescues Fas2G0336 mutant tissue, consistent with the reciprocal finding that UAS-Fas2 rescues Nrg14 mutants (Figure 2A). Together, these data show that Fas2 and Nrg can compensate for one another provided they are expressed at a sufficient level.
Given that the loss of Nrg or Fas2 corresponds to an infrequent number of popped-out cells and our work suggests that reintegration works at low total IgCAM function (Figure 1), we considered whether rescue of a mild phenotype does not equate to a full rescue of protein activity. We tested this possibility by using Inscuteable to increase the severity of a single mutant phenotype. These experiments were feasible with Nrg genomic-rescue transgenes because they rely on their own promoter. Contrastingly, we could not be convinced that the pool of GAL4 was sufficient to drive full expression of both UAS-Fas2 and UAS-Inscuteable.
In the sensitized condition, Nrg incompletely rescues Fas2 (Figure 2C). These results are consistent with the possibility that Nrg and Fas2 have both overlapping and separate functions. Because both Nrg and Fas2 are cell-cell adhesion proteins, we considered whether the basis for their distinction was intracellular. The cytoplasmic C terminus of Nrg/L1CAM includes a highly conserved five-amino-acid subsequence (FIGQY) that helps mediate interaction with ankyrin, a linker between the cytoskeleton and the membrane [18, 19]. Deletion of FIGQY reduces in vitro interaction between ankyrin and the vertebrate L1CAM paralog neurofascin by 80% [19]. The remaining contact requires FIGQY-adjacent amino acids, which are also highly conserved (Figure 2D) [19].
We used NrgΔFIGQY transgenes to test whether FIGQY contributes to reintegration [17]. In both insects and vertebrates, one Nrg/L1CAM isoform is expressed in epithelia (Drosophila Nrg167) and a different isoform in neural tissue (Nrg180) [20]. Both isoforms have a FIGQY sequence, but they use different exons to encode it (Figure S2A). Therefore, a genomic rescue encoding Nrg180ΔFIGQY should express full-length Nrg167 and show the same rescue as full-length Nrg. Consistent with this, Nrg180ΔFIGQY allows for complete rescue of reintegration failure in Nrg14 mutant tissue, even in the presence of Inscuteable (Figures S2A and S2B). Contrastingly, Nrg167ΔFIGQY rescues reintegration failure in Nrg14 mutants but allows only partial rescue in the presence of Inscuteable (Figures 2A and 2C). Furthermore, Nrg167ΔFIGQY promotes substantially less rescue of Nrg14, Fas2G0336 than does full-length Nrg (Figure 2A’). Our mathematical model (Figure 1E) suggests that Nrg167ΔFIGQY provides ~30%–35% of wild-type function.
We considered whether removal of the FIGQY subsequence affects localization of Nrg to cell-cell contacts. Nrg167ΔFIGQY localizes to follicle cell-cell borders in Nrg14 mutant clones, as revealed by immunostaining (Figure 2E). Quantification of these images shows that the relative strength of Nrg immunoreactivity corresponds to genetic dosage; signal decreases by ~25% at the border between a cell expressing two copies of Nrg and a cell expressing only one copy and ~50% at the border between cells expressing one copy each (Figure S2C). Notably, Nrg does not require homotypic adhesion to localize to the membrane, as the signal decreases by only half at the border between wild type and Nrg14 clones. These results are consistent with observations made in other organisms; neither the cytoplasmic domain of L1CAM nor its C. elegans ortholog SAX-7 is required for cell-cell adhesion or membrane localization [21, 22]. Together, our findings indicate that the FIGQY sequence mediates an intracellular function involved in cell reintegration.
Neuroglian Mediates Reintegration through Interaction with Ankyrin
During axon growth, ankyrin stabilizes Nrg/L1CAM at the cell surface. The interaction between them, which is mediated by the FIGQY domain, links Nrg to the spectrin-based membrane skeleton (SBMS), a subcortical scaffold that mechanically supports the plasma membrane [17]. Ankyrin therefore acts as a molecular “clutch” by which new outgrowth is mechanically stabilized [23]. We tested whether this mechanism also participates in cell reintegration.
Like Neuroglian and Fas2, ankyrin localizes along the length of lateral cell-cell junctions in the FE [24]. We used short hairpin RNA (shRNA) to knockdown ankyrin. Though we cannot determine the extent of knockdown directly, this shRNA has been effective in a previous study [25]. Ankyrin knockdown in the FE results in popped-out cells, making it the first non-adhesion protein to give this phenotype (Figures 3A and 3B). This result is not explained by a decrease in the junctional localization of Nrg and Fas2, as both are unchanged in ankyrin-shRNA tissue (Figures S3A–S3C). We also find that ankyrin knockdown potentiates reintegration failure in Fas2G0336, but not Nrg14, tissue (Figure 3B). Together, these findings show that Nrg and ankyrin work together to mediate reintegration in parallel to Fas2.
Figure 3. Nrg and Ankyrin Cooperate to Drive Cell Reintegration.
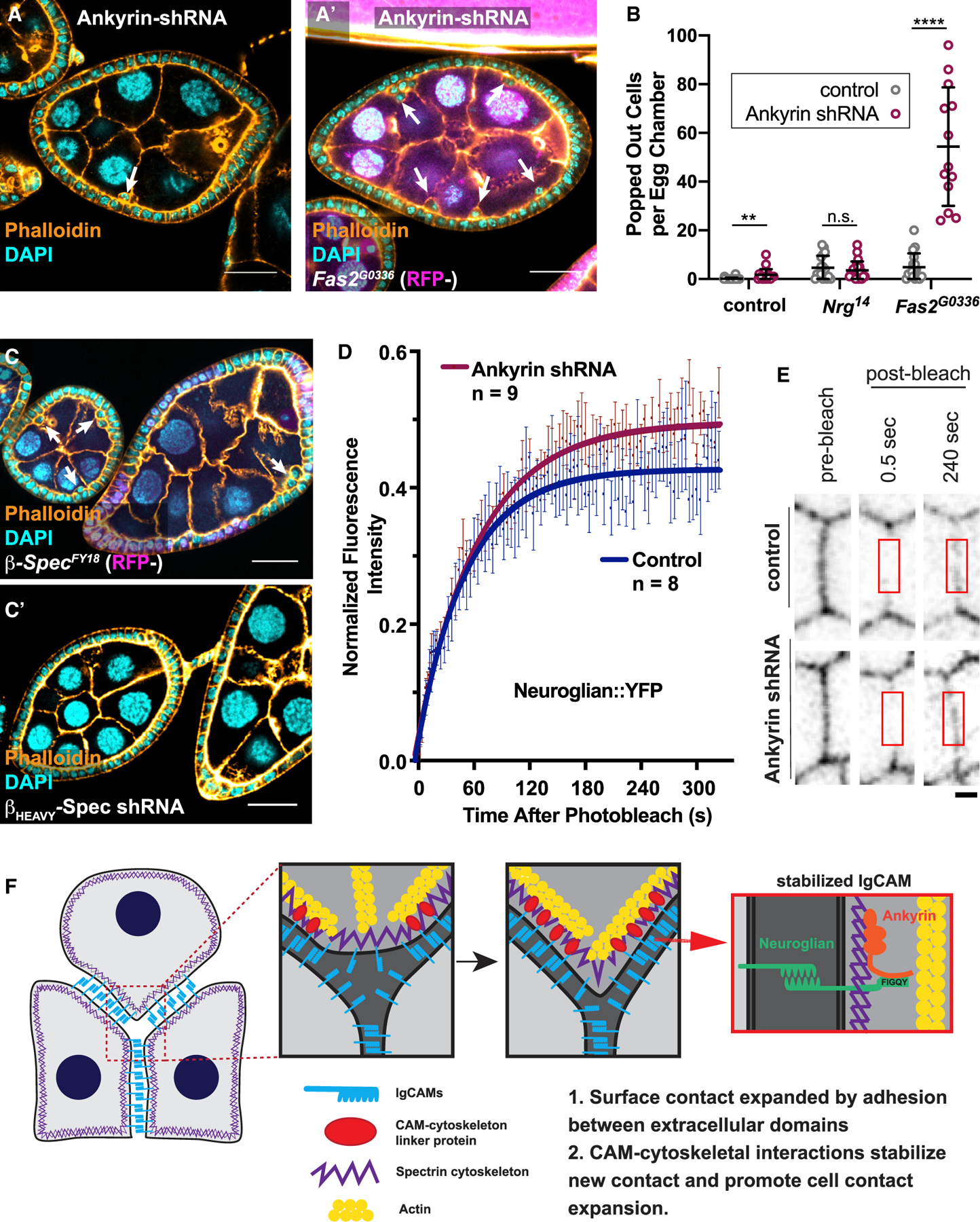
(A and B) Genetic disruption of ankyrin results in failed cell reintegration. Expression of ankyrin shRNA, driven by Traffic Jam-GAL4, results in ~2 popped-out cells per egg chamber. Ankyrin shRNA potentiates the reintegration failure phenotype seen in Fas2-null tissue, but not Nrg-null tissue. Representative images are shown in (A) and quantification in (B). Quantification and statistical tests were performed as in Figure 1. Scale bars in (A) correspond to 20 µm. Average and error bars correspond to mean and SD. p values from left to right: p = 0.0086, p = 0.4663, p < 0.0001.
(C) Popped-out cells, indicating reintegration failure, are apparent in FE tissue mutant for β-spectrinFY18, but not βHEAVY-spectrin. Scale bars correspond to 20 µm.
(D) Fluorescence recovery curves of Nrg::YFP after photobleaching. Lateral FE cell junctions were bleached in egg chambers expressing ankyrin-shRNA driven by Traffic Jam-GAL4 or egg chambers with the driver alone (control). Fluorescence intensity was normalized to account for inherent photobleaching due to imaging. Experiments were performed independently with number of experimental repeats indicated. Curves were calculated by fitting a one-phase association curve. Error bars represent SEM. The 95% confidence interval (CI) of the control plateau is 41–44. This does not overlap with the 95% CI of the ankyrin-shRNA plateau, which is 48–51.
(E) Frames from an example FRAP experiment, showing the fluorescence of Nrg::YFP at a cell junction prior to and post bleaching in control and ankyrin-shRNA-expressing egg chambers. Red boxes indicate photobleached region over which fluorescence intensity was quantified. Scale bars correspond to 1 µm.
(F) We propose a refined model for IgCAM-mediated cell reintegration in epithelia. Both extracellular adhesion and intracellular connection to the cytoskeleton is necessary for the reintegration of cells into epithelial layers. Specifically, we reveal that the spectrin-based cytoskeleton stabilizes Nrg cell-cell adhesion through ankyrin to facilitate reintegration. We propose that the SBMS mechanically stabilizes the trans-interactions of Nrg at the “leading edge” of integrating cells to facilitate the progression of cell reintegration. Nrg-ankyrin interactions thereby provide a traction force (grip).
See also Figure S3.
The SBMS, a lattice composed of ɑ- and β-spectrin tetramers, is polarized in epithelial cells. In the FE, β-spectrin (called β-fodrin or βII-spectrin in vertebrates) and ankyrin line the lateral cortex, whereas βHEAVY-spectrin is along the cell apex, from which ankyrin is excluded [24, 26–29]. Several studies demonstrate disorganization in β-spectrin mutant follicular epithelia [30–32]. In agreement, we observed extensive disorganization phenotypes in large mitotic clones mutant for β-spectrinFY18, particularly after stage 6 of egg chamber development (not shown). However, we also observed popped-out cells in earlier, smaller clones (Figure 3C). Although we cannot exclude the possibility that this phenotype relates to reported defects in Hippo signaling or actomyosin that cause later disorganization, it is consistent with a failure of cell reintegration. The localization of Nrg and Fas2 to cell-cell borders is unchanged in β-spectrinFY18 mutant clones (Figures S3D and S3E). Popped-out cells were not observed after knockdown of βHEAVY-spectrin using an shRNA that has been effective in a previous study [33] (Figure 3C’).
To test whether ankyrin affects Nrg mobility in epithelia, we used fluorescence recovery after photobleaching (FRAP) analysis to measure the effect of ankyrin disruption on Nrg stability at follicle cell-cell junctions. In the control condition, Nrg::YFP signal recovers to only 42% of its pre-bleach intensity, indicating that the majority of Nrg is immobile (Figures 3D and 3E). Ankyrin knockdown increases signal recovery to 50%, indicating that ankyrin binding contributes to some, though not all, of the immobile fraction (Figures 3D and 3E). Conversely, ankyrin knockdown does not change the mobile/immobile fractions of the lateral-junction immunoglobulin membrane protein basigin (Bsg), illustrating that the effect of ankyrin on mobility Nrg is specific (Figure S3F). These findings are consistent with observations made in motor neurons, in which deletion of the FIGQY sequence increases the mobility of Nrg [17]. For technical reasons, our experiment was performed at interphase cell-cell junctions, leaving open the possibility that ankyrin has an even stronger effect in actively reintegrating cells.
Together, these results are consistent with a model in which Nrg cooperates with the lateral SBMS to provide a traction force (grip) during cell reintegration, as it does during axon growth (Figure 3F).
Fas3 Is a Reintegration Factor, but NrxIV Is Not
Like Nrg and Fas2, Fas3 is a homophilic IgCAM found in neurons and in septate junctions [34–38]. Nrg and Fas2 are enriched along follicle cell-cell borders during the developmental time over which follicle cells are dividing, with expression dropping off soon afterward [39, 40]. We observe the same pattern of expression and localization for Fas3, suggesting that it also plays a role in proliferation (Figure 4A) [4, 39, 40].
Figure 4. Fas3 Participates in Reintegration, but NrxIV does Not.
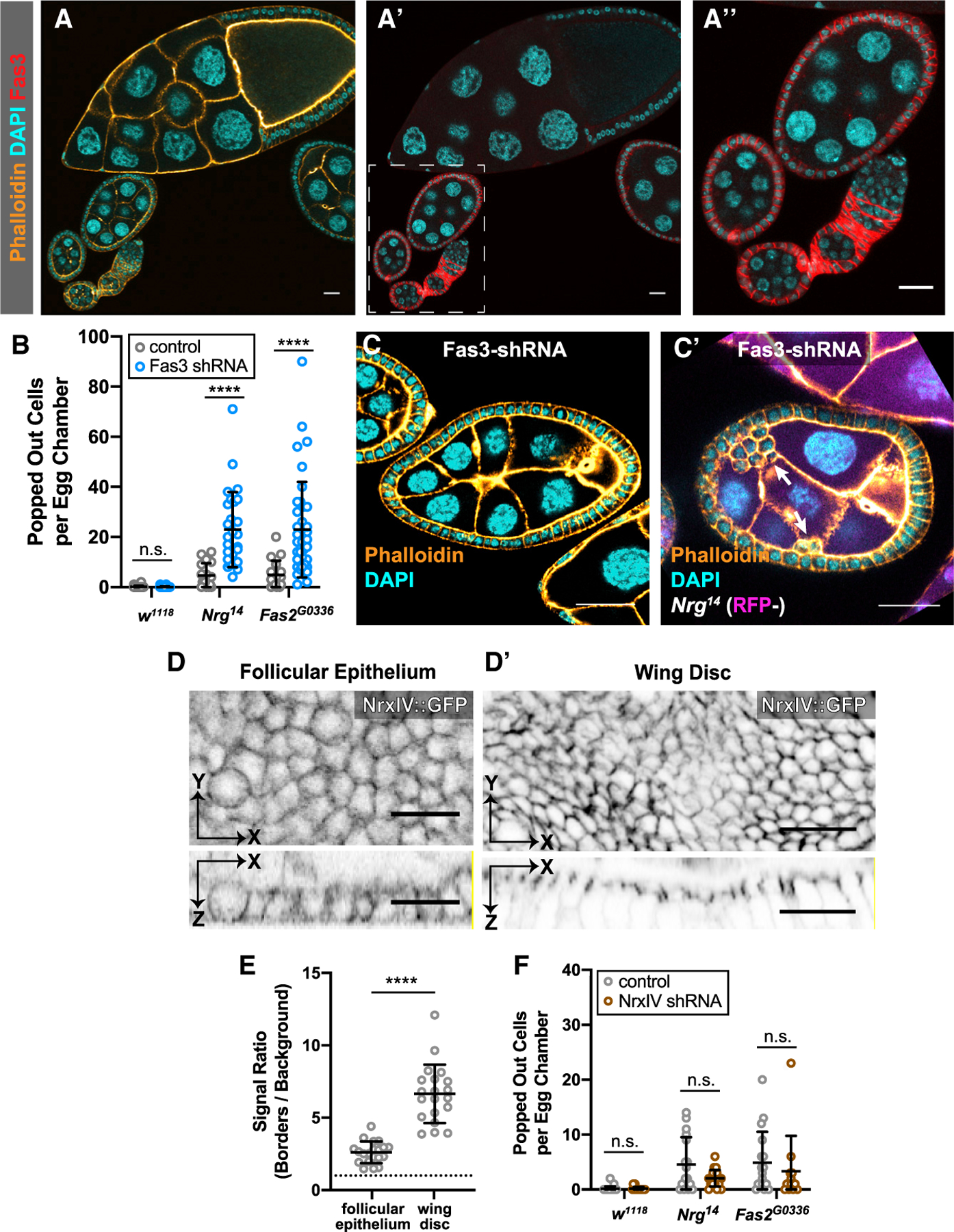
(A) Fas3 localizes to follicle cell-cell borders in egg chambers prior to stage 7. Fas3 immunoreactivity is detected in early egg chambers, in which the follicular epithelium is proliferative (stages 1–6, in dashed box), but drops off substantially by mid-stage (stage 10, top of panel).
(B and C) Fas3 disruption potentiates reintegration failure in Nrg and Fas2 mutant tissue. Fas3 knockdown does not result in misplaced cells (B) unless combined with mutation of either Nrg or Fas2 (B and C). Fas3-shRNA was driven with Traffic Jam-GAL4. Misplaced cells were quantified as in Figure 1. Average and error bars correspond to mean and SD. p values left to right: p = 0.5948, p < 0.0001, p < 0.0001.
(D and E) NrxIV is weakly localized in the early follicular epithelium. NrxIV::GFP, a protein trap, demonstrates weaker junctional localization in the follicular epithelium (D) than in the mature wing disc (D’) relative to local background signal (quantification in E). Signal intensity was measured at septate junctions (wing disc) or along lateral cell-cell contacts (follicular epithelium) and compared with adjacent cytoplasmic signal. Average and error bars represent mean and SD. Significance was determined using an unpaired, two-tailed Student’s t test with Welch’s correction p < 0.0001. (F) NrxIV disruption does not enhance reintegration failure. NrxIV-shRNA, driven with Traffic Jam-GAL4, does not increase the number of misplaced cells in control tissue or in Nrg or Fas2 mutant tissue. Average and error bars represent mean and SD. p values from left to right: p = 0.9166, p = 0.0547, p = 0.5095. Quantification and statistical analysis were performed as in Figure 1. All scale bars correspond to 20 µm.
See also Figure S4.
Fas3 has long been implicated in axon guidance, but mutation of Fas3 alone has no axon guidance phenotype; it is thought to work redundantly with more consequential factors, including Nrg and Fas2 [16, 34]. Consistent with this, we did not observe popped-out cells in Fas3-shRNA follicular epithelia but found that Fas3 knockdown increases popping out in Fas2G0336 or Nrg14 mutant tissue by approximately 5-fold (Figures 4B and 4C). Immunostaining confirmed the efficacy of knockdown (Figure S4A). Because Insc has a similar potentiating effect, we tested whether Fas3-shRNA alters spindle orientation in the FE. Spindle orientation is unaffected by Fas3 knockdown (Figures S4B and S4C). Together, these results suggest that Fas3 plays analogous roles in axon guidance and reintegration.
The follicular epithelium does not have mature septate junctions, but the observation that reintegration involves three septate junction components—Nrg, Fas2, and Fas3—suggests the possibility that reintegration relies on an “immature” septate junction. To test this possibility, we examined the core septate junction molecule neurexin IV (NrxIV) [11, 41, 42]. Like Nrg and Fas2, neurexin-family proteins are found at both synapses and mature septate junctions [11, 43]. However, unlike Nrg and Fas2, neurexins are not implicated in axon guidance. Using both endogenously tagged NrxIV::GFP and immunostaining, we observed NrxIV along follicle cell-cell borders in early-stage egg chambers (Figures 4D and 4E; data not shown). These results confirm NrxIV expression in the FE, though it is significantly weaker (2.6-fold>background; ±0.9) than that observed at mature septate junctions in the imaginal wing disc (6.7-fold>background; ±0.4; Figures 4D and 4E). Knockdown of NrxIV in the FE did not result in reintegration failure, either alone or in combination with Nrg14 or Fas2G0336 (Figure 4F). The efficacy of NrxIV-shRNA was confirmed in the wing disc (Figure S4D). Taken together, these results show that not all septate junction proteins are reintegration factors and instead suggest that reintegration relies on a subset of IgCAMs that participate in axon guidance.
Through genetic manipulations, we have elucidated the molecular mechanism by which the lateral adhesion machinery drives epithelial cell reintegration (Figure 3F). Our work shows that epithelial cell reintegration relies on a distinct junctional complex, previously considered an immature septate junction, that is neither the classical zonula adherens nor the zonula occludens. These junctions are equivalent in composition and function to the axon-axon junctions that support axon growth and pathfinding through fasciculation and parallel the established similarity between mature epithelial pleated septate junctions and synaptic junctions, which have long been proposed to share an evolutionary history [11, 44–48]. We therefore suggest that the maturation of septate junctions is akin to synaptogenesis; both processes (1) preserve IgCAM components of the earlier junction and (2) recruit additional analogous proteins, leading to increased junctional complexity and structural stability. We also suggest the possibility that the epithelial functions of these IgCAMs are ancestral, as both Fas2/NCAM and Nrg/LICAM (Figure 2D) have been identified in Placozoa. These animals have epithelial, but not neural, cell types [49].
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further questions regarding this work or the fly lines and reagents used should be directed toward the Lead Contact, Dan T. Bergstralh (dan.bergstralh@rochester.edu).
Materials Availability
All fly strains can be obtained either through the Lead Contact or through the following Drosophila resource centers: Bloomington Drosophila Stock Center, Kyoto Stock Center, or the Vienna Drosophila Resource Center. Available antibodies are listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-Fas2 (mouse) | DSHB | 1D4 |
| Monoclonal anti-Fas3 (mouse) | DSHB | 7G10 |
| Monoclonal anti-α-tubulin-FITC (mouse) | Sigma | F2168 |
| Polylonal anti-Insc (rabbit) | Jürgen Knoblich, [53] | N/A |
| Monoclonal anti-Nrg (mouse) | Michael Hortsch, [54] | N/A |
| Polyclonal anti-NrxIV (rabbit) | Christian Klämbt, [55] | N/A |
| anti-Centrosomin (rabbit) | Jordan Raff, [56] | N/A |
| Goat-anti-mouse-488 | ThermoFisher | A-11001 |
| Goat-anti-rabbit-488 | ThermoFisher | A-11008 |
| Goat-anti-mouse-633 | ThermoFisher | A-21050 |
| Goat-anti-rabbit-633 | ThermoFisher | A-21071 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FITC Phalloidin | ThermoFisher | F432 |
| Rhodamine Phalloidin | ThermoFisher | R415 |
| Phalloidin 633 | ThermoFisher | A22284 |
| Vectasheild with DAPI | Vector Labs | H-1200 |
| Experimental Models: Organisms/Strains | ||
| D. Melanogaster: Nrg14 | [57] | FBal0013169 |
| D. Melanogaster: Fas2G0336 | [58] | FBal0152502 |
| D. Melanogaster: β-spectrinFY18 | [30] | FBal0325726 |
| D. Melanogaster: RFP-nls, hsflp, FRT19A | N/A | N/A |
| D. Melanogaster: NrgWT on II | [17] | N/A |
| D. Melanogaster: Nrg167ΔFIGQY on II | [17] | FBal0291693 |
| D. Melanogaster: Nrg180ΔFIGQY on II | [17] | FBal0291694 |
| D. Melanogaster: UAS Inscuteable | [53] | FBtp0006279 |
| D. Melanogaster: UAS myristoylated-RFP | [59] | FBtp0017423 |
| D. Melanogaster: UAS Fas2-YFP | [10] | FBtp0051931 |
| D. Melanogaster: UAS Fas2-Extracellular-YFP | [10] | FBtp0051932 |
| D. Melanogaster: UAS Fas2-Intracellular-YFP | [10] | FBtp0051933 |
| D. Melanogaster: UAS βHeavy-spectrin shRNA (HMS00882) | [60] | FBti0140589 |
| D. Melanogaster: UAS NrxIV shRNA (JF03142) | [60] | FBti0127279 |
| D. Melanogaster: UAS Fas3 shRNA (HMC06528) | [60] | FBti0196009 |
| D. Melanogaster: UAS Ank shRNA (GD10431) | [61] | FBal0198724 |
| D. Melanogaster: Traffic Jam-GAL4 | [62] | FBti0034540 |
| D. Melanogaster: GR1-GAL4 | [63] | FBti0040373 |
| D. Melanogaster: nubbin-GAL4 | [64] | FBti0016825 |
| D. Melanogaster: actin5c-FLPout-GAL4 | N/A | N/A |
| D. Melanogaster: Neuroglian::YFP | [65] | N/A |
| D. Melanogaster: Basigin::YFP | [65] | N/A |
| D. Melanogaster: NrxIV::eGFP | [66] | FBti0099828 |
| Software and Algorithms | ||
| FIJI (1.53) | [50] | https://fiji.sc/ |
| Anaconda3 (Python 3.7) | N/A | https://www.anaconda.com/ |
| GraphPad Prism (8) | N/A | https://www.graphpad.com/scientific-software/prism/ |
| TCoffee | N/A | http://tcoffee.crg.cat/ |
Data and Code Availability
The published article includes all datasets generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila Strains
All Drosophila strains used in this study were kept at 22°C and fed with standard cornmeal-agar food. Crosses were carried out at 25°C unless otherwise specified in the Method Details.
METHOD DETAILS
Drosophila Clone Induction and GAL4 Expression
Follicle cell clones of Nrg14, Fas2G0336, and β-spectrinFY18 were induced by incubating larvae or pupae at 37° for two out of every twelve hours over a period of at least two days. Adult females were dissected at least two days after the last heat shock. Ectopic protein expression was accomplished using the UAS-GAL4 system (Brand and Perrimon 1993). UAS-GAL4 flies were maintained at 29° for 48 hours before dissection.
Immunostaining
Ovaries were fixed for 15 minutes in 10% Paraformaldehyde and 0.2% Tween in Phosphate Buffered Saline (PBS). Ovaries were then incubated in 10% Bovine Serum Albumin (in PBS) to block for one hour at room temperature. Primary and secondary immunostainings lasted at least 3 hours in PBS with 0.2% Tween. Three washes (~10 minutes each) in PBS-0.2% Tween were carried out between stainings and after the secondary staining. Primary and secondary antibodies were used at a concentration of 1:150.
Imaging
Fixed imaging was performed using: a Leica SP5 confocal with a 63x/1.4 HCX PL Apo CS oil lens or an Andor Dragonfly Spinning Disk Confocal microscope with a 60x water objective. Images were collected with LAS AF or the Andor Fusion program respectively. Images were processed (Gaussian blur) using ImageJ. Minor drift correction was occasionally applied using a custom Python script that included cv2.filter2D, part of the OpenCV library (https://opencv.org/).
Misplaced Cell Counting
Quantification of extralayer cells was performed on Stage 6–8 egg chambers using at least 3 dissections of at least 5 flies each. For analyses of clonal mutants, the number of extralayer cells was quantified in egg chambers that were at least 60% mutant. Popped-out cells were quantified manually. The entire depth of each egg chamber was examined using confocal microscopy. Images shown are of representative sagittal planes from each condition, while each data point reflects all misplaced cells in a single egg chamber.
FRAP
FRAP experiments were performed on a Nikon A1R HD confocal coupled to a Ti2-E inverted microscope. Images were acquired using a Galvano scanner with a 60x/1.49 Apochromat TIRF oil lens using NIS Elements software. Photobleaching of Nrg::YFP was performed on user-defined 0.5 µm2 region of interest on cell-cell junctions longer than 2 µm using the 488nm laser at 2 x imaging power at 4 x scan speed. Post-bleaching acquisition of Nrg::YFP was performed at 20 s intervals for 2 minutes, followed by 40 s intervals for 5 minutes, or until the sample moved out of the plane of focus. One image of the imaging field of view was acquired pre-bleaching.
Spindle Orientation Measurements
Spindle angle determination was performed using FIJI as previously described in [50]. One line was drawn connecting the centrosomes and another connecting the two apical corners of the cell. The angle between these lines was measured.
QUANTIFICATION AND STATISTICAL ANALYSIS
Mathematical Modeling of Misplaced Cells
Mathematical modeling was performed using Python, making use of the open-source SciPy library (https://scipy.org/scipylib/). To demonstrate the nonlinearity of the number of misplaced cells with respect to loss of function, misplaced cells were plotted against amount of Nrg or Fas2 removed; an arbitrary unit of one was assigned to the loss of either Nrg or Fas2. A simple exponential was used as a preliminary model:
In this model, A and B are arbitrary scale factors. NumPy (https://numpy.org/) was used for array creation and storage. The SciPy optimize.curve_fit tool was used to fit the average and standard deviation of the counts taken for each genotype to our preliminary model. Matplotlib (https://matplotlib.org/), was used to plot the model.
FRAP Quantification
Fluorescence intensity quantification of Nrg::YFP was performed in FIJI [65]. Average fluorescence intensity was quantified over the 0.5 µm2 bleached region for each frame, with xy drift of the ROI corrected manually. FRAP curves were calculated using a double normalization method to correct for imaging acquisition bleaching, analogous to that presented in [66]. The normalized fluorescence intensity of YFP-tagged proteins at photobleached junctions at each time point InormFRAP(t) was calculated as follows:
where Ifrap is the average fluorescence intensity of the photobleached junction region, Iwhole is the average fluorescence intensity of the whole field of view (cropped to include only epithelial tissue where appropriate), Ibase is the average fluorescence of a ‘background’ region manually defined by drawing a 0.5 µm2 region of interest in the cytoplasm of a nearby cell, and Iprebleach is the average fluorescence intensity of the photobleached junction region pre-bleaching. FRAP curves were calculated by calculating a one phase association curve using Prism analysis software (Graphpad).
Nrg Localization Quantification
Signal intensity was measured along a 3-pixel wide ROI perpendicular to the center of a cell-cell border. The normalized intensity ratio was calculated as follows:
where the signal to noise ratio (SNR) refers to pixel intensity at borders divided by the local background (non-cell-cell border) intensity.
Protein Border Enrichment Quantification
Signal intensity was measured along a 4-pixel wide ROI perpendicular to the center of cell-cell borders. An average histogram across the ROI was drawn and border enrichment was calculated as fold enrichment of the peak value of the histogram versus the average signal both sides of the peak, corresponding to the cytoplasmic signal.
Statistical Analyses
An unpaired, two-tailed Student’s t test with Welch’s correction was used to determine significance when comparing the number of misplaced cells in an egg chamber. No statistical method was used to predetermine sample size, the experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. An unpaired, two-tailed Student’s t test with Welch’s correction was also used to compare Nrg localization to cell cell borders. The average Normalized Intensity Ratio for each egg chamber was calculated and the distributions of average fold changes were tested for significance. All statistical tests were carried out using GraphPad Prism.
Sequence Alignments
Protein sequence alignments were performed using T-Coffee (http://tcoffee.crg.cat/).
Supplementary Material
Highlights.
“Neural” IgSF proteins drive epithelial cell reintegration after division
Reintegration relies in part on the interaction between Nrg and ankyrin
The IgSF-mediated reintegration mechanism also mediates axon pathfinding
ACKNOWLEDGMENTS
We are grateful to Holly Lovegrove, Colleen Maillie, Michael Welte and the Welte lab, other members of the Rochester Invertebrate Group, and Nicole Dawney and other Bergstralh lab members for comments and discussion. We thank Kathryn Neville, Philip Bellomio, Naz Ünsal, and Muskaan Vasandani for technical assistance. We thank the Transgenic RNAi Project at Harvard Medical School (NIH/NIGMS R01-GM084947) for fly lines. This work was supported by NIH grants R01GM125839 (PI: D.T.B.) and S10 RR024577-01.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2020.09.061.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Sauer FC (1936). The interkinetic migration of embryonic epithelial nuclei. J. Morphol 60, 1–11. [Google Scholar]
- 2.Spear PC, and Erickson CA (2012). Interkinetic nuclear migration: a mysterious process in search of a function. Dev. Growth Differ 54, 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Packard A, Georgas K, Michos O, Riccio P, Cebrian C, Combes AN, Ju A, Ferrer-Vaquer A, Hadjantonakis A-K, Zong H, et al. (2013). Luminal mitosis drives epithelial cell dispersal within the branching ureteric bud. Dev. Cell 27, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergstralh DT, Lovegrove HE, and St Johnston D (2015). Lateral adhesion drives reintegration of misplaced cells into epithelial monolayers. Nat. Cell Biol 17, 1497–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKinley KL, Stuurman N, Royer LA, Schartner C, Castillo-Azofeifa D, Delling M, Klein OD, and Vale RD (2018). Cellular aspect ratio and cell division mechanics underlie the patterning of cell progeny in diverse mammalian epithelia. eLife 7, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorce B, Escobedo C, Toyoda Y, Stewart MP, Cattin CJ, Newton R, Banerjee I, Stettler A, Roska B, Eaton S, et al. (2015). Mitotic cells contract actomyosin cortex and generate pressure to round against or escape epithelial confinement. Nat. Commun 6, 8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grenningloh G, Rehm EJ, and Goodman CS (1991). Genetic analysis of growth cone guidance in Drosophila: fasciclin II functions as a neuronal recognition molecule. Cell 67, 45–57. [DOI] [PubMed] [Google Scholar]
- 8.Harrelson AL, and Goodman CS (1988). Growth cone guidance in insects: fasciclin II is a member of the immunoglobulin superfamily. Science 242, 700–708. [DOI] [PubMed] [Google Scholar]
- 9.Schuster CM, Davis GW, Fetter RD, and Goodman CS (1996). Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron 17, 641–654. [DOI] [PubMed] [Google Scholar]
- 10.Kohsaka H, Takasu E, and Nose A (2007). In vivo induction of postsynaptic molecular assembly by the cell adhesion molecule Fasciclin2. J. Cell Biol 179, 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harden N, Wang SJH, and Krieger C (2016). Making the connection - shared molecular machinery and evolutionary links underlie the formation and plasticity of occluding junctions and synapses. J. Cell Sci 129, 3067–3076. [DOI] [PubMed] [Google Scholar]
- 12.Mahowald AP (1972). Ultrastructural observations on oogenesis in Drosophila. J. Morphol 137, 29–48. [DOI] [PubMed] [Google Scholar]
- 13.Kristiansen LV, Velasquez E, Romani S, Baars S, Berezin V, Bock E, Hortsch M, and Garcia-Alonso L (2005). Genetic analysis of an overlapping functional requirement for L1- and NCAM-type proteins during sensory axon guidance in Drosophila. Mol. Cell. Neurosci 28, 141–152. [DOI] [PubMed] [Google Scholar]
- 14.Neuert H, Deing P, Krukkert K, Naffin E, Steffes G, Risse B, Silies M, and Klämbt C (2020). The Drosophila NCAM homolog Fas2 signals independently of adhesion. Development 147, dev181479. [DOI] [PubMed] [Google Scholar]
- 15.Halberg KA, Rainey SM, Veland IR, Neuert H, Dornan AJ, Klämbt C, Davies S-A, and Dow JAT (2016). The cell adhesion molecule Fasciclin2 regulates brush border length and organization in Drosophila renal tubules. Nat. Commun 7, 11266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araújo SJ, and Tear G (2003). Axon guidance mechanisms and molecules: lessons from invertebrates. Nat. Rev. Neurosci 4, 910–922. [DOI] [PubMed] [Google Scholar]
- 17.Enneking E-M, Kudumala SR, Moreno E, Stephan R, Boerner J, Godenschwege TA, and Pielage J (2013). Transsynaptic coordination of synaptic growth, function, and stability by the L1-type CAM Neuroglian. PLoS Biol 11, e1001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mualla R, Nagaraj K, and Hortsch M (2013). A phylogenetic analysis of the L1 family of neural cell adhesion molecules. Neurochem. Res 38, 1196–1207. [DOI] [PubMed] [Google Scholar]
- 19.Garver TD, Ren Q, Tuvia S, and Bennett V (1997). Tyrosine phosphorylation at a site highly conserved in the L1 family of cell adhesion molecules abolishes ankyrin binding and increases lateral mobility of neurofascin. J. Cell Biol 137, 703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hortsch M, Bieber AJ, Patel NH, and Goodman CS (1990). Differential splicing generates a nervous system-specific form of Drosophila neuroglian. Neuron 4, 697–709. [DOI] [PubMed] [Google Scholar]
- 21.Pocock R, Bénard CY, Shapiro L, and Hobert O (2008). Functional dissection of the C. elegans cell adhesion molecule SAX-7, a homologue of human L1. Mol. Cell. Neurosci 37, 56–68. [DOI] [PubMed] [Google Scholar]
- 22.Wong EV, Cheng G, Payne HR, and Lemmon V (1995). The cytoplasmic domain of the cell adhesion molecule L1 is not required for homophilic adhesion. Neurosci. Lett 200, 155–158. [DOI] [PubMed] [Google Scholar]
- 23.Suter DM, and Forscher P (2000). Substrate-cytoskeletal coupling as a mechanism for the regulation of growth cone motility and guidance. J. Neurobiol 44, 97–113. [PubMed] [Google Scholar]
- 24.Dubreuil RR, Maddux PB, Grushko TA, and MacVicar GR (1997). Segregation of two spectrin isoforms: polarized membrane-binding sites direct polarized membrane skeleton assembly. Mol. Biol. Cell 8, 1933–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz S, Truglio M, Scott MJ, and Fitzsimons HL (2016). Long-term memory in Drosophila is influenced by histone deacetylase HDAC4 interacting with SUMO-conjugating enzyme Ubc9. Genetics 203, 1249–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubreuil RR, and Yu J (1994). Ankyrin and beta-spectrin accumulate independently of alpha-spectrin in Drosophila. Proc. Natl. Acad. Sci. USA 91, 10285–10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JK, Brandin E, Branton D, and Goldstein LS (1997). alpha-Spectrin is required for ovarian follicle monolayer integrity in Drosophila melanogaster. Development 124, 353–362. [DOI] [PubMed] [Google Scholar]
- 28.Thomas GH, Zarnescu DC, Juedes AE, Bales MA, Londergan A, Korte CC, and Kiehart DP (1998). Drosophila betaHeavy-spectrin is essential for development and contributes to specific cell fates in the eye. Development 125, 2125–2134. [DOI] [PubMed] [Google Scholar]
- 29.Zarnescu DC, and Thomas GH (1999). Apical spectrin is essential for epithelial morphogenesis but not apicobasal polarity in Drosophila. J. Cell Biol 146, 1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong KKL, Li W, An Y, Duan Y, Li Z, Kang Y, and Yan Y (2015). β-spectrin regulates the hippo signaling pathway and modulates the basal actin network. J. Biol. Chem 290, 6397–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fletcher GC, Elbediwy A, Khanal I, Ribeiro PS, Tapon N, and Thompson BJ (2015). The Spectrin cytoskeleton regulates the Hippo signalling pathway. EMBO J 34, 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng BF, Selvaraj GK, Santa-Cruz Mateos C, Grosheva I, Alvarez-Garcia I, Martín-Bermudo MD, and Palacios IM (2016). α-spectrin and integrins act together to regulate actomyosin and columnarization, and to maintain a monolayered follicular epithelium. Development 143, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forest E, Logeay R, Géminard C, Kantar D, Frayssinoux F, Heron-Milhavet L, and Djiane A (2018). The apical scaffold big bang binds to spectrins and regulates the growth of Drosophila melanogaster wing discs. J. Cell Biol 217, 1047–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snow PM, Bieber AJ, and Goodman CS (1989). Fasciclin III: a novel homophilic adhesion molecule in Drosophila. Cell 59, 313–323. [DOI] [PubMed] [Google Scholar]
- 35.Kose H, Rose D, Zhu X, and Chiba A (1997). Homophilic synaptic target recognition mediated by immunoglobulin-like cell adhesion molecule Fasciclin III. Development 124, 4143–4152. [DOI] [PubMed] [Google Scholar]
- 36.Rose D, and Chiba A (1999). A single growth cone is capable of integrating simultaneously presented and functionally distinct molecular cues during target recognition. J. Neurosci 19, 4899–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki E, Rose D, and Chiba A (2000). The ultrastructural interactions of identified pre- and postsynaptic cells during synaptic target recognition in Drosophila embryos. J. Neurobiol 42, 448–459. [DOI] [PubMed] [Google Scholar]
- 38.Wells RE, Barry JD, Warrington SJ, Cuhlmann S, Evans P, Huber W, Strutt D, and Zeidler MP (2013). Control of tissue morphology by Fasciclin III-mediated intercellular adhesion. Development 140, 3858–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szafranski P, and Goode S (2004). A Fasciclin 2 morphogenetic switch organizes epithelial cell cluster polarity and motility. Development 131, 2023–2036. [DOI] [PubMed] [Google Scholar]
- 40.Szafranski P, and Goode S (2007). Basolateral junctions are sufficient to suppress epithelial invasion during Drosophila oogenesis. Dev. Dyn 236, 364–373. [DOI] [PubMed] [Google Scholar]
- 41.Baumgartner S, Littleton JT, Broadie K, Bhat MA, Harbecke R, Lengyel JA, Chiquet-Ehrismann R, Prokop A, and Bellen HJ (1996). A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell 87, 1059–1068. [DOI] [PubMed] [Google Scholar]
- 42.Oshima K, and Fehon RG (2011). Analysis of protein dynamics within the septate junction reveals a highly stable core protein complex that does not include the basolateral polarity protein Discs large. J. Cell Sci 124, 2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Südhof TC (2017). Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell 171, 745–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hortsch M, and Margolis B (2003). Septate and paranodal junctions: kissing cousins. Trends Cell Biol 13, 557–561. [DOI] [PubMed] [Google Scholar]
- 45.Poliak S, and Peles E (2003). The local differentiation of myelinated axons at nodes of Ranvier. Nat. Rev. Neurosci 4, 968–980. [DOI] [PubMed] [Google Scholar]
- 46.Banerjee S, Sousa AD, and Bhat MA (2006). Organization and function of septate junctions: an evolutionary perspective. Cell Biochem. Biophys 46, 65–77. [DOI] [PubMed] [Google Scholar]
- 47.Nans A, Einheber S, Salzer JL, and Stokes DL (2011). Electron tomography of paranodal septate-like junctions and the associated axonal and glial cytoskeletons in the central nervous system. J. Neurosci. Res 89, 310–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLachlan IG, and Heiman MG (2013). Shaping dendrites with machinery borrowed from epithelia. Curr. Opin. Neurobiol 23, 1005–1010. [DOI] [PubMed] [Google Scholar]
- 49.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. (2008). The Trichoplax genome and the nature of placozoans. Nature 454, 955–960. [DOI] [PubMed] [Google Scholar]
- 50.Finegan TM, Na D, Cammarota C, Skeeters AV, Nádasi TJ, Dawney NS, Fletcher AG, Oakes PW, and Bergstralh DT (2019). Tissue tension and not interphase cell shape determines cell division orientation in the Drosophila follicular epithelium. EMBO J 38, 100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraut R, Chia W, Jan LY, Jan YN, and Knoblich JA (1996). Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383, 50–55. [DOI] [PubMed] [Google Scholar]
- 52.Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, Traquina ZR, Schilling J, and Goodman CS (1989). Drosophila neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell 59, 447–460. [DOI] [PubMed] [Google Scholar]
- 53.Stork T, Thomas S, Rodrigues F, Silies M, Naffin E, Wenderdel S, and Klämbt C (2009). Drosophila Neurexin IV stabilizes neuron-glia interactions at the CNS midline by binding to Wrapper. Development 136, 1251–1261. [DOI] [PubMed] [Google Scholar]
- 54.Lucas EP, and Raff JW (2007). Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J. Cell Biol 178, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eeken JCJ, Sobels FH, Hyland V, and Schalet AP (1985). Distribution of MR-induced sex-linked recessive lethal mutations in Drosophila melanogaster. Mutat. Res 150, 261–275. [DOI] [PubMed] [Google Scholar]
- 56.Peter A, Schöttler P, Werner M, Beinert N, Dowe G, Burkert P, Mourkioti F, Dentzer L, He Y, Deak P, et al. (2002). Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep 3, 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersen R, Li Y, Resseguie M, and Brenman JE (2005). Calcium/ calmodulin-dependent protein kinase II alters structural plasticity and cytoskeletal dynamics in Drosophila. J. Neurosci 25, 8878–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, McCall K, Yang-Zhou D, Flockhart I, Binari R, Shim H-S, et al. (2015). The transgenic RNAi project at Harvard Medical School: resources and validation. Genetics 201, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. [DOI] [PubMed] [Google Scholar]
- 60.Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, and Brennecke J (2010). An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29, 3301–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tran DH, and Berg CA (2003). bullwinkle and shark regulate dorsal-appendage morphogenesis in Drosophila oogenesis. Development 130, 6273–6282. [DOI] [PubMed] [Google Scholar]
- 62.Calleja M, Moreno E, Pelaz S, and Morata G (1996). Visualization of gene expression in living adult Drosophila. Science 274, 252–255. [DOI] [PubMed] [Google Scholar]
- 63.Lowe N, Rees JS, Roote J, Ryder E, Armean IM, Johnson G, Drummond E, Spriggs H, Drummond J, Magbanua JP, et al. ; UK Drosophila Protein Trap Screening Consortium (2014). Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library. Development 141, 3994–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, et al. (2007). The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Phair RD, Gorski SA, and Misteli T (2004). Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol 375, 393–414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.


