Abstract
Influenza D is the only type of influenza virus that mainly affects cattle with frequent spillover to other species. Since the initial description of influenza D virus (IDV) in 2011, the virus has been found to circulate among cattle and swine populations worldwide. Research conducted during the past several years has led to an increased understanding of this novel influenza virus with bovines as a reservoir. In this review, we describe the current knowledge of epidemiology and host range of IDV followed by discussion of infection biology and animal model development for IDV. Finally, we review progress towards understanding of the pathogenesis and host response of IDV as well as developing preventive vaccines for IDV.
Introduction
The Orthomyxoviridae has four types of influenza viruses, designated influenza A, B, C, and D. Viruses from influenza A, B, and C are known to cause human respiratory disease [1,2]. Influenza A (IAV) and B (IBV) viruses can cause severe illnesses and seasonal influenza epidemics in humans with IAV having the potential to trigger a pandemic [2]. Clinical manifestations of influenza C virus (ICV) infection are generally mild in healthy adults, but it can be severe with complications of lower respiratory infections in children, especially those under 2 years [1]. Influenza D virus (IDV) circulates in worldwide agricultural animals such as cattle and swine with little known about its impact and pathogenesis on human health [3,4].
Epidemiology and host range of influenza D virus.
IDV was initially isolated from a diseased pig with the severe respiratory symptoms in U.S. Oklahoma in 2011 [4]. Additional serological investigation showed that 9.5% of surveyed pigs in U.S. swine population possessed antibodies specific to IDV, indicating that this previously unidentified influenza virus circulated in U.S. swine [4]. A causal relationship is also evidently supported by the experimental infection of pigs [4,5]. Interestingly, further studies demonstrated that IDV is more prevalent in cattle than in swine in U.S. and the virus likely utilizes cattle as a primary host [3]. IDV is also found in calves or pigs with respiratory disease complex in Europe and Asia [6–14]. In addition to swine and cattle, IDV-specific antibodies have been demonstrated in sheep, goats, horses, camelids, and wild boars [9,10,13,15–25]. Susceptibility to experimental infection by IDV has been established in mice, guinea pigs, and ferrets [4,26–28]. Despite the absence of IDV antibodies in U.S. poultry as demonstrated in a small-scale study [25], a recent study showing the evident detection of IDV genome in Asian poultry farms appears to add this species into a growing list of susceptible hosts for IDV [29]. The results of these studies collectively support a theory that IDV utilizes bovines as a primary reservoir and amplification host with periodical spillover to other animal hosts (Figure 1). Since 2011, IDVs have evolved into two distinctive lineages represented by D/swine/Oklahoma/1334/2011 (D/OK) and D/bovine/Oklahoma/660/2013 (D/660) (Figure 2) [15]. Recently, two new genetic lineage of IDV, designated D/Yama2106 and D/Yama2019, has been reported in cattle herds in Japan (Figure 2) [30]. These four distinctive lineages currently co-circulate in worldwide bovine and pig populations that may facilitate genetic reassortment between different influenza D viruses, which may further affect global animal health.
Figure 1. Infectious ecology and host range of influenza D virus.
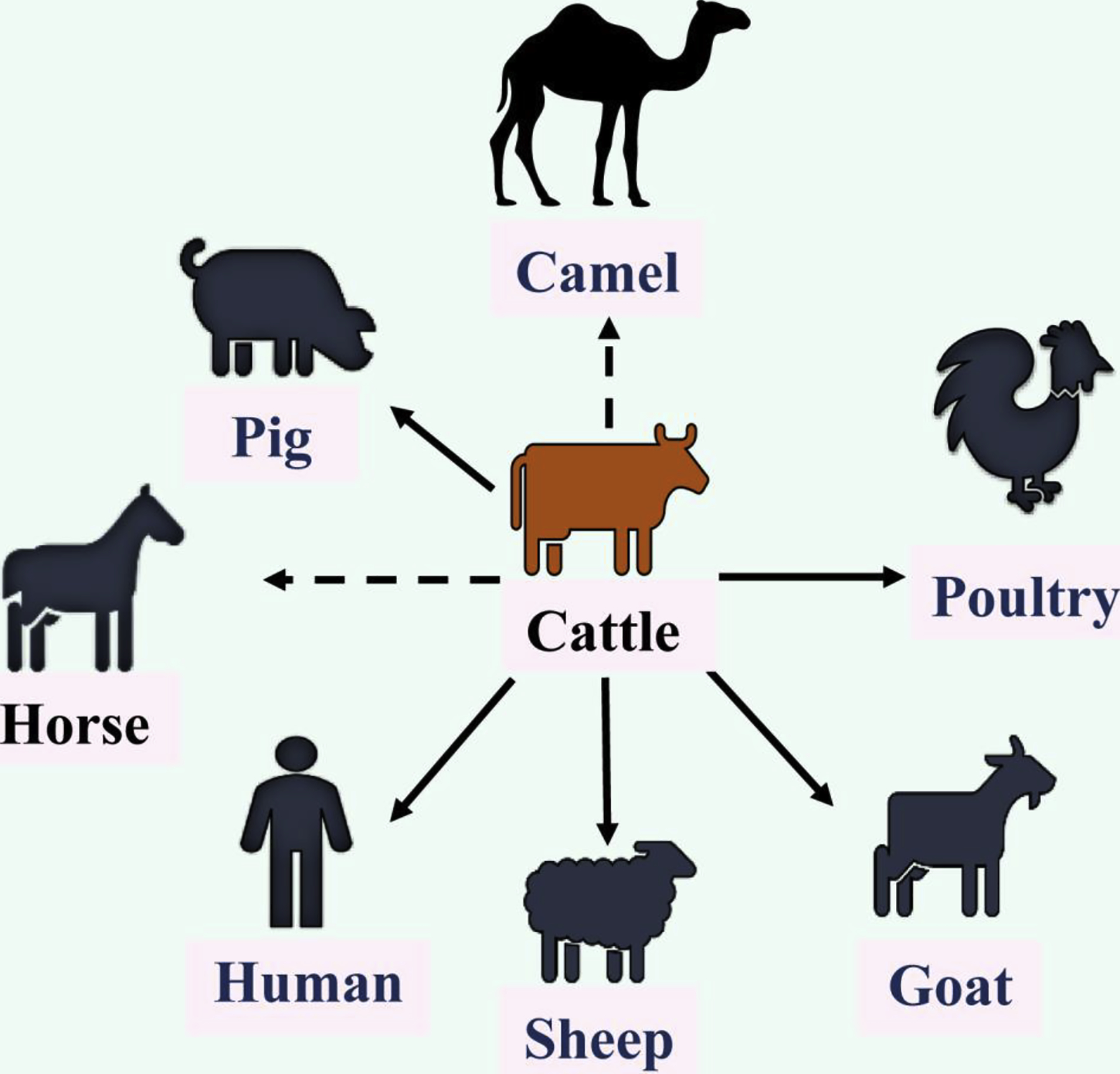
Influenza D virus utilizes bovines as a primary reservoir with frequent spillover to other animals. The bold black line indicates animals or human that derived influenza D virus or viral genome, while the dotted black line represents animals in which viral antibodies were detected without the evidence of virus isolation or the detection of viral genome. Vector graphic images used in the figure were taken from icon pool of the Microsoft Office and Freepik (www.freepik.com).
Figure 2. Maximum-likelihood phylogenetic tree of the Hemagglutinin-esterase (HEF) segment of IDV.
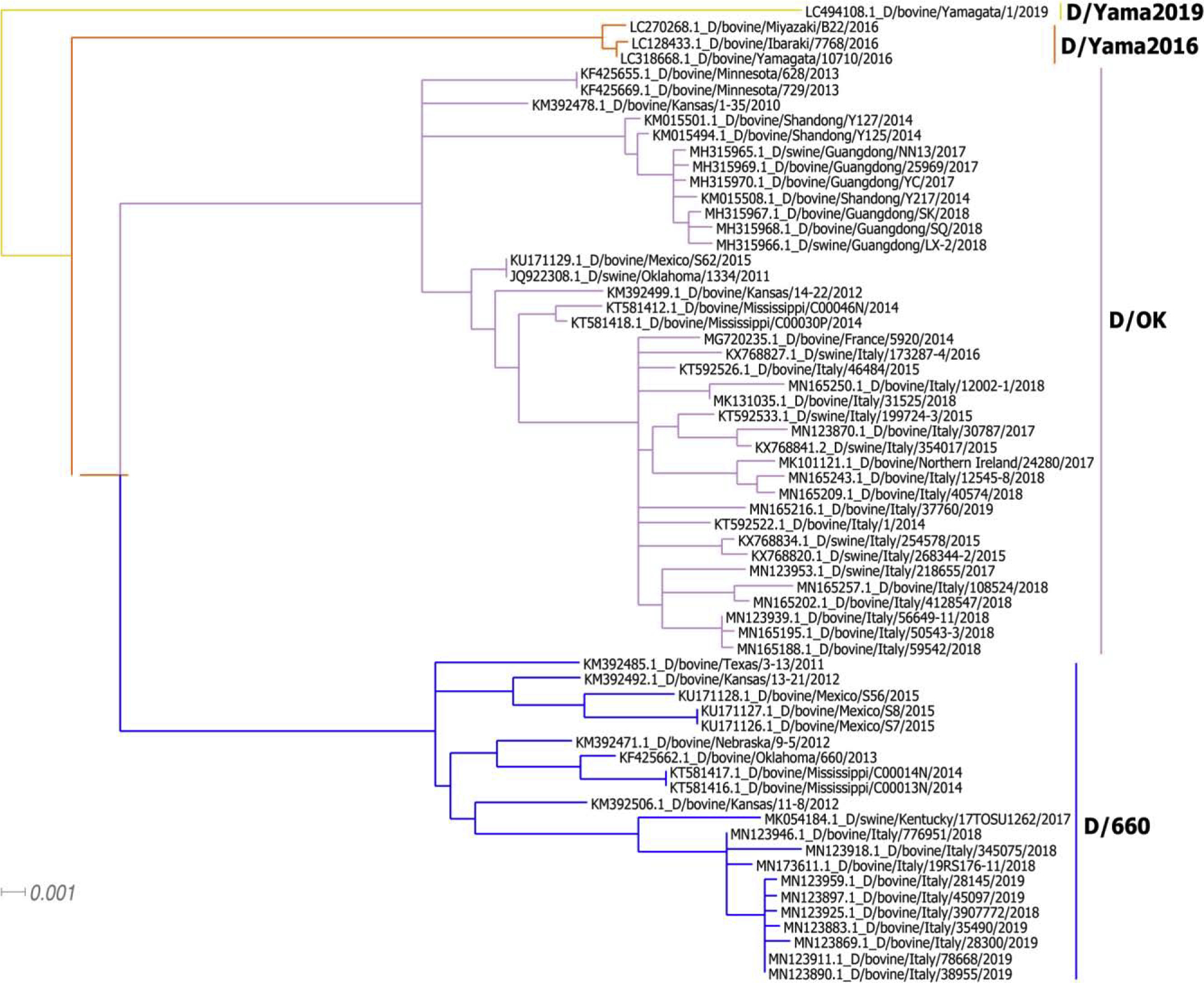
IDV D/660, D/OK, and D/Yama2016, and D/Yama2019 lineages were indicated with different colors, respectively. Total 65 full-length HEF nucleotide sequences available in Genbank were used and the tree was constructed in MEGA 6 using the general time reversible+invariable sites substitution model with 1000 replicates of bootstrap performed.
Impact of influenza D on human health.
Serological evidence of previous exposure to IDV infection in humans has been collected from three independent studies. The first study showed that IDV seroprevalence rate in a cohort aged 60 years and older living in Canada and the eastern US (Connecticut) was about 1.3% (4/312) [4]. The observation of low prevalence of IDV in the general population seems to be in agreement with an etiological investigation in Scotland showing no evidence of IDV infection in archived respiratory samples from hospital-visiting patients [31]. The second study focused on an occupational exposure cohort (cattle-exposed farmers) living in Florida and documented a 97% seroprevalence (34/35), which mirrored the seroprevalence in the cattle population [32]. This study also revealed an 18% seroprevalence (2/11) in non-cattle exposed persons. The third study represents a comprehensively longitudinal study of more than 1,000 human serum samples collected in Italy from 2005 to 2017 for examination of the prevalence of antibodies against influenza D virus [33]. The results of this study showed that the prevalence of IDV antibodies increased in the human population in Italy from 2005 to 2017 with up to 33.9–41.0% of surveyed subjects tested positive in some years. Interestingly, this study revealed a temporal correlation between IDV prevalence peak in humans and epidemics in domestic pigs in Italy [18]. Considering that IDV and its related human influenza C virus share about 50% homology in the hemagglutinin-esterase-fusion protein (HEF), the primary target of the serology assay, the precise nature of human infection with IDV probably needs further investigation by virus-specific antibody assay in future studies.
Significantly, a more recent study showed that IDV genome was detected in nasal washing sample of a swine farm worker in Southeast Asia [34]. Furthermore, molecular surveillance of respiratory viruses with bioaerosol sampling in Raleigh Durham International Airport found that among four (17%) of the 24 samples positive for known respiratory pathogens, one was positive specifically for IDV [35]. It should be noted that none of 24 samples were tested positive for influenza A, B, and C viruses. Using a similar approach, one study has detected IDV genome in hospital visitors in North Carolina [36]. Taken together, these studies collectively demonstrate that IDV is capable of infecting and spreading among humans. The interspecies transmission and the international appearance of IDV in the worldwide animal populations represent a potential risk to global human health, which clearly motivates further investigation of this novel influenza virus.
Biology and tropism of influenza D virus.
IAV and IBV have eight RNA segments with the negative polarity, while ICV and IDV consist of seven negative sense RNAs. Both IAV and IBV contain two major surface glycoproteins: the hemagglutinin (HA) and Neuraminidase (NA). ICV and IDV have only one major surface glycoprotein, hemagglutinin-esterase-fusion (HEF), which performs all entry functions including receptor binding, receptor destroying, and fusion. The overall amino acid sequence of IDV shares approximately 50% identity with that of ICV [4]. It is most closely related to ICV, rather than to IAV and IBV. However, the distance between IDV and ICV is similar to the differences between IAV and IBV for the most of genomic segments [4]. Notably, IDV displays a novel splicing strategy to express the major structural M1 protein [3]. Specifically, IDV M1 protein is translated through a splicing by introduction of a four-residue peptide into the primary transcript, which contrasts with the situation in ICV where a splicing event only introduces a translational termination codon into the primary transcript [37]. In cell culture, IDV is not restricted for replication at 37°C and has a broad cell tropism [4], while ICV is limited for infection at 37°C with a narrow tropism [38,39].
The receptor-binding pocket (RBP) resides in the HEF protein of IDV, which consists of four secondary elements (the 170-loop, 230-helix, 270-loop, and 290-loop) (Figure 3) [40]. Despite the fact that the RBPs between IDV and ICV are similar both in the structure and mode of ligand binding [40,41], IDV RBP possesses an open channel between the 230-helix and 270-loop, whereas this channel is absent in ICV RBP [40]. The open and more accessible RBP may enable IDV readily accommodate diverse extended glycan moieties constituting the cellular receptor, which may explain why the IDV has a broad cell tropism and behaves differentially from ICV in biology and infectious ecology.
Figure 3. 9-O-acetylated SA receptors and HEF structure of IDV.
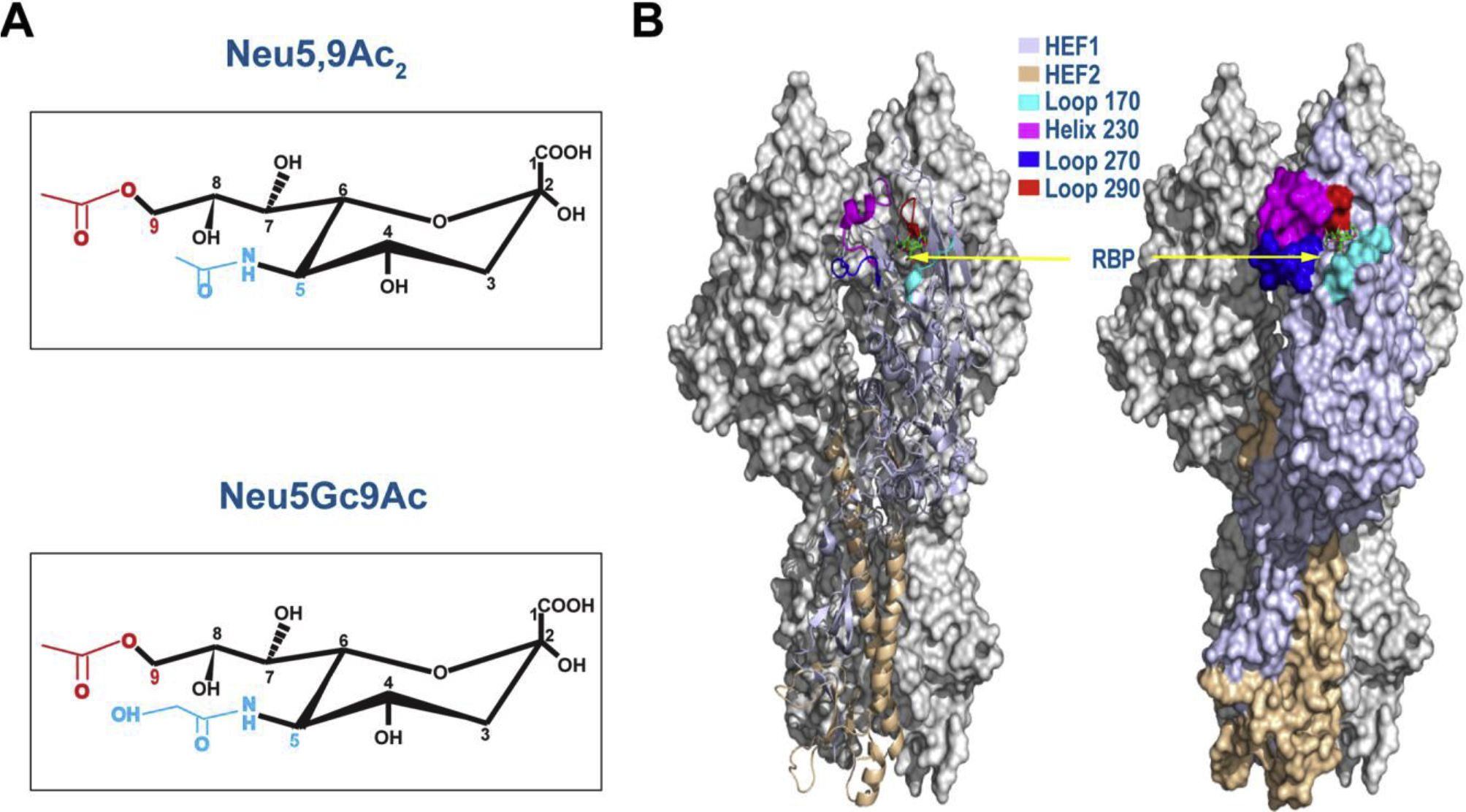
Chemical structures of Neu5,9Ac2 and Neu5Gc9Ac were showed in (A), while structure of IDV D/OK HEF protein trimer (Left, cartoon mode; right, surface mode) (PDB ID:5E65) was presented in the context of binding to an analog of Neu5,9Ac2 Sialic acid (green) (B). Four structural elements involving in the formation of the receptor-binding pocket (RBP) were shown in different colors. Two monomers of a trimer were shown gray with HEF1 and HEF2 subunits of the HEF protein indicated in different colors.
Multiple lines of evidence support that IDV binds to human 9-O-acetylated N-acetylneuraminic acid (Neu5,9Ac2) and non-human N-glycolylneuraminic acid (Neu5Gc9Ac) and utilizes them for viral entry (Figure 3) [40,42]. Several studies demonstrate that IDV is more efficient in recognizing both human Neu5,9Ac2 and nonhuman Neu5Gc9Ac-containing glycans than ICV, and ICV seems to preferentially bind to human Neu5,9Ac2 over non-human Neu5Gc9Ac [40,42]. Neu5Gc and Neu5Ac differ only by a single oxygen atom at the C5 position [43]. Neu5Gc (the precursor of Neu5Gc9Ac) is synthesized by hydroxylation of CMP (cytidine monophosphate)-Neu5Ac to CMP-Neu5Gc through CMP-Neu5Ac hydroxylase (CMAH) [44]. Many mammals contain Neu5Gc/Neu5Gc9Ac in addition to Neu5Ac/Neu5,9Ac2 [45]. In contrast, humans and ferrets can only make Neu5Ac and Neu5,9Ac2, and are deficient in Neu5Gc and Neu5Gc9Ac due to frame-shift mutations occurring in the CMAH that synthesizes Neu5Gc (the Neu5Gc9Ac precursor) [46,47]. Taken together, these results suggest that IDV and ICV diverge in communicating with both O-acetyl group at the C9 and acetyl/glycolyl group at the C5 position in terminal 9-carbon sialic acids (SA). The efficient usage of either human Neu5,9Ac2 or non-human Neu5Gc9Ac gives IDV an advantage over ICV (preferentially binding to human Neu5,9Ac2) in expanding host tropism.
IDV is the most acid stable influenza virus in that only IDV retains approximately 80% of its original infectivity after being treated at pH 3.0 for 30 min, while IAV, IBV, and ICV are completely inactivated under this condition [48]. Importantly, the data from reverse genetics experiment clearly pinpoint that the HEF protein is the primary determinant of IDV acid and thermal stability [48]. The stability of HEF at extremely acidic pH highlights a novel aspect of IDV replication, which warrants further investigation.
Evolution of influenza D virus.
Since 2011, the full genomes of 65 bovine and swine IDV strains have been sequenced in six countries (United States, China, France, Italy, Ireland, and Japan) [3,4,6–11,13,15,16,20,30]. Phylogenetic analyses revealed at least four lineages: D/OK, D/660, D/Yama2016, and D/Yama2019 (Figure 2), with ~4% diversity for the HEF gene [3,4,6–11,13–15,22,30]. Currently, in America and Europe, both D/OK and D/660 lineages are observed [6–9,15]. In China, only strains belonging to the D/OK lineage were reported [11,14], while D/Yama2 recently emerged in Japan [22,30]. Our phylogenetic analysis based on these data showed that IDV diverged from the other three influenza genera about 300 to 1200 years ago [49]. Recently, IDV antibodies were detected in archived US cattle sera dating back at least to 2004 [16]. It indicates that IDV has already existed for a long time.
Evolutionary rate study revealed that IDV evolves with a high substitution rate (1.68×10−3 nucleotide substitutions per site per year for HEF gene), which is about 3-fold faster than ICV HEF and is comparable to some subtypes of IAV and IBV (~1 × 10−3 to 8 × 10−3 nucleotide substitutions per site per year for HA) [50,51] (Figure 4). Thus, unlike ICV, IDV may not be under ‘evolutionarily stasis’. The recent continuous outbreaks of IDV in bovines and other hosts imply that IDV can transmit among hosts with high efficiency. At the same time, the high-affinity attachment to human cells indicates that IDV is superior to ICV in the initiation of viral entry and establishing its infection in humans. Further efforts towards the identification of more IDV lineages and evaluation of their phenotype-and-genotype correlates in replication fitness in in vivo target cells should be useful to locate key genetic and antigenic changes that may modulate its host range and host adaptation.
Figure 4. Evolutionary rate of the HEF gene of influenza D virus.
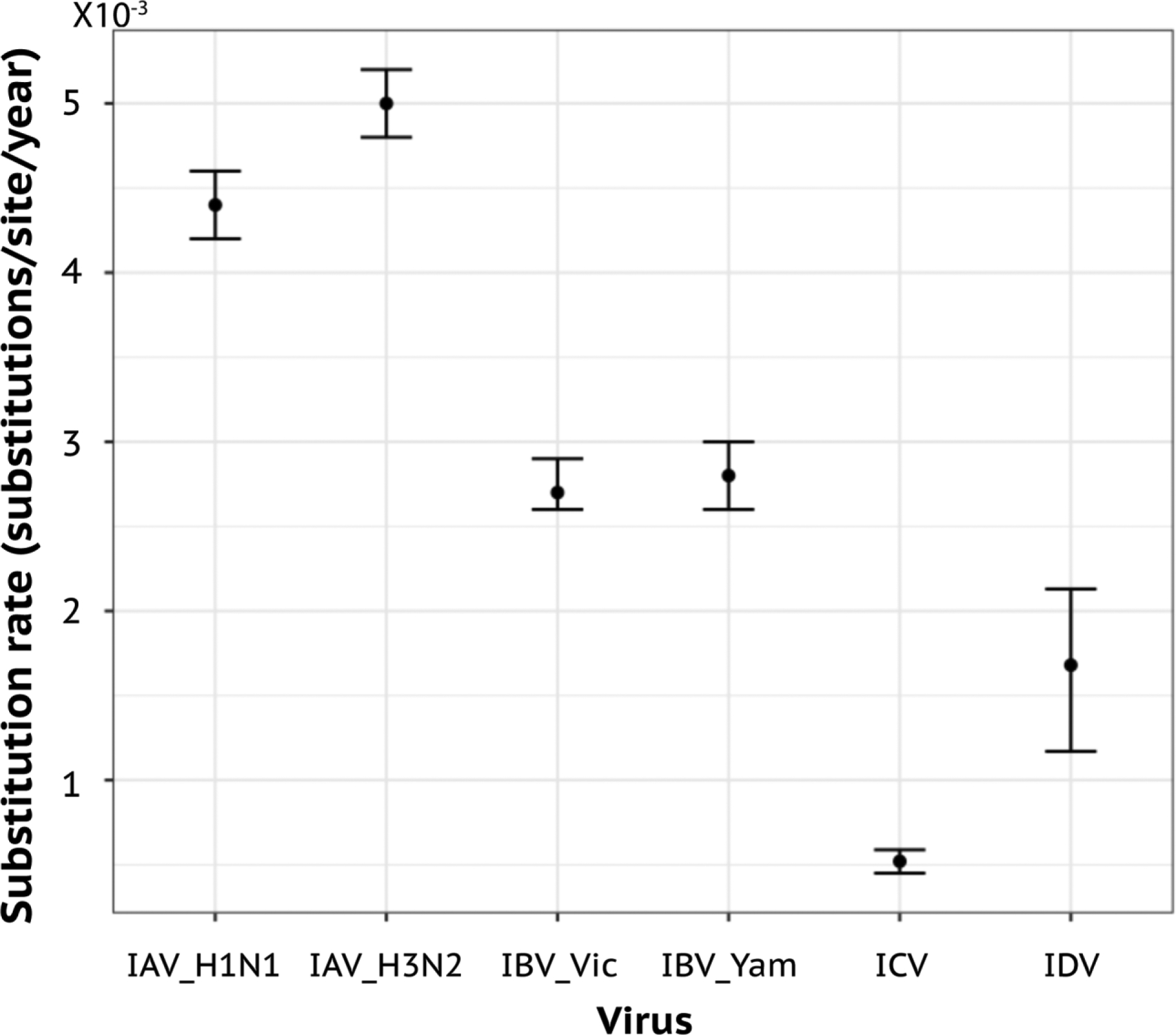
The evolutionary rate of IDV was estimated using BEAST package, with GTR substitution model, restricted molecular clock, and Bayesian Skyline tree prior model. The optimal models were selected using the path sampling approach in BEAST.
Animal models of influenza D virus.
IDV is able to infect and replicate in both ferrets and guinea pigs and transmit to naive animals by direct contact [4,28]. Infected guinea pigs got seroconverted and the virus specific antigens were found in the lung up to 7 days post infection [28]. A notable difference between guinea pig and ferret models is that the virus can be found across the upper and lower respiratory tract including lungs of infected guinea pigs, while detected only in the upper respiratory tract, not in the lungs of infected ferrets [4,28]. In native host, calves supported the replication of IDV in the upper and lower respiratory tract and the efficient virus replication can occur in the lungs of infected animals [9,52]. Virus replication dynamics and transmission in guinea pigs are in a good agreement with those findings obtained in native animals, further strengthening its validity as an animal model to study this newly emerging influenza virus [28]. In addition, mouse model has been developed to study the replication and pathogenesis of IDV [26,27]. Interestingly, in this model, in addition to the respiratory tract, appreciable titers of IDV were found in mice intestines [26]. The observation of IDV presence in the intestines of infected mice seems to be consistent with previous studies showing that IDV shed in fecal samples of animals [14,17], which is supported by the exceptional acid stability of IDV [48].
Pathogenesis and host response of influenza D virus.
Little is known about the pathogenesis of IDV in natural hosts and laboratory animals. Infection in guinea pigs or ferrets was asymptomatic [4,28]. However, lungs from infected guinea pigs showed severe and extensive inflammatory changes in the alveolar space with inflammatory cell infiltration, perivascular cuffing, and destruction of bronchiolar epithelium with exudation [28]. Guinea pigs with IDV infection also demonstrated apoptosis in epithelial cells lining alveoli and bronchioles, as well as in non-epithelial cells of the lungs. In a murine model, neither clinical signs nor weight loss were observed in infected mice [26,27]. Nevertheless, infected mice showed a significant increase of neutrophils and lymphocytes in the lung when compared to mock-infected animals [27]. Additional transcriptomic analyses demonstrated that IDV replication in mice led to activate proinflammatory genes including gamma interferon (IFN-γ) and chemokine CCL2 [26]. These studies also revealed that IDV clearance in mice seemed not to depend on the type I interferon response [26], and IDV infection conferred no appreciable susceptibility to secondary bacterial infection with S. aureus [27].
Limited studies have shown that IDV causes a mild respiratory disease in calves in direct challenge experiments [52,53]. Interestingly, IDV infection can alter the structural integrity of the respiratory epithelium and as a result trigger a significant increase in neutrophils in the trachea of infected animals [53]. This pathological effect seems to suggest an etiological role of IDV in bovine respiratory disease complex (RDC). Further investigation of IDV pathogenesis and host responses in calves showed that IDV infection resulted in moderate bronchopneumonia with restricted lesions of interstitial pneumonia and significant activations of pathogen recognition receptors and chemokines CCL2, CCL3, and CCL4 [52]. Intriguingly, the signaling pathway to activate the type I interferon response was not substantially activated in the lower respiratory tract of IDV-infected calves [52]. This observation seems to be consistent with a recent finding showing that IDV NS1 is capable of suppressing host IFN responses, similar to the NS1 proteins from IAV, IBV, and ICV [54]. Taken together, cattle infection experiments collectively suggest that IDV is a mild respiratory disease pathogen of bovines and acts as an important cofactor of clinically bovine RDC.
Preventive vaccines for influenza D in agricultural animals.
Bovine respiratory disease complex costs the global cattle industry billions per year. As discussed above, IDV has emerged as an important player for this disease complex, thereby indicating a need for an effective and safe vaccine against IDV infection in bovines. Currently, there are no specific vaccines or treatments for influenza D in animals. One candidate vaccine developed for influenza D is a chemical inactivated whole virus particle vaccine [55]. Immunization of cattle with this inactivated vaccine elicited virus-specific immune responses, which provided a partial protection to vaccinated calves from respiratory disease and viral shedding following homologous IDV challenge. In addition, a DNA vaccine expressing the consensus HEF gene of diverse IDV isolates protected guinea pigs from infection by two lineages of IDV [56]. Specifically, guinea pigs received the consensus HEF DNA vaccination developed high-titers neutralizing antibodies. Vaccinated guinea pigs were completely protected against intranasal challenge with two IDV lineage representatives, D/OK and D/660 as viral RNA was not detected in necropsied respiratory tissues of vaccinated animals using quantitative reverse transcription-PCR and in situ hybridization. Although further work is needed to evaluate its protective efficacy against other lineages of IDV in guinea pigs as well as to determine whether the sterilizing protection against diverse lineages of IDV can be observed in bovines or pigs, the encouraging results of this vaccine platform from guinea pigs indicate that a DNA vaccine expressing the consensus HEF has the potential to protect animals from different lineages of IDV infections [56]. As with other types of influenza viruses, IDVs will constantly evolve into more diverse lineages, the IDV vaccine research efforts in the future needs to accommodate for such antigenic drift phenomena and focus on developing a universal vaccine that can effectively protect animals or humans from multiple strains or lineages of IDV. With recently described robust reverse genetics systems for IDV [57,58], rational design of IDV live attenuated vaccine candidates by various mechanisms can be achieved, which represents an alternative platform for the development of safe and efficacious vaccines for IDV.
Conclusions
Novel influenza D virus (IDV) utilizes bovines as a primary reservoir with periodical spillover to other mammalian hosts including pigs. Humans generally have no preexisting immunity against this newly emerging influenza virus. Despite substantial progress made in epidemiology, infection biology, and host range of IDV since its initial appearance in U.S. swine population in 2011, further research on IDV, especially centering on the cross-species transmission and virus binding and fusing into the host cells, is critically needed. Elucidation of IDV entry and adaptation mechanisms to different animal species will be instrumental to develop effective antiviral therapeutics and vaccine strategies against influenza D virus infection in humans and animals.
Acknowledgements:
We thank all the members of the Li and Wang laboratories for their input into this work.
Funding: This work was supported by the National Institutes of Health R01AI141889, SDSU-AES 3AH-673, National Science Foundation/EPSCoR (http://www.nsf.gov/od/iia/programs/epscor/index.jsp) award IIA-1335423, and the SD-CBRC supported by the State of South Dakota’s Governor’s Office of Economic Development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: nothing declared.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Sederdahl BK, Williams JV: Epidemiology and Clinical Characteristics of Influenza C Virus. Viruses 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taubenberger JK, Morens DM: The pathology of influenza virus infections. Annu Rev Pathol 2008, 3:499–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3••.Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, Wang D, Nelson EA, Li F: Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio 2014, 5:e00031–00014. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported on the first isolation of influenza D virus from bovines with respiratory disease symptoms, identified a novel M segment splicing strategy, and proposed a new influenza D genus.
- 4••.Hause BM, Ducatez M, Collin EA, Ran, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, et al. : Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog 2013, 9:e1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study isolated the first influenza D virus from a diseased pig with influenza like symptom and conducted a comprehensive characterization of the novel virus in phylogenetics, biology, and structure modeling. It also demonstrated that ferrets are susceptible to influenza D virus infection.
- 5.Lee J, Wang L, Palinski R, Walsh T, He D, Li Y, Wu R, Lang Y, Sunwoo SY, Richt JA, et al. : Comparison of Pathogenicity and Transmissibility of Influenza B and D Viruses in Pigs. Viruses 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.Chiapponi C, Faccini S, Fusaro A, Moreno A, Prosperi A, Merenda M, Baioni L, Gabbi V, Rosignoli C, Alborali GL, et al. : Detection of a New Genetic Cluster of Influenza D Virus in Italian Cattle. Viruses 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper described the recent introduction of IDV D/660 lineage to Italian cattle.
- 7•.Chiara C, Silvia F, Aurora De M, Laura B, Ilaria B, Carlo R, Arrigo N, Emanuela F: Detection of Influenza D Virus among Swine and Cattle, Italy. Emerging Infectious Disease journal 2016, 22:352. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study described the circulation of influenza D viruses in agricultural animals in Italy.
- 8.Dane H, Duffy C, Guelbenzu M, Hause B, Fee S, Forster F, McMenamy MJ, Lemon K: Detection of influenza D virus in bovine respiratory disease samples, UK. Transbound Emerg Dis 2019, 66:2184–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Ducatez MF, Pelletier C, Meyer G: Influenza D Virus in Cattle, France, 2011–2014. Emerging Infectious Diseases 2015, 21:368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study was the first to show the presence of influenza D viruses in cattle in France.
- 10•.Flynn O, Gallagher C, Mooney J, Irvine C, Ducatez, Hause B, McGrath G, Ryan E: Influenza D Virus in Cattle, Ireland. Emerg Infect Dis 2018, 24:389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrated that influenza D virus circulated in bovines in Ireland.
- 11•.Jiang WM, Wang SC, Peng C, Yu JM, Zhuang QY, Hou GY, Liu S, Li JP, Chen JM: Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes 2014, 49:493–496. [DOI] [PubMed] [Google Scholar]; This study provided the evidence that novel influenza D virus was present in cattle in China.
- 12•.Salem E, Cook EAJ, Lbacha HA, Oliva J, Awoume F, Aplogan GL, Hymann EC, Muloi D, Deem SL, Alali, et al. : Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg Infect Dis 2017, 23:1556–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work provided the serological evience of influenza D infection of Afrcian ruminants and camelids.
- 13.Snoeck CJ, Oliva J, Pauly M, Losch S, Wildschutz F, Muller CP, Hubschen JM, Ducatez MF: Influenza D Virus Circulation in Cattle and Swine, Luxembourg, 2012–2016. Emerg Infect Dis 2018, 24:1388–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai SL, Zhang H, Chen SN, Zhou X, Lin T, Liu R, Lv DH, Wen XH, Wei WK, Wang D, et al. : Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg Infect Dis 2017, 23:1392–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Collin EA, Sheng Z, Lang Y, Ma W, Hause BM, Li F: Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J Virol 2015, 89:1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided the evidence that influenza D viruses have evolved into two different lineages in U.S. animal herds.
- 16.Ferguson L, Eckard L, Epperson WB, Long LP, Smith D, Huston C, Genova S, Webby R, Wan XF: Influenza D virus infection in Mississippi beef cattle. Virology 2015, 486:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Ferguson L, Luo K, Olivier AK, Cunningham FL, Blackmon S, Hanson-Dorr K, Sun H, Baroch J, Lutman MW, Quade B, et al. : Influenza D Virus Infection in Feral Swine Populations, United States. Emerg Infect Dis 2018, 24:1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided the evidence that influenza D virus infection occurred in feral pigs.
- 18.Foni E, Chiapponi C, Baioni L, Zanni I, Merenda M, Rosignoli C, Kyriakis CS, Luini MV, Mandola ML, Bolzoni L, et al. : Influenza D in Italy: towards a better understanding of an emerging viral infection in swine. Sci Rep 2017, 7:11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fusade-Boyer M, Pato PS, Komlan M, Dogno K, Batawui K, Go-Maro E, McKenzie P, Guinat C, Secula A, Paul M, et al. : Risk Mapping of Influenza D Virus Occurrence in Ruminants and Swine in Togo Using a Spatial Multicriteria Decision Analysis Approach. Viruses 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorin S, Fablet C, Queguiner S, Barbier N, Paboeuf F, Herve S, Rose N, Simon G: Assessment of Influenza D Virus in Domestic Pigs and Wild Boars in France: Apparent Limited Spread within Swine Populations Despite Serological Evidence of Breeding Sow Exposure. Viruses 2019, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo J, Ferguson L, Smith DR, Woolums AR, Epperson WB, Wan XF: Serological evidence for high prevalence of Influenza D Viruses in Cattle, Nebraska, United States, 2003–2004. Virology 2017, 501:88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Murakami S, Endoh M, Kobayashi T, Takenaka-Uema A, Chambers JK, Uchida K, Nishihara M, Hause B, Horimoto: Influenza D Virus Infection in Herd of Cattle, Japan. Emerg Infect Dis 2016, 22:1517–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work reported on the first isolation of influenza D virus in cattle herd from Japan.
- 23•.Murakami S, Odagiri T, Melaku SK, Bazartseren B, Ishida H, Takenaka-Uema A, Muraki Y, Sentsui H, Horimoto T: Influenza D Virus Infection in Dromedary Camels, Ethiopia. Emerg Infect Dis 2019, 25:1224–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided the evidence of influenza D infection of African cattle herd.
- 24.Nedland H, Wollman J, Sreenivasan C, Quast M, Singrey A, Fawcett L, Christopher-Hennings J, Nelson E, Kaushik RS, Wang D, et al. : Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health 2018, 65:e148–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quast M, Sreenivasan C, Sexton G, Nedland H, Singrey A, Fawcett L, Miller G, Lauer D, Voss S, Pollock S, et al. : Serological evidence for the presence of influenza D virus in small ruminants. Veterinary microbiology 2015, 180:281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Oliva J, Mettier J, Sedano L, Delverdier M, Bourges-Abella N, Hause B, Loupias J, Pardo I, Bleuart C, Bordignon PJ, et al. : Murine Model for the Study of Influenza D Virus. J Virol 2020, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work established a murine model to study influenza D virus.
- 27.Skelton RM, Shepardson KM, Hatton A, Wilson PT, Sreenivasan C, Yu J, Wang D, Huber VC, Rynda-Apple A: Contribution of Host Immune Responses Against Influenza D Virus Infection Toward Secondary Bacterial Infection in a Mouse Model. Viruses 2019, 11. [Google Scholar]
- 28•.Sreenivasan C, Thomas M, Sheng Z, Hause BM, Collin EA, Knudsen DE, Pillatzki A, Nelson E, Wang D, Kaushik RS, et al. : Replication and Transmission of the Novel Bovine Influenza D Virus in a Guinea Pig Model. J Virol 2015, 89:11990–12001. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that guinea pig model is susceptible to influenza D virus replication and transmission.
- 29.Bailey ES, Fieldhouse JK, Alarja NA, Chen DD, Kovalik ME, Zemke JN, Choi JY, Borkenhagen LK, Toh TH, Lee JSY, et al. : First sequence of influenza D virus identified in poultry farm bioaerosols in Sarawak, Malaysia. Trop Dis Travel Med Vaccines 2020, 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murakami S, Sato R, Ishida H, Katayama M, Takenaka-Uema A, Horimoto T: Influenza D Virus of New Phylogenetic Lineage, Japan. Emerg Infect Dis 2020, 26:168–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith DB, Gaunt ER, Digard P, Templeton K, Simmonds P: Detection of influenza C virus but not influenza D virus in Scottish respiratory samples. J Clin Virol 2016, 74:50–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.White SK, Ma W, McDaniel CJ, Gray GC, Lednicky JA: Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J Clin Virol 2016, 81:31–33. [DOI] [PubMed] [Google Scholar]; This study revealed that influenza D virus-specific antibodies were present in people working in cattle farms in Florida, U.S.
- 33•.Trombetta CM, Marchi S, Manini I, Kistner O, Li F, Piu P, Manenti A, Biuso F, Sreenivasan C, Druce J, et al. : Influenza D Virus: Serological Evidence in the Italian Population from 2005 to 2017. Viruses 2019, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided a longitudinal analysis of influenza D virus antibody prevalence in Italian general population and found that up to 33.9–41.0% of surveyed subjects tested positive to influenza D virus-specific antibodies in some years.
- 34•.Borkenhagen LK, Mallinson KA, Tsao RW, Ha SJ, Lim WH, Toh TH, Anderson BD, Fieldhouse JK, Philo SE, Chong KS, et al. : Surveillance for respiratory and diarrheal pathogens at the human-pig interface in Sarawak, Malaysia. PLoS One 2018, 13:e0201295. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided the first evidence that influenza D virus genome was found in a swine farm worker.
- 35.Bailey ES, Choi JY, Zemke J, Yondon M, Gray GC: Molecular surveillance of respiratory viruses with bioaerosol sampling in an airport. Trop Dis Travel Med Vaccines 2018, 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi JY, Zemke J, Philo SE, Bailey ES, Yondon M, Gray GC: Aerosol Sampling in a Hospital Emergency Room Setting: A Complementary Surveillance Method for the Detection of Respiratory Viruses. Front Public Health 2018, 6:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita M, Krystal M, Palese P: Evidence that the matrix protein of influenza C virus is coded for by a spliced mRNA. J Virol 1988, 62:3348–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu R, Sheng Z, Lin T, Sreenivasan C, Gao R, Thomas M, Druce J, Hause BM, Kaushik RS, Li F, et al. : Genetic and antigenic characteristics of a human influenza C virus clinical isolate. J Med Virol 2020, 92:161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takashita E, Muraki Y, Sugawara K, Asao H, Nishimura H, Suzuki K, Tsuji T, Hongo S, Ohara Y, Kawaoka Y, et al. : Intrinsic temperature sensitivity of influenza C virus hemagglutinin-esterase-fusion protein. J Virol 2012, 86:13108–13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Song H, Qi J, Khedri Z, Diaz S, Yu H, Chen X, Varki A, Shi Y, Gao GF: An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism. PLoS Pathog 2016, 12:e1005411. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study elucidated the structure of an influenza D virus hemagglutinin-esterase-fusion (HEF) protein and revealed the presence of a open receptor binding cavity in the HEF protein that may allow IDV to accommodate diverse extended glycan moieties and have a broad cell tropism.
- 41.Rosenthal PB, Zhang X, Formanowski F, Fitz W, Wong CH, Meier-Ewert H, Skehel JJ, Wiley DC: Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 1998, 396:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Liu R, Sreenivasan C, Yu H, Sheng Z, Newkirk SJ, An W, Smith DF, Chen X, Wang D, Li F: Influenza D virus diverges from its related influenza C virus in the recognition of 9-O-acetylated N-acetyl- or N-glycolyl-neuraminic acid-containing glycan receptors. Virology 2020, 545:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work described the receptor usage of influenza D virus that is different from its related influenza C virus.
- 43.Song X, Yu H, Chen X, Lasanajak Y, Tappert MM, Air GM, Tiwari VK, Cao H, Chokhawala HA, Zheng H, et al. : A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J Biol Chem 2011, 286:31610–31622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawano T, Koyama S, Takematsu H, Kozutsumi Y, Kawasaki H, Kawashima S, Kawasaki T, Suzuki A: Molecular cloning of cytidine monophospho-N-acetylneuraminic acid hydroxylase. Regulation of species- and tissue-specific expression of N-glycolylneuraminic acid. J Biol Chem 1995, 270:16458–16463. [DOI] [PubMed] [Google Scholar]
- 45.Samraj AN, Pearce OM, Laubli H, Crittenden AN, Bergfeld AK, Banda K, Gregg CJ, Bingman AE, Secrest P, Diaz SL, et al. : A red meat-derived glycan promotes inflammation and cancer progression. Proc Natl Acad Sci U S A 2015, 112:542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, Wright KL, Muchmore EA, Nelson DL, Warren ST, Varki A: A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci U S A 1998, 95:11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng PS, Bohm R, Hartley-Tassell LE, Steen JA, Wang H, Lukowski SW, Hawthorne PL, Trezise AE, Coloe PJ, Grimmond SM, et al. : Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors. Nat Commun 2014, 5:5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Yu J, Hika B, Liu R, Sheng Z, Hause BM, Li F, Wang D: The Hemagglutinin Esterase Fusion Glycoprotein Is a Primary Determinant of the Exceptional Thermal and Acid Stability of Influenza D Virus. mSphere 2017, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated a exceptional environmental stability of influenza D virus that is driven by its HEF protein.
- 49.Sheng Z, Ran Z, Wang D, Hoppe AD, Simonson R, Chakravarty S, Hause BM, Li F: Genomic and evolutionary characterization of a novel influenza-C like virus from swine. Arch Virol 2014, 159:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bedford T, Riley S, Barr IG, Broor S, Chadha M, Cox NJ, Daniels RS, Gunasekaran CP, Hurt AC, Kelso A, et al. : Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature 2015, 523:217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuzaki Y, Sugawara K, Furuse Y, Shimotai Y, Hongo S, Oshitani H, Mizuta K, Nishimura H: Genetic Lineage and Reassortment of Influenza C Viruses Circulating between 1947 and 2014. J Virol 2016, 90:8251–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Salem E, Hagglund S, Cassard H, Corre T, Naslund K, Foret C, Gauthier D, Pinard A, Delverdier M, Zohari S, et al. : Pathogenesis, Host Innate Immune Response, and Aerosol Transmission of Influenza D Virus in Cattle. J Virol 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study was a comprehensive investigation of the pathogenesis and virus replication kinetics of influenza D virus in experimentally infected bovines and of the host-mediated anti-IDV innate immune response.
- 53•.Ferguson L, Olivier AK, Genova S, Epperson WB, Smith DR, Schneider L, Barton K, McCuan K, Webby RJ, Wan XF: Pathogenesis of Influenza D Virus in Cattle. J Virol 2016, 90:5636–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work was the first to investigate the pathology and replication and transmission of influenza D virus in bovines.
- 54.Nogales A, Aydillo T, Avila-Perez G, Escalera A, Chiem K, Cadagan R, DeDiego ML, Li F, Garcia-Sastre A, Martinez-Sobrido L: Functional Characterization and Direct Comparison of Influenza A, B, C, and D NS1 Proteins in vitro and in vivo. Front Microbiol 2019, 10:2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Hause BM, Huntimer L, Falkenberg S, Henningson J, Lechtenberg K, Halbur T: An inactivated influenza D virus vaccine partially protects cattle from respiratory disease caused by homologous challenge. Vet Microbiol 2017, 199:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work described a traditionally inactivated vaccine that provided a partial protection of bovines against influenza D virus challenge.
- 56•.Wan Y, Kang G, Sreenivasan C, Daharsh L, Zhang J, Fan W, Wang D, Moriyama H, Li F, Li Q: A DNA Vaccine Expressing Consensus Hemagglutinin-Esterase Fusion Protein Protected Guinea Pigs from Infection by Two Lineages of Influenza D Virus. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided the evidence that a DNA vaccine expressing the consensus Hemagglutinin-Esterase-Fusion protein can protect guinea pigs from infection by two lineages of influenza D virus
- 57.Ishida H, Murakami S, Kamiki H, Matsugo H, Takenaka-Uema A, Horimoto T: Establishment of a Reverse Genetics System for Influenza D Virus. J Virol 2020, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu J, Liu R, Zhou B, Chou TW, Ghedin E, Sheng Z, Gao R, Zhai SL, Wang D, Li F: Development and Characterization of a Reverse-Genetics System for Influenza D Virus. J Virol 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]


