Abstract
Multiple hosts and various life cycle stages prompt the human malaria parasite, Plasmodium falciparum, to acquire sophisticated molecular mechanisms to ensure its survival, spread, and transmission to its next host. To face these environmental challenges, increasing evidence suggests that the parasite has developed complex and complementary layers of regulatory mechanisms controlling gene expression. Here, we discuss the recent developments on the discovery of molecular components that contribute to cell replication and differentiation, and highlight the major contribution of epigenetics, transcription factors, and nuclear architecture in controlling gene regulation and life cycle progression in Plasmodium spp.
Keywords: Malaria, Plasmodium, epigenetics, transcription factor, chromatin architecture
Malaria and the Plasmodium parasite
Despite significant progress in the last decade, malaria remains the deadliest parasitic disease in the world with 228 million cases and 405,000 deaths reported in 2018 [1]. Most malaria cases occur in African and South East Asian region. This disease is caused by the protozoan Plasmodium, which belongs to the phylum Apicomplexa (see Glossary). The genus Plasmodium is composed of more than 200 species infecting various metazoans. Five species can infect humans: P. vivax, P. malariae, P. ovale, P. knowlesi, and P. falciparum, the last being the deadliest because of its ability to escape the host immune system and sequester in capillaries and venules of vital organs including the brain.
These Plasmodium species possess a complex life cycle involving various stages and two hosts: female Anopheles and humans (Figure 1). To survive in such different conditions, a tight regulation of gene expression is crucial for coordinated and controlled transition between the different parasitic stages. Over the past decades, microarray and RNA-seq experiments have examined the transcriptional dynamic at the population level and highlight a simple cascade of gene regulation that directs the parasite development across the different life cycle stages [2–8]. More recently single-cell RNA-seq (scRNA-seq) produced high resolution mapping of transcriptomic profiles across the complete Plasmodium life cycle to not only identify signature of sexual commitment in Plasmodium [9,10] but also discrete and abrupt transition of transcriptional signatures suggesting that expression over development is not as continuous and plastic as commonly thought [10–13]. Deciphering the mechanisms controlling gene expression is an important step in malaria research to identify novel therapeutic strategies to hamper parasite transmission. In this review, we discuss our current understanding of the mechanisms controlling gene regulation in Plasmodium spp. through layers of interconnected biological processes including nucleosome landscape, epigenetic features, transcription factors, and chromatin organization, which together control the parasite’s ability to survive and spread.
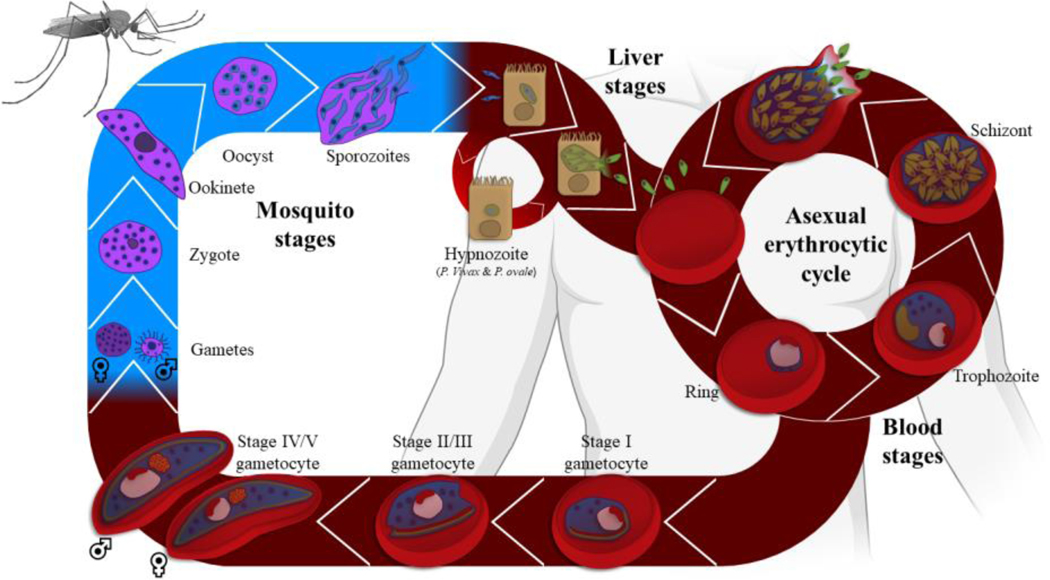
Figure 1. Life cycle of P. falciparum.
An infected mosquito injects sporozoites into the human during a blood meal. After invading the hepatocytes, parasites replicate multiple times to generate thousands of merozoites, which then invade red blood cells and initiate the intraerythrocytic developmental cycle (IDC). In P. vivax and P. ovale, a dormant stage, hypnozoite, is able to persist in the liver for months, sometime years, before being reactivated and released into the blood stream. During IDC, a parasite multiplies asexually and undergoes several rounds of DNA replication resulting in the formation of 16 to 32 new merozoites. These merozoites reinvade new RBCs after bursting out of the host cells. During this cycle, a number of these parasites differentiate into male and female gametocytes and are taken up by a mosquito. The formed zygote develops into an ookinete, then an oocyst in the Anopheles midgut. The oocyst grows and divides into thousands of sporozoites which migrate to the salivary glands to ensure a new full cycle. The mosquito stages are represented in blue and human stages in red.
Histone and nucleosome landscape in Plasmodium
Inside the nucleus of all eukaryotic species, the genome is packed and organized into chromatin. Principles of chromatin organization and nucleosome occupancy are generally well conserved in eukaryotes including Plasmodium spp. (Figure 2A). First, most promoters of active and highly expressed genes share a nucleosome-depleted region (NDR) in the promoter near the transcription start site (TSS) flanked by a strong positioning of nucleosomes in the coding regions. This NDR allows the access and binding of the general transcription machinery to actively transcribe the genes needed [14–17]. Alternatively, while still displaying an NDR in their promoters near the TSS, silenced or lowly expressed genes display a higher level of nucleosome occupancy indicating the presence of a transcription barrier in their promoter regions [14–16]. This was particularly true in nucleosome landscape studies completed in asexual Plasmodium blood stages where genes known to be significantly upregulated during sexual and transmission stages (gametocytes and sporozoites) had a high level of nucleosome occupancy observed in their promoters [15,18]. As a whole, results from these initial studies indicated that nucleosome architecture undergoes drastic changes throughout the parasite cell cycle following through silent and active transcriptional stages [8,15,18–20]. Nucleosomes loosen after the invasion of red blood cells to allow massive transcription at the trophozoite stage, and then repack prior to the next cycle. Changes in nucleosome occupancy within promoter regions follow this genome-wide pattern, with a few exceptions such as genes involved in invasion, virulence (var genes), sexual differentiation, and transmission [15,18–21]. In addition to their positioning, nucleosome subunits can be exchanged with histone variants H2A.Z, H2B.Z, CenH3, and H3.3 to confer complementary and alternative functions (Figure 2A) [19,22,23]. Interestingly, the positioning of the histone variant H2A.Z seems to have a parasite-specific feature as it was shown to be enriched in P. falciparum intergenic AT-rich regions, irrespective of their active or inactive stage [24,25]. This was specifically true for most genes with the exception of clonally variant genes (var genes) (Box 1) expressing the Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1), proteins involved in mechanism regulating antigenic variation that allow the parasite to evade the human host immune system for successful proliferation and transmission. Exploring nucleosome landscape in other life cycle stages or in other Plasmodium species such as P. vivax or rodent parasites, could highlight specific occupancy or composition features to validate preliminary results and further confirm the importance of nucleosome occupancy in Plasmodium gene regulation.
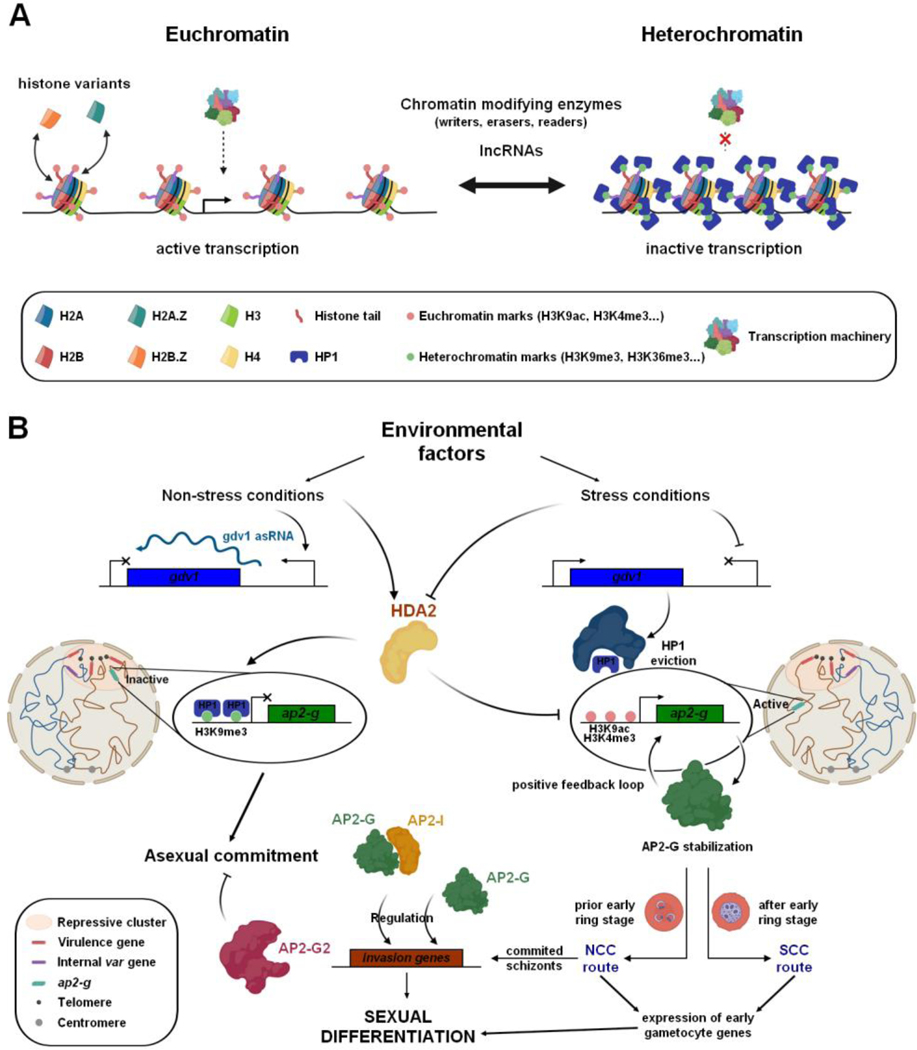
Figure 2. Chromatin remodeling and sexual commitment in Plasmodium.
(A) Nucleosome occupancy, histone variants, and epigenetic marks participate in the chromatin remodeling at a given promoter and lead to the accessibility of the transcription machinery. LncRNAs and chromatin modifying enzymes are also involved in the regulation of gene expression. (B) Cellular and molecular mechanisms implied in the gametocyte commitment. Dynamic chromatin structure and epigenetic factors are crucial in the control of the master regulator of gametocytogenesis, AP2-G.
Box 1: Epigenetic regulation of var genes expression.
The most studied epigenetics regulation mechanism in P. falciparum include the var multi-gene family that encode for the PfEMP1 antigen, critical to control antigenic variation and virulence factors, and of which only one gene is transcribed at a time. The expression of PfEMP1 is highly regulated in asexual blood stages with all repressed var genes enriched in H3K9me3, H2A and HP1 [26,27,56,91,92,102] and present in the nuclear periphery [92,95,96,103], while the promoter of the expressed gene is enriched in H3K9ac and double-variant histones H2A.Z/H2B.Z [25] together with the histone methyltransferase PfSET10 required for H3K4me3 modification [104]. PfSir2, a histone deacetylase, as well as the methyltransferase, PfSETvs, required for H3K36me3, have been shown to actively participate in var gene silencing [53,55,56,91,92]. H3.3 occupancy may also serve as a potential memory mark for virulence gene expression [105]. Other antigen families such as eba and clag exhibit similar epigenetic mechanisms with H3K9me3 and H3K9ac enriched in repressed and active states, respectively [106]. Oocyst and sporozoite stage data have demonstrated the presence of a reset mechanism in the regulation of var family with the presence of repression markers such as PfHP1 and H3K9me3 [107], and active marks H3K9ac and H3K27ac correlating with gene regulation [108]. Hi-C analysis of var genes in different strains of P. falciparum suggests that transcriptional switching is more dependent on local changes in the region adjacent to the active locus than on long-range interactions [97]. In addition to histone PTMs, sense and antisense lncRNAs, expressed from var introns, have been identified as essential for the activation and var gene switching [42–44]. Likewise, GC-rich genes, adjacent to internal virulence genes, are transcribed to control in trans- and cis- the mutually exclusive expression of var genes [45,46]. CRISPR interference of this family affected the steady state of var genes as well as the clonally variant gene family Pfmc-2TM [47].
Epigenetic regulation in Plasmodium
Although this dynamic rearrangement of the histone core and histone variants provides a crucial regulatory mechanism, various post-translational modifications (PTMs) of the histone tails such as methylation, acetylation, and phosphorylation, also described as the “histone code”, can significantly alter the degree of interaction of the nucleosome with the DNA. Such changes in the interaction play an important role in chromatin remodeling and, thus, in gene expression as it enables the recruitment or inhibition of the transcriptional machinery in the promoter regions. Several quantitative mass spectrometry approaches have identified a large amount of histone PTMs in Plasmodium [22,26–28]. The most recent and complete study detected over 230 PTMs during the intraerythrocytic developmental cycle (IDC) of P. falciparum [29]. Some of these epigenetic modifications are considered universal marks of active promoters or characteristics of euchromatin. This is specifically true for the histones H3K9me3, shown to interact with the heterochromatin protein 1 (HP1), and H3K36me3, which are associated with silenced promoters and heterochromatin regions [26,27,30,31], and are mutually exclusive of histones H3K9ac and H3K4me3 (Figure 2A) [19,20,32]. In P. falciparum, the importance and the role of histone marks in gene expression have been extensively studied for the regulation of var genes involved in antigenic variation and pathogenesis of severe malaria (Box 1). A recent machine learning algorithm using most histone modification and nucleosome positioning studies published in P. falciparum specifically suggests that H3K9me3, H4K20me3, and H3K9ac have a schizont-specific regulatory role in the erythrocytic cycle of Plasmodium [33]. Histone PTMs are also associated with the regulation of genes involved in sexual differentiation or gametocytogenesis and these changes could be influenced by environmental stimuli. We describe below and in Figure 2 some of the modifications involved in the expression of two major contributors of the sexual commitment, P. falciparum gametocyte development protein 1 (PfGDV1) and AP2-G. Furthermore, a subset of gametocyte-specific histone PTMs have been identified by mass spectrometry and contains H3K36me2, H3K36me3, and H3K79me3 marks specific to early or late gametocytes, while H3K9me2 and H3K18me1 were only detected during IDC [28]. While a significant number of experiments have been performed in P. falciparum, recent studies have generated additional epigenetic maps in other Plasmodium species. In P. vivax sporozoites, a high level of H3K9me3 marks was observed in subtelomeric regions, probably due to the richness of these regions in virulence and invasion genes [34]. Meanwhile, H3K9ac and H3K4me3 were reported within 1000 kb up and downstream of highly expressed genes, rather than in the coding region itself [34]. In the rodent malaria parasite P. berghei, a recent epigenomics map generated across asexual stages, male and female gametocytes, as well as ookinetes, confirmed, as already described in all other Plasmodium species, the restricted presence of H3K9me3 mark and HP1 in telomeric and subtelomeric regions [35]. H3K9ac marks were also broadly distributed across whole chromosomes in all stages studied. This later study reported that H3K9ac enrichment in the 5’ UTR was not positively associated with transcripts known to be upregulated in female gametocytes, most likely because these mRNAs have already been produced and stored at this particular stage. In P. cynomolgi, a recent study using a histone methyltransferase inhibitor was able to accelerate the rate of hypnozoite activation in primary hepatocyte cultures [36]. Whether the effect of the compound was directly mediated through the parasite or the host histone methylation status remains to be determined. However, these results indicate that epigenetic mechanisms are implicated in the control of gene expression in all major stages of the malaria parasite life cycles. These histone PTMs could also be subject to changes induced by environmental and stress conditions.
Another aspect of the epigenetic regulation concerns the involvement of noncoding RNAs (ncRNAs), conventionally divided into small (sncRNA) and long ncRNA (lncRNA) based on the RNA length (± 200 nucleotides). Although the existence and function of these RNAs remains unclear, thousands of lncRNAs have been reported in eukaryotes including Plasmodium. In higher eukaryotes, several lncRNAs have been implicated in gene regulation, such as chromosome dosage compensation, imprinting, transcription, translation, splicing, epigenetic regulation, and cell differentiation [37]. They often serve as a molecular signal in response to various stimuli and act as signal, decoy, guide, and scaffold for some of the key regulatory proteins. In P. falciparum, hundreds of ncRNAs have been identified as expressed in the genome in a stage-specific manner [38]. One subset of these lncRNAs was identified as transcribed from the telomere-associated repetitive elements (TAREs) and telomeres, and could contribute to the stability of these regions and the maintenance of heterochromatin state [39–41]. The involvement of lncRNAs [42–44] and GC-rich ncRNAs in var gene activation [45–47] (Box 1) or an antisense RNA in regulation of the P. falciparum gametocyte development protein 1 (PfGDV1) [38,48] shows that ncRNAs may have an important impact on all stages. Given the crucial and diverse functions of lncRNAs in higher eukaryotes [37], their involvement in controlling Plasmodium epigenetic features and gene regulation remains to be investigated at the mechanistic level [49].
Chromatin-associated proteins
In view of the fundamental role of epigenetic modifications on gene regulation, these reversible PTMs are under control of wide range of proteins identified as histone writers, erasers, and readers which respectively deposit, remove, and interpret these histone marks. The histone acetylome is regulated by histone acetyltransferases (HATs), including GNAT and MYST proteins, and their antagonists, histone deacetylases (HDACs), composed of HDAC and sirtuin proteins [50]. These proteins are crucial to maintain correct epigenetic marks at the time, such as PfGCN5 and HDACs, whose respective inhibition leads to deregulation of gene expression in P. falciparum [32,51]. Histone lysine methylations are regulated by methyltransferases (HKMTs) and demethylases (HKDMs), but their role has been less studied in the parasite [52]. Disruption of Sir2A and B [53,54], and PfSETvs [55,56], a HKMT enzyme, showed an increase of the transcription of a larger number of var genes indicating their involvement in virulence genes silencing (Box 1). Considering the critical role of these enzymes, the use of inhibitors that could target these enzymes has been intensively investigated to disrupt the parasite life cycle progression with high specificity [57]. While writers and erasers are critical for the parasite development, histone marks are also recognized by readers to promote recruitment or stabilization of protein complexes mediating diverse biological processes. A recent proteomic approach identified several readers in P. falciparum [58] including HP1 or PfGCN5-PfADA2, two members of the SAGA complex, well-known in eukaryotes to be involved in multiple transcriptional steps such as transcription initiation and elongation [59]. Another identified reader from this study is the bromodomain protein PfBDP1, in association with PfBDP2 and PfAP2-I, that binds to acetylated histones present in promoters of invasion genes and has been shown to be involved in the expression of these parasite specific genes to ensure the proper invasion of erythrocytes [60,61].
In addition to writers, erasers and readers, numerous non-histone proteins are associated to chromatin. They participate in multiple biological processes such as DNA maintenance and replication as well as transcription. Several of these proteins including SMCs and CRWN-like proteins were recently identified in P. falciparum by chromatin enrichment for proteomics [62]. The latter are believed to have a function similar to the lamina protein in plants [63], and seem to be associated to heterochromatin regions in Plasmodium [62]. SMC proteins are more conserved across eukaryotes and are involved in chromosome condensation, segregation, and DNA repair. In P. falciparum, using ChiP-seq experiment, PfSMC3, a subunit of the cohesin complex was shown to be localized to the centromere region on all 14 chromosomes [62]. This result is similar to what is observed in yeast [64] and suggest that in Plasmodium, cohesin has a possible role in sister chromatid separation and cell cycle regulation. In P. berghei, SMC2 and SMC4, two core members of the condensin complex, were localized in centromeres and demonstrated to be, not only essential to parasite development, but also to participate in chromosome organization during schizogony and endoreduplication of male gametocytes [65]. While additional experiments are needed to validate the function of many other proteins identified in this recent study [62], preliminary results indicate that these chromatin-associated proteins have a crucial role in parasite development and validate the importance of chromatin structure to various biological processes.
Transcription factors repertory
As described above, transcriptional activity is highly influenced by the nucleosome occupancy and epigenetic state at the promoter region to modulate accessibility of the transcription machinery to the chromatin. Transcription of protein coding genes is carried out by RNA polymerase II (RNA Pol II) and associated subunits. Recruitment of this complex to the promoter region requires the general transcription factors that include TFII subunits [66]. In eukaryotes, the TATA-binding protein (TBP) and TBP associated factors (TAFs) constitute the TFIID complex. However, some of the TAFs identified in Plasmodium do not have the histone fold domain responsible for the heterodimerization of TAFs [67]. Alternative mechanisms of transcriptional regulation have been demonstrated in the parasite, such as the binding of PfTBP and PfTFIIE to promoters independently of their transcriptional activity and presence of acetylated histones [68]. Analysis of RNA Pol II occupancy showed two phases of binding during IDC which do not fully correlate with to the mRNA levels detected, indicating the existence of regulatory mechanisms downstream of transcription initiation [69]. In a more recent study, that compared genome wide nascent RNA to mRNA steady state profiling, the pausing of RNA Pol II observed at the early ring stage was significantly higher than in all other blood stages analysed validating that RNA Pol II pausing mechanism is important for regulating gene expression [70].
Another critical component of the transcriptional machinery is the specific transcription factors (TFs). They are required to facilitate the recruitment of the chromatin remodelling and chromatin modifying enzymes necessary to modulate nucleosome positioning and occupancy. Once nucleosomes are removed from the promoter regions upstream of the genes, the transcription preinitiation complex, that includes the RNA polymerase, can form and bind to the promoter to activate transcription. In Plasmodium, the discovery of TFs has been quite challenging. In addition to the very rare representatives of common TFs present in Plasmodium [71], an apicomplexan-specific TF family, designated ApiAP2, has been identified in the parasite genome [72]. These ~27 AP2s are analogous to plant Apetala-2 and each protein contains one to four AP2 DNA binding domains [73]. Despite the identification of this family, TFs remain extremely low in number to regulate the ~5,500 genes in the parasite genome compared to other eukaryotes [71], and researchers remain puzzled as to how such a small number of TFs can control a tight chromatin remodeling program and a complex gene expression pattern. Several AP2 TFs have been reported as potential master regulators to mediate stage specific transitions including gametocyte (AP2-G and AP2-G2) [74,75], ookinete (AP2-O) [76], sporozoite (AP2-SP) [77], and liver stages (AP2-L) of either in P. falciparum or P. berghei [78]. Recent knockout screenings of ApiAP2 family members in P. berghei and P. yoelii points toward their complex pattern of controlling the gene expression to act as repressors or activators in the stage transitions [79,80]. RNA-seq analysis of AP2-exp mutant parasites has also revealed transcriptional changes in the subset of exported proteins associated with clonally variant gene families, RIFIN and STEVOR [81]. While the presence of AP2 motifs upstream of genes whose expression varied across the erythrocytic cycle led to the hypothesis that these AP2 factors could be the missing reservoir of sequence specific TFs in Plasmodium, however machine learning algorithm did not identify these motifs as critical in regulating gene expression during the Plasmodium erythrocytic cycle [33]. It is tempting to speculate that AP2 DNA-binding requires both a TF-specific motif and a permissive epigenetic state at a given locus to regulate gene expression. DNA accessibility and epigenetic state are also known to play a critical role in restricting TF binding in eukaryotes, in general [82]. Additional experiments would be essential to determine the specificity of these TFs throughout the parasite life cycle progression.
Regulation of gametocytogenesis
A key gap in information regarding these TFs is our understanding of their activation, their precise mode of action, and how they interact with other factors of the transcriptional machinery, including chromatin remodelling and modifying enzymes. For years, researchers have focused their interest on the transcriptional master regulator of sexual commitment in Plasmodium spp., AP2-G (see Figure 2B) [74,83,84]. In the asexual stages, the AP2-G locus is enriched in HP1 and heterochromatin marks leading to its silencing [48,85,86]. Stress conditions such as high parasitemia or lysophosphatidylcholine restriction [87] promote sexual differentiation indicating that environmental signals are crucial determinants for this commitment. How these signals act early on the chromatin remains to be determined, but these factors counteract the repression of GDV1 expression by its own antisense RNA, triggering HP1 expulsion on the ap2-g gene [48]. With this eviction, the heterochromatin surrounding the AP2-G locus is destabilized and facilitates the binding of transcription factors to activate the first wave of AP2-G expression [9,48]. After an initial AP2-G peak, a drop in its transcripts is observed temporarily and correlate with increased level of PF3D7_1222400 expression, an adjacent ApiAP2, which could play a role in AP2-G regulation [9]. The stabilization of AP2-G is required for the second wave of expression in a positive transcriptional feedback loop. Depending on when AP2-G is stabilized during IDC, two pathways of sexual commitment were described [88]. The classical route corresponds to the Next Cycle Conversion (NCC) and implies schizogony and reinvasion while the Same Cycle Conversion (SCC) route requires PfAP2-G stabilization in early ring stage allowing a direct sexual conversion (Figure 2B). Hundreds of genes across the genome have been reported to be transcriptionally activated by AP2-G [9,83,84,89], which binds specific binding motifs on their promoter regions [73,74,84,89]. These AP2-G targets correspond mainly to early gametocyte genes including its own promoter and secreted proteins in P. falciparum [9,83], which is involved in its atypical erythrocyte remodeling during gametocytogenesis and not present in P. berghei [84]. Some invasion genes (eba175, ron5 and sera) are also targeted by AP2-G, sometimes in association with AP2-I in committed schizonts [9,89], while another ApiAP2, AP2-G2, plays an essential function in gametocyte development by repressing asexual proliferation [75]. Conditional knock-down of all of the identified TFs as well as pull down and ChIP-seq experiments would be valuable tools in determining how they interact with chromatin and regulate gene expression precisely throughout the parasite life cycle progression [33].
Nuclear architecture in Plasmodium
Along with the nucleosome landscape and other layers of regulation such as histone PTMs and TFs, the way the genome is organized and compartmentalized at the 3-dimensional level inside the nucleus represents another layer of complexity in our understanding of chromatin structure and gene regulation in the parasite life cycle. Initial results using Fluorescence In Situ Hybridization (FISH) and fluorescence microscopy approaches have shown a precise arrangement of the chromatin inside the nucleus of P. falciparum. During intra‐erythrocytic development, parasite centromeres were identified as demarcated by the presence of the histone variant PfCENH3 and sequestered to a single nuclear location from ring to mature schizont stages [90]. Clonally variant genes, associated with antigenic variation and localized mostly in subtelomeric regions, were detected at the periphery of the nucleus in a heterochromatin state marked by H3K9me3 and HP1 [91,92]. An electron microscopy technique was able to construct a 3D model of the nucleus at distinct stages of parasite development [93]. These results suggest that change in the patterns of gene expression observed during the erythrocytic cycle correlate with modifications in the number and distribution of nuclear pores as well as chromatin organization, and confirm changes in nucleosome positioning observed using complementary genome-wide approaches [8,15,16,18]. The number of nuclear pores increase in the trophozoite stage and correlate with active gene expression patterns associated with this particular stage [93,94]. Their localization is consistent with euchromatin clusters, probably to ensure an adequate export of mRNAs into the cytoplasm from the transcriptionally active territory.
Recently, chromosome conformation capture based on incidences of physical interactions between loci and coupled with next-generation sequencing (Hi-C) (Box 2) were used on various Plasmodium genomes to model, at the genome wide level, the chromatin spatial organization and how it relates to gene expression [30,95–97]. In Plasmodium genomes, Hi-C data and 3D modeling have shown a generally simple nuclear organization throughout different stages of the parasite life cycle. The genome architecture does not seem to contain well-defined topologically associating domains like those found in human, mouse, and Drosophila nuclei [97–99]. However, the P. falciparum nucleus does exhibit complex features that are largely associated with virulence genes. Similar to FISH, Hi-C experiments confirmed that the var genes and heterochromatin cluster colocalize at the periphery of the nucleus and demonstrated that chromatin looping brings internal var and repressed genes physically close to subtelomeric chromosomal regions (Figure 2B). Clustering of the SICAvar genes was also observed in P. knowlesi, most likely to ensure control of their mutually exclusive gene expression [95]. Hi-C results also established that centromeres and telomeres form two clusters at opposite sides of the nucleus (Figure 3) [95,96]. Moreover, the maximum size and volume of the genome is detected at trophozoite stage, confirming an open and accessible chromatin to the transcription machinery, in agreement with the intense transcriptional activity described for this stage [96]. Clustering of A-type ribosomal DNAs (rDNA), transcriptionally active in the asexual and gametocyte stages but not in sporozoite, were localized near the nuclear periphery [30,96,97]. This observation strongly suggests the importance of genome architecture and loop formation in gene regulation.
Box 2: Overview of chromosome conformation capture techniques.
Identifying chromatin interactions is necessary to fully understand how chromatin folding and the resulting 3-dimensional genome architecture regulate transcription. Analysis of the interactions between two distinct loci within the genome was first performed using the Chromosome Conformation Capture (3C) technique [109]. However, this method is limited in throughput because the use of locus-specific primers only allows investigation of a single pair of interacting loci at a time. To overcome this limitation, the 3C technique was adapted using microarrays and chromosome conformation capture-on-chip (4C) was developed to screen an entire genome for all interactions between all loci and any region of interest [110,111]. Simultaneously another method was being developed called chromosome conformation capture carbon copy (5C), which uses multiplexed ligation-mediated amplification (LMA) to amplify fragments within a 3C library prior to microarray or DNA sequencing analysis [112,113]. There are also limitations using 4C and 5C techniques because although throughput is increased over traditional 3C techniques, it is still necessary to have a target locus of interest and therefore cannot scan the genome in an unbiased manner.
An unbiased snapshot of interactions across an entire genome (Hi-C) was constructed using massively parallel next-generation sequencing to identify interactions in an “all-vs-all” manner [114,115]. Although exact protocols differ between experiments—most prominently in the restriction endonuclease used to digest the cross-linked chromatin [96,98,115–117], which affects the data resolution [118]—the general Hi-C protocol in various studies remains the same. Limitations in resolution of the data are due to sequencing depth and linear separation of interacting loci [119].
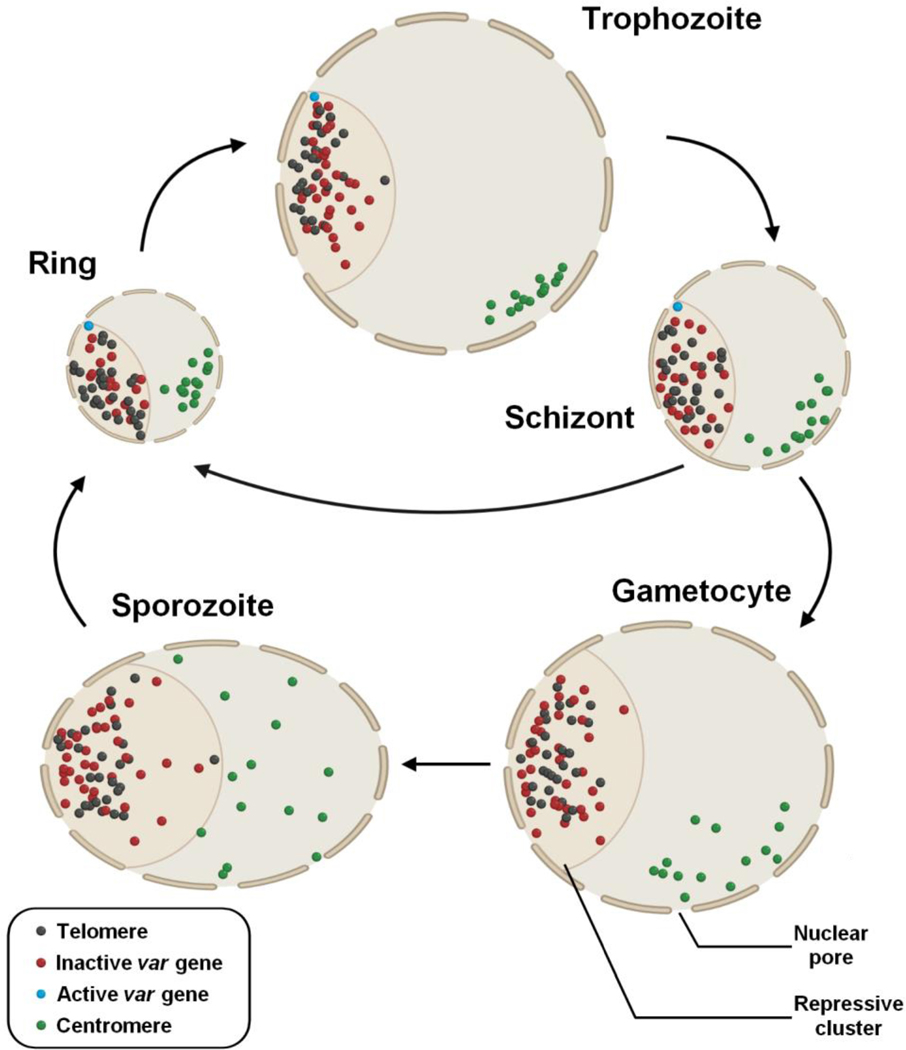
Figure 3. Dynamic chromatin organization across different stages of P. falciparum.
Telomere regions (black spheres) as well as inactive var genes (red spheres) are located within the repressive cluster at the periphery of the nucleus, while centromeres (green spheres) form a cluster on the opposite side. The active var gene is represented by a blue sphere. Throughout the life cycle, extensive remodeling of the chromatin occurs and provides essential modulation of gene expression during parasite development.
During the sexual transition, Hi-C data demonstrates an important reorganization of the genome while maintaining the telomeric and centromeric clusters (Figure 3) [30]. Extension of the repressive cluster was observed and associated not only with var genes but genes exported at the surface of the red blood cell and involved in erythrocyte remodeling, as well as genes involved in invasion (e.g. clag and eba), as none of these genes are required in gametocytogenesis. Additional features were also observed during sexual differentiation. The transcription factor pfap2-g, the master regulator of sexual differentiation, no longer colocalizes with the heterochromatin cluster in early stage II/III gametocytes, suggesting that pfap2-g physically leaves the repressive center during early gametocytogenesis to follow the transcriptional activation needed at that stage for this particular TF (Figure 2B) [30]. Furthermore, large chromatin rearrangements were also observed on chromosome 14. Two superdomains were reported in gametocytes, most likely to facilitate the expression of ptpa and ap2–03, two genes essential during sexual development. Altogether these data support the idea that large chromatin rearrangements are essential to modulate gene expression during parasite development [30,95].
At the sporozoite stage, Hi-C experiments confirmed a strong association between the repressive cluster and invasion and virulence genes in P. falciparum and P. vivax (Figure 3) [30]. Additional long-range interactions and loops are also detected for genes involved in the liver stages. This is especially true for csp, trap, and gest, required for cell traversal and hepatocyte invasion [100,101]. Finally, a gradient of gene expression with the lowest expression on the side of the telomeres and high expression near the centromere was observed in P. falciparum and P. knowlesi suggesting a strong association between genome organization and gene expression [95]. Taken together, these results highlight the importance of the dynamic nuclear architecture in gene expression across the different life stages.
Concluding Remarks
Altogether, the studies discussed above demonstrate that transcriptional regulation in the parasite is controlled at multiple levels and that nucleosome positioning, epigenetics, TFs, and chromatin structure must participate collectively in the dynamics of nuclear organization to coordinate differentiation, multiplication, and transmission of the parasite. Because chromatin structure and epigenetics have such an impact on gene expression, the discovery of molecular components, including lncRNAs that regulating genome architecture and gene expression (see Outstanding Questions), could serve as novel drug targets that can disrupt parasite development and transmission.
Outstanding Questions Box
What mechanisms control expression of the master transcription factors across the life cycle of the malarial parasite?
Which chromatin regulating proteins and lncRNAs are crucial for stage transitions and gene expression in malaria parasites?
How important are the specificities observed in 3D models of Plasmodium species?
Highlights
Nucleosome occupancy and configuration affect gene expression.
Epigenetic modifications regulate the parasite genome and the mutually exclusive expression of virulence genes.
ApiAP2 are master regulators in the stage transitions.
The stereotypical organization of chromosomes in the parasite nucleus facilitates gene expression.
Acknowledgments
This work was supported by the National Institutes of Allergy and Infectious Diseases and the National Institutes of Health (grants R01 AI136511, R01 AI142743-01 and R21 AI142506-01 to KLR) and the University of California, Riverside (NIFA-Hatch-225935 to KLR).
Glossary
- A-type ribosomal DNA (rDNA):
28S, 5.8S, and 18S ribosomal units on chromosomes 1 and 7 of P. falciparum are transcribed predominantly during IDC
- AP2-O3:
member of the ApiAP2 transcription factor family. This protein is detected in the nucleus of female gametocytes and its disruption leads to the formation of aberrant ookinetes
- Apicomplexa:
large phylum of parasitic protists including Plasmodium and Toxoplasma. They possess an apical complex and a second plastid, the apicoplast.
- Chromatin enrichment for proteomics:
method to enrich chromatin and identify cross-linked proteins that bind to DNA under stringent conditions.
- Chromosome conformation capture (3C):
technique to reconstruct the spatial organization of chromatin within nucleus (Box 2).
- ChIP-seq:
method for genome-wide profiling of DNA-binding proteins. Proteins are cross-linked to the chromatin then immunoprecipitated allowing the identification of their DNA binding sites by next-generation sequencing
- Eraser:
proteins removing the marks laid down by the writers. They include among others, histone demethylases, histone deacetylases and sirtuins.
- Fluorescence In Situ Hybridization (FISH):
technique to visualize specific chromosomal regions using complementary fluorescent probes
- Gametocytogenesis:
sexual development of male and female gametocytes through which the parasite is transmitted from human to mosquitoes
- GDV1:
Gametocyte development protein 1 plays an essential role in the sexual commitment and gametocyte production in Plasmodium
- Hi-C:
genome-wide sequencing technique used to identify long-range interactions on a “all-vs-all” manner inside the nucleus (Box 2)
- Intraerythrocytic Developmental Cycle (IDC):
correspond to the asexual blood cycle and is responsible for the symptoms of malaria. The periodicity of this cycle is dependent of the Plasmodium species
- Post-translational modifications (PTMs):
correspond to the enzymatic modification of proteins and alter their respective cellular functions. Variety of these PTMs are reported as phosphorylation, methylation, acetylation, glycosylation, ubiquitination and proteolysis
- PTPA:
Protein Phosphatase 2 Phosphatase Activator is a regulator of one of the most important Ser/Thr protein phosphatase, PP2A, and are involved in the regulation of the cell cycle
- Reader:
proteins able to recognize the epigenetic marks and mediate their effects. Diverse protein domains are associated such as bromodomain and PHD finger
- SICAvar:
family of virulent genes in P. knowlesi playing an essential role for antigenic variation and are comparable to var genes in P. falciparum
- Sporozoite:
transmission stage injected by mosquitoes to humans. Sporozoites migrate from mosquito salivary glands and during a blood meal, penetrate human skin, blood vessels, and then invade hepatocytes
- Topologically associating domains:
correspond to genomic regions interacting more frequently within the domain than regions outside. They are involved in regulation of gene expression
- Trophozoite:
transcriptionally active blood stage in which the parasite matures before dividing during schizogony
- Writer:
proteins including mainly histone methyltransferases, histone acetyltransferases and kinases. These proteins catalyze modifications of specific residues on the exposed histone tail
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization (2019) World Malaria Report 2019. World Health [Google Scholar]
- 2.Le Roch KG et al. (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508 [DOI] [PubMed] [Google Scholar]
- 3.Otto TD et al. (2010) New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol 76, 12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lasonder E. et al. (2016) Integrated transcriptomic and proteomic analyses of P. falciparum gametocytes: molecular insight into sex-specific processes and translational repression. Nucleic Acids Res. 44, 6087–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López-Barragán MJ et al. (2011) Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otto TD et al. (2014) A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Med. 12, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozdech Z. et al. (2003) The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toenhake CG et al. (2018) Chromatin Accessibility-Based Characterization of the Gene Regulatory Network Underlying Plasmodium falciparum Blood-Stage Development. Cell Host Microbe 23, 557–569.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poran A. et al. (2017) Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature 551, 95–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howick VM et al. (2019) The malaria cell atlas: Single parasite transcriptomes across the complete Plasmodium life cycle. Science 365, eaaw2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reid AJ et al. (2018) Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. Elife 7, e33105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walzer KA et al. (2018) Single-Cell Analysis Reveals Distinct Gene Expression and Heterogeneity in Male and Female Plasmodium falciparum Gametocytes. mSphere 3, 130–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sà JM et al. (2020) Single-cell transcription analysis of Plasmodium vivax blood-stage parasites identifies stage- and species-specific profiles of expression. PLoS Biol. 18, e3000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponts N. et al. (2011) Nucleosome occupancy at transcription start sites in the human malaria parasite: A hard-wired evolution of virulence? Infect. Genet. Evol 11, 716–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bunnik EM et al. (2014) DNA-encoded nucleosome occupancy is associated with transcription levels in the human malaria parasite Plasmodium falciparum. BMC Genomics 15, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kensche PR et al. (2015) The nucleosome landscape of Plasmodium falciparum reveals chromatin architecture and dynamics of regulatory sequences. Nucleic Acids Res. 44, 2110–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westenberger SJ et al. (2009) Genome-wide nucleosome mapping of Plasmodium falciparum reveals histone-rich coding and histone-poor intergenic regions and chromatin remodeling of core and subtelomeric genes. BMC Genomics 10, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponts N. et al. (2010) Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 20, 228–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bártfai R. et al. (2010) H2A.Z demarcates intergenic regions of the Plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog. 6, e1001223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz JL et al. (2018) Characterization of the accessible genome in the human malaria parasite Plasmodium falciparum. Nucleic Acids Res. 46, 9414–9431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu XM et al. (2015) Analysis of nucleosome positioning landscapes enables gene discovery in the human malaria parasite Plasmodium falciparum. BMC Genomics 16, 1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao J. et al. (2006) The malaria parasite Plasmodium falciparum histones: Organization, expression, and acetylation. Gene 369, 53–65 [DOI] [PubMed] [Google Scholar]
- 23.Sullivan WJ (2003) Histone H3 and H3.3 variants in the protozoan pathogens Plasmodium falciparum and Toxoplasma gondii. DNA Seq. - J. DNA Seq. Mapp 14, 227–231 [DOI] [PubMed] [Google Scholar]
- 24.Hoeijmakers WAM et al. (2013) H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the Plasmodium falciparum genome. Mol. Microbiol 87, 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petter M. et al. (2013) H2A.Z and H2B.Z double-variant nucleosomes define intergenic regions and dynamically occupy var gene promoters in the malaria parasite Plasmodium falciparum. Mol. Microbiol 87, 1167–1182 [DOI] [PubMed] [Google Scholar]
- 26.Trelle MB et al. (2009) Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum. J. Proteome Res 8, 3439–3450 [DOI] [PubMed] [Google Scholar]
- 27.Salcedo-Amaya AM et al. (2009) Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A 106, 9655–9660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coetzee N. et al. (2017) Quantitative chromatin proteomics reveals a dynamic histone post-translational modification landscape that defines asexual and sexual Plasmodium falciparum parasites. Sci. Rep 7, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saraf A. et al. (2016) Dynamic and combinatorial landscape of histone modifications during the intraerythrocytic developmental cycle of the malaria parasite. J. Proteome Res 15, 2787–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunnik EM et al. (2018) Changes in genome organization of parasite-specific gene families during the Plasmodium transmission stages. Nat. Commun 9, 1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraschka SA et al. (2018) Comparative Heterochromatin Profiling Reveals Conserved and Unique Epigenome Signatures Linked to Adaptation and Development of Malaria Parasites. Cell Host Microbe 23, 407–420.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui L. et al. (2007) PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryot. Cell 6, 1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read DF et al. (2019) Predicting gene expression in the human malaria parasite Plasmodium falciparum using histone modification, nucleosome positioning, and 3D localization features. PLoS Comput. Biol 15, e1007329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vivax Sporozoite Consortium (2019) Transcriptome and histone epigenome of Plasmodium vivax salivary-gland sporozoites point to tight regulatory control and mechanisms for liver-stage differentiation in relapsing malaria. Int. J. Parasitol 49, 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witmer K. et al. (2020) An epigenetic map of malaria parasite development from host to vector. Sci. Rep 10, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dembélé L. et al. (2014) Persistence and activation of malaria hypnozoites in long-term primary hepatocyte cultures. Nat. Med 20, 307–312 [DOI] [PubMed] [Google Scholar]
- 37.Rinn JL et al. (2020) Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu. Rev. Biochem 89, 283–308 [DOI] [PubMed] [Google Scholar]
- 38.Broadbent KM et al. (2015) Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics 16, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sierra-Miranda M. et al. (2012) Two long non-coding RNAs generated from subtelomeric regions accumulate in a novel perinuclear compartment in Plasmodium falciparum. Mol. Biochem. Parasitol 185, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broadbent KM et al. (2011) A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol. 12, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raabe CA et al. (2009) A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res. 38, 608–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jing Q. et al. (2018) Plasmodium falciparumvar Gene Is Activated by Its Antisense Long Noncoding RNA. Front. Microbiol 9, 3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amit-Avraham I. et al. (2015) Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A 112, E982–E991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Epp C. et al. (2009) Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA 15, 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guizetti J. et al. (2016) Trans-acting GC-rich non-coding RNA at var expression site modulates gene counting in malaria parasite. Nucleic Acids Res. 44, 9710–9718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei G. et al. (2015) Dual regulatory effects of non-coding GC-rich elements on the expression of virulence genes in malaria parasites. Infect. Genet. Evol 36, 490–499 [DOI] [PubMed] [Google Scholar]
- 47.Barcons-Simon A. et al. (2020) CRISPR interference of a clonally variant GC-rich noncoding RNA family leads to general repression of var genes in Plasmodium falciparum. MBio 11, e03054–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filarsky M. et al. (2018) GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science 359, 1259–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y. et al. (2020) Noncoding RNAs in Apicomplexan Parasites: An Update. Trends Parasitol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanyal A. et al. (2018) Genome‐wide survey and phylogenetic analysis of histone acetyltransferases and histone deacetylases of Plasmodium falciparum. FEBS J. 285, 1767–1782 [DOI] [PubMed] [Google Scholar]
- 51.Chaal BK et al. (2010) Histone Deacetylases Play a Major Role in the Transcriptional Regulation of the Plasmodium falciparum Life Cycle. PLoS Pathog. 6, e1000737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui L. et al. (2008) Histone lysine methyltransferases and demethylases in Plasmodium falciparum. Int. J. Parasitol 38, 1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duraisingh MT et al. (2005) Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121, 13–24 [DOI] [PubMed] [Google Scholar]
- 54.Tonkin CJ et al. (2009) Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 7, 0771–0788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang L. et al. (2013) PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499, 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ukaegbu UE et al. (2014) Recruitment of PfSET2 by RNA Polymerase II to Variant Antigen Encoding Loci Contributes to Antigenic Variation in P. falciparum. PLoS Pathog. 10, e1003854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Coetzee N. et al. (2020) Epigenetic inhibitors target multiple stages of Plasmodium falciparum parasites. Sci. Rep 10, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoeijmakers WAM et al. (2019) Epigenetic reader complexes of the human malaria parasite, Plasmodium falciparum. Nucleic Acids Res. 47, 11574–11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheon Y. et al. (2020) Dynamic modules of the coactivator SAGA in eukaryotic transcription. Exp. Mol. Med [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Josling GA et al. (2015) A Plasmodium Falciparum Bromodomain Protein Regulates Invasion Gene Expression. Cell Host Microbe 17, 741–751 [DOI] [PubMed] [Google Scholar]
- 61.Santos JM et al. (2017) Red Blood Cell Invasion by the Malaria Parasite Is Coordinated by the PfAP2-I Transcription Factor. Cell Host Microbe 21, 731–741.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Batugedara G. et al. (2020) The chromatin bound proteome of the human malaria parasite. Microb. Genomics 6, e000327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H. et al. (2013) Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol. 13, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glynn EF et al. (2004) Genome-Wide Mapping of the Cohesin Complex in the Yeast Saccharomyces cerevisiae. PLoS Biol. 2, e259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pandey R. et al. (2020) Plasmodium Condensin Core Subunits SMC2/SMC4 Mediate Atypical Mitosis and Are Essential for Parasite Proliferation and Transmission. Cell Rep. 30, 1883–1897.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coulson RMR et al. (2004) Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 14, 1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Callebaut I. et al. (2005) Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: Conserved features and differences relative to other eukaryotes. BMC Genomics 6, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gopalakrishnan AM et al. (2009) Plasmodium falciparum: Preinitiation complex occupancy of active and inactive promoters during erythrocytic stage. Exp. Parasitol 121, 46–54 [DOI] [PubMed] [Google Scholar]
- 69.Rai R. et al. (2014) Genome-wide analysis in Plasmodium falciparum reveals early and late phases of RNA polymerase II occupancy during the infectious cycle. BMC Genomics 15, 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu XM et al. (2017) Nascent RNA sequencing reveals mechanisms of gene regulation in the human malaria parasite Plasmodium falciparum. Nucleic Acids Res. 45, 7825–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Templeton TJ et al. (2004) Comparative analysis of apicomplexa and genomic diversity in eukaryotes. Genome Res. 14, 1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balaji S. et al. (2005) Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33, 3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campbell TL et al. (2010) Identification and Genome-Wide Prediction of DNA Binding Specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 6, e1001165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sinha A. et al. (2014) A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature 507, 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuda M. et al. (2015) Global transcriptional repression: An initial and essential step for Plasmodium sexual development. Proc. Natl. Acad. Sci. U. S. A 112, 12824–12829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuda M. et al. (2009) Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol. Microbiol 71, 1402–1414 [DOI] [PubMed] [Google Scholar]
- 77.Yuda M. et al. (2010) Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol. Microbiol 75, 854–863 [DOI] [PubMed] [Google Scholar]
- 78.Iwanaga S. et al. (2012) Identification of an AP2-family Protein That Is Critical for Malaria Liver Stage Development. PLoS One 7, e47557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Modrzynska K. et al. (2017) A Knockout Screen of ApiAP2 Genes Reveals Networks of Interacting Transcriptional Regulators Controlling the Plasmodium Life Cycle. Cell Host Microbe 21, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C. et al. (2017) Systematic CRISPR-Cas9-mediated modifications of plasmodium yoelii ApiAP2 genes reveal functional insights into parasite development. MBio 8, e01986–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martins RM et al. (2017) An ApiAP2 member regulates expression of clonally variant genes of the human malaria parasite Plasmodium falciparum. Sci. Rep 7, 14042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.MacQuarrie KL et al. (2011) Genome-wide transcription factor binding: Beyond direct target regulation. Trends Genet. 27, 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kafsack BFC et al. (2014) A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kent RS et al. (2018) Inducible developmental reprogramming redefines commitment to sexual development in the malaria parasite Plasmodium berghei. Nat. Microbiol 3, 1206–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brancucci NMB et al. (2014) Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe 16, 165–176 [DOI] [PubMed] [Google Scholar]
- 86.Coleman BI et al. (2014) A Plasmodium falciparum Histone Deacetylase regulates antigenic variation and gametocyte conversion. Cell Host Microbe 16, 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brancucci NMB et al. (2017) Lysophosphatidylcholine Regulates Sexual Stage Differentiation in the Human Malaria Parasite Plasmodium falciparum. Cell 171, 1532–1544.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bancells C. et al. (2019) Revisiting the initial steps of sexual development in the malaria parasite Plasmodium falciparum. Nat. Microbiol 4, 144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Josling GA et al. (2020) Dissecting the role of PfAP2-G in malaria gametocytogenesis. Nat. Commun 11, 1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoeijmakers WAM et al. (2012) Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell. Microbiol 14, 1391–1401 [DOI] [PubMed] [Google Scholar]
- 91.Freitas LH et al. (2005) Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121, 25–36 [DOI] [PubMed] [Google Scholar]
- 92.Lopez-Rubio JJ et al. (2009) Genome-wide Analysis of Heterochromatin Associates Clonally Variant Gene Regulation with Perinuclear Repressive Centers in Malaria Parasites. Cell Host Microbe 5, 179–190 [DOI] [PubMed] [Google Scholar]
- 93.Weiner A. et al. (2011) 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum. Cell. Microbiol 13, 967–977 [DOI] [PubMed] [Google Scholar]
- 94.Dahan-Pasternak N. et al. (2013) Pfsec13 is an unusual chromatin-associated nucleoporin of plasmodium falciparum that is essential for parasite proliferation in human erythrocytes. J. Cell Sci 126, 3055–3069 [DOI] [PubMed] [Google Scholar]
- 95.Bunnik EM et al. (2019) Comparative 3D genome organization in apicomplexan parasites. Proc. Natl. Acad. Sci. U. S. A 116, 3183–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ay F. et al. (2014) Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 24, 974–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lemieux JE et al. (2013) Genome-wide profiling of chromosome interactions in Plasmodium falciparum characterizes nuclear architecture and reconfigurations associated with antigenic variation. Mol. Microbiol 90, 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dixon JR et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sexton T. et al. (2012) Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 [DOI] [PubMed] [Google Scholar]
- 100.Kumar H and Tolia NH (2019) Getting in: The structural biology of malaria invasion. PLOS Pathog. 15, e1007943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Talman AM et al. (2011) PbGEST mediates malaria transmission to both mosquito and vertebrate host. Mol. Microbiol 82, 462–474 [DOI] [PubMed] [Google Scholar]
- 102.Chookajorn T. et al. (2007) Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. U. S. A 104, 899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ralph SA et al. (2005) Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc. Natl. Acad. Sci. U. S. A 102, 5414–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Volz JC et al. (2012) PfSET10, a Plasmodium falciparum Methyltransferase, Maintains the Active var Gene in a Poised State during Parasite Division. Cell Host Microbe 11, 7–18 [DOI] [PubMed] [Google Scholar]
- 105.Fraschka SAK et al. (2016) H3.3 demarcates GC-rich coding and subtelomeric regions and serves as potential memory mark for virulence gene expression in Plasmodium falciparum. Sci. Rep 6, 31965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Crowley VM et al. (2011) Heterochromatin formation in bistable chromatin domains controls the epigenetic repression of clonally variant Plasmodium falciparum genes linked to erythrocyte invasion. Mol. Microbiol 80, 391–406 [DOI] [PubMed] [Google Scholar]
- 107.Zanghì G. et al. (2018) A Specific PfEMP1 Is Expressed in P. falciparum Sporozoites and Plays a Role in Hepatocyte Infection. Cell Rep 22, 2951–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gómez-Díaz E. et al. (2017) Epigenetic regulation of Plasmodium falciparum clonally variant gene expression during development in Anopheles gambiae. Sci. Rep 7, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dekker J. et al. (2002) Capturing chromosome conformation. Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 110.Simonis M. et al. (2007) An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods 4, 895–901 [DOI] [PubMed] [Google Scholar]
- 111.Simonis M. et al. (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet 38, 1348–1354 [DOI] [PubMed] [Google Scholar]
- 112.Ferraiuolo MA et al. (2012) From cells to chromatin: Capturing snapshots of genome organization with 5C technology. Methods 58, 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dostie J. et al. (2006) Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res 16, 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van Berkum NL et al. (2010) Hi-C: A Method to Study the Three-dimensional Architecture of Genomes. J. Vis. Exp 39, 1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lieberman-Aiden E. et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rao SSP et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Belaghzal H. et al. (2017) Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123, 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pal K. et al. (2019) Hi-C analysis: from data generation to integration. Biophys. Rev 11, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Belton JM et al. (2012) Hi-C: A comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]