Abstract
Viruses are obligate intracellular parasites that usurp cellular signaling networks to promote pathogen spread and disease progression. Signaling through extracellular vesicles (EVs) is an emerging field of study in the virus-host interaction network. EVs relay information both locally and distally through incorporated contents, typically without tripping innate immune sensors. Therefore, this extracellular signaling axis presents itself as a tantalizing target for promoting a favorable niche for the pathogen(s) takeover of the host, particularly for chronic infections. From the incorporation of virus-encoded molecules such as micro RNAs and proteins/enzymes to the envelopment of entire infectious particles, evolutionary distinct viruses have shown a remarkable ability to converge on this means of communication. In this review, we will cover the recent advances in this field and explore how EV can be used as potential biomarkers for chronic, persistent, or latent virus infections.
Keywords: Viruses, exosomes, microvesicles, extracellular vesicles, HIV, KSHV, EBV, HCV, HAV, HSV, latency, pathogenesis
Graphical abstract
Extracellular vesicles are realeased by all cell types and transfer internal contents from one cell to the next. Viruses can mocify these internal contents, thereby influrncing recipient cell physiology without infection.
Introduction
As a part of the extracellular signaling network, cells release extracellular vesicles (EVs). The current model posits that EVs exist as three major subgroups: (i) apoptotic bodies, (ii) microvesicles, and (iii) exosomes. Apoptotic bodies are produced when a cell is undergoing apoptosis, and contain materials from every cellular subcompartment, and have a broad size range (100 nanometers (nm) - >1 μm in diameter). Microvesicles originate from the pinching off of the plasma membrane and are enriched for surface membrane proteins, and usually have a diameter of 80 – 500 nm). Strictly speaking, exosomes originate from the invagination of the late endosome into the multivesicular body (MVB) and are enriched for members of the endosomal sorting complex required for transport (ESCRT) machinery. Exosomes have a size range of 40 – 150 nm in diameter [1,2]. In cell supernatant or biological fluids, it is difficult to identify origin for any individual EV.
EVs function similarly to a virion in their ability to transfer active materials from one cell to another, either locally or distally. In primary fluids such as blood, EVs exist at higher concentrations (up to 1010 particles/mL) than even high titer viruses such as flaviviruses, filoviruses, or human immunodeficiency virus (HIV) [3,4]. Similar to a virus, contents delivered by an EV influence cell and tissue physiology such as transcription, differentiation, migration, signaling, and metabolic state [1,5–10]. EVs from tumor cells have been linked to metastasis [10–13]. Given their function in steering local and systemic equilibrium, it is unsurprising that viruses have evolved to co-opt an EV to facilitate pathogen spread and disease progression (Figure 1).
Figure 1.
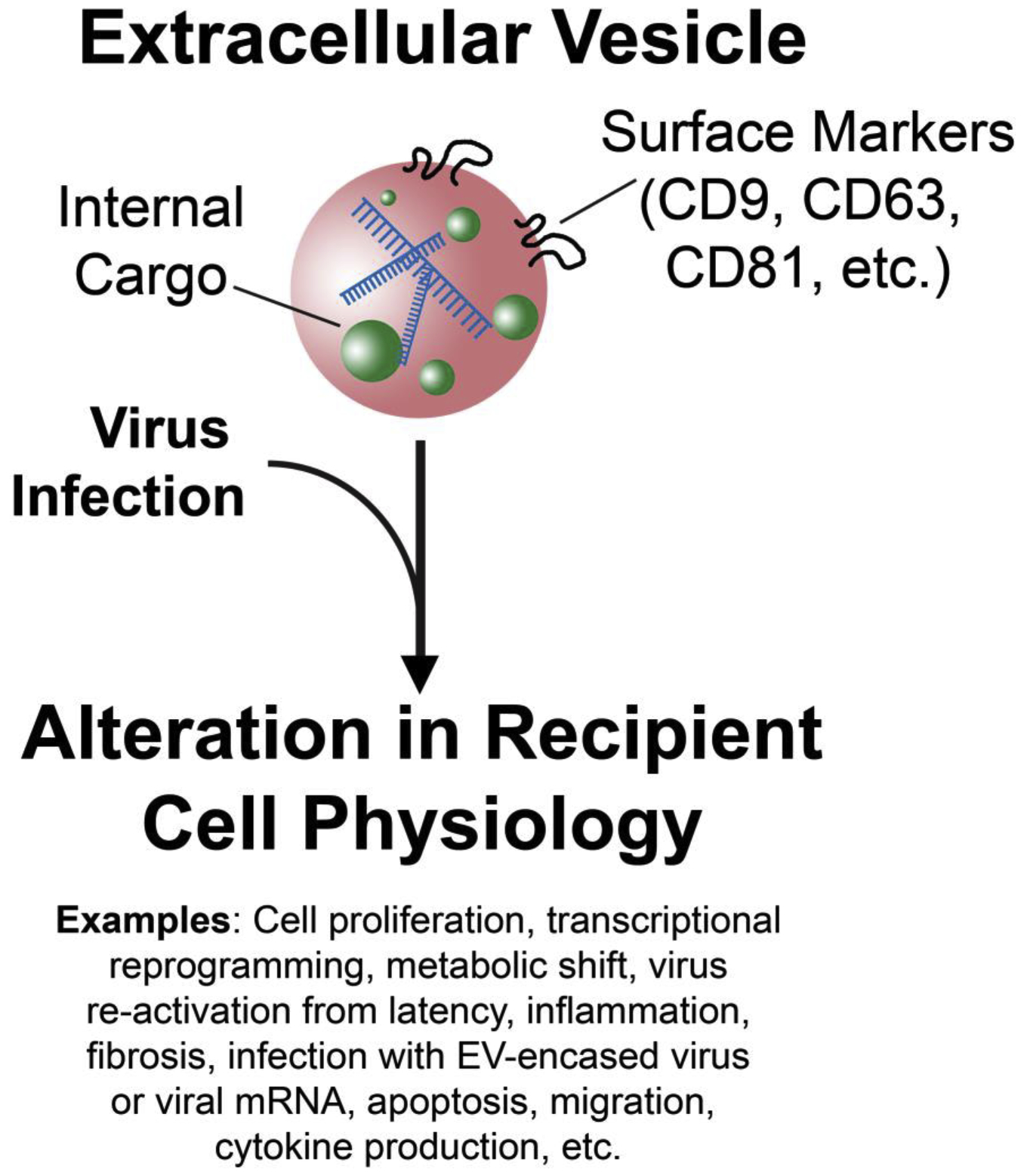
Virus infection alters the cargo of an extracellular vesicle (EV) in vivo. The virus-modified EV is used to establish a more favorable niche for pathogen takeover through the alterations of recipient cell physiology and signaling networks such as those listed.
Much of our current understanding of EVs comes from studies using viruses that deliberately manipulate EV cargo through the incorporation of viral nucleic acids, proteins, and even entire virions into EV [14–23]. Delivery of these virus-modified EVs to naïve cells alters their physiology, demonstrating that EVs can functionally deliver pathogenic materials. Given that EVs exist naturally (i.e. in the absence of virus infection) they transport these materials unbeknownst to the host’s immune response. This phenomenon is believed to have occurred for millennia but has only become appreciated in recent years as a means by which viruses usurp cellular resources.
This review will focus on established models of EV hijacking by viruses, as well as recent advances in this field. We will highlight the work done on EV-virus interactions in the context of viruses that can induce chronic infections. Finally, the potential of using EV as a biomarker for chronic, persistent, and latent infection will be discussed.
Herpesviruses
A shared trait among the Herpesviridae family is that they persist at an equilibrium between latent and lytic replication phases. Both phases are required for viral spread and disease, but only the lytic is responsible for the production of infectious particles. Herpesviruses are some of the oldest mammalian viruses known, with estimates of their origin being hundreds of million years ago, and have host ranges extending from humans (Homo sapiens) to turtles (Chelonia mydas) [24,25].
Herpes simplex virus (HSV-1) has been shown to export viral miRNAs and the innate immune regulator stimulator of interferon genes (STING) protein into EVs post-infection [19,26]. EV transfer of STING has prompted the development of a model in which HSV-1 attenuates its shift from latency into the lytic phase through EVs. Multiple interferon stimulatory genes and pro-inflammatory regulators such as tumor necrosis factor-alpha (TNF-α) were upregulated in cells treated with EVs from HSV-1 infected cells. This led to a decrease in viral transcripts, pushing HSV-1’s equilibrium towards latency [27,28]. This shift would allow the existing, chronically-infected cells to avoid detection through innate sensors triggered by cytokines like TNF-α. Other work has shown that in oligodendrocytic cells, knockdown of the MVB protein Rab27a attenuated HSV-1 infection [29,30]. This work was bolstered by the findings that HSV-1 hijacks a microvesicle-like body to transfer the virus to receptor-negative, neighboring cells. These HSV-1 particles were resistant to neutralizing antibodies and could package multiple virions into one body. Interestingly, these virus-filled vesicles contained markers of the autophagosome [31]. This phenomenon resembles poliovirus exiting through the autophagosome-mediated exit without lysing (AWOL) pathway [32].
Studies on Epstein-Barr virus (EBV) have shown an intricate subversion of EV-signaling. The viral latent membrane protein 1 (LMP1) is a driver for the transformation of B-cells [33–35]. In a B-cell lymphoma, LMP1 is expressed during the latency-II and latency-III phases in germinal center B cells. LMP1 is incorporated into exosomes and delivered to non-infected cells through LMP1’s association with CD63 and lipid rafts [36–39]. LMP1 is primarily expressed in the terminal type III latency, which has a mRNAseq profile distinct from transient latency-IIb phases [40]. Exosome-mediated export of LMP1 in an EBV-transformed B-cell during latency-III to exert effects on non-infected cells or even receptor-negative cells is supported by other findings in which EVs play a role in cancer metastasis (reviewed in [10]). In addition to LMP1, EBV transfers viral miRNAs through EVs. Lymphoblastoid cells express high concentrations of EBV BHRF1 and BART cluster viral miRNAs and are trafficked into an EV. EBV miRNAs could be transferred to naïve dendritic cells and act on their known cellular target mRNA [41].
Kaposi’s Sarcoma-associated herpesvirus (KSHV) is the causative agent of primary effusion lymphoma (PEL) and Kaposi’s Sarcoma (KS) [42–44]. Post-infection, the KSHV genome is maintained in a mostly transcriptionally quiescent state, but a handful of latent proteins and noncoding RNAs are expressed [45–47]. During latency, KSHV packages high concentrations of viral miRNAs into EVs from infected cells (KSHV-EVs) [9,48]. Delivery of KSHV-EVs reprograms recipient, non-infected endothelial cells transcriptionally, and alters their metabolic profile into a distinct, cancer-cell defining state termed the “Warburg state” or “Warburg effect” [7,8]. The Warburg effect, named after Otto Warburg, describes a cell with a metabolic state with largely glycolytic-based energy production as opposed to oxidative phosphorylation through the electron transport chain [49]. These metabolic and transcriptome shifts occur without the transfer of the virus itself. These alterations in cellular physiology by KSHV-EV do not activate innate immune sensors such as interferon regulatory factor 3 (IRF3), STING, and nuclear factor kappa b (NF-κB). In contrast, extracellular signal-regulated kinase (ERK) is activated by KSHV-EVs. Upon chronic exposure of naïve endothelial cells to KSHV-EVs, cells transition into a hyper-proliferative state, with Ki67 positive staining similar to that of a bona fide KS tumor. These complex reprogramming events occur during latency and infectious particle production is low [7]. Therefore, the detection of KSHV miRNAs in blood has been proposed as a biomarker for virus infection [8,9].
Retroviruses
The human immunodeficiency virus (HIV) emerged into the human population in the early 20th century from spillover events of the simian immunodeficiency virus (SIV) [43,50,51]. HIV chronically infects T-lymphocytes and macrophages, depleting them over the course of years, ultimately leading to a collection of clinical symptoms referred to as AIDS.
The HIV-encoded Tat potently transactivates the viral genome and forms a positive feedback loop [52–54]. Early HIV transcripts are fully spliced, and encode for Tat, Rev, and Nef. In addition to Tat’s role at the HIV promoter, a portion of Tat is enriched on membranes through its interaction with phosphatidylinositol (4–5) bisphosphate (PI(4,5)P2) [55,56]. Tat’s membrane affinity is largely controlled by the first few amino acids of its RNA binding domain (RBD). This particular domain of Tat has also doubled as a peptide transduction sequence, allowing for the cellular penetration of membrane-impermeable drugs [57,58]. The Tat-PI(4,5)P2 interaction has been proposed to allow for Tat to be exported from the cell via EV [59]. Experimentally, even particle-free Tat proteins and Tat-fusion moieties retain the ability to cross-target cell membranes and enter the cytosol directly.
In addition to Tat, the HIV-encoded 5’ RNA element TAR has also been found in EVs [60–63]. TAR is transcribed at the HIV promoter by RNA Polymerase II, and Tat binds to the RNA with elongation factors with a remarkable affinity [64]. The finding of both Tat and TAR inside of EVs further highlights this unique RNA-protein binding couple. Interestingly, the delivery of Tat and/or TAR-enriched EV has been shown to activate HIV from resting CD4+T lymphocytes [65] and activate cellular migration/proliferation in an ERK-dependent manner [66].
Nef was identified as the first detectable viral protein expressed post-HIV-infection, yet the nef open reading frame is dispensable for in vitro growth [67,68]. Nef is nevertheless a critical driver for HIV and SIV disease progression in human and non-human primates, respectively [69–74]. Nef downregulates surface CD4 and MHC-I presumably to prevent super-infection of a cell and avoid immune detection [75–84]. A large fraction of Nef is secreted from cells through EVs in vivo. This is likely owed to its interactions with the endosomal recycling pathway [85,86] and its anchoring onto the inner leaflet of the plasma membrane [87–93]. The ability of this viral protein to be incorporated into EV is evolutionarily conserved between HIV and SIV. The Nef-EV can be uptaken by naïve CD4+ T cells and can induce their apoptosis [94], metabolic and lipid raft reorganization, and enhance HIV infection through inflammatory signaling [95]. Nef-EVs can also be uptaken by macrophages and endothelial cells which line the vasculature where Nef localizes to punctate intracellular structures [4,96]. Nef’s ability to interact with and/or modify lipid rafts may be the mechanism by which the viral protein is trafficked into EVs [96–98]. Both HIV and SIV Nef can be visualized in complex with CD81, a known lipid raft component, by super-resolution microscopy (Figure 2) [1,4]. This has lead to a model that Nef exerts pathogenic effects via EV, contributing to co-morbidities frequently associated with long-term HIV infection and late stage AIDS. Supporting this hypothesis, Nef can be detected in EV during treatment with antiretroviral therapy (ART), which effectively abrogates virus production [4,93,99].
Figure 2.
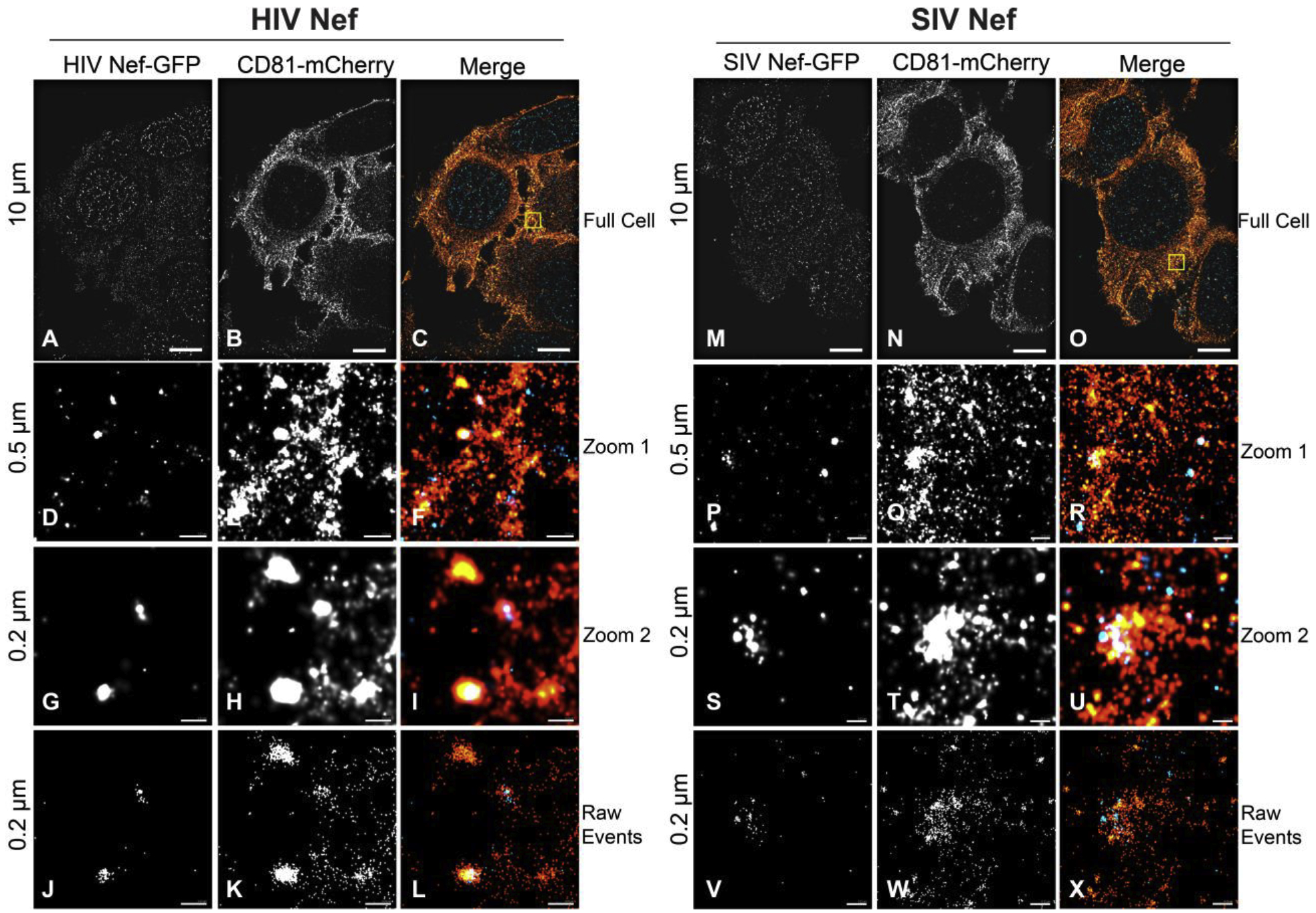
Super-resolution microscopy of SIV and HIV Nef with the EV marker CD81. A-C. dSTORM of a CD81-mCherry expressing cell transfected with HIV Neg-GFP (scale = 10 μm). D-F. Zoomed in view of box in (C) (scale = 0.5 μm). G-I Further zoomed-in image of CD81-mCherry and HIV Nef-GFP co-localizing events (scale = 0.2 μm). J-L. Raw photoswitching events of -(G-I). M-X. Same as (A-L), but for SIV Nef-GFP.
Hepatitis viruses
There are several taxonomically distinct hepatitis viruses that infect humans. Vaccinations exist for hepatitis A virus (HAV, a picornavirus) and hepatitis B virus (HBV, a hepadnavirus), and successful antiviral therapy regimens for hepatitis C virus (HCV, a flavivirus). Yet, these three viruses continue to infect millions of people worldwide each year. Other viruses cause viral hepatitis as well, but we will focus on HAV and HCV.
HAV belongs to the Picornaviridae family of viruses, which are positive-sense RNA viruses without an envelope [100,101]. However, early work showed that HAV exists on a spectrum of buoyancies, indicating interaction with lipid moieties [102]. It was later discovered that HAV acquires a cellular envelope through the endosomal recycling pathway. This “quasi-enveloped HAV” (eHAV) is resistant to neutralizing antibodies [103–106]. Uptaken HAV and eHAV are trafficked through the endosomal pathway, but eHAV ultimately goes to the lysosome where the envelope is degraded [107]. HAV represent the prominent species in feces, and source of person-to-person transmission whereas eHAV circulates in the bloodstream and liver. Other members of the Picornaviridae family have also shown the ability to acquire cell membranes despite being defined as “non-enveloped” viruses [108,109]. This envelope appears to be from the late endosome due to its enrichment in phosphatidylserine (PS) [9,110]. PS binds to TIM-family of receptors on the cell surface, notably TIM1 and TIM4, allowing for phagocytosis of apoptotic cells, EV, or a virion coated with PS [111–114]. Interestingly, CRISPR-mediated knockout of TIM1 reduced eHAV infection in Vero cells but was not essential for HAV or eHAV virus infection [115]. Instead, both HAV and eHAV appear to be uptaken through a β 1 integrin-mediated clathrin and dynamin endocytic processes [107], in addition to TIM1.
HCV is a unique member of the Flaviviridae family in that it is transmitted from human-to-human, in contrast to other flaviviruses such as Dengue virus, West Nile virus, and Zika virus that are transmitted by arthropods [116,117]. Post-entry (see review [118]), the positive-strand viral RNA genome is sufficient to initiate an infection state in a susceptible cell. EV transfer of HCV RNA has been observed in hepatocytes, and virus constructs lacking functional structural (virion) proteins could traffic viral RNA into EVs and promote an infection state [119,120]. HCV RNA can be transferred to receptor-negative plasmacytoid dendritic cells, triggering IFN-α production [121]. This occurs through the ESCRT machinery proteins chromatin-modifying protein 4B (CHMP4B) and the tumor susceptibility gene 101 (TSG101) [122–124]. EVs from HCV-infected cells promote activation and fibrosis in hepatic stellate cells, which themselves cannot be infected with HCV. This phenomenon is believed to occur via the EV-mediated transfer of miR19a and miR192 from HCV-infected cells. These two miRNAs lead to higher levels of transforming growth factor β (TGF-β) signaling and activation of liver fibrosis [125,126].
Concluding Remarks
Extracellular signaling is paramount for organism homeostasis, and unsurprisingly viruses evolved to hijack this network. By manipulating EVs, viruses can establish a niche favorable for pathogen takeover without de novo infection or activation of innate immune cascades. Importantly, this can occur in receptor-negative cells. Additionally, the transfer of viral contents through EVs is mostly unaffected by neutralization by immunoglobulins (Figure 3). In many ways, EVs are akin to platelets, which unlike organs or cells, can be transplanted across HLA-mismatched individuals [127].
Figure 3.
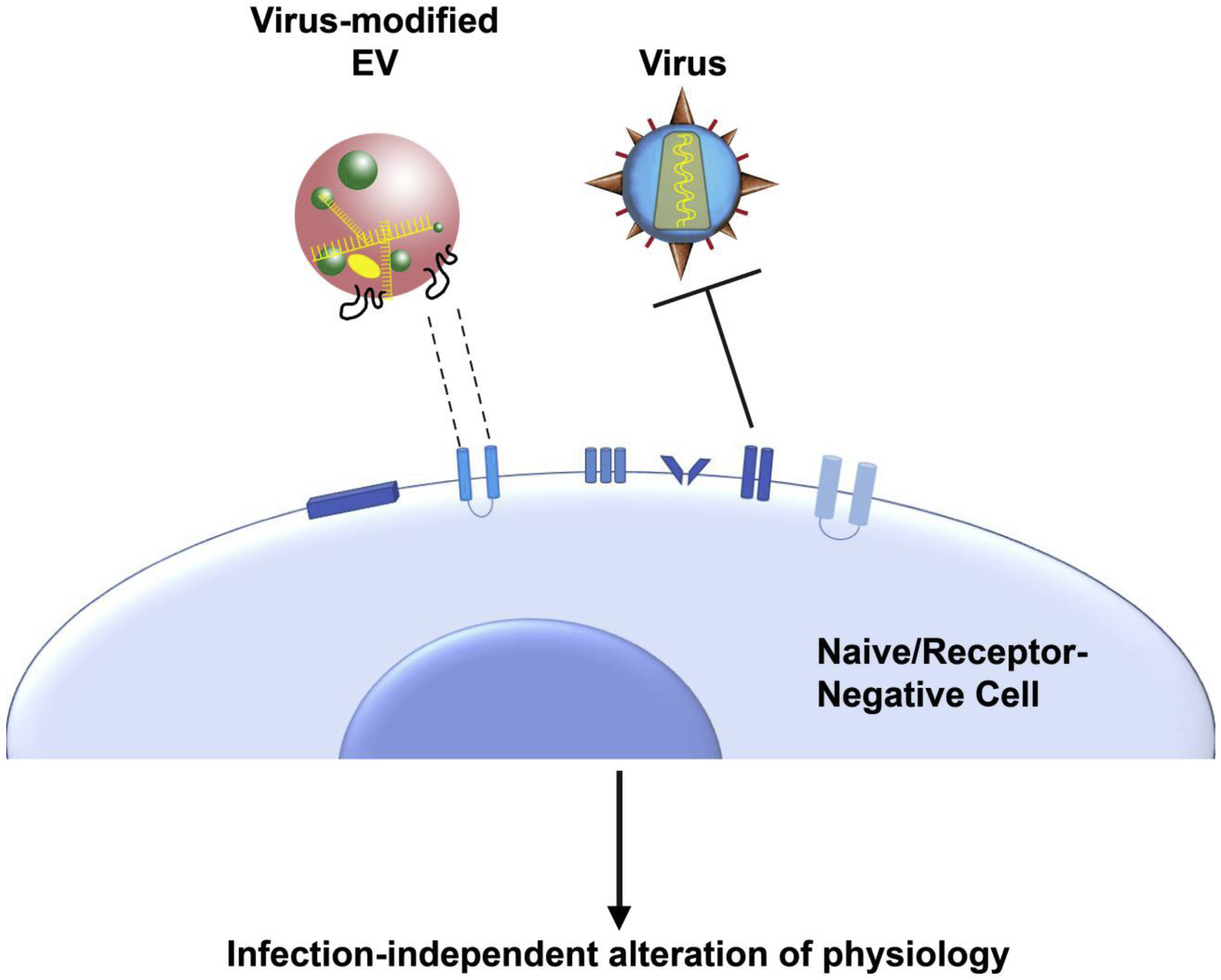
Virus-modified EV alter the physiology of naïve and non-infectable (receptor-negative) cells. Cells lacking receptors for specific viruses can still be targeted by viruses through the incorporation of virus-modified cargo into EV (colored in yellow) emanating from an infected cell. This allows the virus to reprogram cellular signaling, gene expression, and metabolic state without de novo infection.
The phenomenon of reshaping the environment can occur during latency and antiviral treatment. For example, HIV and SIV Nef have been detected in EVs even during ART [4,99]. Nef-EVs may originate from leaky latent reservoirs expressing early HIV genes but not functional virions. Eradication of HIV latent reservoirs represent the single greatest hurdle towards a curative strategy for HIV, and Nef-EV may provide some utility as a biomarker for disease. Most strategies of HIV detection and quantitation rely on viral RNA genome amplification or p24 detection, which are made in the context of full viral genome activation, and thus cells with leaky early gene expression can be missed. It has been proposed that both SIV and HIV latency is an intentional strategy by the virus to promote virus survival [128–130]. Current strategies of activating this latent reservoir to purge out SIV/HIV, particularly in resting CD4+ T lymphocytes, have seen a marked improvement in recent years [131–133]. Studies examining how EVs fit into HIV latency, curative strategies, and biomarkers for disease state seem warranted.
Given their ability to vehicle viral factors during latency, using EVs as biomarkers for infection has been explored [21]. For the oncogenic EBV and KSHV, viral genomes remain remarkably low during tissue transformation. This is in sharp contrast to viral miRNAs, which are robustly transcribed during latency. These miRNAs are trafficked into exosomes and are present at high concentrations in cultured cell supernatant and primary fluids [7–9,41,47,48,122,134,135]. This is analogous to certain tumors that have a unique exosome RNA profile [136,137]. Therefore EVs can have clinical utility in the form of liquid biopsy.
While this review focused on chronically infecting viruses, there is considerable literature showing a dynamic relationship between EVs and acutely infecting viruses. In many of these cases, EVs play an integral anti-viral role. Examples of this include infection with influenza virus [138,139], dengue virus [140], zika virus [141], and hepatitis B virus (which can elicit both acute and chronic infection) [142]. Similar to examples listed in this review, these responses can be induced through the transfer of specific factors to a naïve cell. Comparisons and contrasts in the interplay between EVs and acutely and chronically infecting viruses will be of considerable interest in the coming years as our understanding of this signaling axis continues expanding.
Other viruses that elicit chronic infections utilize EV pathogenesis, and we were unable to cover these in the depth they deserve. Examples include CMV, HPV, and HTLV-1 [15,20,143–147]. Evolutionarily distinct viruses converged to utilize EVs for the functional transfer of materials. This allows viruses to modify the local area of infection and avoid detection by the immune system and may play a previously underappreciated role in maintaining virus latency.
Highlights.
Distinct viruses converged to usurp extracellular vesicle signaling.
Extracellular vesicles transfer virus-encoded factors during latency.
Extracellular vesicles can be used for biomarker detection.
Acknowledgments
We would like to thank Dr. Y. Zhou for constructive comments and feedback on this review. This review was funded through the AIDS Malignancy Consortium Fellowship (part of the 5UM1CA121947-10) awarded to R.P.M. and the National Institutes of Health grant 1R01DA040394 awarded to D.P.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interests
This review was funded through the AIDS Malignancy Consortium Fellowship (part of the 5UM1CA121947–10) awarded to R.P.M. and the National Institutes of Health grant 1R01DA040394 awarded to D.P.D. The funding sources had no involvement in the interpretations or writing of this review.
References
- 1.Pegtel DM, Gould SJ: Exosomes. Annu Rev Biochem 2019, 88:487–514. [DOI] [PubMed] [Google Scholar]
- 2.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, et al. : Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018, 7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNamara RP, Dittmer DP: Modern Techniques for the Isolation of Extracellular Vesicles and Viruses. J Neuroimmune Pharmacol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNamara RP, Costantini LM, Myers TA, Schouest B, Maness NJ, Griffith JD, Damania BA, MacLean AG, Dittmer DP: Nef Secretion into Extracellular Vesicles or Exosomes Is Conserved across Human and Simian Immunodeficiency Viruses. MBio 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meckes DG, Gunawardena HP, Dekroon RM, Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B, Raab-Traub N: Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc Natl Acad Sci U S A 2013, 110:E2925–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, Marini JC, Tudawe T, Seviour EG, San Lucas FA, et al. : Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016, 5:e10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yogev O, Henderson S, Hayes MJ, Marelli SS, Ofir-Birin Y, Regev-Rudzki N, Herrero J, Enver T: Herpesviruses shape tumour microenvironment through exosomal transfer of viral microRNAs. PLoS Pathog 2017, 13:e1006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNamara RP, Chugh PE, Bailey A, Costantini LM, Ma Z, Bigi R, Cheves A, Eason AB, Landis JT, Host KM, et al. : Extracellular vesicles from Kaposi Sarcoma-associated herpesvirus lymphoma induce long-term endothelial cell reprogramming. PLoS Pathog 2019, 15:e1007536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chugh PE, Sin SH, Ozgur S, Henry DH, Menezes P, Griffith J, Eron JJ, Damania B, Dittmer DP: Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog 2013, 9:e1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wortzel I, Dror S, Kenific CM, Lyden D: Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell 2019, 49:347–360. [DOI] [PubMed] [Google Scholar]
- 11.Brzozowski JS, Bond DR, Jankowski H, Goldie BJ, Burchell R, Naudin C, Smith ND, Scarlett CJ, Larsen MR, Dun MD, et al. : Extracellular vesicles with altered tetraspanin CD9 and CD151 levels confer increased prostate cell motility and invasion. Sci Rep 2018, 8:8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ: Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol 2018, 15:617–638. [DOI] [PubMed] [Google Scholar]
- 13.Zomer A, Maynard C, Verweij FJ, Kamermans A, Schafer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SIJ, et al. : In Vivo Imaging Reveals Extracellular Vesicle-Mediated Phenocopying of Metastatic Behavior. Cell 2015, 161:1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson M, Kashanchi F, Jacobson S: Role of Exosomes in Human Retroviral Mediated Disorders. J Neuroimmune Pharmacol 2018, 13:279–291. [DOI] [PubMed] [Google Scholar]
- 15.Anderson MR, Kashanchi F, Jacobson S: Exosomes in Viral Disease. Neurotherapeutics 2016, 13:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raab-Traub N, Dittmer DP: Viral effects on the content and function of extracellular vesicles. Nat Rev Microbiol 2017, 15:559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Z, Hirai-Yuki A, McKnight KL, Lemon SM: Naked Viruses That Aren’t Always Naked: Quasi-Enveloped Agents of Acute Hepatitis. Annu Rev Virol 2014, 1:539–560. [DOI] [PubMed] [Google Scholar]
- 18.Meckes DG, Raab-Traub N: Microvesicles and viral infection. J Virol 2011, 85:12844–12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bello-Morales R, López-Guerrero JA: Extracellular Vesicles in Herpes Viral Spread and Immune Evasion. Front Microbiol 2018, 9:2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schorey JS, Harding CV: Extracellular vesicles and infectious diseases: new complexity to an old story. J Clin Invest 2016, 126:1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigues M, Fan J, Lyon C, Wan M, Hu Y: Role of Extracellular Vesicles in Viral and Bacterial Infections: Pathogenesis, Diagnostics, and Therapeutics. Theranostics 2018, 8:2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orefice NS, Souchet B, Braudeau J, Alves S, Piguet F, Collaud F, Ronzitti G, Tada S, Hantraye P, Mingozzi F, et al. : Real-Time Monitoring of Exosome Enveloped-AAV Spreading by Endomicroscopy Approach: A New Tool for Gene Delivery in the Brain. Mol Ther Methods Clin Dev 2019, 14:237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maguire CA, Balaj L, Sivaraman S, Crommentuijn MH, Ericsson M, Mincheva-Nilsson L, Baranov V, Gianni D, Tannous BA, Sena-Esteves M, et al. : Microvesicle-associated AAV vector as a novel gene delivery system. Mol Ther 2012, 20:960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA: Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol 1995, 247:443–458. [DOI] [PubMed] [Google Scholar]
- 25.McGeoch DJ, Dolan A, Ralph AC: Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J Virol 2000, 74:10401–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bello-Morales R, Ripa I, López-Guerrero JA: Extracellular Vesicles in Viral Spread and Antiviral Response. Viruses 2020, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalamvoki M, Du T, Roizman B: Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci U S A 2014, 111:E4991–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deschamps T, Kalamvoki M: Extracellular Vesicles Released by Herpes Simplex Virus 1-Infected Cells Block Virus Replication in Recipient Cells in a STING-Dependent Manner. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bello-Morales R, Crespillo AJ, Fraile-Ramos A, Tabarés E, Alcina A, López-Guerrero JA: Role of the small GTPase Rab27a during herpes simplex virus infection of oligodendrocytic cells. BMC Microbiol 2012, 12:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et al. : Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 2010, 12:19–30; sup pp 11–13. [DOI] [PubMed] [Google Scholar]
- 31.Bello-Morales R, Praena B, de la Nuez C, Rejas MT, Guerra M, Galán-Ganga M, Izquierdo M, Calvo V, Krummenacher C, López-Guerrero JA: Role of Microvesicles in the Spread of Herpes Simplex Virus 1 in Oligodendrocytic Cells. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor M, Burgon T, Kirkegaard K, Jackson W: Role of Microtubules in Extracellular Release of Poliovirus. Journal of Virology 2009, 83:6599–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang LW, Jiang S, Gewurz BE: Epstein-Barr Virus LMP1-Mediated Oncogenicity. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu HP, Chen CC, Wu CC, Huang YC, Liu SC, Liang Y, Chang KP, Chang YS: Epstein-Barr virus-encoded LMP1 interacts with FGD4 to activate Cdc42 and thereby promote migration of nasopharyngeal carcinoma cells. PLoS Pathog 2012, 8:e1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tellam J, Connolly G, Webb N, Duraiswamy J, Khanna R: Proteasomal targeting of a viral oncogene abrogates oncogenic phenotype and enhances immunogenicity. Blood 2003, 102:4535–4540. [DOI] [PubMed] [Google Scholar]
- 36.Rider MA, Cheerathodi MR, Hurwitz SN, Nkosi D, Howell LA, Tremblay DC, Liu X, Zhu F, Meckes DG: The interactome of EBV LMP1 evaluated by proximity-based BioID approach. Virology 2018, 516:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurwitz SN, Cheerathodi MR, Nkosi D, York SB, Meckes DG: Tetraspanin CD63 Bridges Autophagic and Endosomal Processes To Regulate Exosomal Secretion and Intracellular Signaling of Epstein-Barr Virus LMP1. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurwitz SN, Nkosi D, Conlon MM, York SB, Liu X, Tremblay DC, Meckes DG: CD63 Regulates Epstein-Barr Virus LMP1 Exosomal Packaging, Enhancement of Vesicle Production, and Noncanonical NF-κB Signaling. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, van der Kant R, Neefjes J, et al. : LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J 2011, 30:2115–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messinger JE, Dai J, Stanland LJ, Price AM, Luftig MA: Identification of Host Biomarkers of Epstein-Barr Virus Latency IIb and Latency III. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM: Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 2010, 107:6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.(CDC) CfDC: Kaposi’s sarcoma and Pneumocystis pneumonia among homosexual men--New York City and California. MMWR Morb Mortal Wkly Rep 1981, 30:305–308. [PubMed] [Google Scholar]
- 43.Sharp PM, Hahn BH: Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 2011, 1:a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dittmer DP, Damania B: Kaposi’s Sarcoma-Associated Herpesvirus (KSHV)-Associated Disease in the AIDS Patient: An Update. Cancer Treat Res 2019, 177:63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sin SH, Dittmer DP: Viral latency locus augments B-cell response in vivo to induce chronic marginal zone enlargement, plasma cell hyperplasia, and lymphoma. Blood 2013, 121:2952–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sin SH, Fakhari FD, Dittmer DP: The viral latency-associated nuclear antigen augments the B-cell response to antigen in vivo. J Virol 2010, 84:10653–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pearce M, Matsumura S, Wilson AC: Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi’s sarcoma-associated herpesvirus originate from a common promoter. J Virol 2005, 79:14457–14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoshina S, Sekizuka T, Kataoka M, Hasegawa H, Hamada H, Kuroda M, Katano H: Profile of Exosomal and Intracellular microRNA in Gamma-Herpesvirus-Infected Lymphoma Cell Lines. PLoS One 2016, 11:e0162574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liberti MV, Locasale JW: The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci 2016, 41:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worobey M, Telfer P, Souquière S, Hunter M, Coleman CA, Metzger MJ, Reed P, Makuwa M, Hearn G, Honarvar S, et al. : Island biogeography reveals the deep history of SIV. Science 2010, 329:1487. [DOI] [PubMed] [Google Scholar]
- 51.Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Prosser AT, Torimiro JN, Wright A, Mpoudi-Ngole E, et al. : Naturally acquired simian retrovirus infections in central African hunters. Lancet 2004, 363:932–937. [DOI] [PubMed] [Google Scholar]
- 52.Feinberg MB, Baltimore D, Frankel AD: The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci U S A 1991, 88:4045–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D’Orso I, Frankel AD: RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol 2010, 17:815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J, Tamilarasu N, Hwang S, Garber ME, Huq I, Jones KA, Rana TM: HIV-1 TAR RNA enhances the interaction between Tat and cyclin T1. J Biol Chem 2000, 275:34314–34319. [DOI] [PubMed] [Google Scholar]
- 55.Rayne F, Debaisieux S, Yezid H, Lin YL, Mettling C, Konate K, Chazal N, Arold ST, Pugnière M, Sanchez F, et al. : Phosphatidylinositol-(4,5)-bisphosphate enables efficient secretion of HIV-1 Tat by infected T-cells. EMBO J 2010, 29:1348–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Debaisieux S, Rayne F, Yezid H, Beaumelle B: The ins and outs of HIV-1 Tat. Traffic 2012, 13:355–363. [DOI] [PubMed] [Google Scholar]
- 57.Joliot A, Prochiantz A: Transduction peptides: from technology to physiology. Nat Cell Biol 2004, 6:189–196. [DOI] [PubMed] [Google Scholar]
- 58.Koren E, Apte A, Sawant RR, Grunwald J, Torchilin VP: Cell-penetrating TAT peptide in drug delivery systems: proteolytic stability requirements. Drug Deliv 2011, 18:377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mele AR, Marino J, Chen K, Pirrone V, Janetopoulos C, Wigdahl B, Klase Z, Nonnemacher MR: Defining the molecular mechanisms of HIV-1 Tat secretion: PtdIns(4,5)P. Traffic 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narayanan A, Iordanskiy S, Das R, Van Duyne R, Santos S, Jaworski E, Guendel I, Sampey G, Dalby E, Iglesias-Ussel M, et al. : Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J Biol Chem 2013, 288:20014–20033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patters BJ, Kumar S: The role of exosomal transport of viral agents in persistent HIV pathogenesis. Retrovirology 2018, 15:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sampey GC, Saifuddin M, Schwab A, Barclay R, Punya S, Chung MC, Hakami RM, Zadeh MA, Lepene B, Klase ZA, et al. : Exosomes from HIV-1-infected Cells Stimulate Production of Pro-inflammatory Cytokines through Trans-activating Response (TAR) RNA. J Biol Chem 2016, 291:1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeMarino C, Pleet ML, Cowen M, Barclay RA, Akpamagbo Y, Erickson J, Ndembe N, Charurat M, Jumare J, Bwala S, et al. : Antiretroviral Drugs Alter the Content of Extracellular Vesicles from HIV-1-Infected Cells. Scientific Reports 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH: Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 2010, 465:747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang X, Lu H, Dooner M, Chapman S, Quesenberry PJ, Ramratnam B: Exosomal Tat protein activates latent HIV-1 in primary, resting CD4+ T lymphocytes. JCI Insight 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L, Feng Z, Yue H, Bazdar D, Mbonye U, Zender C, Harding CV, Bruggeman L, Karn J, Sieg SF, et al. : Exosomes derived from HIV-1-infected cells promote growth and progression of cancer via HIV TAR RNA. Nat Commun 2018, 9:4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim S, Ikeuchi K, Byrn R, Groopman J, Baltimore D: Lack of a negative influence on viral growth by the nef gene of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 1989, 86:9544–9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SY, Byrn R, Groopman J, Baltimore D: Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol 1989, 63:3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chakrabarti LA, Metzner KJ, Ivanovic T, Cheng H, Louis-Virelizier J, Connor RI, Cheng-Mayer C: A truncated form of Nef selected during pathogenic reversion of simian immunodeficiency virus SIVmac239Deltanef increases viral replication. J Virol 2003, 77:1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Connor RI, Montefiori DC, Binley JM, Moore JP, Bonhoeffer S, Gettie A, Fenamore EA, Sheridan KE, Ho DD, Dailey PJ, et al. : Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J Virol 1998, 72:7501–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kirchhoff F, Schindler M, Specht A, Arhel N, Muench J: Role of Nef in primate lentiviral immunopathogenesis. Cellular and Molecular Life Sciences 2008, 65:2621–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rhodes DI, Ashton L, Solomon A, Carr A, Cooper D, Kaldor J, Deacon N: Characterization of three nef-defective human immunodeficiency virus type 1 strains associated with long-term nonprogression. Australian Long-Term Nonprogressor Study Group. J Virol 2000, 74:10581–10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang Y, Zhang L, Ho DD: Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. J Virol 1995, 69:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC: Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med 1995, 332:228–232. [DOI] [PubMed] [Google Scholar]
- 75.Pereira EA, daSilva LL: HIV-1 Nef: Taking Control of Protein Trafficking. Traffic 2016, 17:976–996. [DOI] [PubMed] [Google Scholar]
- 76.Lindwasser OW, Chaudhuri R, Bonifacino JS: Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr Mol Med 2007, 7:171–184. [DOI] [PubMed] [Google Scholar]
- 77.Wildum S, Schindler M, Münch J, Kirchhoff F: Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J Virol 2006, 80:8047–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piguet V, Gu F, Foti M, Demaurex N, Gruenberg J, Carpentier JL, Trono D: Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes. Cell 1999, 97:63–73. [DOI] [PubMed] [Google Scholar]
- 79.Aiken C, Konner J, Landau NR, Lenburg ME, Trono D: Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 1994, 76:853–864. [DOI] [PubMed] [Google Scholar]
- 80.Dirk BS, Pawlak EN, Johnson AL, Van Nynatten LR, Jacob RA, Heit B, Dikeakos JD: HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling. Sci Rep 2016, 6:37021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Collins DR, Collins KL: HIV-1 accessory proteins adapt cellular adaptors to facilitate immune evasion. PLoS Pathog 2014, 10:e1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G: HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 2002, 111:853–866. [DOI] [PubMed] [Google Scholar]
- 83.Manrique S, Sauter D, Horenkamp FA, Lülf S, Yu H, Hotter D, Anand K, Kirchhoff F, Geyer M: Endocytic sorting motif interactions involved in Nef-mediated downmodulation of CD4 and CD3. Nat Commun 2017, 8:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schaefer MR, Wonderlich ER, Roeth JF, Leonard JA, Collins KL: HIV-1 Nef targets MHC-I and CD4 for degradation via a final common beta-COP-dependent pathway in T cells. PLoS Pathog 2008, 4:e1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Del Río-Iñiguez I, Vázquez-Chávez E, Cuche C, Di Bartolo V, Bouchet J, Alcover A: HIV-1 Nef Hijacks Lck and Rac1 Endosomal Traffic To Dually Modulate Signaling-Mediated and Actin Cytoskeleton-Mediated T Cell Functions. J Immunol 2018, 201:2624–2640. [DOI] [PubMed] [Google Scholar]
- 86.Madrid R, Janvier K, Hitchin D, Day J, Coleman S, Noviello C, Bouchet J, Benmerah A, Guatelli J, Benichou S: Nef-induced alteration of the early/recycling endosomal compartment correlates with enhancement of HIV-1 infectivity. J Biol Chem 2005, 280:5032–5044. [DOI] [PubMed] [Google Scholar]
- 87.Doms RW, Trono D: The plasma membrane as a combat zone in the HIV battlefield. Genes Dev 2000, 14:2677–2688. [DOI] [PubMed] [Google Scholar]
- 88.Boeske A, Schwarten M, Ma P, Tusche M, Mötter J, Möller C, Neudecker P, Hoffmann S, Willbold D: Direct binding to GABARAP family members is essential for HIV-1 Nef plasma membrane localization. Sci Rep 2017, 7:5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pužar Dominkuš P, Ferdin J, Plemenitaš A, Peterlin BM, Lenassi M: Nef is secreted in exosomes from Nef.GFP-expressing and HIV-1-infected human astrocytes. J Neurovirol 2017, 23:713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sami Saribas A, Cicalese S, Ahooyi TM, Khalili K, Amini S, Sariyer IK: HIV-1 Nef is released in extracellular vesicles derived from astrocytes: evidence for Nef-mediated neurotoxicity. Cell Death Dis 2017, 8:e2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raymond AD, Diaz P, Chevelon S, Agudelo M, Yndart-Arias A, Ding H, Kaushik A, Jayant RD, Nikkhah-Moshaie R, Roy U, et al. : Microglia-derived HIV Nef+ exosome impairment of the blood-brain barrier is treatable by nanomedicine-based delivery of Nef peptides. J Neurovirol 2016, 22:129–139. [DOI] [PubMed] [Google Scholar]
- 92.Khan MB, Lang MJ, Huang MB, Raymond A, Bond VC, Shiramizu B, Powell MD: Nef exosomes isolated from the plasma of individuals with HIV-associated dementia (HAD) can induce Aβ(1–42) secretion in SH-SY5Y neural cells. J Neurovirol 2016, 22:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee JH, Schierer S, Blume K, Dindorf J, Wittki S, Xiang W, Ostalecki C, Koliha N, Wild S, Schuler G, et al. : HIV-Nef and ADAM17-Containing Plasma Extracellular Vesicles Induce and Correlate with Immune Pathogenesis in Chronic HIV Infection. EBioMedicine 2016, 6:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lenassi M, Cagney G, Liao M, Vaupotic T, Bartholomeeusen K, Cheng Y, Krogan NJ, Plemenitas A, Peterlin BM: HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 2010, 11:110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dubrovsky L, Ward A, Choi SH, Pushkarsky T, Brichacek B, Vanpouille C, Adzhubei AA, Mukhamedova N, Sviridov D, Margolis L, et al. : Inhibition of HIV Replication by Apolipoprotein A-I Binding Protein Targeting the Lipid Rafts. mBio 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mukhamedova N, Hoang A, Dragoljevic D, Dubrovsky L, Pushkarsky T, Low H, Ditiatkovski M, Fu Y, Ohkawa R, Meikle PJ, et al. : Exosomes containing HIV protein Nef reorganize lipid rafts potentiating inflammatory response in bystander cells. PLoS Pathog 2019, 15:e1007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cui HL, Grant A, Mukhamedova N, Pushkarsky T, Jennelle L, Dubrovsky L, Gaus K, Fitzgerald ML, Sviridov D, Bukrinsky M: HIV-1 Nef mobilizes lipid rafts in macrophages through a pathway that competes with ABCA1-dependent cholesterol efflux. J Lipid Res 2012, 53:696–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alexander M, Bor YC, Ravichandran KS, Hammarskjöld ML, Rekosh D: Human immunodeficiency virus type 1 Nef associates with lipid rafts to downmodulate cell surface CD4 and class I major histocompatibility complex expression and to increase viral infectivity. J Virol 2004, 78:1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ferdin J, Goričar K, Dolžan V, Plemenitaš A, Martin JN, Peterlin BM, Deeks SG, Lenassi M: Viral protein Nef is detected in plasma of half of HIV-infected adults with undetectable plasma HIV RNA. PLoS One 2018, 13:e0191613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tapparel C, Siegrist F, Petty TJ, Kaiser L: Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol 2013, 14:282–293. [DOI] [PubMed] [Google Scholar]
- 101.Wang X, Ren J, Gao Q, Hu Z, Sun Y, Li X, Rowlands DJ, Yin W, Wang J, Stuart DI, et al. : Hepatitis A virus and the origins of picornaviruses. Nature 2015, 517:85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lemon SM, Jansen RW, Newbold JE: Infectious hepatitis A virus particles produced in cell culture consist of three distinct types with different buoyant densities in CsCl. J Virol 1985, 54:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM: A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 2013, 496:367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McKnight KL, Xie L, González-López O, Rivera-Serrano EE, Chen X, Lemon SM: Protein composition of the hepatitis A virus quasi-envelope. Proc Natl Acad Sci U S A 2017, 114:6587–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hirai-Yuki A, Hensley L, Whitmire JK, Lemon SM: Biliary Secretion of Quasi-Enveloped Human Hepatitis A Virus. mBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.González-López O, Rivera-Serrano EE, Hu F, Hensley L, McKnight KL, Ren J, Stuart DI, Fry EE, Lemon SM: Redundant Late Domain Functions of Tandem VP2 YPX. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rivera-Serrano EE, González-López O, Das A, Lemon SM: Cellular entry and uncoating of naked and quasi-enveloped human hepatoviruses. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, et al. : Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 2015, 160:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, Williams W, Kha N, Cruz C, Hancock BM, et al. : Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog 2014, 10:e1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zakharova L, Svetlova M, Fomina AF: T cell exosomes induce cholesterol accumulation in human monocytes via phosphatidylserine receptor. Journal of Cellular Physiology 2007, 212:174–181. [DOI] [PubMed] [Google Scholar]
- 111.Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S: Identification of Tim4 as a phosphatidylserine receptor. Nature 2007, 450:435–439. [DOI] [PubMed] [Google Scholar]
- 112.Kobayashi N, Karisola P, Peña-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, Butte MJ, Nagumo H, Chernova I, Zhu B, et al. : TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 2007, 27:927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A: The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 2012, 12:544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Feng Z, Li Y, McKnight KL, Hensley L, Lanford RE, Walker CM, Lemon SM: Human pDCs preferentially sense enveloped hepatitis A virions. J Clin Invest 2015, 125:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Das A, Hirai-Yuki A, González-López O, Rhein B, Moller-Tank S, Brouillette R, Hensley L, Misumi I, Lovell W, Cullen JM, et al. : TIM1 (HAVCR1) Is Not Essential for Cellular Entry of Either Quasi-enveloped or Naked Hepatitis A Virions. mBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weaver SC, Reisen WK: Present and future arboviral threats. Antiviral Research 2010, 85:328–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weaver SC, Barrett AD: Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol 2004, 2:789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhu YZ, Qian XJ, Zhao P, Qi ZT: How hepatitis C virus invades hepatocytes: the mystery of viral entry. World J Gastroenterol 2014, 20:3457–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G: Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog 2014, 10:e1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin Raj V, Jenster G, et al. : Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A 2013, 110:13109–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dreux M, Garaigorta U, Boyd B, Décembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV: Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe 2012, 12:558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McNamara RP, Caro-Vegas CP, Costantini LM, Landis JT, Griffith JD, Damania BA, Dittmer DP: Large-scale, cross-flow based isolation of highly pure and endocytosis-competent extracellular vesicles. J Extracell Vesicles 2018, 7:1541396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernández-Delgado I, Torralba D, Moreno-Gonzalo O, Baldanta S, Enrich C, Guerra S, Sánchez-Madrid F: ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun 2016, 7:13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kuang Z, Seo EJ, Leis J: Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15. J Virol 2011, 85:7153–7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB: Exosome-Mediated Intercellular Communication between Hepatitis C Virus-Infected Hepatocytes and Hepatic Stellate Cells. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim JH, Lee CH, Lee SW: Exosomal Transmission of MicroRNA from HCV Replicating Cells Stimulates Transdifferentiation in Hepatic Stellate Cells. Mol Ther Nucleic Acids 2019, 14:483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Saha P, Sharma S, Korutla L, Datla SR, Shoja-Taheri F, Mishra R, Bigham GE, Sarkar M, Morales D, Bittle G, et al. : Circulating exosomes derived from transplanted progenitor cells aid the functional recovery of ischemic myocardium. Sci Transl Med 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Razooky BS, Pai A, Aull K, Rouzine IM, Weinberger LS: A hardwired HIV latency program. Cell 2015, 160:990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rouzine IM, Weinberger AD, Weinberger LS: An evolutionary role for HIV latency in enhancing viral transmission. Cell 2015, 160:1002–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, et al. : A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019, 566:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nixon CC, Mavigner M, Sampey GC, Brooks AD, Spagnuolo RA, Irlbeck DM, Mattingly C, Ho PT, Schoof N, Cammon CG, et al. : Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature 2020, 578:160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McBrien JB, Mavigner M, Franchitti L, Smith SA, White E, Tharp GK, Walum H, Busman-Sahay K, Aguilera-Sandoval CR, Thayer WO, et al. : Robust and persistent reactivation of SIV and HIV by N-803 and depletion of CD8. Nature 2020, 578:154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Herzig E, Kim KC, Packard TA, Vardi N, Schwarzer R, Gramatica A, Deeks SG, Williams SR, Landgraf K, Killeen N, et al. : Attacking Latent HIV with convertibleCAR-T Cells, a Highly Adaptable Killing Platform. Cell 2019, 179:880–894.e810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sin SH, Kim Y, Eason A, Dittmer DP: KSHV Latency Locus Cooperates with Myc to Drive Lymphoma in Mice. PLoS Pathog 2015, 11:e1005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Meckes DG, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N: Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A 2010, 107:20370–20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.He C, Zheng S, Luo Y, Wang B: Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8:237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jia Y, Chen Y, Wang Q, Jayasinghe U, Luo X, Wei Q, Wang J, Xiong H, Chen C, Xu B, et al. : Exosome: emerging biomarker in breast cancer. Oncotarget 2017, 8:41717–41733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Maemura T, Fukuyama S, Sugita Y, Lopes TJS, Nakao T, Noda T, Kawaoka Y: Lung-Derived Exosomal miR-483–3p Regulates the Innate Immune Response to Influenza Virus Infection. J Infect Dis 2018, 217:1372–1382. [DOI] [PubMed] [Google Scholar]
- 139.Bedford JG, Infusini G, Dagley LF, Villalon-Letelier F, Zheng MZM, Bennett-Wood V, Reading PC, Wakim LM: Airway Exosomes Released During Influenza Virus Infection Serve as a Key Component of the Antiviral Innate Immune Response. Front Immunol 2020, 11:887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sung PS, Huang TF, Hsieh SL: Extracellular vesicles from CLEC2-activated platelets enhance dengue virus-induced lethality via CLEC5A/TLR2. Nat Commun 2019, 10:2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Müller JA, Harms M, Krüger F, Groß R, Joas S, Hayn M, Dietz AN, Lippold S, von Einem J, Schubert A, et al. : Semen inhibits Zika virus infection of cells and tissues from the anogenital region. Nat Commun 2018, 9:2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li J, Liu K, Liu Y, Xu Y, Zhang F, Yang H, Liu J, Pan T, Chen J, Wu M, et al. : Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat Immunol 2013, 14:793–803. [DOI] [PubMed] [Google Scholar]
- 143.Jaworski E, Narayanan A, Van Duyne R, Shabbeer-Meyering S, Iordanskiy S, Saifuddin M, Das R, Afonso PV, Sampey GC, Chung M, et al. : Human T-lymphotropic virus type 1-infected cells secrete exosomes that contain Tax protein. J Biol Chem 2014, 289:22284–22305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Walker JD, Maier CL, Pober JS: Cytomegalovirus-infected human endothelial cells can stimulate allogeneic CD4+ memory T cells by releasing antigenic exosomes. J Immunol 2009, 182:1548–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zicari S, Arakelyan A, Palomino RAÑ, Fitzgerald W, Vanpouille C, Lebedeva A, Schmitt A, Bomsel M, Britt W, Margolis L: Human cytomegalovirus-infected cells release extracellular vesicles that carry viral surface proteins. Virology 2018, 524:97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kannan A, Hertweck KL, Philley JV, Wells RB, Dasgupta S: Genetic Mutation and Exosome Signature of Human Papilloma Virus Associated Oropharyngeal Cancer. Sci Rep 2017, 7:46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sadri Nahand J, Moghoofei M, Salmaninejad A, Bahmanpour Z, Karimzadeh M, Nasiri M, Mirzaei HR, Pourhanifeh MH, Bokharaei-Salim F, Mirzaei H, et al. : Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: A review. Int J Cancer 2020, 146:305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]


