Abstract
Background
Circulating microRNAs that post‐transcriptionally regulate gene expressions have been reported as promising biomarkers in cancer monitoring. This study was to identify the potential role of circulating miR‐212 in gastric cancer and whether it could serve as a novel biomarker for gastric cancer.
Methods
We detected the serum levels of miR‐212 in 100 health people and 110 gastric cancer patients and analyzed the relationships of the serum level of miR‐212 with gastric cancer. We detected the expression of miR‐212 in human gastric mucosal epithelial cell line (GES‐1) and human gastric cancer cell lines (NCI‐N87 and SNU‐16) using qRT‐PCR. Then, we detected the role of 5‐aza‐deoxycytidine on the epigenetic regulation of miR‐212 in human gastric cancer cell lines. Furthermore, luciferase reporter assay was used to detect binding activity of miR‐212 on SOX4 mRNA, and their functions on the cell proliferation and apoptosis.
Results
The expression of miR‐212 was higher in health people than that in gastric cancer patients, higher in gastric mucosal epithelial cell line than that in gastric cancer cells. miR‐212 can be a circulating biomarker and an independent prognostic factor of gastric cancer. Moreover, miR‐212 can directly regulate the 3′UTR of SOX4 mRNA to suppress p53 and Bax, resulting gastric cancer cells proliferation inhibition and apoptosis induction.
Conclusion
Our study demonstrated that miR‐212 was epigenetically downregulated in gastric cancer, and resulting low level of miR‐212 can be a potential circulating biomarker and poor prognosis predicator of gastric cancer.
Keywords: 5‐aza‐deoxycytidine, circulation microRNA, gastric cancer, miR‐212, SOX4
miR‐212 can be served as gastric cancer diagnostic indicator and inhibits cell apoptosis.
Abbreviations
- 5‐aza
5‐aza‐2′‐deoxycytidine
- GC
gastric cancer
1. INTRODUCTION
Gastric cancer (GC) is the second most common death cause of cancer. It is characterized by resistance and failure in chemotherapy and surgery, especially the advanced gastric cancer. 1 , 2 Nowadays, endoscopic biopsy is the gold standard for diagnosis of gastric cancer. 3 However, biopsy is invasive and requires proficient surgeons, experienced pathologists, and expensive equipment. Many known diagnostic serum markers, such as CEA (carcinoembryonic antigen), CA19‐9 (carbohydrate antigen 19‐9), and CA72‐4 (carbohydrate antibody 72‐4), 4 were used in the clinical diagnosis. However, these markers have low sensitivity, specificity, and reproducibility and cannot satisfy for accurate detection of gastric cancer. 5 Therefore, it is in urgent need of a non‐invasiveness marker with high sensitivity and specificity for early diagnosis of GC.
miRNAs which comprise a large group of endogenous non‐coding RNAs can regulate mRNA translation and stability and play an important role in regulating gene expression. 6 , 7 miR‐212, which gene locating in the chromosomal region 17p13, plays important roles in many normal developmental processes such as epithelial‐stromal interactions and expansion of mammary progenitor cell populations and serves as a tumor suppressor in mammary cancer and lung cancer. 8 , 9 , 10 , 11
Circulating microRNAs that post‐transcriptionally regulate gene expressions have been reported as promising biomarkers in cancer monitoring. This study was to identify the potential role of circulating miR‐212 in gastric cancer and to evaluate its clinical application. The serum levels of miR‐212 in 100 health people and 110 gastric cancer patients were detected and analyzed the relationships of the serum level of miR‐212 with gastric cancer. We detected the expression of miR‐212 in human gastric mucosal epithelial cell line and human gastric cancer cell lines (NCI‐N87 and SNU‐16). Then, 5‐aza‐deoxycytidine was used to detect the roles of miR‐212 in epigenetic regulation in human gastric cancer cell lines. The expression of miR‐212 was higher in health people than that in gastric cancer patients, higher in gastric mucosal epithelial cell line than that in gastric cancer cells, especially in poorly differentiated gastric cancer cells. Furthermore, the molecular mechanism of miR‐212 was investigated, which indicated that SOX4 was a direct target of miR‐212, and overexpression of SOX4 can rescue miR‐212 mediated GC cells proliferation inhibition and apoptosis induction. Therefore, this study demonstrated that miR‐212 serum level could be served as circulating biomarkers of GC and indicate the prognosis of GC.
2. MATERIALS AND METHODS
2.1. Clinical samples
Serum samples were obtained from 100 health people and 110 gastric cancer patients in the Fifth Central Hospital of Tianjin, China. Health people included 63 men and 37 women; the median age was 58.5 years (range: 40‐80 years). The gastric cancer patients included 62 men and 48 women; the median age of the patients was 61.5 years (range: 36‐84 years); the median follow‐up time was 38.1 months; contain 51 stage I/stage II, 19 stage III and 40 stage IV patients. The pathological diagnosis was counterchecked by two senior pathologists; follow‐ups were conducted by telephone, which were sent to obtain information on the patients’ outcomes. The clinical‐pathological data were collected. Post‐treatment samples were collected at least 2 weeks after the last dose of the fluorouracil, and before second‐line chemotherapy was started, in order to avoid any acute drug effects on influencing the expression profile. This assay was approved by the ethics committee of the Fifth Central Hospital of Tianjin, China.
2.2. Cell lines and 5‐Aza‐deoxycytidine treatment
Human gastric mucosal epithelial cell line GES‐1 was purchased from Beijing Institute of Cancer Research (Beijing, China) and cultured in DMEM medium with 10% calf serum (CS). Human gastric cancer cell lines NCI‐N87 and SNU‐16 were purchased from American Type Cell culture Collection (ATCC). Cells were cultured in RPMI1640 medium (Invitrogen) supplemented with 10% FBS and incubated at 37°C and 5% CO2. Gastric cancer cells were cultured with or without 2.5 µmol/L 5‐Aza‐dC for 48 hours. After that, total RNA was extracted using miRNeasy Mini Kit (Qiagen) for the miRNAs in the cells and miRNeasy Serum/Plasma Kit (Qiagen) for the miRNAs secreted into the culture medium according to the manufacturer's protocol.
2.3. miRNA extraction
About 400 μL of plasma was used to extract the total RNAs containing small RNA by miRNeasy Serum/Plasma Kit (Qiagen) according to the manufacturer's protocol. And we extracted the total RNAs in 106 cells of each cell line by miRNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. The concentration and quality of the RNA samples were measured by NanoDrop 1000 (Nanodrop).
2.4. Quantitive real‐time polymerase chain reaction
Quantitive real‐time polymerase chain reaction (qRT‐PCR) was used to measure the expression of miRNA. Maxima SYBR Green qPCR master mix (Fermentas) was used according to the manufacturer's instruction. Spiked‐in cel‐miR‐39 was analyzed as a normalization control. The experiments were repeated at least three times.
2.5. MTT assay
SNU‐16 cell viability was analyzed with 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT). Briefly, the cells were seeded in 96‐well plates (4 × 103 cells per well), followed by transfection with NC, miR‐212 mimics, pcDNA3‐SOX4, miR‐212 + pcDNA3‐SOX4 for 24 hours. Then, the optical density (OD) was measured at 570 nm and analyzed in comparison to the control group (NC group).
2.6. LDH assay
Lactic dehydrogenase (LDH) activity was detected using the LDH assay kit (Jiancheng) according to the manufacturer's instructions.
2.7. Caspase 3/7 assay
Caspase activity was detected as a mean to evaluate apoptosis in cells expressing miRNAs or miRNA inhibitors. SNU‐16 cells transfected with NC, miR‐212 mimics, pcDNA3‐SOX4, miR‐212 + pcDNA3‐SOX4 were analyzed for Caspase 3/7 activity using Caspase 3/7 Glo substrate solution (Promega), followed by measuring the luminescence using a Glomax luminometer (Promega). The reading values were shown as RLU/cell number per mL.
2.8. Luciferase reporter assay
The 3′‐UTR sequence of SOX4 which was predicted to interact with miR‐212 or a mutant sequence with the predicted target sites were synthesized and inserted into the pGL3 promoter vector (Invitrogen) named pGL3‐SOX4‐wt and pGL3‐SOX4‐mut. Cells were cultured in a 24‐well plate and then co‐transfected with miRNA control or miR‐212 mimics and pGL3‐SOX4‐wt or pGL3‐SOX4‐mut using Lipofectamine 3000 reagent (Invitrogen). After 48 hours, the cells were collected and analyzed with the Dual‐Luciferase Reporter Assay kit (Promega). All transfection experiments were repeated three times independently.
2.9. Statistical analysis
Data were analyzed by SPSS 19.0 software (SPSS). One‐way analysis of variance and Mann‐Whitney test between the groups were used to determine the statistical significance. ROC (Receiver operating characteristic curve) analysis and AUC (area under the ROC curve) were performed to determine the diagnostic accuracy of each parameter. Cox regression analysis was performed to reveal the correlation between serum miR‐212 expression and the age, gender, and stages of GC patients. The relationship of overall survival with serum miR‐212 levels was analyzed by the Kaplan‐Meier method and log‐rank test. P < .05 was considered as significance.
3. RESULTS
3.1. miR‐212 serum expression is lower in GC patients and related to its stage
To detect the expression of miR‐212, we collected the serums from 110 gastric cancer patients and 100 controls (non‐cancer donors). Mann‐Whitney test showed that serum levels of miR‐212 in health people is significantly higher than that in GC patients (Figure 1A). Moreover, the level of miR‐212 was significantly related to the stages of GC. miR‐212 serum level is higher in GC stage I‐II than that in GC stage III and IV. The results showed that the miR‐212 serum level is significantly negative related to the GC stage (Figure 1B). We analyzed the relationship of miR‐212 with the age and gender of GC. The results showed that the serum level of miR‐212 had no relationship with the patients' age or gender (Table 1). Further analysis indicated that miR‐212 was upregulated after therapy (Figure 1C, D, and E, Table 2).
FIGURE 1.
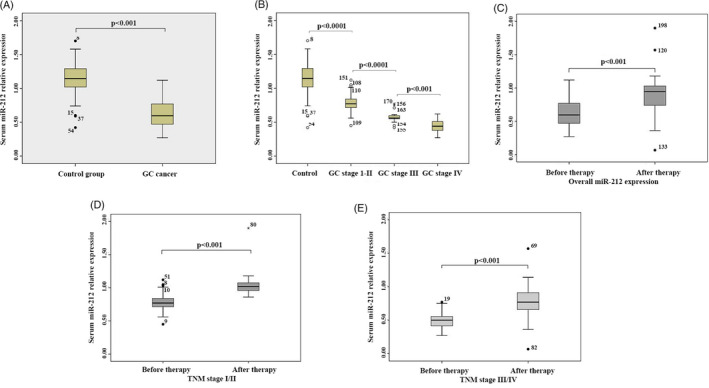
Serum levels of miR‐212 were significantly upregulated in gastric patients and related to GC stages. A, Mann‐Whitney test showed that serum levels of miR‐212 in non‐cancer donors was significantly higher than that in GC patients. B, miR‐212 serum level was higher in GC stage I‐II than that in GC stage III‐IV. C, miR‐212 serum level was higher after therapy in overall GC stages. D, miR‐212 serum level was higher after therapy in GC stage I‐II. E, miR‐212 serum level was higher after therapy in GC stage III‐IV
TABLE 1.
The distribution of serum miR‐212
| Variables | Control | Patients | ||||
|---|---|---|---|---|---|---|
| Number |
miR‐212 Median (95% CI) |
P value | Number |
miR‐212 Median (95% CI) |
P value | |
| Age (y) | ||||||
| ≤60 | 46 | 1.09 (0.98‐1.48) | .494 | 45 | 1.55 (0.98‐3.83) | .489 |
| >60 | 54 | 1.10 (0.51‐2.03) | 65 | 1.64 (0.43‐2.76) | ||
| Gender | ||||||
| Male | 63 | 1.09 (0.51‐2.03) | .175 | 62 | 1.35 (0.98‐3.18) | .093 |
| Female | 37 | 1.13 (0.74‐1.42) | 48 | 1.65 (0.59‐3.7) | ||
TABLE 2.
Comparison between change in microRNA‐212 expression with respect to therapy
| Clinical parameter |
Before therapy miR‐212 expression mean ± SD |
After therapy miR‐212 expression mean ± SD |
P value |
|---|---|---|---|
| Overall miR‐212 expression | 0.62 ± 0.19 | 0.89 ± 0.23 | <.001 |
| TNM stage | |||
| I/II | 0.78 ± 0.78 | 1.03 ± 0.14 | <.001 |
| III/IV | 0.49 ± 0.11 | 0.77 ± 0.22 | <.001 |
3.2. miR‐212 can be a circulating biomarker and an independent prognostic factor of GC
To further explore whether the miR‐212 serum level could be served as circulating biomarkers of GC, ROC analysis was performed and the results showed that the AUC of miR‐212 serum level (GC vs control) was 0.960, the specificity was 0.787, and the sensitivity was 0.951 (Figure 2A). ROC analysis comparing stages I‐II and control revealed that miR‐212 had a sensitivity of 0.946, a specificity of 0.608 and an AUC of 0.951 (Figure 2B). ROC analysis of stage III vs control showed a sensitivity of 0.954 and specificity of 0.385 with an AUC of 0.984 (Figure 2C). Comparison between stage IV and control showed a sensitivity of 0.949 and specificity of 0.598 with an AUC of 0.993 (Figure 2D). These results indicated that miR‐212 serum level could be considered as a circulating biomarker of GC.
FIGURE 2.
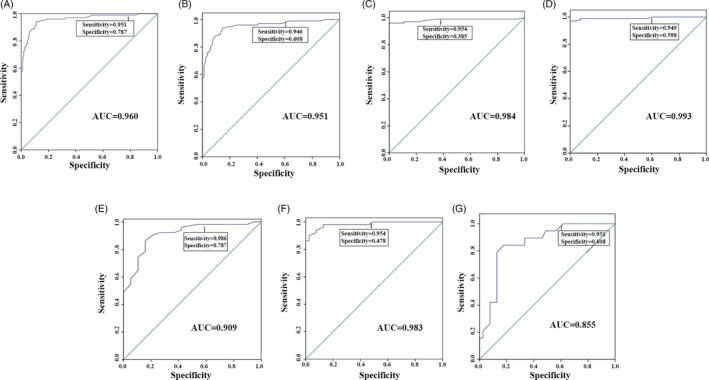
ROC analysis of miR‐212 serum level and different stages of Gastric cancer patients (GC patients) and health persons (Control). A, All of GC patients vs Control. B, stages I‐II of GC patients vs Control. C, Stage III of GC patients vs Control. D, Stage IV of GC patients vs Control. ROC analysis of miR‐212 serum level of different stage of Gastric cancer patients (GC patients). E, Stage I‐II vs stage III. F, Stage I‐II vs stage IV. G, Stage III of GC patients vs stage IV
To further detect whether the miR‐212 serum level could clarify the stage of GC, ROC analysis was performed and the results displayed that the AUC, sensitivity, and specificity of stage I‐II VS stage III were 0.909, 0.986, and 0.787, respectively (Figure 2E). As for stage I‐II vs stage IV, the AUC, sensitivity, and specificity were 0.983, 0.954, and 0.478, respectively (Figure 2F). Comparison between stage III and IV showed sensitivity of 0.951 and specificity of 0.608 with an AUC of 0.855 (Figure 2G). The results suggested that miR‐212 serum level can significantly distinguish the different stage of GC and serve as an independent prognostic factor analyzed by Cox regression analysis (Table 3). Furthermore, Kaplan‐Meier method and log‐rank test showed that lower expression of miR‐212 was significantly related to the shorter survival rate of GC (P < .0001, Figure 3).
TABLE 3.
Serum miR‐212 is an independent prognostic factor by Cox regression analysis
| Variables | Number | Median (mo) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P1 value | Hazard ratio (95% CI) | P2 value | |||
| Age (y) | ||||||
| ≤60 | 45 | 31 | 1.52 (0.997‐2.320) | .052 | 1.33 (0.410‐4.360) | .640 |
| >60 | 65 | 30 | ||||
| Gender | ||||||
| Male | 62 | 34 | 0.759 (0.502‐1.149) | .192 | 0.347 (0.056‐2.14) | .255 |
| Female | 48 | 28 | ||||
| Stages | ||||||
| I/II | 51 | 45 | 1.907 (1.403‐2.593) | <.0001 | 3.66 (0.830‐16.080) | <.0001 |
| III | 19 | 40 | ||||
| IV | 40 | 34 | ||||
FIGURE 3.
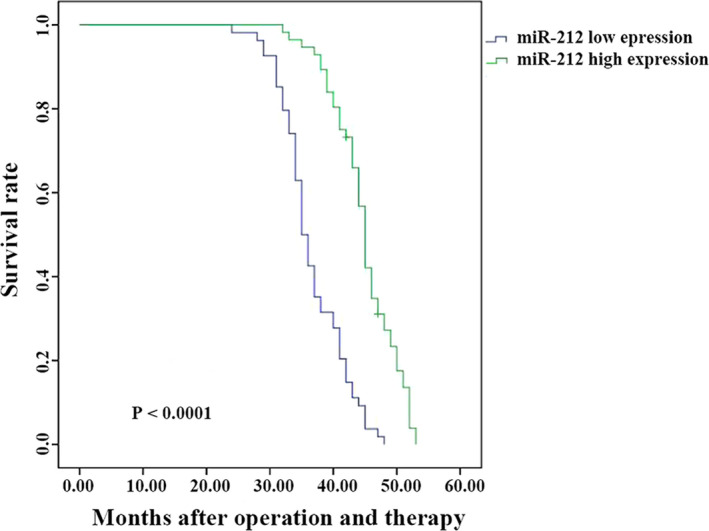
Lower expression of miR‐212 was related to the poor prognosis of GC. Kaplan‐Meier method and log‐rank test showed that lower expression of miR‐212 was significantly related to the poor prognosis of GC (P < .0001)
3.3. Epigenetic regulation of miR‐212 expression in human gastric cancer cell lines
To further detect the expression of miR‐212 in GC cells, the intracellular and culture medium level of miR‐212 was detected, which was reduced in GC cell lines (human gastric mucosal epithelial cell line GES‐1 was used as control). However, when we cultured the GC cells with 5‐Aza‐dC, the intracellular and medium levels of miR‐212 were increased (Figure 4C,D). Therefore, we inferred that the transcriptional activity of mature miR‐212 is silenced by DNA methylation in GC cells.
FIGURE 4.
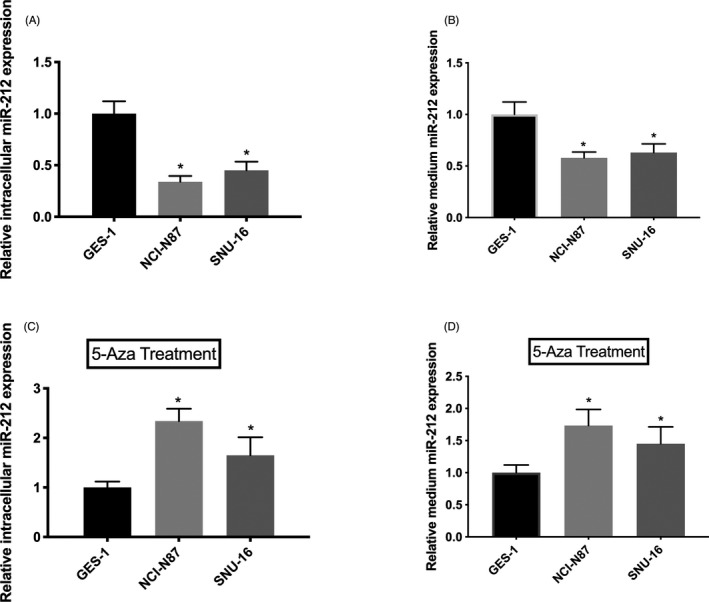
Transcriptional activity of mature miR‐212 was silenced by DNA methylation in GC cells. A, The intracellular expression level of miR‐212 was reduced in GC cell lines compared with human gastric mucosal epithelial cell line GES‐1. B, The culture medium expression level of miR‐212 was reduced in GC cell lines compared with human gastric mucosal epithelial cell line GES‐1. C, The intracellular expression level of miR‐212 was increased in GC cell lines compared with human gastric mucosal epithelial cell line GES‐1 after cultured with 5‐Aza‐dC. D, The culture medium expression level of miR‐212 was increased in GC cell lines compared with human gastric mucosal epithelial cell line GES‐1
3.4. SOX4 is a direct target of miR‐212
To further investigate the downstream molecular mechanisms of miR‐212, TargetScan was used to predict potential target gene of miR‐212. We found that the 3′‐UTR of SOX4 mRNA contained a potential target site for miR‐212 (Figure 5A). The transfection efficiency showed that miR‐212 mimics significantly increased the level of miR‐212 (Figure 5B). To confirm SOX4 as a direct target of miR‐212, luciferase reporter assay was performed. Our results showed that miR‐212 significantly suppressed the luciferase activity of the wild type (wt) (P < .05) but not the mutant (mut) 3′‐UTR of SOX4 (P > .05, Figure 5C). Moreover, qRT‐PCR and Western blot analyses showed that overexpression of miR‐212 significantly decreased the expression of SOX4 in SNU‐16 cells (P < .05, Figure 5D,E,F). Previous study found that SOX4 overexpression regulates the p53/Bax‐mediated apoptosis in hepatocellular carcinoma 12 ; therefore, we further investigated whether miR‐212 mediated SOX4 downregulation can affect p53/Bax pathway. Interestingly, the results identified that miR‐212 mediated downregulation of p53 and Bax, which was rescued by the overexpression of SOX4 (P < .05, Figure 5E,F). Taken together, these results indicated that miR‐212 directly targets SOX4 to repress the expression of p53 and Bax.
FIGURE 5.
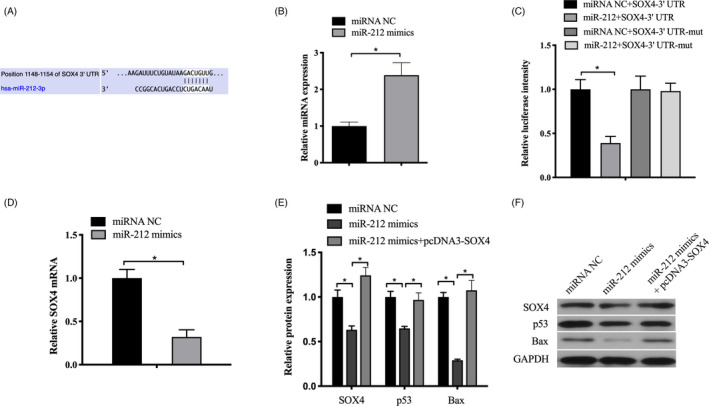
SOX4 was identified as a direct target of miR‐212. A, Computer prediction of the 3′‐UTR of SOX4 mRNA contained a target site for miR‐212. B, Transfection efficiency of miR‐212 mimics. C, Luciferase activity assay revealed that miR‐212 suppressed SOX4 3′‐UTR‐wt luciferase activity, while it had no effect on SOX4 3′‐UTR luciferase activity compared to control in SNU‐16 cells. D, The mRNA expression of SOX4 was examined by qRT‐PCR analysis in SNU‐16 cells. E and F, The protein level of SOX4 was detected by western blot after transfected with miR‐212 mimics or control in SNU‐16 cells. GAPDH was chosen as a loading control. *P < .05
3.5. miR‐212 regulated gc cells proliferation inhibition and apoptosis induction was mediated by SOX4
We further determined the role of SOX4 on GC cell proliferation and apoptosis by overexpressing SOX4 in SNU‐16 cells, and the results showed that overexpression of miR‐212 inhibited the proliferation (Figure 6A) and promoted the apoptosis in SNU‐16 cells (Figure 6B,C), while co‐transfection of miR‐212 mimics and pcDNA3‐SOX4 rescued the function of miR‐212 mediated effects (Figure 6A‐C). These results suggested that SOX4 played as a downstream regulator in miR‐212 mediated GC cells proliferation inhibition and apoptosis induction.
FIGURE 6.
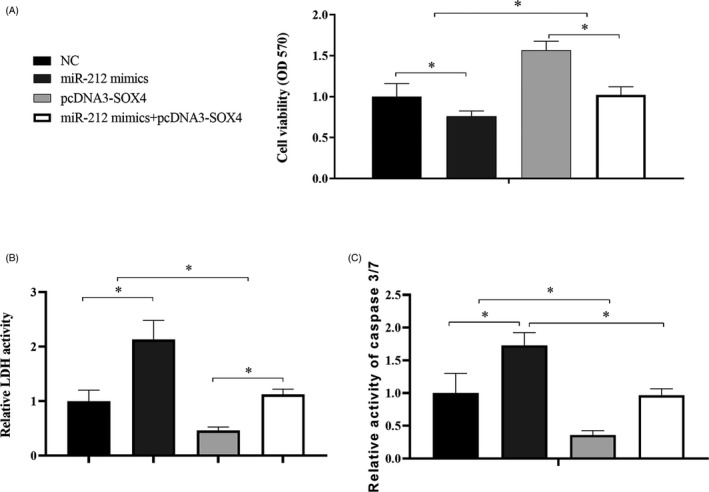
Overexpression of SOX4 rescued miR‐212 mediated GC cells functions. SNU‐16 cells were transfected with NC, miR‐212 mimics, pcDNA3‐SOX4, and miR‐212 mimics + pcDNA3‐SOX4 and detected the cell proliferation with MTT (A) and apoptosis with LDH activity (B) and caspase3/7 activity (C). *P < .05
4. DISCUSSION
GC is no longer the most common cancer worldwide, but it remains the second leading cause of cancer‐related mortality. 13 Moreover, prognosis of GC remains poor 14 and the 5‐year survival rates of GC in stages I and II can reach up to 70%. 15
As we all know that, upper gastrointestinal endoscopy is the gold standard and widely used for the diagnosis of GC. However, it was reported that endoscopic screening is cost‐effective only in high‐incidence areas, not the average risk populations. 16 , 17 Moreover, endoscopy is an invasive procedure, may lead people to die. 12 Therefore, it is urgent to find a new specific and reliable non‐invasiveness method and biomarker for the diagnosis of GC.
Accumulating studies have certified that miRNAs play an important role in the carcinogenesis and progression of gastric cancer. Some reports showed that miR‐212 promotes the malignant biological behaviors of pancreatic ductal adenocarcinoma cells by targeting the hedgehog signaling pathway receptor patched‐1. 18 Also, some researchers found that miR‐212 can constitute the silencing moiety of chimera by negatively modulating PED expression in non‐small‐cell lung cancer. 19 In this study, qRT‐PCR results and statistical analysis indicated that miR‐212 was expressed lower in serum of GC patients and GC cell lines, especially in the high stage of GC. Kaplan‐Meier method and log‐rank test indicated that the lower serum expression of miR‐212 was related to the poor prognosis of GC. ROC analysis suggested that the miR‐212 serum level could be served as circulating biomarkers of GC, while the serum expression of miR‐212 elevated noticeably in GC patients after the chemotherapy with fluorouracil, which was correlated with the TNM stages, and the higher serum level of miR‐212 related with higher survival rate. Previously, a study evaluated the miRNAs alteration in GC treated with SOX chemotherapeutic regimen including Oxaliplatin and S‐1, and they found lower serum expressions of miR‐145 and miR‐185 were elevated drastically, and the higher level of increase indicates better relief outcomes, while the lower level of miR‐145 after SOX neoadjuvant chemotherapy may predict myelotoxicity. These results suggested miRNAs would be potential treatment outcome and adverse effects predictors of different chemotherapy regimens for GC, 20 further study of circulating miR‐212 with different chemotherapy regimens and related adverse effects, including bone marrow suppression, severe gastrointestinal reactions, and significant peripheral nerve damages may better illustrate the value of miR‐212 as the circulating markers of GC.
Evidences indicated that the silencing of tumor suppressor miRNAs in cancer cells is tightly linked to epigenetic mechanism, such as genetic mutation, epigenetic aberration, and deregulated transcriptional activity. 21 To evaluate this possible role of methylation, we treated GC cells by DNA demethylating agent 5‐aza‐2′‐dexoxycytidine. 22 Our results showed that culturing GC cell lines with 5‐aza‐2′‐deoxycytidine (5‐aza) could induce the endogenous level of miR‐212; therefore, we speculated that the lower expression of miR‐212 in GC was caused by methylation.
To further investigated the downstream mediator of miR‐212, bioinformatic analysis was performed using online databases TargetScan, which showed that SOX4, a member of the SOX (SRY‐related HMG‐box) family of transcription factors, might be a direct target of miR‐212. In vitro, we demonstrated that miR‐212 inhibited SOX4 mRNA and protein expression through binding to its 3’‐UTR in SNU‐16 cells. Recent studies demonstrated that SOX4 may contribute to the tumor progression. The expression of SOX4 was upregulated in many types of human cancers, including non‐small cell lung cancer, prostate cancer, colon cancer, and hepatocellular carcinoma. 23 Song et al found that SOX4 overexpression was an unfavorable prognostic factor in breast cancer patients. 24 Moreover, amount of studies showed miRNAs could play an important role in the regulation of SOX4. Liu et al demonstrated that miRNA‐132 inhibited cell growth and metastasis in osteosarcoma cell lines by targeting SOX4. 25 Yeh et al 26 also reported that miRNA‐138 suppressed ovarian cancer cell invasion and metastasis by targeting SOX4. In this study, we confirmed miR‐212 can target SOX4 to repress p53 and Bax in GC cells. Furthermore, miR‐212 mediated GC cells proliferation inhibition and apoptosis induction can be rescued by the overexpression of SOX4.
In conclusion, our study demonstrated that miR‐212 was epigenetically downregulated in GC, re‐expression of miR‐212 suppressed the proliferation and induced cell apoptosis via directly targeting SOX4. Furthermore, low level of miR‐212 was identified as a potential circulating biomarker and poor prognosis predicator for GC.
ACKNOWLEDGMENT
The study was supported by the Science and Technology Project of Health and Family Planning Commission of Tianjin Binhai New District (grant no. 2018BWKZ004) and Medical Research Project of the Fifth Central Hospital of Tianjin (grant no. wzx202001).
Shao J‐P, Su F, Zhang S‐P, et al. miR‐212 as potential biomarker suppresses the proliferation of gastric cancer via targeting SOX4. J Clin Lab Anal. 2020;34:e23511 10.1002/jcla.23511
REFERENCES
- 1. Song EJ, Chan MWY, Shin JW, Chen CC. Hard clam extracts induce atypical apoptosis in human gastric cancer cells. Exp Therap Med. 2017;14:1409‐1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng L, Chen Y, Ye L, et al. miRNA‐584‐3p inhibits gastric cancer progression by repressing Yin Yang 1‐ facilitated MMP‐14 expression. Sci Rep. 2017;7:8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leake PA, Cardoso R, Seevaratnam R, et al. A systematic review of the accuracy and indications for diagnostic laparoscopy prior to curative‐intent resection of gastric cancer. Gastric Cancer. 2012;15(Suppl 1):S38‐S47. [DOI] [PubMed] [Google Scholar]
- 4. Marrelli D, Pinto E, De Stefano A, Farnetani M, Garosi L, Roviello F. Clinical utility of CEA, CA 19–9, and CA 72–4 in the follow‐up of patients with resectable gastric cancer. Am J Surg. 2001;181:16‐19. [DOI] [PubMed] [Google Scholar]
- 5. Carpelan‐Holmstrom M, Louhimo J, Stenman UH, Alfthan H, Haglund C. CEA, CA 19–9 and CA 72–4 improve the diagnostic accuracy in gastrointestinal cancers. Anticancer Res. 2002;22:2311‐2316. [PubMed] [Google Scholar]
- 6. Sun K, Lai EC. Adult‐specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350‐355. [DOI] [PubMed] [Google Scholar]
- 8. Zhao JL, Zhang L, Guo X, et al. miR‐212/132 downregulates SMAD2 expression to suppress the G1/S phase transition of the cell cycle and the epithelial to mesenchymal transition in cervical cancer cells. IUBMB Life. 2015;67:380‐394. [DOI] [PubMed] [Google Scholar]
- 9. Liu H, Li C, Shen C, et al. MiR‐212‐3p inhibits glioblastoma cell proliferation by targeting SGK3. J Neurooncol. 2015;122:431‐439. [DOI] [PubMed] [Google Scholar]
- 10. Jiang X, Chen X, Chen L, et al. Upregulation of the miR‐212/132 cluster suppresses proliferation of human lung cancer cells. Oncol Rep. 2015;33:705‐712. [DOI] [PubMed] [Google Scholar]
- 11. Damavandi Z, Torkashvand S, Vasei M, Soltani BM, Tavallaei M, Mowla SJ. Aberrant expression of breast development‐related microRNAs, miR‐22, miR‐132, and miR‐212, in breast tumor tissues. J Breast Cancer. 2016;19:148‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hur W, Rhim H, Jung CK, et al. SOX4 overexpression regulates the p53‐mediated apoptosis in hepatocellular carcinoma‐ clinical implication and functional analysis in vitro. Carcinogenesis. 2010;31(7):1298‐1307. [DOI] [PubMed] [Google Scholar]
- 13. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700‐713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wagner AD, Syn NL, Moehler M, et al. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2017;8:CD004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Isobe Y, Nashimoto A, Akazawa K, et al. Gastric cancer treatment in Japan: 2008 annual report of the JGCA nationwide registry. Gastric Cancer. 2011;14:301‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ricci C, Holton J, Vaira D. Diagnosis of Helicobacter pylori: invasive and non‐invasive tests. Best Pract Res Clin Gastroenterol. 2007;21:299‐313. [DOI] [PubMed] [Google Scholar]
- 17. Choi KS, Kwak MS, Lee HY, Jun JK, Hahm MI, Park EC. Screening for gastric cancer in Korea: population‐based preferences for endoscopy versus upper gastrointestinal series. Cancer Epidemiol Biomarkers Prev. 2009;18:1390‐1398. [DOI] [PubMed] [Google Scholar]
- 18. Ma C, Nong K, Wu B, et al. miR‐212 promotes pancreatic cancer cell growth and invasion by targeting the hedgehog signaling pathway receptor patched‐1. J Exp Clin Cancer Res. 2014;33:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iaboni M, Russo V, Fontanella R, et al. Aptamer‐miRNA‐212 conjugate sensitizes NSCLC cells to TRAIL. Mol Ther Nucleic Acids. 2016;5:e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tan B, Li Y, Di Y, et al. Clinical value of peripheral blood microRNA detection in evaluation of SOX regimen as neoadjuvant chemotherapy for gastric cancer. J Clin Lab Anal. 2018;32(4):e22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma J, Hong L, Chen Z, Nie Y, Fan D. Epigenetic regulation of microRNAs in gastric cancer. Dig Dis Sci. 2014;59:716‐723. [DOI] [PubMed] [Google Scholar]
- 22. Li D, Li Z, Xiong J, et al. MicroRNA‐212 functions as an epigenetic‐silenced tumor suppressor involving in tumor metastasis and invasion of gastric cancer through down‐regulating PXN expression. Am J Cancer Res. 2015;5(10):2980‐2997. [PMC free article] [PubMed] [Google Scholar]
- 23. Wang D, Hao T, Pan Y, Qian X, Zhou D. Increased expression of SOX4 is a biomarker for malignant status and poor prognosis in patients with non‐small cell lung cancer. Mol Cell Biochem. 2015;402:75‐82. [DOI] [PubMed] [Google Scholar]
- 24. Song GD, Sun Y, Shen H, Li W. SOX4 overexpression is a novel biomarker of malignant status and poor prognosis in breast cancer patients. Tumour Biol. 2015;36:4167‐4173. [DOI] [PubMed] [Google Scholar]
- 25. Liu Y, Li Y, Liu J, Wu Y, Zhu Q. MicroRNA‐132 inhibits cell growth and metastasis in osteosarcoma cell lines possibly by targeting Sox4. Int J Oncol. 2015;47:1672‐1684. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Yeh YM, Chuang CM, Chao KC, Wang LH. MicroRNA‐138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF‐1alpha. Int J Cancer. 2013;133:867‐878. [DOI] [PubMed] [Google Scholar]


