ABSTRACT
Recent studies on recombinant adeno‐associated viral (rAAV) vector production demonstrated the generation of infectious viral particles in Saccharomyces cerevisiae. Proof‐of‐concept results showed low vector yields that correlated with low AAV DNA encapsidation rates. In an attempt to understand the host cell response to rAAV production, we profiled proteomic changes throughout the fermentation process by mass spectrometry. By comparing an rAAV‐producing yeast strain with a respective non‐producer control, we identified a subset of yeast host proteins with significantly different expression patterns during the rAAV induction period. Gene ontology enrichment and network interaction analyses identified changes in expression patterns associated mainly with protein folding, as well as amino acid metabolism, gluconeogenesis, and stress response. Specific fold change patterns of heat shock proteins and other stress protein markers suggested the occurrence of a cytosolic unfolded protein response during rAAV protein expression. Also, a correlative increase in proteins involved in response to oxidative stress suggested cellular activities to ameliorate the effects of reactive oxygen species or other oxidants. We tested the functional relevance of the identified host proteins by overexpressing selected protein leads using low‐ and high‐copy number plasmids. Increased vector yields up to threefold were observed in clones where proteins SSA1, SSE1, SSE2, CCP1, GTT1, and RVB2 were overexpressed. Recombinant expression of SSA1 and YDJ insect homologues (HSP40 and HSC70, respectively) in Sf9 cells led to a volumetric vector yield increase of 50% relative to control, which validated the importance of chaperone proteins in rAAV‐producing systems. Overall, these results highlight the utility of proteomic‐based tools for the understanding and optimization of rAAV‐producing recombinant strains.
Keywords: adeno‐associated virus, gene therapy, mass spectrometry, Saccharomyces
Proteomic changes in yeast recombinant and control strains were profiled to identify cellular events associated with adeno‐associated viral (AAV) vector production. Bioinformatics data analysis suggested cytoplasmic unfolded protein response was primarily taking place in the AAV‐producing yeast strain. Additional changes related to response to stress and amino acid and glucose metabolism were identified. Host protein overexpression was hypothesized as a means for molecular optimization. Overexpression of SSA1, YDJ, SSE1, and other selected proteins in AAV‐producing yeast strains and insect cells resulted in increased vector yield.
1. INTRODUCTION
Recombinant adeno‐associated viral vectors (rAAV) are emerging drugs for gene therapy applications. Ongoing animal and human trials are showing promising data for a variety of clinical conditions. The FDA approvals of Luxturna (Spark Therapeutics) and Zolgensma (Novartis) open regulatory and commercialization pathways for new drugs, motivating other companies to pursue the development of rAAV‐based products.
Vector production at a laboratory scale requires the interaction of several biological inputs (e.g., plasmids, viral inoculum, auxiliary helper genes, and host cells) within a controlled cell culture environment (Aponte‐Ubillus et al., 2018). This process is currently carried out using mammalian cells (HEK293, BHK) or insect cells (Sf9) genetically modified to express AAV proteins. Expression of the capsid proteins VP1, VP2, and VP3 and an assembly activating protein (AAP) leads to the formation of viral capsids in the nucleolus (Samulski & Muzyczka, 2014). The expression of the non‐structural proteins Rep68/78 and Rep52/40 triggers rAAV DNA replication and encapsidation of the generated single‐stranded sequence (Balakrishnan & Jayandharan, 2014). In mammalian cells, the expression of auxiliary adenovirus or herpesvirus proteins is necessary to complement rAAV production; the identified helper genes participate as trans‐activating agents of AAV promoters, or modifiers of the host cell milieu (Geoffroy & Salvetti, 2005).
As an alternative to complex eukaryotic production host cells, a few groups have studied rAAV production in simpler organisms such as yeast. The potential development of a microbial platform would bring simple, lower‐cost, and scalable means for rAAV vector biomanufacturing. Proof‐of‐concept results showed efficient rAAV DNA replication and capsid formation, but low volumetric rAAV2 yield (Barajas et al., 2017; Galli et al., 2017). The low vector yields obtained in yeast could come as a result of an unbalanced expression of rAAV proteins, or a negative effect of host cell response to expression of foreign proteins and multiple forms of rAAV DNA. In the present study, we aimed at increasing our understanding of yeast host cell response to rAAV expression by profiling proteomic changes throughout the production process. Bioinformatics analysis of change patterns helped us build secondary hypotheses regarding potential production constraints in the producer cell line. A final protein overexpression approach confirmed the importance of selected yeast host proteins in vector production, proved by up to a threefold increase in rAAV2 yield for selected overexpression strains.
2. MATERIALS AND METHODS
2.1. Strain and culture media
Saccharomyces cerevisiae strain YPH501 (MATa/MATα ura3‐52/ura3‐52 lys2‐801/lys2‐801 ade2‐101/ade2‐101 trp1‐Δ63/trp1‐Δ63 his3‐Δ200/his3‐Δ200 leu2‐Δ1/leu2‐Δ1) was obtained from Agilent Technologies. 20% glycerol stocks were maintained at −80°C. YPD broth (1% yeast extract, 2% peptone, 2% dextrose) was used for culture start‐up. Synthetic complete (SC) media lacking the appropriate amino acids was used for yeast transformation. SC media supplemented with 0.1 M Na2HPO4/NaH2PO4 phosphate buffer and 2% glucose or 3% galactose was used for fermentation of rAAV‐producing strains. Flasks were incubated in a MaxQ orbital shaker (Thermo Fisher) at 30°C and 250 rpm agitation.
Spodoptera furgiperda (Sf9) cells were cultured in Sf900III‐SFM (Life Technologies). Cell passage was carried out every 4 days in 250‐ml shake flasks at 40% working volume, using an inoculation cell density of 5 × 105 cells/ml. Flasks were incubated in a Multitron orbital shaker (Infors HT) at 28°C temperature and 125 rpm agitation.
2.2. Plasmid design
Coding sequences for rAAV2 capsid and replication proteins were amplified from a pAAV RC2 plasmid and inserted into pESC 2‐micron plasmids under the control of galactose‐induced promoters, as described in Barajas et al. (2017). Briefly, all plasmids were generated using a pESC plasmid (Agilent Technologies) as a vector. DB046 contains a His3 selection marker and VP3 and AAP expression cassettes controlled by GAL1/10 bidirectional promoter. DB228 and DB138 plasmids contain a Leu2 selection marker and GAL‐based Rep52 and VP1 expression cassettes. DB029 plasmid contains a Trp1 selection marker and GAL‐based Rep78 and VP2 expression cassettes. JA001 plasmid consolidated the aforementioned AAV coding sequences into one plasmid. Plasmid DB040 is a pAAV‐GFP‐based plasmid (Cell Biolabs) containing Ura3 and 2‐micron sequences. JA002 plasmid resembles DB040, with the difference that a Leucine marker was placed instead of the original Uracil marker. Protein overexpression plasmids were generated using DB327 (pESC(U)‐GAL10‐), as a backbone vector. Specific primer sets (see Appendix 1, https://doi.org/10.6084/m9.figshare.13040591.v1) were designed to amplify coding sequences from yeast genomic DNA. SmaI‐digested DB327 plasmid and amplified sequences were ligated by Gibson assembly. DB327 2‐micron and CEN variants were generated to promote high and low gene copy number, respectively.
The plasmid pFastBac (ThermoFisher) was modified to include a blasticidin resistance gene and the baculovirus HR5 region in the plasmid backbone, outside of the Tn7 transposable cassette, generating pFB‐HR5‐BSD. S. frugiperda HSC70 and HSP40 were PCR amplified with primers 1010/1011 and 1008/1009, respectively, and inserted into XhoI‐linearized pFB‐HR5‐BSD under the control of a polh promoter, generating plasmids pFB‐HR5‐BSD‐HSC70 and pFB‐HR5‐BSD‐HSP40. To generate the plasmid pFB‐HR5‐BSD‐HSC70‐HA‐HSP40‐HA, expressing the two HA‐tagged proteins from a polh and a p10 promoter, respectively, the HSC70 gene was amplified with primers 1150/1140; a SV40 terminator‐p10 promoter DNA fragment was amplified with primers 1141/1142 from pFB‐inCap‐inRep (Chen, 2008), and HSP40 was amplified with primers 1143/1151. The three DNA fragments were inserted by Gibson assembly into BamHI/XhoI‐digested pFB‐HR5‐BSD. The resulting plasmids were transformed into E. coli DH10Bac strain to generate recombinant bacmids following the Bac‐to‐bac system (Thermo Fisher Scientific).
2.3. Baculovirus viral stocks
Three baculovirus strains were developed in the laboratory using the Bac‐to‐bac system (Thermo Fisher Scientific). rBV‐GFP contains the green fluorescent protein gene controlled by a CMV promoter. The cassette is flanked by AAV inverted terminal repeats (ITRs). rBV‐RepCap contains the Rep and Cap genes controlled by p10 and polh promoters, respectively. rBV‐HSP is used to overexpressed specific insect cell proteins. Viral stocks were generated in Sf9 cells growing in the Sf900III‐SFM medium, with a mixture of bacmid DNA and Cellfectin (Thermo Fisher Scientific). After four days of cell culture, the baculovirus stock was harvested by centrifugation at 230 g × 10 min. The stocks were maintained at 4°C in the dark.
2.4. Proteomic profiling study design
The experimental design was aimed at identifying protein expression differences between rAAV‐producing and non‐producing YPH501 strains. The YPH501‐rAAV strain was developed by transformation with plasmids DB046, DB138, DB029, and DB040. A control strain was generated by transformation with 4 pESC empty plasmids. Fermentation was performed in 250‐ml shake flasks containing 50 ml of SC medium +2% glucose. Each strain was inoculated at an approximated cell density of 0.2 OD 600 nm. An orbital shaker was used to agitate flasks at a rate of 240 rpm. After 24 hours of culture, the medium was exchanged with fresh SC medium +0.1 M phosphate buffer +2% galactose to induce rAAV protein expression (Figure 1a). The culture was extended for four days after the media exchange. Cell growth, pH, and AAV protein expression were monitored throughout the culture. This experiment was performed in triplicate, at three different times for both strains (n = 9). Yeast biomass samples were taken on days 0, 2, and 3 post‐induction to analyze their proteomic profile.
FIGURE 1.
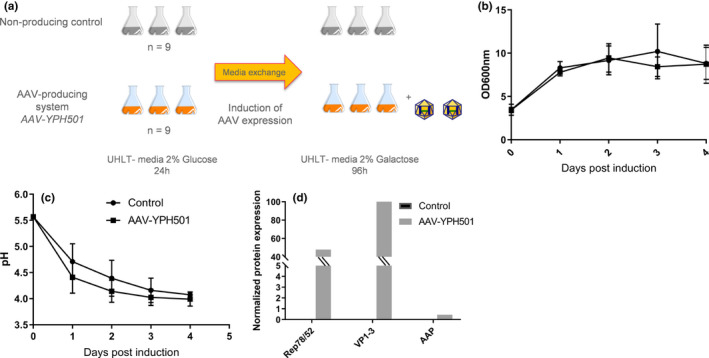
Proteomic profiling study. Outline of experiment (a). Time‐course yeast biomass (b) and pH monitoring (c) of all shake flasks conditions. Normalized rAAV protein expression in yeast samples on day 3 post‐galactose induction, detected by mass spectrometry (d)
2.5. Sample preparation
The samples were prepared for MS analysis following the methodology suggested by Paulo et al. (2015). Cells were washed twice with water and suspended in a buffer containing 50 mM HEPES (pH 8.5), 8 M urea, 75 mM NaCl, and protease inhibitors Complete Mini (EDTA‐free) and PhosStop (Roche). The cell suspension was concentrated to approximately 1.5‐2 OD 600 nm. Glass beads were added to the suspension in a cell:buffer:beads ratio of approximately 1:2:2. Cell suspensions were submitted to three homogenization cycles of 30 seconds each with 30‐second rest intervals, using a Maxiprep 24 homogenizer. Lysates were centrifuged at 100 g × 5 min, and the supernatant was aliquoted for further treatment. Sample aliquots were reduced by incubation in 5 mM tris 2‐carboxyethyl phosphine (TCEP) for 25 min at room temperature. Alkylation was subsequently performed by 30‐min incubation with 10 mM iodoacetamide. Immediately after, samples were incubated in 15 mM Dithiothreitol (DTT), and protein fractions were separated by methanol chloroform precipitation. The protein concentration of all samples was monitored by using the BCA assay kit (Thermo Fisher Scientific). Protein fractions were dissolved in 50 mM HEPES +0.05% Rapigest and digested with trypsin (EMD Millipore) at 100:1 protein to protease ratio, for 6 hours at 37°C. Enzymatic digestion was stopped by the addition of 1% formic acid. Prepared samples were flash‐frozen for further analysis.
2.6. Liquid chromatography‐mass spectrometry
Chromatographic separation was performed using Acquity M‐Class UPLC fitted with an Acquity HSS T3 column (1.8 µm, 1.0 × 150 mm, 100 Å; Waters Corporation). Peptides were separated with a reversed‐phase gradient elution running 0.1% formic acid and 0.1% formic acid in acetonitrile (Burdick and Jackson) from 3% B to 40% B over 80 min at 25 µl/min. Before injection on the column, 2 µg peptide sample solutions were spiked with 500 fmol of Hi3 E. coli peptide internal standard mixture (Waters Corporation) for subsequent quantitation by the “Hi3” method (Silva et al., 2006). Proteomics data were acquired on a Synapt G2‐Si mass spectrometer (Waters Corporation) operating in HDMSE mode. Raw mass spectrometry data were processed for proteomics analysis with Progenesis QI for Proteomics software (version 3.0, Nonlinear Dynamics). Chromatograms were aligned and normalized using the “all proteins” approach. Peptides were identified within Progenesis from a search of the Uniprot Saccharomyces cerevisiae database (UP000002311) appended to include key AAV protein amino acid sequences and Hi3 internal standard peptides. Peptide search criteria included 10 ppm mass measuring accuracy, fixed carbamidomethylation, variable methionine oxidation, and a 4% false discovery rate. Ion matching requirements were two or more fragments/peptide, three or more fragments/protein, and one or more peptides/protein.
2.7. Bioinformatics data analysis
Preliminary Progenesis protein expression data were refined by establishing a cutoff confidence ID value of 15, and the presence of at least two unique peptides per hit. The principal component analysis was performed in JMP 12 software (SAS) to visualize sample clustering and to identify potential run outliers. The analysis was run under the default estimation method. Basic statistical analysis (mean, standard deviation, p‐value) of MS data was performed using Progenesis QI software and Microsoft Excel. An adjusted ANOVA p‐value of 5 × 10−5 was used to determine the statistical difference in protein expression among conditions and/or time points.
Log2 protein expression changes were determined per strain using Microsoft Excel, and heat map analyses were performed using GraphPad Prism to visualize protein expression changes within the control strain between days 0 and 3 post‐induction (column A), within the rAAV‐producing strain between days 0 and 3 post‐induction (column B), and between strains based on days 2‐3 post‐induction data (column C). To systematically identify the proteins showing the biggest expression changes, we compared changes in columns A and B and quantified the difference among them. A 25% change was used as the minimum threshold for selection (ABS [log2(B) − log2(A)] > 0.32). Column C values were used to confirm upregulation or downregulation events linked to rAAV production. DAVID bioinformatics database (https://david.ncifcrf.gov/home.jsp) was used for gene ontology enrichment analysis. A Benjamini‐corrected p‐value of 0.05 was established for the analysis. Additional bioinformatics analysis was performed using the STRING network interaction database (https://string‐db.org/). A high confidence interaction score (0.7) was selected for predicted interaction analysis.
2.8. Protein overexpression study
The effect of protein overexpression on vector yield was assessed. Results from protein profiling led to the identification of protein candidates that could potentially improve rAAV expression. Nineteen candidates were screened by using an rAAV 2‐plasmid yeast system (JA001 and JA002) on which a third plasmid containing the coding sequence of a protein of interest controlled by GAL10 promoter was transformed. Four clones per strain were isolated and grown in 24‐deep well plates following the fermentation strategy described above. 500 µl samples were taken on day 4 post‐induction, and benzonase‐resistant vector yield from each yeast lysate was determined. Paired, one‐tailed t tests were performed between each variable and control strains to determine whether the average yield of the variable strain was significantly higher.
2.9. rAAV production in Sf9 cells
rAAV production using insect cells was performed using standard 3‐rBV, Sf9 cell culture process parameters as reported in the literature(Aucoin et al., 2006; Negrete & Kotin, 2008). Briefly, Sf9 cells were inoculated at a cell density of 1 × 106 cells/ml in a 250‐ml shake flask containing 50 ml of Sf900III‐SFM medium. Cells were incubated at 28°C and 135 rpm agitation. At 24 h of growth, three rBV stocks (rBV‐GFP, rBV‐RepCap, and rBV‐HSP) were added to the culture at an individual MOI of 3. Viable cell density, viability, and cell diameter were monitored with the use of the ViCell analyzer (Beckman Coulter). The culture progressed over five days until viability percent is approximately 40% or lower. Crude harvest was centrifuged at 4300 g × 15 min and then filtered with a 0.22 µm, PVDF‐based syringe filter. Harvest was treated with benzonase to degrade DNA impurities. Inorganic salts with high ionic strength were added to prevent rAAV particle aggregation (Wright et al., 2005). rAAV vg titer was determined by ddPCR analysis of the clarified harvest.
2.10. rAAV analytical testing
Digital droplet PCR (ddPCR) was performed as described in Barajas et al. (2017) to quantify benzonase‐resistant rAAV DNA. Yeast‐treated material was diluted 100:1000‐fold to target the ddPCR dynamic range. Five µL of diluted material was mixed with 20 µl of Taqman‐based master mix (BioRad) including GFP primers DB307/DB309 and a FAM dye‐labeled probe DB308. Droplets were generated by an automated droplet generator (Biorad), and amplified material was analyzed in a QX200 droplet reader (Biorad) using Quantasoft software (Biorad).
3. RESULTS
To evaluate yeast proteome changes occurring because of rAAV expression, fermentation runs using a yeast rAAV‐producing strain, and a non‐producing control was performed in triplicate at three different times. The results from fermentation runs are presented in Figure 1b–d. Most conditions showed a consistent growth profile throughout the 5 days of fermentation. There was no significant difference in growth rate trends after galactose induction, suggesting that rAAV protein expression did not significantly impact cell growth. pH trends were also comparable between strains, showing a subsequent mild decrease over the four days of growth in galactose. rAAV protein expression was tracked in both strains, and mass spectrometry was used for their detection on day 3 post‐induction. Figure 1d displayed the detection of Rep, Cap, and AAP proteins only in the rAAV‐producing strain and not in the control strain.
We used Progenesis QI for mass spectrometry raw data processing. A total of 925 yeast proteins were identified, covering protein IDs present in several cellular structures such as cell wall, cytoplasm, and nucleus. Out of this total, approximately 70% met the confidence ID and unique peptide cutoff requirements for our profiling analysis (See Appendix 2 for details, https://doi.org/10.6084/m9.figshare.13040591.v1). Principal component analysis (PCA) of samples from days 0, 2, and 3 post‐induction was performed to visualize the clustering of proteomic data among yeast samples. Analysis of day 0 post‐induction samples showed no separation among control and recombinant strain samples (Figure 2a). This expected trend corroborated that, before induction, both strains shared basic metabolic features common to early logarithmic growth. A secondary PCA analysis of day 2 and day 3 post‐induction samples evidenced a clear clustering of the control samples and the recombinant strain samples, being principal component 2 a transformed variable that explained 20.6% of proteome sample differences (Figure 2b). We can infer that the identity of the variables that comprise component 2 might be implicated in significant biological processes associated with rAAV production.
FIGURE 2.
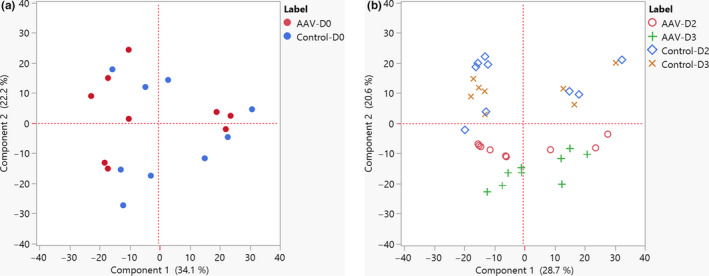
Principal component analysis. Dynamic profile analysis of recombinant (rAAV) and control yeast strains at day 0 post‐galactose induction (a) and at days 2 and 3 post‐galactose induction (b)
Mass spectrometry analysis identified 304 proteins that showed statistically significant changes in protein accumulation throughout the fermentation process (adjusted p‐value <5 × 10−5). Heat map analysis of the proteome subset was performed in three sets that compared induction day and strain type (Figure 3). Our categorization criteria helped us select a subset of 148 proteins with the biggest protein expression changes in the rAAV‐producing strain relative to the control strain. We rationalized that this subset would be more representative of the changes associated with rAAV production; therefore, subsequent gene ontology and network analysis were performed with this protein list. Gene ontology enrichment analysis was performed using DAVID software. Table 1a displays biological processes identified from the original 304 protein subset. In addition to broad processes such as amino acid/carbon metabolism and oxidation‐reduction processes, this list also comprised specific events like protein folding and refolding, translation, ribosomal subunit assembly, and gluconeogenesis. GO analysis of the highest‐changing 148 proteins displayed four hits, which included protein folding, protein refolding, de novo protein folding, and metabolic process (Table 1b). Additional biological processes identified but with higher p‐value included gluconeogenesis, response to heat, and cellular amino acid biosynthetic process. These results suggest the previously mentioned events might be directly or indirectly associated with rAAV expression in yeast. Further analysis performed with STRING software contributed to the generation of a prediction‐based interaction network, based on the protein set ID and protein–protein interactions reported in the literature. Network results corroborated the presence of four important clusters of proteins identified during bioinformatic analysis: protein folding/refolding, gluconeogenesis, amino acid biosynthesis, and carbohydrate metabolic processes (Figure 4).
FIGURE 3.
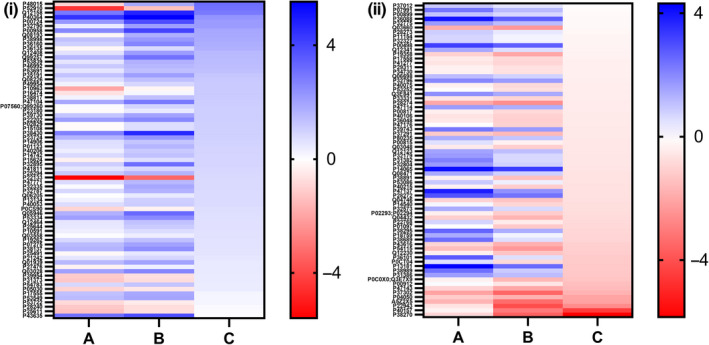
Heat map analyses of yeast host protein expression changes between day 0 and day 3 post‐induction within the control strain (column A) and between day 0 and day 3 post‐induction within the rAAV‐producing recombinant strain (column B), and between control strain and AAV‐producing strain on days 2‐3 post‐induction (column C). Subset I comprises 72 proteins showing the biggest overexpression levels, determined by log2 expression change at least 25% higher in B than A. Subset II comprises 73 proteins showing the biggest downregulation levels, determined by log2 expression change at least 25% lower in B than A. The upregulation/downregulation trends were confirmed by column C values. Proteins P39079, P00004, and Q06177 are not included in this graph. Additional information can be found in Appendix 3 (https://doi.org/10.6084/m9.figshare.13040591.v1).
TABLE 1.
Gene ontology enrichment analysis focused on biological processes was performed using the DAVID bioinformatics database. A first evaluation used a subset of 304 host cell proteins that showed statistically significant change among conditions (A), whereas the second evaluation used the previously mentioned subset of 148 proteins that showed the biggest upregulation/downregulation changes during recombinant AAV expression (B)
| Term | Gene count | Benjamini |
|---|---|---|
| (A) | ||
| Cytoplasmic translation | 38 | 6.2E‐11 |
| Metabolic process | 38 | 1.2E‐10 |
| Translation | 46 | 4.4E‐7 |
| Gluconeogenesis | 9 | 6.0E‐4 |
| Oxidation‐reduction process | 41 | 6.1E‐4 |
| Ribosomal small subunit assembly | 10 | 6.6E‐4 |
| Protein refolding | 8 | 1.1E‐3 |
| Cellular amino acid biosynthetic process | 18 | 1.5E‐3 |
| Carbohydrate metabolic process | 17 | 9.4E‐3 |
| rRNA export from nucleus | 7 | 2.0E‐2 |
| Glycolytic process | 8 | 2.4E‐2 |
| Protein folding | 15 | 3.7E‐2 |
| Pyruvate metabolic process | 6 | 3.8E‐2 |
| (B) | ||
| Protein refolding | 7 | 6.3E‐4 |
| Metabolic process | 20 | 1.5E‐2 |
| Protein folding | 11 | 1.8E‐2 |
| “de novo” protein folding | 4 | 3.2E‐2 |
| Gluconeogenesis | 5 | 9.7E‐2 |
| Response to heat | 5 | 1.4E‐1 |
| Cellular amino acid biosynthetic process | 9 | 1.4E‐1 |
| Carbohydrate metabolic process | 9 | 2.1E‐1 |
| One‐carbon metabolic process | 4 | 4.7E‐1 |
FIGURE 4.
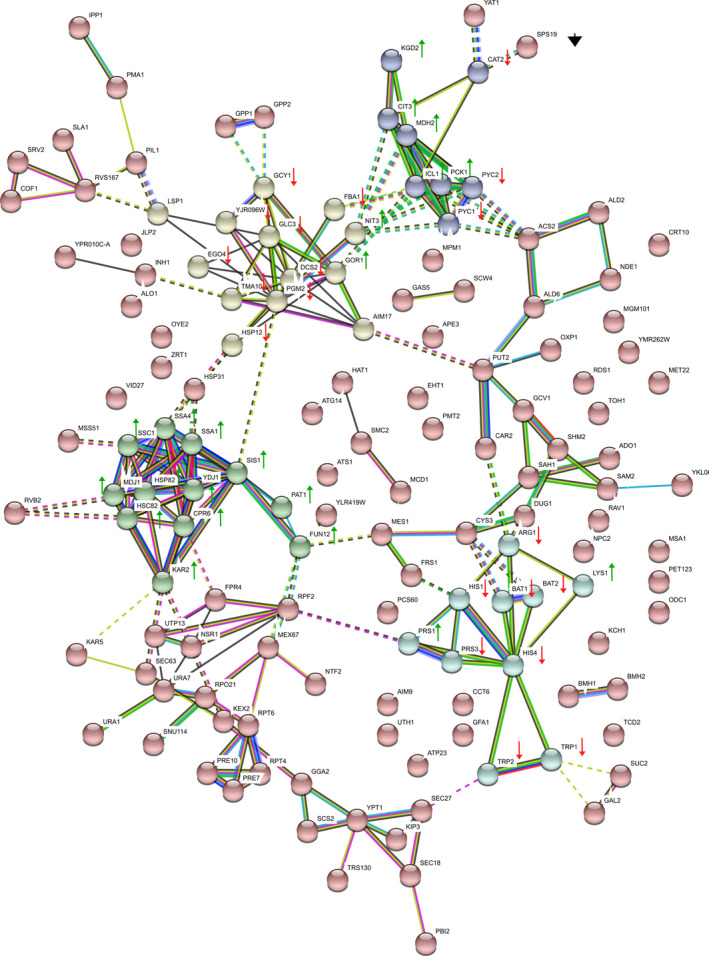
Predicted protein interaction network performed in STRING software. The analysis was done based on a previously mentioned subset of 148 proteins with differential protein expression. A high confidence factor (0.7) was used for the analysis. Proteins were clustered based on their participation in relevant biological processes: Protein refolding (green), amino acid biosynthesis (sky blue), carbohydrate metabolism/response to stress (yellow), and gluconeogenesis(purple). Non‐clustered proteins are presented in red. Each protein group displays green and red arrows next to its protein components to denote upregulation or downregulation in the rAAV‐producing strain relative to the control strain
After examining the potential implications of expression change of these proteins, we postulated that the overexpression of specific host cell proteins involved in protein folding, response to stress, proteasome activity, and some carbon metabolic processes could provide a stress‐relief activity to the yeast cell, and potentially contribute to vector yield improvement. Most of the nineteen proteins selected for overexpression studies were part of the protein subset mentioned in Figures 3 and 4, and they were all upregulated during the AAV expression phase. We generated a set of low‐copy and high‐copy number plasmids containing expression cassettes for 19 yeast host cell proteins. We overexpressed these proteins in a yeast strain transformed with two rAAV plasmids (JA001 and JA002), using a total of three plasmids for each yeast strain variant. As shown in Figure 5, the expression of some of these proteins using high‐copy and low‐copy number plasmids resulted in an improvement in the rAAV vector yield. Variant strains SSA1, RVB2, SSE1, SSE2, CCP1, and GTT1 showed significant increases in vector yield that go as high as threefold and twofold when 2‐micron and CEN‐based plasmids were used, respectively. These results support the significance of the aforementioned biological processes and the functional relevance of selected host cell proteins on rAAV vector production when using S. cerevisiae as host.
FIGURE 5.
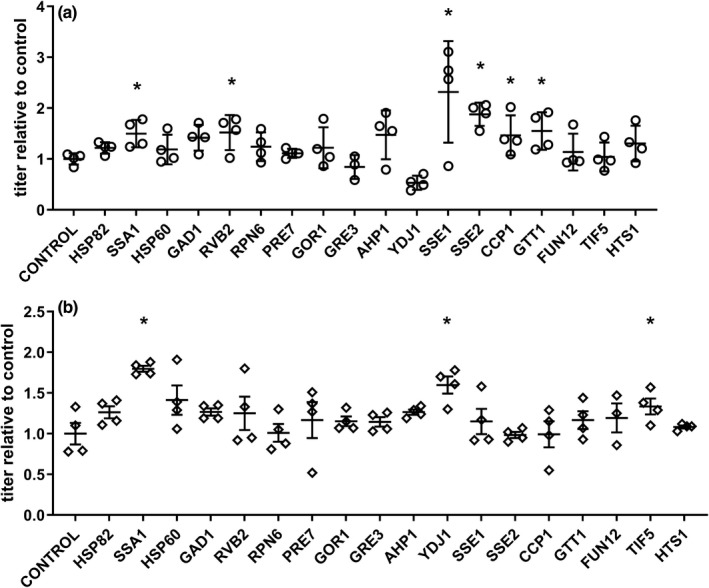
rAAV2 vector yield results from yeast protein overexpression strains. Nineteen strains were generated by transforming the control rAAV‐producing strain with an additional 2‐micron (a) or CEN (b) plasmid containing a GAL10‐X host protein expression cassette. Benzonase‐resistant vector yield results from each clone are presented as vector titer relative to the control mean value. Bars represent the mean and standard deviation (n = 4). The asterisk represents conditions that are significantly higher than the control values, based on a paired, one‐tailed t test (p < 0.05)
Overexpression of selected insect cell homologues was carried out in Sf9 cells to validate some of the findings found in the yeast model. Since bioinformatics and protein analysis highlighted protein folding and refolding as events potentially associated with rAAV expression, we decided to test overexpressed cytosolic chaperone proteins with foldase activity. S. frugiperda HSC70 and HSP40 were overexpressed during rAAV‐GFP production, and the yield was compared against a no‐overexpression control. Overexpression of HSP40 and HSC70 led to a vector yield of 7 × 1010 vg/ml, which represents a 50% volumetric yield improvement (Figure 6). This increase is associated with higher per‐cell productivity, likely due to an enhanced folding and expression capacity of the tested cells.
FIGURE 6.
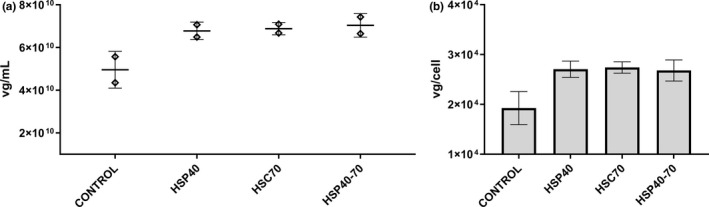
rAAV‐GFP vector production in Sf9 cells overexpressing selected protein folding chaperones. Three recombinant baculovirus strains (rBV‐GFP, rBV‐RepCap, and rBV‐HSP) were used to infect Sf9 cells at an individual MOI of 3. Each condition was run in duplicates. In the HSP40‐70 condition, rBV‐HSP40 and rBV‐HSC70 were combined at an individual MOI of 1.5. Crude supernatant was harvested at 120 h post‐infection and analyzed by ddPCR after benzonase digestion (a). Per‐cell productivity was calculated by the ratio of vg titer on harvest day by peak cell density (b)
4. DISCUSSION
Time‐course mass spectrometry analysis of yeast samples allowed us to survey changes in the proteomic profile of the rAAV‐producing yeast strain. Bioinformatics and statistical tools played an important role in highlighting expression changes and clustering them as part of biological processes. With those results, we identified processes that significantly varied when rAAV2 proteins were expressed. We built a secondary hypothesis that could potentially link these few biological processes to bottlenecks in rAAV vector production.
Our results from proteomic profiling highlighted events related to protein folding and conformational stress. Protein folding/refolding comprises cellular activities aimed at shaping the native conformation of proteins (Gasser et al., 2008). To keep cellular homeostasis, cell responses are focused on correcting the conformation of misfolded proteins, either by refolding sequestration, or degradation (Chen et al., 2011). Protein folding takes place in the endoplasmic reticulum (ER) and cytoplasm, and each compartment has its arsenal of folding proteins capable of doing a variety of modifications to the target protein. In recombinant strains, protein expression and processing machinery are mostly allocated in the ER, where the nascent protein is modified and prepared for secretion (Gasser et al., 2008; Mattanovich et al., 2004). For our case, the rAAV expression cassettes encode 6 non‐glycosylated viral proteins that undergo processing in the cytoplasmic compartment. Our proteomic profiling results highlighted the overexpression of several cytoplasmic heat shock proteins (Hsp) and chaperones during the last days of galactose induction. This correlation implies a link between protein folding activities and the expression of rAAV proteins. No differences in expression of foldases and other chaperones associated with ER processing were seen, which gives support to the hypothesis that saturation of the protein processing machinery might be taking place, potentially leading to protein misfolding at the cytoplasmic compartment.
Table 2 shows MS results regarding the fold change activity of the principal heat shock proteins during induction time. Results showed a twofold increase in KAR2 protein (also known as BiP). This protein is a stress marker and its upregulation is usually linked to unfolded protein response (Hohenblum et al., 2004). This protein was one of the few ER‐related proteins that changed in concentration after galactose induction of rAAV expression. The cytoplasmic proteins SSA1, SSA2, and SSA4 increased their expression levels more than 25%, and the vector yield increase seen after protein overexpression of SSA/SSE chaperones confirmed their functional relevance for rAAV vector production. These chaperones belong to the HSP70 and HSP110 families and are implicated in protein folding activity at the cytoplasm (Bush & Meyer, 1996; Dragovic et al., 2006). Big and small heat shock proteins showed a different change in their expression patterns, which aligned with results reported by Geiler‐Samerotte et al. (2011). The authors referred to this particular phenomenon as cytoplasmic unfolded protein response, and it has been reported on other occasions when surface viral proteins are expressed in yeast (Čiplys et al., 2011). It is believed that chaperone action is crucial to mitigate negative impacts related to protein misfolding. Valaviciute et al. (2016) evaluated the effect of overexpression and mild downregulation of HSP90, HSP70, and HSP40 chaperones and co‐chaperones during recombinant expression of VP1 hamster polyomavirus protein in yeast. Downregulation of cytosolic chaperones such as SSA1/SSA2, SSA3/SSA4, HSP82, and HSC82 had a negative effect on VP1‐EGFP levels. Also, mild overexpression of these proteins translated into a surplus of VP1 yield. Their results suggested that these sub‐group of proteins had a direct impact on protein processing, and by extension on active recombinant protein yield.
TABLE 2.
Expression fold change of important chaperones and other host proteins implicated in cytosolic unfolded protein response and response to cellular stress. Function descriptions were obtained from Yeastmine (https://yeastmine.yeastgenome.org/)
| Protein name | Fold change | Function |
|---|---|---|
| SSA4 | +2.04 | Protein folding, cellular response to heat, SRP‐dependent co‐translational protein‐membrane targeting, and translocation |
| HSP82 | +2.02 | protein refolding, proteasome assembly, box C/D RNP assembly |
| HSC82 | +2.02 | Protein refolding, proteasome assembly box C/D RNP assembly |
| CPR6 | +2.54 | Protein folding, protein refolding |
| SSA1 | +1.53 | Translation, protein refolding, proteasome‐mediated catabolic process, protein polyubiquitination |
| YDJ1 | +1.48 | “de novo” protein folding, ER‐associated ubiquitin‐dependent catabolic process, tRNA import into nucleus |
| SSC1 | +1.63 | Protein refolding, protein unfolding, protein import into the mitochondrial matrix |
| HSP60 | +1.57 | Protein refolding, protein maturation, chaperone‐mediated complex protein assembly |
| KAR2 | +2.20 | Unfolded protein binding, participates in ubiquitin‐dependent ERAD pathway, protein import into the ER |
| AHP1 | +1.06 | Cell redox homeostasis, response to oxidative stress, response to metal ion |
| ZUO1 | +1.02 | Protein folding, regulation of translational fidelity, ribosomal subunit export from the nucleus |
| PDI1 | +1.03 | Unfolded protein binding, protein disulfide isomerase activity |
| HSP12 | −6.25 | Lipid binding protein; involved in plasma membrane organization and response to oxidative, osmotic, and heat stress |
We also found multiple changes in proteins associated with gluconeogenesis such as ENO1, ENO2, MDH2, PCK1, and PCK2. It is unclear what the major driver for this change is; however, we speculate that the formation of secondary metabolic products like ethanol and trehalose could have shifted this expression pattern. Ethanol is a common byproduct during yeast consumption of glucose. This component tends to be used right after yeast diauxic shift, once glucose or the primary fermentable carbon source is depleted (Peng et al., 2015). It has been reported that ethanol consumption drives the expression of factors that reduces gluconeogenesis activity (Soontorngun et al., 2007). Moreover, yeast accumulates trehalose during recombinant protein production, a mechanism that is believed to mitigate stress (D'Amore et al., 1991). An increment in protein TPS1, directly involved in trehalose production, influences gluconeogenesis as part of the general stress response (Deroover et al., 2016).
An additional function of the host cell proteins highlighted in our analysis is related to protein degradation. During the profiling analysis, changes in proteins that are components of the proteasome subunits such as RPN6 and PRE7 were identified. Many cytoplasmic chaperones participate in ubiquitin‐dependent and independent degradation processes, which include proteasome activity (Ben‐Nissan & Sharon, 2014; Santamaria et al., 2003). The increased expression of this set of proteins suggests that protein degradation activities might have taken place during rAAV protein expression, likely by proteasome activity on misfolded rAAV proteins such as capsids. Empirical knowledge from Western blot analysis showed VP capsid proteins of multiple sizes besides the three expected sizes (data not shown), which would support the notion of potential protein degradation events occurring in the cytosol. We also identified host cell proteins that get upregulated due to oxidative stress. Thioredoxins, catalases, superoxide dismutases, and glutathione transferases usually play an important role during oxidative stress (Gasch, 2003). GAD1 and GTT1 showed an increase in protein levels after galactose induction. These two proteins participate in the metabolism of glutamate and glutathione, respectively; which indirectly impacts the intracellular redox potential and modulates the stress generated by toxic oxidants (Coleman et al., 2001; Collinson & Grant, 2003; Grant, 2001). Other overexpressed proteins were CCP1, GRE3, and AHP1. These proteins, present in cytosolic and mitochondrial compartments, are expressed under stress conditions and are implicated in different metabolic routes that protect cells against oxidant damage (Aguilera & Prieto, 2001; Charizanis et al., 1999; Lee et al., 1999). Increased expression of some of these antioxidant proteins, however, has been associated with a diauxic shift in yeast strains. Since this metabolic event is common in strains that grow in glucose‐galactose media transitions (Murphy et al., 2015), it is plausible to think that the specific increase of some of these proteins might have been triggered by metabolic events different from rAAV production.
We performed a protein overexpression strategy based on the analysis of proteome changes and their potential implications on rAAV production. We hypothesized that additional expression of host cell proteins would benefit stress‐free cell metabolism. The results shown in our yeast model using low‐copy and high‐copy number plasmids supported the utility of the rAAV‐producing yeast strain for proteomics‐guided optimization. In few cases such as YDJ1, protein overexpression using high‐copy number plasmids led to a vector yield lower than the one obtained with the control strain, suggesting that modulation of chaperone/host protein levels is required to achieve optimal yields. Vector titer improvement was evidenced after overexpression of proteins related to protein folding, response to oxidative stress, and regulation of gene expression. More importantly, improved results translated into insect cells when the respective protein folding homologues were overexpressed. These results might indicate that protein folding takes place during rAAV production among different systems and that the extent of foldase protein activity might lead to more efficient vector production at a cellular level.
5. CONCLUSIONS
The present study provided a molecular snapshot of proteomic changes related to rAAV expression in yeast. Data analysis suggested that cytoplasmic unfolded protein response might be taking place primarily in our rAAV‐producing yeast strain, and secondary events such as proteasome degradation and cellular stress might be associated with it. We used this information to look for potential molecular strategies for optimization, and a host protein overexpression strategy resulting in increased yields supported this effort. We validated the rAAV‐enhancing activity of protein folding chaperones in Sf9 cells, which sustains the use of the yeast model for the study and optimization of rAAV‐producing systems. Further steps will include combined overexpression of proteins involved in multiple mechanisms, in the hope that this strategy shows a synergistic effect on vector yield improvement.
CONFLICTS OF INTEREST
JAU, DB, JP, DG, MR, and HS are current employees of Biomarin Pharmaceutical Inc.
AUTHOR CONTRIBUTION
Juan Aponte‐Ubillus: Conceptualization (lead); Data curation (equal); Formal analysis (lead); Investigation (lead); Writing‐original draft (lead). Daniel Barajas: Conceptualization (equal); Investigation (supporting); Supervision (supporting); Writing‐review & editing (equal). Harry Sterling: Conceptualization (equal); Data curation (equal); Formal analysis (supporting); Investigation (supporting); Supervision (supporting); Writing‐review & editing (equal). Ali Aghajanirefah: Formal analysis (supporting); Investigation (supporting); Writing‐review & editing (equal). Cameron Bardliving: Conceptualization (equal); Supervision (supporting); Writing‐review & editing (equal). Joseph Peltier: Conceptualization (equal); Formal analysis (supporting); Supervision (supporting); Writing‐review & editing (equal). Parviz Shamlou: Conceptualization (equal); Supervision (supporting); Writing‐review & editing (equal). Mimi Roy: Conceptualization (equal); Project administration (supporting); Writing‐review & editing (equal). Daniel Gold: Conceptualization (equal); Funding acquisition (lead); Writing‐review & editing (equal).
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
The authors would like to thank Yvette Tang, Tomas Cinek, and Marc‐Andre Robert at BioMarin Pharmaceutical Inc for their valuable suggestions during study data analysis.
DATA AVAILABILITY STATEMENT
The data sets generated and/or analyzed during the current study plus supplementary information are available in the figshare repository at https://doi.org/10.6084/m9.figshare.13040591.v1: Appendix 1. List of primers sets used for amplification of sequences from yeast genomic DNA and further Gibson assembly of overexpression plasmids; Appendix 2. MS data set of identified proteins; Appendix 3. Table and heatmap showing protein expression changes per strain.
REFERENCES
- Aguilera, J. , & Prieto, J. A. (2001). The Saccharomyces cerevisiae aldose reductase is implied in the metabolism of methylglyoxal in response to stress conditions. Current Genetics, 39, 273–283. [DOI] [PubMed] [Google Scholar]
- Aponte‐Ubillus, J. J. , Barajas, D. , Peltier, J. , Bardliving, C. , Shamlou, P. , & Gold, D. (2018). Molecular design for recombinant adeno‐associated virus (rAAV) vector production. Applied Microbiology and Biotechnology, 102, 1045–1054. 10.1007/s00253-017-8670-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aucoin, M. G. , Perrier, M. , & Kamen, A. A. (2006). Production of adeno‐associated viral vectors in insect cells using triple infection: optimization of baculovirus concentration ratios. Biotechnology and Bioengineering, 95, 1081–1092. 10.1002/bit.21069 [DOI] [PubMed] [Google Scholar]
- Balakrishnan, B. , & Jayandharan, G. R. (2014). Basic biology of adeno‐associated virus (AAV) vectors used in gene therapy. Current Gene Therapy, 14, 86–100. [DOI] [PubMed] [Google Scholar]
- Barajas, D. , Aponte‐Ubillus, J. J. , Akeefe, H. , Cinek, T. , Peltier, J. , & Gold, D. (2017). Generation of infectious recombinant Adeno‐associated virus in Saccharomyces cerevisiae . PLoS One, 12, e0173010 10.1371/journal.pone.0173010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Nissan, G. , & Sharon, M. (2014). Regulating the 20S proteasome ubiquitin‐independent degradation pathway. Biomolecules, 4, 862–884. 10.3390/biom4030862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, G. L. , & Meyer, D. I. (1996). The refolding activity of the yeast heat shock proteins Ssa1 and Ssa2 defines their role in protein translocation. Journal of Cell Biology, 135, 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis, C. , Juhnke, H. , Krems, B. , & Entian, K. D. (1999). The mitochondrial cytochrome c peroxidase Ccp1 of Saccharomyces cerevisiae is involved in conveying an oxidative stress signal to the transcription factor Pos9 (Skn7). Molecular and General Genetics, 262, 437–447. [DOI] [PubMed] [Google Scholar]
- Chen, B. , Retzlaff, M. , Roos, T. , & Frydman, J. (2011). Cellular strategies of protein quality control. Cold Spring Harbor Perspectives in Biology, 3, a004374 10.1101/cshperspect.a004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. (2008). Intron splicing‐mediated expression of AAV Rep and Cap genes and production of AAV vectors in insect cells. Molecular Therapy, 16, 924–930. 10.1038/mt.2008.35 [DOI] [PubMed] [Google Scholar]
- Čiplys, E. , Samuel, D. , Juozapaitis, M. , Sasnauskas, K. , & Slibinskas, R. (2011). Overexpression of human virus surface glycoprotein precursors induces cytosolic unfolded protein response in Saccharomyces cerevisiae. Microbial Cell Factories, 10, 37 10.1186/1475-2859-10-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, S. T. , Fang, T. K. , Rovinsky, S. A. , Turano, F. J. , & Moye‐Rowley, W. S. (2001). Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. Journal of Biological Chemistry, 276, 244–250. 10.1074/jbc.M007103200 [DOI] [PubMed] [Google Scholar]
- Collinson, E. J. , & Grant, C. M. (2003). Role of yeast glutaredoxins as glutathione S‐transferases. Journal of Biological Chemistry, 278, 22492–22497. 10.1074/jbc.M301387200 [DOI] [PubMed] [Google Scholar]
- D'Amore, T. , Crumplen, R. , & Stewart, G. G. (1991). The involvement of trehalose in yeast stress tolerance. Journal of Industrial Microbiology, 7, 191–195. 10.1007/BF01575882 [DOI] [Google Scholar]
- Deroover, S. , Ghillebert, R. , Broeckx, T. , Winderickx, J. , & Rolland, F. (2016). Trehalose‐6‐phosphate synthesis controls yeast gluconeogenesis downstream and independent of SNF1. FEMS Yeast Research, 16(4), fow036 10.1093/femsyr/fow036 [DOI] [PubMed] [Google Scholar]
- Dragovic, Z. , Broadley, S. A. , Shomura, Y. , Bracher, A. , & Hartl, F. U. (2006). Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO Journal, 25, 2519–2528. 10.1038/sj.emboj.7601138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli, A. , Della Latta, V. , Bologna, C. , Pucciarelli, D. , Cipriani, F. , Backovic, A. , & Cervelli, T. (2017). Strategies to optimize capsid protein expression and single‐stranded DNA formation of adeno‐associated virus in Saccharomyces cerevisiae . Journal of Applied Microbiology, 123, 414–428. 10.1111/jam.13511 [DOI] [PubMed] [Google Scholar]
- Gasch, A. P. (2003). The environmental stress response: A common yeast response to diverse environmental stresses In Hohmann S., & Mager W. H. (Eds.), Yeast stress responses (pp. 11–70). Springer. [Google Scholar]
- Gasser, B. , Saloheimo, M. , Rinas, U. , Dragosits, M. , Rodríguez‐Carmona, E. , Baumann, K. , Giuliani, M. , Parrilli, E. , Branduardi, P. , Lang, C. , Porro, D. , Ferrer, P. , Tutino, M. , Mattanovich, D. , & Villaverde, A. (2008). Protein folding and conformational stress in microbial cells producing recombinant proteins: A host comparative overview. Microbial Cell Factories, 7, 11 10.1186/1475-2859-7-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler‐Samerotte, K. A. , Dion, M. F. , Budnik, B. A. , Wang, S. M. , Hartl, D. L. , & Drummond, D. A. (2011). Misfolded proteins impose a dosage‐dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proceedings of the National Academy of Sciences of the United States of America, 108, 680–685. 10.1073/pnas.1017570108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy, M.‐C. , & Salvetti, A. (2005). Helper functions required for wild type and recombinant adeno‐associated virus growth. Current Gene Therapy, 5, 265–271. [DOI] [PubMed] [Google Scholar]
- Grant, C. M. (2001). Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Molecular Microbiology, 39, 533–541. [DOI] [PubMed] [Google Scholar]
- Hohenblum, H. , Gasser, B. , Maurer, M. , Borth, N. , & Mattanovich, D. (2004). Effects of gene dosage, promoters, and substrates on unfolded protein stress of recombinant Pichia pastoris . Biotechnology and Bioengineering, 85, 367–375. 10.1002/bit.10904 [DOI] [PubMed] [Google Scholar]
- Lee, J. , Spector, D. , Godon, C. , Labarre, J. , & Toledano, M. B. (1999). A new antioxidant with alkyl hydroperoxide defense properties in yeast. Journal of Biological Chemistry, 274, 4537–4544. [DOI] [PubMed] [Google Scholar]
- Mattanovich, D. , Gasser, B. , Hohenblum, H. , & Sauer, M. (2004). Stress in recombinant protein producing yeasts. Journal of Biotechnology, 113, 121–135. 10.1016/j.jbiotec.2004.04.035 [DOI] [PubMed] [Google Scholar]
- Murphy, J. P. , Stepanova, E. , Everley, R. A. , Paulo, J. A. , & Gygi, S. P. (2015). Comprehensive temporal protein dynamics during the diauxic shift in Saccharomyces cerevisiae . Molecular & Cellular Proteomics: MCP, 14, 2454–2465. 10.1074/mcp.M114.045849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete, A. , & Kotin, R. M. (2008). Large‐scale production of recombinant adeno‐associated viral vectors. Methods in Molecular Biology, 433, 79–96. 10.1007/978-1-59745-237-3_5 [DOI] [PubMed] [Google Scholar]
- Paulo, J. A. , O'Connell, J. D. , Gaun, A. , & Gygi, S. P. (2015). Proteome‐wide quantitative multiplexed profiling of protein expression: Carbon‐source dependency in Saccharomyces cerevisiae . Molecular Biology of the Cell, 26, 4063–4074. 10.1091/mbc.E15-07-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, B. , Williams, T. C. , Henry, M. , Nielsen, L. K. , & Vickers, C. E. (2015). Controlling heterologous gene expression inyeast cell factories on different carbon substrates and across the diauxic shift: Acomparison of yeast promoter activities. Microbial Cell Factories, 14, 91 10.1186/s12934-015-0278-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski, R. J. , & Muzyczka, N. (2014). AAV‐mediated gene therapy for research and therapeutic purposes. Annual Review of Virology, 1, 427–451. 10.1146/annurev-virology-031413-085355 [DOI] [PubMed] [Google Scholar]
- Santamaria, P. G. , Finley, D. , Ballesta, J. P. G. , & Remacha, M. (2003). Rpn6p, a proteasome subunit from Saccharomyces cerevisiae, is essential for the assembly and activity of the 26 S proteasome. Journal of Biological Chemistry, 278, 6687–6695. 10.1074/jbc.M209420200 [DOI] [PubMed] [Google Scholar]
- Silva, J. C. , Gorenstein, M. V. , Li, G.‐Z. , Vissers, J. P. C. , & Geromanos, S. J. (2006). Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Molecular & Cellular Proteomics: MCP, 5, 144–156. 10.1074/mcp.M500230-MCP200 [DOI] [PubMed] [Google Scholar]
- Soontorngun, N. , Larochelle, M. , Drouin, S. , Robert, F. , & Turcotte, B. (2007). Regulation of gluconeogenesis in Saccharomyces cerevisiae is mediated by activator and repressor functions of Rds2. Molecular and Cellular Biology, 27, 7895–7905. 10.1128/MCB.01055-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valaviciute, M. , Norkiene, M. , Goda, K. , Slibinskas, R. , & Gedvilaite, A. (2016). Survey of molecular chaperone requirement for the biosynthesis of hamster polyomavirus VP1 protein in Saccharomyces cerevisiae . Archives of Virology, 161, 1807–1819. 10.1007/s00705-016-2846-3 [DOI] [PubMed] [Google Scholar]
- Wright, J. F. , Le, T. , Prado, J. , Bahr‐Davidson, J. , Smith, P. H. , Zhen, Z. , Sommer, J. M. , Pierce, G. F. , & Qu, G. (2005). Identification of factors that contribute to recombinant AAV2 particle aggregation and methods to prevent its occurrence during vector purification and formulation. Molecular Therapy, 12, 171–178. 10.1016/j.ymthe.2005.02.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and/or analyzed during the current study plus supplementary information are available in the figshare repository at https://doi.org/10.6084/m9.figshare.13040591.v1: Appendix 1. List of primers sets used for amplification of sequences from yeast genomic DNA and further Gibson assembly of overexpression plasmids; Appendix 2. MS data set of identified proteins; Appendix 3. Table and heatmap showing protein expression changes per strain.


