Abstract
Background
Circular RNAs (circRNAs) may act as biomarkers of coronary artery disease (CAD). However, the relationship between expression characteristics of circRNAs and coronary atherosclerosis has not been fully explored. The aim of this study was to determine and characterize the circRNAs from human coronary artery.
Methods
The coronary artery segments were obtained from an 81‐year‐old male patient with sudden death of myocardial infarction at autopsy. The coronary stenosis and atherosclerosis were evaluated by hematoxylin and eosin (H&E) staining, and the circRNAs expression profile was characterized by RNA sequencing (RNA‐seq). The differentially expressed circRNAs were validated by qRT‐PCR.
Results
The analysis of H&E staining indicated that coronary atherosclerosis grade and extent in the LM was more serious than that in other coronary arteries. Twenty‐seven circRNAs were selected for expression validation in coronary artery. CircRNAs corresponding cyclization sites of 3 circRNAs (hsa_circ_0016868, hsa_circ_0001364, hsa_circ_0006731) have been verified by Sanger sequencing.
Conclusion
The 3 circRNAs are suggested to play a pathological role underlying the coronary arteries atherosclerosis and may serve as a valuable resource as diagnostic or therapeutic targets against CAD.
Keywords: CircRNAs, coronary artery samples, coronary heart disease, qRT‐PCR
The overall technology roadmap of the present study (A) Coronary artery samples preparation (B) RNA‐Seq library preparation, and sequencing (C) qRT‐PCR forward and reverse prime (D) Sanger sequencing samples preparation, and steps

1. INTRODUCTION
Coronary artery disease (CAD) is one of the major forms of atherosclerosis and becomes a heavy burden on social economy and public health worldwide. 1 The morbility and mortality of CAD have maintained dismal, and prognosis has remained ineligible in spite of the development of diagnosis and treatment strategies of CAD, including medications, cardiac interventional therapy, and surgery operation. 2 The early diagnosis and treatment may reduce major mortality of CAD 3 ; therefore, highly sensitive and specific biomarkers are urgently needed for the earlier diagnosis and treatment of CAD. Recently, circular RNAs (circRNAs) have been suggested to become a potential class of biomarkers for CAD. 4
CircRNAs are genetic regulators that have covalently linked ends with no polyadenylated tails in contrast to linear RNAs. In addition, circRNAs regulate gene expression via multiple mechanisms including sponging miRNA, binding RNA‐binding proteins, sequestering agents, regulating transcript, and transforming into functional proteins. 5 , 6 Due to the characteristics of circRNAs including highly stable and detectable in body fluids, abundantly expressed, evolutionarily conserved in humans, circRNAs have been suggested as better biomarkers than linear RNAs in clinical diagnosis, treatment, and prognosis of CAD. 7 , 8 , 9 , 10 However, the total amount of circRNAs is low in blood to be easily detected. 11 Therefore, the expression characteristics of circRNAs and association with coronary atherosclerosis remain elusive.
In the present study, the transcriptome‐wide circRNAs expression in coronary artery segments was profiled, and the differential expression circRNAs were validated in coronary artery segments via qRT‐PCR method in order to explore the relationship between the circular RNA expression content and severity of coronary atherosclerosis in coronary artery.
2. MATERIALS AND METHODS
2.1. Study subjects
The coronary artery samples were obtained from an autopsy case at Department of Human Anatomy in Nanjing Medical University. Informed consent was obtained from the bereaved family that pathological samples were used for the research only, and the autopsy was conducted according to the guideline of the university. The methods were performed in accordance with the approved guiding principles, and all experimental protocols were approved by the ethics committee of the Nanjing Medical University and the First Affiliated Hospital of Nanjing Medical University.
The overall research technology roadmap of the present study is shown in Figure S1A‐D.
2.2. Coronary artery segments preparation
The study included one autopsy performed at 2018 at the Department of Human Anatomy in Nanjing Medical University. The 81‐year‐old male subject died from heart attack, and the postmortem delay was about 1 hour. At autopsy, epicardial coronary arteries were removed from the hearts. The epicardial coronary artery of the autopsy was divided into 10 segments: the proximal segment, the midsegment, the distal segment of the left anterior descending (LAD), left circumflex (LCX), right coronary artery (RCA), respectively, and the left main trunk (LM). And, every coronary artery segment was divided into two groups: RNA group and pathological group. The segments at the RNA group were snap‐frozen in liquid nitrogen and stored at −80˚C for RNA‐seq and validated by qRT‐PCR. In addition, the segments at the pathological group were fixed overnight in 10% formalin and embedded in paraffin for histological analysis.
2.3. Pathological analysis
The pathological analysis of the coronary arteries was conducted in Department of Human Anatomy in Nanjing Medical University. The specific steps of the pathological examination have been previously described. 12
Coronary atherosclerosis grade and extent of the coronary artery segments were analyzed by independent pathologists who attempted to relate them to the current histological grading according to American Heart Association (AHA) classification guidelines. 13
2.4. RNA isolation, RNA‐Seq library preparation, and sequencing
Total RNA content in the coronary artery samples was isolated by TRIzol (15 596 018, Invitrogen) following the manufacturer's instructions and detected by an Agilent Bioanalyzer 2100 (Agilent technologies, US) for a RIN number to inspect RNA integration. RNA degradation and contamination were detected using a 1% agarose gel. In addition, NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) was used to measure RNA purity and concentration. The input RNA sample material was prepared as a total of 3 μ g RNA per sample. The steps for RNA‐Seq library preparation and sequencing are the same as we previously described. 12
2.5. Selection of circular RNAs
The differentially expressed circular RNAs that showed a two‐fold or greater change in the coronary artery samples were screened, and the overlapping among the data sets was illustrated using Venn diagrams. To further assess the association of circular RNA expression content with severity of coronary atherosclerosis, the expression levels of the differentially expressed circulars were validated by qRT‐PCR in coronary artery samples.
2.6. qRT‐PCR
Total RNA (1 µg) was used as a template to prepare cDNA (ReverTra Ace qPCR RT Kit, TOYOBO, FSQ‐101, Japan). The differentially expressed circRNAs were quantified using SYBR Green Realtime PCR Master Mix (TOYOBO, QPK‐212) on the ABI 7900HT sequence detection system (Applied Biosystems, 7900HT). PCR was performed with the following thermocycling conditions: An initial 1 minutes at 95°C, followed by 40 cycles of 95°C for 15 seconds, 60°C for 30 seconds. PCR products were gel‐extracted and Sanger‐sequenced. The primers were obtained from GeneCreate Bio Co. (Wuhan, China). Housekeeping gene β‐actin was used as an internal reference to normalize the results. All experiments were conducted in triplicate. Finally, the 2−ΔΔCt method was performed to calculate the relative expression. 14
2.7. Statistical analysis
All statistical analyses were performed using SPSS Software 16.0 (Chicago, USA). Results are given as means ± standard deviation. t test was used to compare the differences between two groups. GraphPad Prism 5 software was used to analyze data and create graphs. p values < 0.05 were considered statistically significant.
3. RESULTS
3.1. Natural history and histological classification of atherosclerotic lesions of the coronary artery samples
This research is part of our series of studies, the results of which have been published in an our previously published article. 12
3.2. Expression validation of selected differentially expressed circRNAs by qRT‐PCR analysis
The overlapping circRNAs of the different clusters of coronary artery including LAD, LCX, and RCA were analyzed via using Venn diagram analysis by online software at the following URL: http://bioinformatics.psb.ugent.be/webtools/Venn/. Furthermore, Venn diagrams were adopted to illustrate the overlapping of differentially expressed circRNAs in coronary artery samples among the three data sets including LAD, LCX, and RCA (Figure S2).
In this study, a total of 7 circRNAs (Table 1) were differentially expressed in both the LAD and LCX coronary artery samples, while in Table 2, we found that a total of 12 circRNAs were differentially expressed in both the LAD and RCA coronary artery samples, and 11 (Table S1) differentially expressed circRNAs were found both the LCX and RCA coronary artery samples. Furthermore, the hsa_circ_0004672 expressed circRNA was found in the LAD, LCX, and RCA coronary artery samples. Among the differentially expressed circRNAs, 10 circRNAs were not recruited for expression validation due to their fragments are short; thus, 27 circRNAs were selected for expression validation in coronary artery and the data are listed in Table S2.
Table 1.
List of 7 cirRNAs that were co‐overexpressed in the left anterior descending and left circumflex coronary artery samples
| circRNAs | LAD‐p | LAD‐m | LAD‐d | LCX‐p | LCX‐m | LCX‐d | log2.F C | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAD‐d vs. LAD‐m | LAD‐d vs. LAD‐p | LAD‐m vs. LAD‐p | LCX‐d vs. LCX‐m | LCX‐d vs. LCX‐p | LCX‐m vs. LCX‐p | |||||||
| has_circ_0004672 | 0 | 474.2547425 | 195.1600312 | 824.0626288 | 0 | 343.9085203 | –1.2409 | 8.6876 | 9.9328 | 9.428 | –1.2719 | –10.706 |
| hsa_circ_0003244 | 535.188654 | 0 | 195.1600312 | 206.0156572 | 0 | 429.8856504 | 8.6286 | –1.2971 | –10.021 | 9.7499 | 1.0501 | –8.7063 |
| hsa_circ_0006693 | 0 | 135.501355 | 487.9000781 | 274.6875429 | 0 | 773.7941708 | 1.8884 | 10.01 | 8.1255 | 10.598 | 1.483 | –9.1214 |
| hsa_circ_0006848 | 178.396218 | 474.2547425 | 975.8001561 | 618.0469716 | 0 | 171.9542602 | 1.081 | 2.6098 | 1.4972 | 8.428 | –1.8568 | –10.291 |
| hsa_circ_0008068 | 356.792436 | 135.501355 | 0 | 137.3437715 | 413.2914531 | 0 | –8.0621 | –9.3998 | –1.3102 | –9.6889 | –8.1072 | 1.55 |
| hsa_circ_0016868 | 445.990545 | 203.2520325 | 0 | 137.3437715 | 495.9497438 | 0 | –8.6471 | –9.7217 | –1.0472 | –9.952 | –8.1072 | 1.813 |
| hsa_circ_0037886 | 178.396218 | 406.504065 | 0 | 137.3437715 | 0 | 343.9085203 | –9.6471 | –8.3998 | 1.2748 | 9.428 | 1.3131 | –8.1214 |
Abbreviations: FC, fold change; LAD‐d, distal segment of the left anterior descending; LAD‐m, midsegment of the left anterior descending; LAD‐p, proximal segment of the left anterior descending; LCX‐d, distal segment of the left circumflex; LCX‐m, midsegment of the left circumflex; LCX‐p, proximal segment of the left circumflex.
Table 2.
List of 12 cirRNAs that were co‐overexpressed in the left anterior descending and right coronary artery samples
| circRNAs | LAD‐p | LAD‐m | LAD‐d | RCA‐p | RCA‐m | RCA‐d | log2.F C | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAD‐d vs. LAD‐m | LAD‐d vs. LAD‐p | LAD‐m vs. LAD‐p | RCA‐d vs. RCA‐m | RCA‐d vs. RCA‐p | RCA‐m vs. RCA‐p | |||||||
| hsa_circ_0001364 | 178.396218 | 338.7533875 | 0 | 553.1483359 | 239.2344498 | 0 | –9.3841 | –8.3998 | 1.0117 | –8.9983 | –10.121 | –1.0429 |
| hsa_circ_0002500 | 624.386763 | 135.501355 | 0 | 184.3827786 | 478.4688995 | 0 | –8.0621 | –10.207 | –2.1176 | –9.9983 | –8.5358 | 1.542 |
| hsa_circ_0002607 | 0 | 135.501355 | 292.7400468 | 460.9569466 | 159.4896332 | 0 | 1.1514 | 9.2726 | 8.1255 | –8.4133 | –9.8577 | –1.3649 |
| hsa_circ_0003632 | 535.188654 | 1016.260163 | 0 | 737.5311146 | 239.2344498 | 0 | –10.969 | –9.9848 | 1.0117 | –8.9983 | –10.536 | –1.458 |
| hsa_circ_0006041 | 178.396218 | 338.7533875 | 0 | 553.1483359 | 0 | 259.235256 | –9.3841 | –8.3998 | 1.0117 | 8.9221 | –1.1119 | –10.028 |
| hsa_circ_0006731 | 0 | 880.7588076 | 390.3200625 | 460.9569466 | 877.1929825 | 0 | –1.134 | 9.6876 | 10.826 | –10.873 | –9.8577 | 1.0946 |
| hsa_circ_0008664 | 0 | 135.501355 | 585.4800937 | 276.574168 | 1036.682616 | 0 | 2.1514 | 10.273 | 8.1255 | –11.114 | –9.1208 | 2.0725 |
| hsa_circ_0008925 | 535.188654 | 135.501355 | 0 | 460.9569466 | 159.4896332 | 0 | –8.0621 | –9.9848 | –1.8952 | –8.4133 | –9.8577 | –1.3649 |
| hsa_circ_0008985 | 891.98109 | 338.7533875 | 0 | 276.574168 | 558.2137161 | 0 | ‐9.3841 | ‐10.722 | ‐1.3102 | ‐10.221 | ‐9.1208 | 1.1794 |
| hsa_circ_0014174 | 445.990545 | 135.501355 | 0 | 184.3827786 | 0 | 518.470512 | ‐8.0621 | ‐9.7217 | ‐1.6321 | 9.9221 | 1.4731 | ‐8.4434 |
Among them, 10 circRNA (has_circ_0004672, hsa_circ_0016868, hsa_circ_0001364, hsa_circ_0003632, hsa_circ_0006731, hsa_circ_0008925, hsa_circ_0008985, hsa_circ_0001551, hsa_circ_0006987, and hsa_circ_0049434) can be plotting with the expression validation data by means of qRT‐PCR analysis, and the 2^‐△△ct of the 10 circRNAs is shown in Table S3. The comparisons of expression levels of circRNAs by qRT‐PCR in the other eight coronary artery segments versus LM are shown in Table S4.
We validated that several of these circRNAs were indeed differentially expressed in the artery samples (Figure 1A‐J). In addition, circRNA corresponding cyclization sites of 3 circRNAs (hsa_circ_0016868, hsa_circ_0001364, and hsa_circ_0006731) have been verified by Sanger sequencing (Figure 2A‐C). Therefore, the objective circRNA validation rate was 3/27 in the present study.
Figure 1.
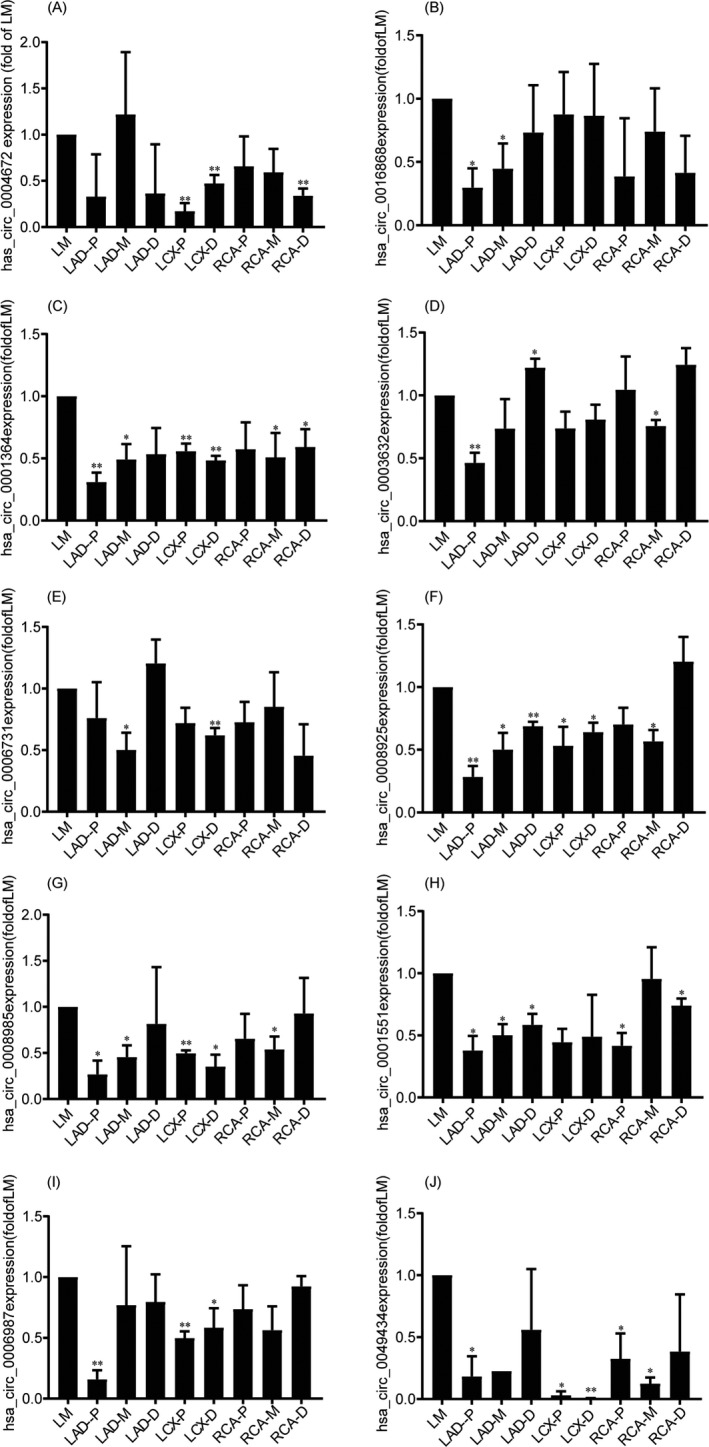
qRT‐PCR analysis of expression of 10 circRNAs in the coronary artery samples. A, has_circ_0004672. B, hsa_circ_0016868. C, hsa_circ_0001364. D, hsa_circ_0003632. E, hsa_circ_0006731. F, hsa_circ_0008925. G, hsa_circ_0001551. H, hsa_circ_0006987. I, hsa_circ_0049434. J, hsa_circ_0049434. *P < .05, **P < .01 vs. LM (two‐tailed t test)
Figure 2.
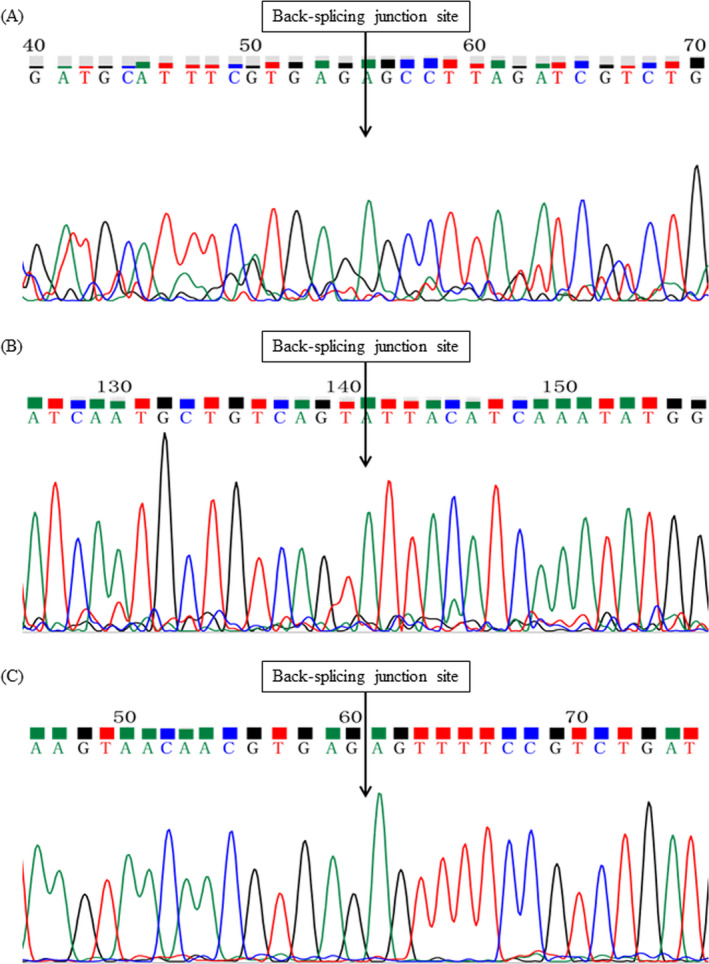
The cyclization site of qRT‐PCR products confirmed by Sanger sequencing "A, hsa_circ_0016868, B, hsa_circ_0001364, C, hsa_circ_0006731"
4. DISCUSSION
In this study, we reported the circRNAs validation results by qRT‐qPCR measurements in coronary artery samples based on the RNA‐seq profile, and the results revealed that 3 circRNAs (hsa_circ_0016868, hsa_circ_0001364, and hsa_circ_0006731) have been verified by qRT‐PCR measurements and Sanger sequencing among 27 differentially expressed circRNAs. Furthermore, the association between the expression content of 3 validated circular RNA and severity of coronary atherosclerosis in coronary artery was explored.
As endogenous transcripts, circRNAs display differential expression among species, developmental stages, and various pathologies. Due to the characteristics of increased stability compared with the linear transcripts, circRNAs have been proposed as potential candidates for biomarker of diagnostic and therapeutic interventions. 15 Recently, circRNAs have been explored in CAD and represented to act important roles in CAD. Wang et al have identified that two circular RNAs (Hsa_circ_0001879 and Hsa_circ_0004104) as novel biomarkers for CAD. 16 Mao et al demonstrated that the circ‐SATB2 regulates vascular smooth muscle cell proliferation and differentiation through upregulates STIM1 expression via miR‐939. 17 Li X et al reported that hsa‐circRNA11783‐2 level in peripheral blood is associated with CAD and type 2 diabetes mellitus. 18 However, in the above studies, due to the absence of experimental confirmation of the circular nature of the validated circRNAs, the false‐positive results by detection of linear RNA molecules with sequences closely resembled to the specific backsplice junction have not been avoided. 19 In the present study, among 27 differentially expression circRNAs, 10 circRNAs have been validated successfully by means of qRT‐PCR assays. And they have been shown to be expressed differently in the coronary artery samples. However, only 3 circRNAs (hsa_circ_0016868, hsa_circ_0001364, and hsa_circ_0006731) circular nature of the identified transcripts have been verified by Sanger sequencing. Because circRNAs are subtly expressed in circulating blood, it is difficult to accurately measure their contents in peripheral blood by qRT‐PCR. But in our study, circRNAs were highly expressed in coronary arteries, and the expression levels have shown notable difference in different segments of coronary artery. Hsa_circ_0016868 showed significant subtler expression levels in LAD‐p and LAD‐m than LM. Hsa_circ_0001364 had a significantly subtler expression in LAD‐p, LAD‐m, LCX‐p, RCA‐m, and RAC‐d compared to LM. Hsa_circ_0006731 was significant subtler expression in LAD‐m and LCX‐d than LM. And with the results of hematoxylin and eosin (H&E), different degrees of coronary artery stenosis were precisely presented. Moreover, some circRNAs showed tissue‐specific expression patterns in human. This will help to further explore the relationship between the progression of coronary atherosclerosis and circRNAs.
It has been reported that the mechanisms involved in cardiovascular aging and the potential for targeting novel pathways implicate in endothelial dysfunction, mitochondrial oxidative stress, chromatin remodeling and genomic instability. 20 Likewise, it has been reported that cardiovascular diseases are closely related to the expression of circRNA and that they point to a high abundance of specific cardiac‐expressed circRNA. 21 Besides, upregulation of atherosclerosis‐susceptible genes, such as indoleamine 2,3‐dioxygenase1 (IDO1), MMP8, and CD40, and downregulation of anti‐atherosclerosis genes, such as apolipoprotein AI (ApoA I), RNASE1, by overexpression of circRNAs in CAD patients might contribute to the pathogenesis of atherosclerosis and CAD. 11 CircRNAs have been demonstrated to play important roles in a variety of inflammation‐related diseases by acting as miRNA sponges and thus may influence the progression of coronary atherosclerosis by regulating inflammatory response. 22 Therefore, the mechanism of the effect of differentially expressed circRNAs in coronary arteries on coronary atherosclerosis still needs to be further explored.
There are still some limitations in this study. The human coronary artery samples of the present study are only from one patient, and the sample size is a little small; Besides, the verification of the circRNA profile by qRT‐PCR needs to be conducted in another independent large‐size sample cohort. Additionally, since this study does not include functional validation assays, it has not been speculated that circRNA may be involved in the specific mechanisms of atherosclerosis development. The case died from heart attack, and postmortem delay was about 1 hour. Therefore, we do not have any information of the possible effects of death on the degradation of circRNAs.
In conclusion, in this work, we found 27 differentially expression circRNAs and successfully validated 10 circRNAs by qRT‐PCR assays. However, we only verified 3 circRNAs (hsa_circ_0016868, hsa_circ_0001364, and hsa_circ_0006731) circular nature of the identified transcripts by Sanger sequencing. The results indicate that the identified circRNAs may play an important role in the development of human atherosclerosis.
AUTHORS' CONTRIBUTIONS
As the guarantor, Enzhi Jia conceived the study. Can Hou initially drafted the manuscript. Lingfeng Gu, Yi Guo, Yaqing Zhou, Lei Hua, Jiaxin Chen, and Shu He enrolled participants and collected data under the supervision of Qiaowei Jia and Sheng Zhang. Chenhui Zhao, Guangxu Xu, and Jing Zhang coordinated the study.
CONFLICT OF INTEREST
There was not any conflict of interest existing in this manuscript.
Supporting information
Figure S1
Figure S2
Table S1
Table S2
Table S3
Table S4
ACKNOWLEDGMENTS
This study received support from the National Natural Science Foundations of China (No. 81970302, 81170180, 30400173, and 30971257) and the Priority Academic Program Development of Jiangsu Higher Education Institutions. Dr En‐Zhi Jia is an Assistant Fellow at the Collaborative Innovation Center for Cardiovascular Disease Translational Medicine.
Hou C, Gu L, Guo Y, et al. Association between circular RNA expression content and severity of coronary atherosclerosis in human coronary artery. J Clin Lab Anal. 2020;34:e23552 10.1002/jcla.23552
DATA AVAILABILITY STATEMENT
All data and materials have been made available.
REFERENCES
- 1. Negi S, Anand A. Atherosclerotic coronary heart disease‐epidemiology, classification and management. Cardiovasc Hematol Disord Drug Targets. 2010;10(4):257‐261. [DOI] [PubMed] [Google Scholar]
- 2. Waldo SW, Secemsky EA, O’Brien C, et al. Surgical ineligibility and mortality among patients with unprotected left main or multivessel coronary artery disease undergoing percutaneous coronary intervention. Circulation. 2014;130(25):2295‐2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim MC, Ahn Y, Cho KH, et al. Early statin therapy within 48 hours decreased one‐year major adverse cardiac events in patients with acute myocardial infarction. Int Heart J. 2011;52(1):1‐6. [DOI] [PubMed] [Google Scholar]
- 4. Geng HH, Li R, Su YM, et al. The Circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR‐7a on its target genes expression. PLoS One. 2016;11(3):e0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333‐338. [DOI] [PubMed] [Google Scholar]
- 6. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384‐388. [DOI] [PubMed] [Google Scholar]
- 7. Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2017;14(8):978‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang PL, Bao Y, Yee MC, et al. Circular RNA is expressed across the eukaryotic tree of life [published correction appears in PLoS One. 2014;9(4):e95116]. PLoS One. 2014;9(6):e90859. Published 2014 Mar 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang YU, Sui X, Zhao H, et al. Decreased circular RNA hsa_circ_0001649 predicts unfavorable prognosis in glioma and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:117‐122. [DOI] [PubMed] [Google Scholar]
- 10. Zhang Y, Liu B, Shao C, et al. Evaluation of the inclusion of circular RNAs in mRNA profiling in forensic body fluid identification. Int J Legal Med. 2018;132(1):43‐52. [DOI] [PubMed] [Google Scholar]
- 11. Wang W, Wang Y, Piao H, et al. Circular RNAs as potential biomarkers and therapeutics for cardiovascular disease. PeerJ. 2019;7:e6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan RY, Zhao CH, Yuan JX, et al. Circular RNA profile in coronary artery disease. Am J Transl Res. 2019;11(11):7115‐7125. [PMC free article] [PubMed] [Google Scholar]
- 13. Stary HC. Natural history and histological classification of atherosclerotic lesions: an update. Arterioscler Thromb Vasc Biol. 2000;20(5):1177‐1178. [DOI] [PubMed] [Google Scholar]
- 14. Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101‐1108. [DOI] [PubMed] [Google Scholar]
- 15. Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588‐5600. [DOI] [PubMed] [Google Scholar]
- 16. Wang L, Shen C, Wang Y, et al. Identification of circular RNA Hsa_circ_0001879 and Hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis. 2019;286:88‐96. [DOI] [PubMed] [Google Scholar]
- 17. Mao YY, Wang JQ, Guo XX, Bi Y, Wang CX. Circ‐SATB2 upregulates STIM1 expression and regulates vascular smooth muscle cell proliferation and differentiation through miR‐939. Biochem Biophys Res Commun. 2018;505(1):119‐125. [DOI] [PubMed] [Google Scholar]
- 18. Li X, Zhao Z, Jian D, Li W, Tang H, Li M. Hsa‐circRNA11783‐2 in peripheral blood is correlated with coronary artery disease and type 2 diabetes mellitus. Diab Vasc Dis Res. 2017;14(6):510‐515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siede D, Rapti K, Gorska AA, et al. Identification of circular RNAs with host gene‐independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol. 2017;109:48‐56. [DOI] [PubMed] [Google Scholar]
- 21. Tan WL, Lim BT, Anene‐Nzelu CG, et al. A landscape of circular RNA expression in the human heart [published correction appears in Cardiovasc Res. 2017 May 1;113(6):704]. Cardiovasc Res. 2017;113(3):298‐309. [DOI] [PubMed] [Google Scholar]
- 22. Wang F, Chen X, Han Y, Xi S, Wu G. circRNA CDR1as regulated the proliferation of human periodontal ligament stem cells under a lipopolysaccharide‐induced inflammatory condition. Mediators Inflamm. 2019;2019:1625381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Table S1
Table S2
Table S3
Table S4
Data Availability Statement
All data and materials have been made available.


