Abstract
Background
The emergence and transmission of tigecycline‐ and carbapenem‐resistant Klebsiella pneumoniae (TCRKP) have become a major concern to public health globally. Here, we investigated the molecular epidemiology and mechanisms of tigecycline resistance in carbapenem‐resistant K pneumoniae (CRKP) isolates.
Methods
Forty‐five non‐duplicate CRKP isolates were collected from January 2017 to June 2019. We performed antimicrobial susceptibility tests, multilocus sequence typing (MLST), and pulsed‐field gel electrophoresis (PFGE). PCR and DNA sequencing were performed for the detection and mutation analysis of acrR, oqxR, ramR, rpsJ, tet(A), and tet(X) genes, which are related to tigecycline resistance. The expression levels of efflux pump genes acrB and oqxB and their regulator genes rarA, ramA, soxS, and marA were assessed by quantitative real‐time PCR.
Results
The resistance rate to tigecycline in CRKP isolates was 37.8% (17/45). K pneumoniae ST307 was a predominant clone type (70.6%, 12/17) among the TCRKP isolates. The expression levels of acrB (P < .001) and marA (P = .009) were significantly higher in the tigecycline‐resistant group than in the tigecycline‐intermediate and tigecycline‐susceptible groups. Increased expression of acrB was associated with marA expression (r = 0.59, P = .013).
Conclusions
We found that the activated MarA‐induced overexpression of AcrAB efflux pump plays an important role in the emergence of tigecycline resistance in CRKP isolates.
Keywords: AcrAB‐TolC efflux pump, carbapenem‐resistant Klebsiella pneumoniae, tigecycline resistance
We introduce the mechanisms of tigecycline resistance in carbapenem‐resistant K pneumoniae (CRKP) isolates. The transcriptional activator MarA‐mediated overexpression of AcrAB efflux pump plays an important role in the emergence of tigecycline resistance in CRKP isolates.
1. INTRODUCTION
As the emergence and dissemination of carbapenem‐resistant Klebsiella pneumoniae (CRKP) are being increasingly reported worldwide, multidrug‐resistant K pneumoniae is considered a serious global health threat. 1 , 2 , 3 , 4 Tigecycline, a glycylcycline antibiotic, has become one of the few available last resort antibiotics for the treatment of carbapenem‐resistant Enterobacteriaceae (CRE) infections such as CRKP. 5 , 6 , 7 , 8 , 9 According to an international ESCMID cross‐sectional survey, tigecycline monotherapy is the most commonly used treatment for patients with intra‐abdominal infections and skin and soft‐tissue infections caused by CRE. 10 However, CRKP isolates with tigecycline resistance have been reported. 11 , 12 , 13 According to Taiwan's national surveillance study, the resistance rate of tigecycline (minimum inhibitory concentration, MIC > 2 mg/L) in carbapenem non‐susceptible K pneumoniae was reported to be 9%. 14 In South Korea, the resistance rate of tigecycline (MIC > 2 mg/L) among carbapenemase‐producing K pneumoniae was found to be approximately 14.5%. 15 Tigecycline resistance in CRKP has thus become a serious problem that can eventually lead to treatment failure. 11 , 12 , 13
To date, there are several known mechanisms associated with resistance to tigecycline in K pneumoniae 11 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 The most common mechanism is the overproduction of non‐specific active resistance‐nodulation‐cell division (RND) efflux pumps such as AcrAB‐TolC 11 , 19 and OqxAB. 17 Expression of the acrAB efflux pump genes is regulated by the global AraC‐family transcriptional activators such as RamA, MarA, and SoxS and the local TetR‐family transcriptional repressor AcrR. 16 , 19 The transcription of ramA, marA, and soxS is repressed by RamR, MarR, and SoxR, respectively. 13 , 26 RamA is also regulated by Lon protease. 27 , 28 As RamR directly represses the expression of ramA, loss‐of‐function mutations in ramR can cause the overexpression of ramA. 18 , 21 The expression of the oqxAB efflux pump genes is also regulated by the global activators RarA, MarA, and SoxS and the local repressor OqxR. 17 Mutations in the rpsJ gene, which encodes the ribosomal protein S10, are associated with reduced tigecycline susceptibility. 24 Moreover, mutations in tet(A), which encodes one of the major facilitator superfamily (MFS) efflux pumps, and tet(X), which encodes a tigecycline‐modifying enzyme, are associated with decreased tigecycline susceptibility. 20 , 22 , 23 , 25
The aim of this study was to investigate the phenotypic characteristics, molecular epidemiology, and mechanisms of tigecycline resistance in CRKP isolates from a tertiary care hospital in South Korea.
2. MATERIALS AND METHODS
2.1. Bacterial strains
A total of 3461 non‐duplicate K pneumoniae isolates were collected from Chungnam National University Hospital in South Korea from January 2017 to June 2019. The VITEK 2 ID‐GNB cards (bioMérieux SA, Marcy l’Étoile, France) were used for the identification of isolates. We retrospectively reviewed the clinical data for each isolate.
2.2. Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of cefepime, cefotaxime, ceftazidime, gentamicin, amikacin, ciprofloxacin, trimethoprim/sulfamethoxazole, and chloramphenicol were determined using the E‐test (Biomerieux, Marcy l'Etoile, France) on Mueller‐Hinton (MH) agar (Difco Laboratories, Detroit, MI, USA) according to the Clinical and Laboratory Standards Institute (CLSI) guideline. 29 Antimicrobial susceptibility to the carbapenems (ertapenem, imipenem, and meropenem) was determined by the agar dilution method, in accordance with the CLSI guidelines. 30 The MICs for tigecycline (Sigma‐Aldrich, USA) were assessed by the broth microdilution method with MH broth (Difco Laboratories) in accordance with the European Committee on Antimicrobial Susceptibility Testing (2016 EUCAST) criteria (1.0 mg/L is susceptible, 2.0 mg/L is intermediate, and >2.0 mg/L is resistant). 31 Both Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains for antimicrobial susceptibility testing. Tigecycline‐resistant and carbapenem‐resistant K pneumoniae (TCRKP) was defined if K pneumoniae was resistant to both at least one carbapenem [ertapenem (MIC ≥ 2.0 mg/L) and/or imipenem (MIC ≥ 4.0 mg/L) and/or meropenem (MIC ≥ 4.0 mg/L)] and tigecycline (MIC ≥ 4.0 mg/L).
2.3. Resistance gene detection
The following resistance genes were detected by PCR: (a) carbapenemase genes (bla NDM, bla IMP, bla VIM, bla KPC, bla OXA‐48‐like, and bla GES), (b) extended‐spectrum‐β‐lactamase (ESBL) genes (bla CTX−M−1−like, bla CTX−M−9−like, bla TEM, and bla SHV), (c) ampC β‐lactamase genes (bla CIT, bla MOX, bla DHA, bla ACC, bla EBC, and bla FOX), (d) quinolone resistance‐determinant genes (qnrA, qnrB, qnrS, aac(6’)‐Ib‐cr, and qepA), and (e) aminoglycoside resistance‐determinant resistance genes (armA, rmtA, rmtB, and rmtC). 32 The amplicons were determined by DNA sequencing. Primers used for PCR are shown in the Table S1.
2.4. Multilocus sequence typing (MLST) and pulsed‐field gel electrophoresis (PFGE)
MLST and PFGE were used to determine the genetic relatedness among the 45 CRKP isolates. PCR and sequencing for MLST were carried out for seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) for K pneumoniae and the sequences were compared in the MLST database, so that the allelic numbers and sequence types (STs) could be determined. 33 The allelic profiles and STs were assigned using an online database (https://pubmlst.org/). For PFGE, bacterial DNA was cleaved with XbaI endonuclease (Roche, Penzberg, Germany), and the XbaI‐digested genomic DNA was subjected to PFGE using a CHEF‐DR® III Variable Angle System (Bio‐Rad, USA). 34 The PFGE patterns were compared using BioNumerics software (Applied Maths, Kortrijk, Belgium) with dice correlation for band matching at a 1.5% position tolerance and the unweighted pair group method with an arithmetic average (UPGMA). Clusters were defined as DNA patterns sharing >80% similarity.
2.5. Quantitative real‐time PCR (qRT‐PCR)
The expression levels of the efflux pump genes acrB and oqxB and their regulator genes rarA, ramA, soxS, and marA were assessed by qRT‐PCR. Total RNA of CRKP isolates was extracted with the MagListo™ 5M Cell Total RNA Extraction Kit (Bioneer, Daejeon, Korea) according to the manufacturer's instructions. Reverse transcription of RNA to cDNA was performed using AccuPower® CycleScript RT Premix (Bioneer, Daejeon, Korea). Quantitative real‐time PCR performance using Solg™ 2X Real‐Time PCR Smart mix with EvaGreen™ 500 (SolGent Co., Ltd., Daejeon, Korea) was run on a Exicycler™ 96 Real‐time Quantitative Thermal Block (Bioneer, Daejeon, Korea). All experiments were performed in triplicate. The mRNA expression level of each gene was normalized based on an endogenous reference gene (rrsE). Relative expression of each gene was calculated based on tigecycline‐susceptible isolate CRKP 87 (tigecycline MIC 0.5 mg/L, expression = 1) as the control strain. The level of expression of each gene was calculated using 2−ΔΔCt method. The primers used in the experiments are listed in the Table S2.
2.6. Detection and mutation analysis of the acrR, oqxR, ramR, rpsJ, tet(A), and tet(X) genes and pI andpII promoter regions
We performed PCR to detect acrR, oqxR, ramR, rpsJ, tet(A), and tet(X) genes and pI (upstream of the romA controlling romA‐ramA operon transcription) and pII (located in the open reading frame of romA) promoter regions, which are transcriptional start sites of the ramA. 11 , 35 The amplicons were sequenced. For mutation analysis, we compared each gene sequence with that of the wild‐type reference strains, K pneumoniae MGH78578 (GenBank accession number CP000647) in case of acrR, oqxR, ramR, and rpsJ genes and pI and pII promoter regions and E coli plasmid RP1 (GenBank accession number X00006) for the tet(A) gene. Primers used for PCR are shown in the Table S2.
2.7. Statistical analysis
All statistical analyses were performed using MedCalc statistical software 14.12.0 (MedCalc Software, Mariakerke, Belgium). Data are presented as the mean ± standard deviation (SD) unless otherwise stated. In groups with a non‐normal distribution, we evaluated the intergroup comparisons using either a Mann‐Whitney rank sum test or Kruskal‐Wallis test followed by Dunn's multiple comparison test. To assess the correlations between the expression levels of each gene, linear regressions were calculated. A P value < .05 was considered statistically significant.
3. RESULTS
3.1. Clinical characteristics of CRKP isolates
Of the 3416 non‐duplicate K pneumoniae strains, CRKP accounted for 1.3% (45/3416). The isolates were obtained from various clinical specimens, including rectal swab (25/45, 55.6%), urine (6/45, 13.3%), blood (5/45, 11.1%), bile fluid (4/45, 8.9%), sputum (4/45, 8.9%), and cerebrospinal fluid (CSF) (1/45, 2.2%), from hospitalized patients aged 0 to 86, with a median age of 70 years (Figure 1). Two clinical K pneumoniae strains (CRKP4 and CRKP60) were isolated from patients who had previously been treated with tigecycline.
Figure 1.
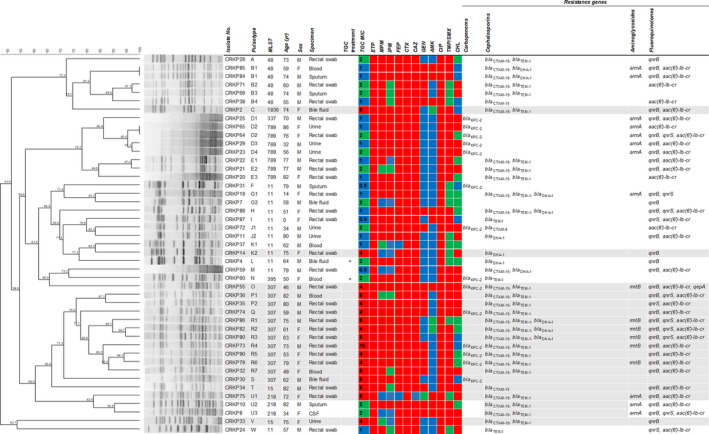
Dendrogram based on pulsed‐field gel electrophoresis patterns, multilocus sequence typing, characteristics, antibiotic susceptibility profile, and resistance gene profile of 45 carbapenem‐resistant Klebsiella pneumoniae isolates. The red, green, and blue squares indicate resistant, intermediate, and susceptible to each antibiotic, respectively. The gray background indicates 17 tigecycline‐ and carbapenem‐resistant Klebsiella pneumoniae (TCRKP) isolates. AMK, amikacin; CAZ, ceftazidime; CHL, chloramphenicol; CIP, ciprofloxacin; CTX, cefotaxime; ETP, ertapenem; FEP, cefepime; GEN, gentamicin; IPM, imipenem; MIC, minimum inhibitory concentration; MLST, multilocus sequence typing; MPM, meropenem; TGC, tigecycline; TMP/SMX, trimethoprim/sulfamethoxazole
3.2. Antibiotic resistance profile and distribution of resistance genes in CRKP isolates
All CRKP isolates were resistant to ertapenem, cefotaxime, and ciprofloxacin. These isolates were also co‐resistant to ceftazidime (44/45, 97.8%), cefepime (43/45, 95.6%), meropenem (36/45, 80.0%), trimethoprim/sulfamethoxazole (32/45, 71.1%), imipenem (29/45, 64.4%), chloramphenicol (22/45, 48.9%), gentamicin (18/45, 40.0%), and amikacin (9/45, 20.0%) (Figure 1). Fourteen of 45 CRKP isolates (31.1%) harbored the carbapenemase genes bla KPC‐2. Except for two strains (CRKP7 and CRKP37), the remaining 29 carbapenemase non‐producing CRKP isolates had ESBL genes (27/29, 93.1%) such as bla CTX‐M‐14, bla CTX‐M‐15, and bla TEM‐1 and/or ampC lactamase such as bla DHA‐1 (14/29, 48.3%). Among the other resistance genes, the most commonly accompanied gene in CRKP was aac(6')‐Ib‐cr (34/45, 75.6%), followed by qnrB (32/45, 71.1%), armA (11/45, 24.4%), qnrS (11/45, 24.4%), rmtB (6/45, 13.3%), and qepA (1/45, 2.2%). The other resistant genes that were evaluated, such as carbapenemase genes (bla NDM, bla IMP, bla VIM, bla OXA‐48‐like, and bla GES), bla SHV, ampC β‐lactamase genes (bla CIT, bla MOX, bla ACC, bla EBC, and bla FOX), qnrA, rmtA, and rmtC, were not detected in the CRKP isolates. Among CRKP isolates, the prevalence of tigecycline non‐susceptible (MICs ≥ 2 mg/L) strains was 64.4% (29/45) and that of tigecycline‐resistant (MICs ≥ 4 mg/L) strains was 37.8% (17/45). Among the 17 TCRKP isolates, six (35.3%) harbored bla KPC‐2.
3.3. Molecular epidemiology based on MLST and PFGE
Nine distinct STs were observed among the 45 CRKP isolates as follows: ST11 (12/45, 26.7%), ST307 (12/45, 26.7%), ST789 (7/45, 15.6%), ST48 (6/45, 13.3%), ST218 (3/45, 6.7%), ST15 (2/45, 4.4%), ST337 (1/45, 2.2%), ST395 (1/45, 2.2%), and ST1836 (1/45, 2.2%) (Figure 1). For PFGE analysis, 43 different pulsotypes and 23 clonal groups were observed among the 45 CRKP isolates. Among 17 TCRKP isolates, five distinct STs, which were differentiated into 17 pulsotypes and 10 clonal groups, were observed as follows: ST307 (12/17, 70.6%, PFGE pulsotype O, P1‐P2, Q, R1‐R7, S), ST15 (2/17, 11.8%, PFGE pulsotype T, V), ST11 (1/17, 5.9%, PFGE pulsotype K2), ST1836 (1/17, 5.9%, PFGE pulsotype C), and ST218 (1/17, 5.9%, PFGE pulsotype U1).
3.4. Gene expression level analysis of efflux pumps acrB and oqxB and their regulators rarA, ramA, soxS, and marA in the tigecycline‐susceptible, intermediate, and resistant CRKP groups
The expression levels of the efflux pump‐encoding acrB gene (P < .001) and its transcriptional activator‐encoding marA gene (P = .009) in TCRKP isolates were significantly higher than those in both the tigecycline‐intermediate and tigecycline‐susceptible groups (Figure 2). Moreover, correlation analysis between the expression of acrB and marA in the TCRKP isolates indicated a moderate correlation (r = 0.59, P = .013; Figure 3). There was no significant difference in the expression levels of ramA, soxS, oqxB, and rarA genes among the three groups.
Figure 2.
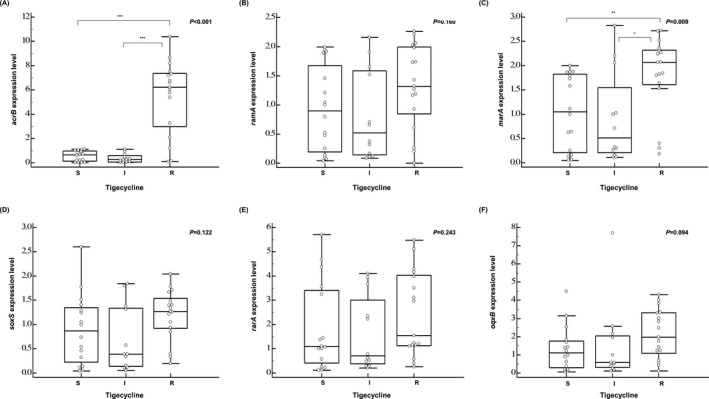
The relative expression level of each gene in the tigecycline‐susceptible (N = 16), tigecycline‐intermediate (N = 12), and tigecycline‐resistant (N = 17) groups. *P < .05; **P < .01; ***P < .001; Kruskal‐Wallis test followed by Dunn's multiple comparison test. I, intermediate; R, resistant; S, susceptible
Figure 3.
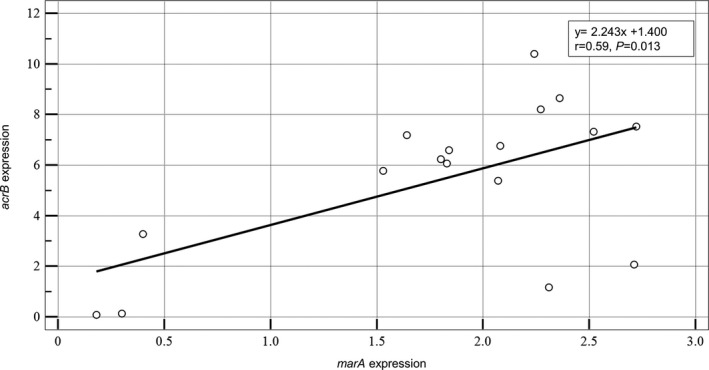
Correlation between expression of the transcriptional activator‐encoding marA gene and efflux pump‐encoding acrB gene in tigecycline‐ and carbapenem‐resistant Klebsiella pneumoniae. P value; Mann‐Whitney rank sum test
3.5. Mutation analysis of acrR, oqxR, ramR, rpsJ, tet(A), and tet(X) genes and pI and pII promoter regions, and their relationship with gene expression levels
The relative expression levels of acrB, ramA, marA, soxS, rarA, and oqxB genes and mutation analysis of TCRKP isolates are presented in Table 1. The acrR, oqxR, ramR, and rpsJ genes and pI and pII promoter regions were detected in all 17 TCRKP isolates. The tet(A) gene was detected in 52.9% (9/17) of the TCRKP isolates. In contrast, the tet(X) gene was not detected in any of the TCRKP isolates.
Table 1.
Mutation analysis of acrR, ramR,oqxR, and tet(A)genes and pI and pII promoter regions and the relative expression levels of acrB, ramA, marA, soxS, rarA, and oqxB in TCRKP isolates
|
TGC MIC (mg/L) |
Isolates | Mutation analysis a | Relative expression level b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| acrR | pI promoter | pII promoter | ramR | oqxR | tet(A) | acrB | ramA | marA | soxS | rarA | oqxB | ||
| 4 | CRKP14 | △396‐415, g441a | a54g | 919_920insa, a1106g, c1133t, t1139a, △1316 | None | t389c (V130A) | ‐ | 0.13 ± 0.02 | 0.20 ± 0.01 | 0.30 ± 0.02 | 0.34 ± 0.02 | 0.58 ± 0.03 | 0.56 ± 0.14 |
| CRKP33 | None | a54g | 919_920insa, △1316 | c56t (A19V)*, △343‐351* | None | ‐ | 1.17 ± 0.37 | 2.22 ± 0.74 | 2.31 ± 0.35 | 1.71 ± 0.31 | 5.11 ± 0.87 | 3.82 ± 0.72 | |
| CRKP35 | None | a54g | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | △332‐342* | None | ‐ | 7.33 ± 0.67 | 1.73 ± 0.17 | 2.52 ± 0.36 | 1.41 ± 0.20 | 4.29 ± 1.46 | 4.00 ± 0.49 | |
| CRKP55 | None | a54g, g133a* | 919_920insa, a1106g, c1133t, t1139a, △1316 | None | t389c (V130A) | ‐ | 8.21 ± 0.82 | 0.00 ± 0.00 | 2.27 ± 0.24 | 2.04 ± 0.48 | 3.99 ± 0.25 | 3.35 ± 0.30 | |
| CRKP74 | None | a54g | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | None | None | △1928, 1936_1937insc | 6.60 ± 0.54 | 1.19 ± 0.05 | 1.84 ± 0.31 | 0.85 ± 0.13 | 1.23 ± 0.17 | 1.43 ± 0.20 | |
| CRKP79 | None | a54g | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | t563a (L188Q)* | None | ‐ | 7.19 ± 0.68 | 0.93 ± 0.16 | 1.64 ± 0.08 | 1.22 ± 0.05 | 1.14 ± 0.09 | 1.26 ± 0.18 | |
| CRKP82 | None | a54g | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | t563a (L188Q)* | None |
△1928 1936_1937insc |
6.06 ± 0.52 | 1.32 ± 0.36 | 1.83 ± 0.18 | 1.27 ± 0.04 | 1.54 ± 0.12 | 1.76 ± 0.05 | |
| CRKP90 | None | a54g | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | t563a (L188Q)* | None | ‐ | 3.28 ± 0.48 | 0.61 ± 0.09 | 0.40 ± 0.07 | 0.40 ± 0.10 | 0.41 ± 0.26 | 0.70 ± 0.20 | |
| 8 | CRKP2 | None | a54g | 919_920insa, △1316 | △44‐46* | t92a (A30Q)* |
△1928 1936_1937insc |
0.09 ± 0.05 | 0.26 ± 0.06 | 0.18 ± 0.04 | 0.20 ± 0.08 | 0.26 ± 0.07 | 0.42 ± 0.20 |
| CRKP30 | None | a54g | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | △103* | None | ‐ | 10.41 ± 3.45 | 2.26 ± 0.36 | 2.24 ± 0.33 | 1.34 ± 0.22 | 2.97 ± 1.31 | 4.32 ± 0.75 | |
| CRKP32 | None | a54g, △163* | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | None | None |
△1928 1936_1937insc |
7.53 ± 0.30 | 1.98 ± 0.15 | 2.72 ± 0.06 | 1.49 ± 0.21 | 3.51 ± 0.21 | 3.29 ± 0.16 | |
| CRKP34 | None | a54g | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | △332* | None |
△1928 1936_1937insc |
6.77 ± 0.86 | 2.06 ± 0.22 | 2.08 ± 0.19 | 1.78 ± 0.45 | 4.18 ± 0.34 | 2.84 ± 0.34 | |
| CRKP36 | None | a54g | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | c364t (Q122stop) | None |
△1928 1936_1937insc |
8.65 ± 0.72 | 2.03 ± 0.24 | 2.36 ± 0.30 | 1.67 ± 0.13 | 5.48 ± 0.48 | 3.02 ± 0.24 | |
| CRKP75 | None | a54g | 919_920insa, c992t, a1106g, △1316 | △332‐344*, g474a* | t389c (V130A) | ‐ | 2.07 ± 0.86 | 1.27 ± 0.12 | 2.71 ± 0.35 | 1.26 ± 0.09 | 3.12 ± 0.29 | 0.11 ± 0.02 | |
| CRKP80 | None | a54g | g929a*, c992t, c999t*, a1106g, △1316 | t563a (L188Q)* | None |
△1928 1936_1937insc |
5.39 ± 0.23 | 1.43 ± 0.13 | 2.07 ± 0.22 | 0.94 ± 0.06 | 1.20 ± 0.25 | 1.23 ± 0.23 | |
| CRKP86 | None | a54g | g929a*, c992t, c999t*, a1106g, △1316 | g554t (W185L)*, t563a (L188Q)* | None |
△1928 1936_1937insc |
5.78 ± 0.48 | 1.17 ± 0.05 | 1.53 ± 0.21 | 1.40 ± 0.011 | 1.15 ± 0.18 | 1.96 ± 0.13 | |
| 16 | CRKP73 | △385‐395* | a54g | 919_920insa, g929a*, c992t, c999t*, a1106g, △1316 | None | None |
△1928 1936_1937insc |
6.23 ± 0.71 | 1.74 ± 0.26 | 1.80 ± 0.24 | 1.05 ± 0.12 | 1.12 ± 0.13 | 2.47 ± 0.42 |
The numbers in bold indicate at least 2‐fold higher transcriptional level of the gene compared to that in the tigecycline‐susceptible control CRKP87 isolate containing the wild‐type gene.
Abbreviations: ‐, the gene was not confirmed in PCR; △, deletion; ins, insertion; MIC, minimum inhibitory concentration; None, the gene was confirmed in PCR, but no mutation was detected; TCRKP, tigecycline‐resistant and carbapenem‐resistant Klebsiella pneumoniae; TGC, tigecycline.
In mutation analysis, uppercase letters indicate amino acids and lowercase letters indicate the nucleotide bases.
Data are expressed as the mean ± standard deviation (SD).
Mutations observed only in tigecycline‐resistant CRKP isolates.
Mutations in the efflux pump‐encoding acrR and oqxR genes were identified in 11.8% (2/17) and 23.5% (4/17) of the TCRKP isolates, respectively. Of isolates harboring the mutations only identified in TCRKP isolates, except for mutations identified in tigecycline‐susceptible and/or tigecycline‐intermediate CRKP isolates, CRKP 73, harboring the deletion 385‐395gcccagcggca in acrR, showed increased expression levels of acrB (6.23 ± 0.71). There was no increase in the expression level of the oqxB gene (0.42 ± 0.20) in the CRKP2 isolate harboring the point mutation t92a (A30Q) in the oqxR gene. Mutants of RamR, the repressor of ramA, were detected in 12 TCRKP isolates (12/17, 70.6%). Some TCRKP isolates (CRKP30, CRKP33, and CRKP34) harboring frameshift mutations in ramR tended to express slightly higher transcriptional levels of ramA than the tigecycline‐susceptible control CRKP87 isolate. The expression level of ramA was not sufficiently increased by mutations such as the deletion 163g or point mutation g133a in the pI promoter region and point mutations g929a and c999t in the pII promoter region. All nine TCRKP isolates harboring the tet(A) gene had the same mutation, deletion 1928a, and insertion 1936_1937c, changing the 201‐203rd amino acids in the sequence from serine, phenylalanine, and valine (SFV) to alanine, serine, and proline (ASP). However, this mutation was also detected in tigecycline‐susceptible CRKP isolates (Table S3). There were no mutations in the rpsJ gene.
4. DISCUSSION
In the present study, we analyzed phenotypic characteristics including tigecycline susceptibility, molecular epidemiology, and mechanisms of tigecycline resistance in CRKP isolates from a tertiary care hospital in South Korea. According to a multicenter study in the United States, the resistance rate of tigecycline in CRKP isolates was reported to be 18.0%. 36 However, in this study, tigecycline non‐susceptible CRKP and TCRKP accounted for 64.4% and 37.8% of strains, respectively, with a higher tigecycline resistance rate in CRKP isolates when analyzing tigecycline MIC results according to the EUCAST criteria. 36
Some studies have reported that treatment with tigecycline could lead to the development of tigecycline resistance. 11 , 37 However, in this study, all TCRKP isolates were collected from patients who had not been exposed to tigecycline treatment previously. These findings revealed that tigecycline resistance might occur even without exposure to this antibiotic. According to another study, exposure to other antibiotics that are effluxed by non‐specific pumps, such as AcrAB, could indirectly contribute to reduced tigecycline susceptibility. 38
The MLST and PFGE analyses revealed that K pneumoniae ST307, which was divided into five clonal groups based on PFGE, was the predominant clone among the TCRKP isolates in this study. Moreover, some of the K pneumonia ST307 isolates in this study harbored bla KPC‐2. K pneumoniae ST307 has been internationally reported as a high‐risk pathogen associated with high resistance to fluoroquinolones, third generation cephalosporins, and carbapenem. 39 , 40 Moreover, K pneumoniae ST307 is one of the dominant clonal types, along with ST11, ST768, ST15, ST23, and ST48, among the tigecycline‐resistant K pneumoniae isolates in South Korea. 9 In this study, we confirmed that high‐risk TCRKP isolates such as K pneumoniae ST307 had already emerged and are disseminating in this area. Therefore, we should thoroughly monitor these high‐risk pathogens to prevent their transmission.
Regarding the mechanisms of tigecycline resistance, we found that tigecycline resistance in most of the CRKP isolates was associated with increased expression of the efflux pump‐encoding acrB gene. The upregulation of acrB could be mediated by a local repressor AcrR and/or transcriptional activators such as MarA, RamA, and SoxS. 38 , 39 Among them, we found that increased acrB expression correlated with overexpression of the transcriptional activator marA in the TCRKP isolates. However, the overexpression of acrB was not detected in the three TCRKP isolates. These findings suggest that tigecycline resistance in these isolates might be due to an alternative pathway or efflux pumps other than AcrAB or OqxAB.
In previous studies, it has been reported that mutations in the ramR gene could contribute to ramA overexpression and subsequent acrAB upregulation. 13 , 18 , 21 , 22 However, the expression levels of ramA in TCRKP isolates harboring mutations in ramR were not significantly increased compared to those in the control strain with a wild‐type ramR gene in the present study. Moreover, based on mutation analysis of the transcriptional start sites (pI and pII promoter regions) of the ramA gene, there were no specific mutations that could affect the expression level of ramA. This implied that ramA overexpression might not be required to upregulate acrB and to confer tigecycline resistance. Among mutations, a Q122stop mutant in RamR 13 and V130A mutant of OqxR 22 were reported to confer resistance to tigecycline in previous studies; however, in this study, these mutants were also observed in the tigecycline‐susceptible CRKP isolate indicating that it has little impact on tigecycline resistance.
There is one limitation to this study. Our study suggested that the upregulation of acrB mediated by the transcriptional activator MarA plays an important role in the mechanisms of tigecycline resistance. However, the expression of marA could also be regulated by a transcriptional regulator such as MarR. Therefore, further studies on additional regulators such as MarR, SoxR, and Lon protease, which affect the expression of marA, soxS, and ramA genes, respectively, and subsequent acrAB expression will be needed to assess the possible role in tigecycline resistance. In conclusion, although the mechanisms of tigecycline resistance are complex and have not been fully understood, our study indicates that the main mechanisms of tigecycline resistance in the CRKP isolates can be attributed to transcriptional activator MarA‐mediated overexpression of AcrAB efflux pump.
Supporting information
Supplementary Material
Park Y, Choi Q, Kwon GC, Koo SH. Molecular epidemiology and mechanisms of tigecycline resistance in carbapenem‐resistant Klebsiella pneumoniae isolates. J Clin Lab Anal. 2020;34:e23506 10.1002/jcla.23506
REFERENCES
- 1. David S, Reuter S, Harris SR, et al. Epidemic of carbapenem‐resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4(11):1919‐1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vaez H, Sahebkar A, Khademi F. Carbapenem‐resistant Klebsiella pneumoniae in Iran: a systematic review and meta‐analysis. J Chemother. 2019;31(1):1‐8. [DOI] [PubMed] [Google Scholar]
- 3. Lee H‐J, Lee D‐G. Carbapenem‐resistant Enterobacteriaceae: recent updates and treatment strategies. J Korean Med Assoc. 2018;61(4):281‐289. [Google Scholar]
- 4. Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem‐resistant enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53(1):60‐67. [DOI] [PubMed] [Google Scholar]
- 5. Osei Sekyere J, Govinden U, Bester LA, Essack SY. Colistin and tigecycline resistance in carbapenemase‐producing gram‐negative bacteria: emerging resistance mechanisms and detection methods. J Appl Microbiol. 2016;121(3):601‐617. [DOI] [PubMed] [Google Scholar]
- 6. van Duin D, Cober ED, Richter SS, et al. Tigecycline therapy for carbapenem‐resistant Klebsiella pneumoniae (CRKP) bacteriuria leads to tigecycline resistance. Clin Microbiol Infect. 2014;20(12):1117‐1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Townsend ML, Pound MW, Drew RH. Tigecycline in the treatment of complicated intra‐abdominal and complicated skin and skin structure infections. Ther Clin Risk Manag. 2007;3(6):1059‐1070. [PMC free article] [PubMed] [Google Scholar]
- 8. Sader HS, Castanheira M, Flamm RK, Mendes RE, Farrell DJ, Jones RN. Tigecycline activity tested against carbapenem‐resistant Enterobacteriaceae from 18 European nations: results from the SENTRY surveillance program (2010–2013). Diagn Microbiol Infect Dis. 2015;83(2):183‐186. [DOI] [PubMed] [Google Scholar]
- 9. Ahn C, Yoon SS, Yong TS, Jeong SH, Lee K. The resistance mechanism and clonal distribution of tigecycline‐nonsusceptible Klebsiella pneumoniae isolates in Korea. Yonsei Med J. 2016;57(3):641‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Papst L, Beović B, Pulcini C, et al. Antibiotic treatment of infections caused by carbapenem‐resistant gram‐negative bacilli: an international ESCMID cross‐sectional survey among infectious diseases specialists practicing in large hospitals. Clin Microbiol Infect. 2018;24(10):1070‐1076. [DOI] [PubMed] [Google Scholar]
- 11. Yoon E‐J, Oh Y, Jeong SH. Development of tigecycline resistance in carbapenemase‐producing Klebsiella pneumoniae sequence type 147 via AcrAB overproduction mediated by replacement of the ramA promoter. Ann Lab Med. 2020;40(1):15‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiu S‐K, Chan M‐C, Huang L‐Y, et al. Tigecycline resistance among carbapenem‐resistant Klebsiella pneumoniae: clinical characteristics and expression levels of efflux pump genes. PLoS One. 2017;12(4):e0175140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He F, Fu Y, Chen Q, et al. Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC‐producing Klebsiella pneumoniae . PLoS One. 2015;10(3):e0119064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wi YM, Kang C‐I. Antimicrobial therapy for infections caused by carbapenem‐resistant gram‐negative bacteria. Korean J Med. 2018;93(5):439‐446. [Google Scholar]
- 15. Jeong SH, Kim HS, Kim JS, et al. Prevalence and molecular characteristics of carbapenemase‐producing enterobacteriaceae from five hospitals in Korea. Ann Lab Med. 2016;36(6):529‐535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blanco P, Hernando‐Amado S, Reales‐Calderon JA, et al. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms. 2016;4(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Veleba M, Higgins PG, Gonzalez G, Seifert H, Schneiders T. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae . Antimicrob Agents Chemother. 2012;56(8):4450‐4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X, Chen H, Zhang Y, et al. Genetic characterisation of clinical Klebsiella pneumoniae isolates with reduced susceptibility to tigecycline: role of the global regulator RamA and its local repressor RamR. Int J Antimicrob Agents. 2015;45(6):635‐640. [DOI] [PubMed] [Google Scholar]
- 19. Ruzin A, Visalli MA, Keeney D, Bradford PA. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae . Antimicrob Agents Chemother. 2005;49(3):1017‐1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leski TA, Bangura U, Jimmy DH, et al. Multidrug‐resistant tet(X)‐containing hospital isolates in Sierra Leone. Int J Antimicrob Agents. 2013;42(1):83‐86. [DOI] [PubMed] [Google Scholar]
- 21. Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob Agents Chemother. 2010;54:2720‐2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chiu S‐K, Huang L‐Y, Chen H, et al. Roles of ramR and tet(A) mutations in conferring tigecycline resistance in carbapenem‐resistant Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2017;61(8):e00391‐e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moore IF, Hughes DW, Wright GD. Tigecycline is modified by the flavin‐dependent monooxygenase TetX. Biochemistry. 2005;44(35):11829‐11835. [DOI] [PubMed] [Google Scholar]
- 24. He F, Shi Q, Fu Y, Xu J, Yu Y, Du X. Tigecycline resistance caused by rpsJ evolution in a 59‐year‐old male patient infected with KPC‐producing Klebsiella pneumoniae during tigecycline treatment. Infect Genet Evol. 2018;66:188‐191. [DOI] [PubMed] [Google Scholar]
- 25. Du X, He F, Shi Q, et al. The rapid emergence of tigecycline resistance in blaKPC‐2 harboring Klebsiella pneumoniae, as mediated in vivo by mutation in tetA during tigecycline treatment. Front Microbiol. 2018;9:648.29675006 [Google Scholar]
- 26. Hladicz A, Kittinger C, Zarfel G. Tigecycline resistant Klebsiella pneumoniae isolated from Austrian River Water. Int J Environ Res Public Health. 2017;14(10):1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ricci V, Blair JMA, Piddock LJV. RamA, which controls expression of the MDR efflux pump AcrAB‐TolC, is regulated by the Lon protease. J Antimicrob Chemother. 2013;69(3):643‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fang L, Chen Q, Shi K, et al. Step‐Wise Increase in Tigecycline resistance in Klebsiella pneumoniae associated with mutations in ramR, lon and rpsJ. PLoS One. 2016;11(10):e0165019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. CLSI . Performance Standards for Antimicrobial Susceptibility Testing; Twenty‐fifth Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 30. CLSI . Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 31. EUCAST . European Committee on Antimicrobial Susceptibility Testing 2016 [Available from: http://www.eucast.org/clinical_breakpoints/]. Accessed July 10, 2019.
- 32. Park Y, Choi Q, Kwon GC, Koo SH. Emergence and transmission of New Delhi metallo‐beta‐lactamase‐5‐producing Escherichia coli sequence type 361 in a tertiary hospital in South Korea. J Clin Lab Anal. 2020;34:e23041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178‐4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han H, Zhou H, Li H, et al. Optimization of pulse‐field gel electrophoresis for subtyping of Klebsiella pneumoniae . Int J Environ Res Public Health. 2013;10:2720‐2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosenblum R, Khan E, Gonzalez G, Hasan R, Schneiders T. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae . Int J Antimicrob Agents. 2011;38(1):39‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Duin D, Cober E, Richter SS, et al. Residence in skilled nursing facilities is associated with tigecycline nonsusceptibility in carbapenem‐resistant Klebsiella pneumoniae . Infect Control Hosp Epidemiol. 2015;36(8):942‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang R, Dong N, Huang Y, et al. Evolution of tigecycline‐ and colistin‐resistant CRKP (carbapenem‐resistant Klebsiella pneumoniae) in vivo and its persistence in the GI tract. Emerg Microbes Infect. 2018;7(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy S, Datta S, Viswanathan R, Singh AK, Basu S. Tigecycline susceptibility in Klebsiella pneumoniae and Escherichia coli causing neonatal septicaemia (2007–10) and role of an efflux pump in tigecycline non‐susceptibility. J Antimicrob Chemother. 2013;68(5):1036‐1042. [DOI] [PubMed] [Google Scholar]
- 39. Rojas R, Macesic N, Tolari G, Guzman A, Uhlemann A‐C. Multidrug‐resistant Klebsiella pneumoniae ST307 in traveler returning from Puerto Rico to Dominican Republic. Emerg Infect Dis. 2019;25(8):1583‐1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Villa L, Feudi C, Fortini D, et al. Diversity, virulence, and antimicrobial resistance of the KPC‐producing Klebsiella pneumoniae ST307 clone. Microbial genomics. 2017;3(4):e000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


