Abstract
Objective
To develop and test computer software to detect, quantify, and monitor progression of pneumonia associated with COVID-19 using chest CT scans.
Methods
One hundred twenty chest CT scans from subjects with lung infiltrates were used for training deep learning algorithms to segment lung regions and vessels. Seventy-two serial scans from 24 COVID-19 subjects were used to develop and test algorithms to detect and quantify the presence and progression of infiltrates associated with COVID-19. The algorithm included (1) automated lung boundary and vessel segmentation, (2) registration of the lung boundary between serial scans, (3) computerized identification of the pneumonitis regions, and (4) assessment of disease progression. Agreement between radiologist manually delineated regions and computer-detected regions was assessed using the Dice coefficient. Serial scans were registered and used to generate a heatmap visualizing the change between scans. Two radiologists, using a five-point Likert scale, subjectively rated heatmap accuracy in representing progression.
Results
There was strong agreement between computer detection and the manual delineation of pneumonic regions with a Dice coefficient of 81% (CI 76–86%). In detecting large pneumonia regions (> 200 mm3), the algorithm had a sensitivity of 95% (CI 94–97%) and specificity of 84% (CI 81–86%). Radiologists rated 95% (CI 72 to 99) of heatmaps at least “acceptable” for representing disease progression.
Conclusion
The preliminary results suggested the feasibility of using computer software to detect and quantify pneumonic regions associated with COVID-19 and to generate heatmaps that can be used to visualize and assess progression.
Key Points
• Both computer vision and deep learning technology were used to develop computer software to quantify the presence and progression of pneumonia associated with COVID-19 depicted on CT images.
• The computer software was tested using both quantitative experiments and subjective assessment.
• The computer software has the potential to assist in the detection of the pneumonic regions, monitor disease progression, and assess treatment efficacy related to COVID-19.
Electronic supplementary material
The online version of this article (10.1007/s00330-020-07156-2) contains supplementary material, which is available to authorized users.
Keywords: COVID-19, Biomarkers, Pneumonia, Neural network
Introduction
Since its outbreak in December 2019, the novel coronavirus (COVID-19) has evolved from its epicenter of Wuhan City (Hubei Province, China) into a worldwide pandemic. Recent reports describe characteristic pulmonary infiltrative patterns on chest computed tomography (CT) including progressive ground-glass opacities (GGO), crazy paving, and consolidations occurring in the majority of patients [1–8]. In fact, such findings on CT exam have demonstrated a greater sensitivity than real-time polymerase chain reaction (RT-PCR) (98% vs 71%) [9] in diagnosing longitudinally confirmed COVID-19 infection. At this time, it is critical to develop efficient and effective tools to detect the disease and reliably assess its progress and response to therapy.
CT image findings associated with COVID-19, which include subtle boundaries, pleural-based location, and variations in size, density, location, and texture [1, 4, 8, 10], present challenges to automated identification and quantification using traditional lung segmentation algorithms. While automated detection of pneumonia on chest radiography [11–14] has been reported, limited effort has been directed toward CT images. One effort to detect ground-glass opacities using a neural network was associated with a significant false-positive rate [15].
Our objective was to develop and test computer software to automatically detect pneumonia associated with COVID-19, quantify the extent of disease, and assess the progression of the disease using chest CT images. To accomplish our objective, we needed to segment the lung in the presence of severe disease and to reliably detect and quantify pneumonic regions in the lung, which required the integration of deep learning and computer vision technologies. The performance of the software was evaluated using the agreement between visual and automated computer assessment of lung infiltrates associated with COVID-19.
Materials and methods
A. Image datasets
Two datasets were retrospectively collected: Dataset 1 consisted of 120 chest CT scans and was used to develop, train, and test deep learning algorithms to segment the lung boundaries and main lung vessels. A variety of lung disease, including atelectasis (n = 47), interstitial lung disease (ILD) (n = 31), tuberculosis (n = 13), pneumonia (n = 17), and others (n = 12) (including five CT scans with emphysema and seven CT scans that were negative for lung disease based on visual interpretation by a thoracic radiologist). Dataset 1 was randomly split into three groups: (1) training set (n = 80), (2) interval validation set (n = 20), and (3) independent test set (n = 20). These CT scans were collected from various sources and acquired using different protocols (e.g., manufacturers, radiation dose, and slice thickness).
Dataset 2 consisted of 72 serial chest CT scans from 24 subjects with a confirmed COVID-19 diagnosis and used development and test the algorithm to detect and quantify the pneumonic regions. Each subject had at least three consecutive CT scans performed at (1) T0—baseline CT scan; (2) T1—first follow-up scan; and (3) T2—second follow-up scan, which were performed at 3.4 days ± 1.8, 9.7 days ± 1.9, and 15.8 days ± 3.6 after symptom onset, respectively. Only the first three consecutive CT scans were included in this study. The cohort represents all the subjects identified with three consecutive CT scans and no other criteria were used to exclude subjects. All subjects had close contact with individuals from Wuhan and were later confirmed to have COVID-19 by RT-PCR. The chest CT exams were performed on a 64-row spiral CT (Siemens) without radiopaque contrast with the participants in a supine position and holding their breath. The scan parameters were tube voltage of 120 kVp, tube current modulation of 100 mA, and spiral pitch factor of 1. The image slice thickness ranged from 1.0 to 2.0 mm. Subject ages ranged from 15 to 74 years with a mean of 44.8 ± 15.6 with 13 of the 24 subjects male (Table 1). There were no reported or obvious comorbidities reported in the medical records of the Dataset 2 cohort. The CT scans from four subjects were used to develop the algorithm (training set), and the remaining CT scans from 20 subjects were used to independently test the algorithm (test set). All the CT scans from the subjects (i.e., T0, T1, and T2) were used in the development and testing process.
Table 1.
Demographics of the COVID-19 subjects (n = 24)
| Characteristics | Value |
|---|---|
| Male, n (%) | 13 (54.2) |
| Age, mean years (SD) | 44.8 (15.6) |
| Exposure history | |
| Exposure to infected patient from Wuhan, n (%) | 19 (79.2) |
| Recent travel to Wuhan, China, n (%) | 5 (20.8) |
| Time from symptom onset to CT scans | |
| T0 | 3.4 days ± 1.8 |
| T1 | 9.7 days ± 1.9 |
| T2 | 15.8 days ± 3.6 |
| Comorbidities, n (%) | 0 (0) |
SD standard deviation, T0 baseline CT scan, T1 first follow-up CT scan, T2 second follow-up CT scan
The protected health information was removed from all data and was re-identified with a unique study ID. This study was approved by both the Ethics Committee at the Xian Jiaotong University The First Affiliated Hospital (XJTU1AF2020LSK-012) and the University of Pittsburgh Institutional Review Boards (IRB) (# STUDY20020171).
B. The computerized scheme
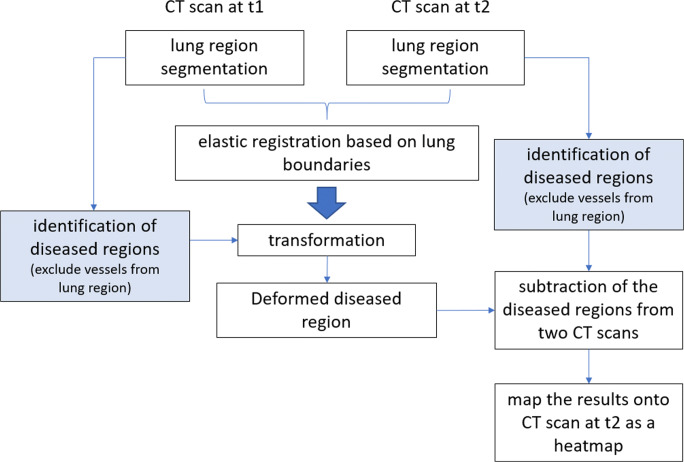
There are four primary components to our approach (Fig. 1): (1) automated segmentation of the lung boundary and major vessels, (2) elastic registration of the CT scans acquired at two time points, (3) computerized identification of the pneumonitis regions, and (4) assessment of disease progression. See Supplemental power point file demonstrating the 3-dimensional visualization of the lung, vascular, and pneumonia segmentations (slide 2) as well as the heatmap visualization of disease progression (slides 3 and 4).
Fig. 1.
The scheme flowchart of the developed algorithm
Automated lung segmentation
A deep learning approach based on the U-Net framework [16–18] was developed to ensure the automated segmentation of the lung boundary when there is pneumonia or consolidation adjacent to the chest wall. It is well-known that deep learning approaches are data-hungry. The 120 CT scans in Dataset 1 used to develop the lung and vessel segmentation algorithm had the lung boundaries delineated and other types of lung diseases labeled by an experienced thoracic radiologist (D.P.). Our computational geometric approach [19] used to identify the intrapulmonary vessels often failed to identify the vessels near the hilum due to the entanglement of the arteries and veins. Therefore, the U-Net framework was used to identify the main extrapulmonary vessels and vessels near the hilum. When training the U-Net framework, the CT images were transformed into an isotropic format and used 3D patches with a size of 96 × 96 × 96 mm. The Adam optimizer was used with an initial learning rate of 0.001 on a batch size of 2 and set the maximum number of epochs as 100. The voxel-wise cross-entropy loss function was minimized for the optimization, and the model with the smallest validation loss was saved as the final inference model.
Elastic lung registration
Our previously developed bidirectional elastic registration algorithm [20] was used to register two CT scans at different time points. The registration procedure produced a deformation field, by which we could elastically transform the CT images from one CT scan to another. We computed the intensity of the deformed voxels by performing a linear interposition based on the eight neighboring voxels of the new locations in the initial CT images.
Automated detection of COVID-19 disease
Pneumonia depicted on CT scans typically has a higher density compared with the lung parenchyma, but the density of pneumonia can vary widely. On chest CT scans, the lung vessels, fissures, and airway walls have a higher HU value compared with the surrounding parenchyma. The pulmonary fissures and the airway walls are small relative to the parenchyma and can largely be ignored or easily filtered from the image. The vessels are larger, thus intrapulmonary vessels and extrapulmonary vessels in the mediastinum were segmented (or filtered) and excluded from the images during the detection of the diseased regions, specifically pneumonia. The average density of the images in the middle of the lungs was used to compute a threshold (the lowest density) to detect regions associated with pneumonia. An experienced thoracic radiologist (J.S.) labeled the pneumonic regions associated with COVID-19 in the 72 CT scans from the 24 subjects in Dataset 1. As stated above, the 12 CT scans from four subjects were used in the development of the algorithm, which were not part of the test CT scans.
Quantitative assessment of disease progression
One approach to assess disease progression would be to independently quantify the volumes of the diseased regions in the lungs depicted on two chest CT scans and then compute the volume differences. For pneumonia caused by COVID-19, which most often includes multiple infected regions (Fig. 4), this method provides an overall estimation of the disease progress but lacks information regarding specific regional disease differences. A more robust approach to evaluating disease progression includes the independent assessment of longitudinal changes in each diseased region. To compare progression across individual regions, the paired regions of disease need to be identified on serial CT scans (e.g., T0 and T1). To automatically pair the diseased regions on two different CT scans, we used our previously developed bidirectional elastic registration algorithm [20] to register the two CT scans. Given two chest CT scans, the registration procedure produced a deformation field by which we could elastically transform the CT images with the identified disease at an early time point to a deformed version that is expected to be aligned with the CT images obtained at a later time point. We calculated the intensity of the deformed voxels by performing a linear interposition based on the eight neighboring voxels of the new locations from the initial CT images. The CT images from T0 and T1 were registered as described above, and based on the alignment of the regions of disease, which are automatically aligned, the difference between the regions of disease was used to quantify disease progression between T0 and T1 in terms of volume and density. A simple subtraction was also performed, based on the image registration between the aligned regions of disease between T0 and T1, to visualize the longitudinal changes by creating a heatmap. The voxel values on the subtraction images could be either positive or negative. A positive value indicates that the density increases, and a negative value indicates that the density decreases from T0 to T1.
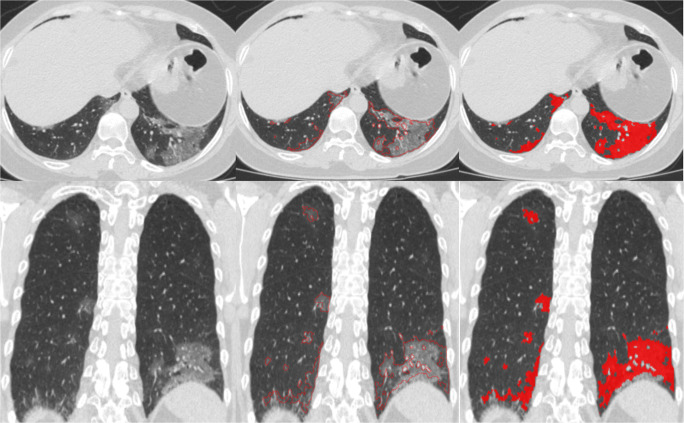
Fig. 4.
Example of automated segmentation of lung abnormalities depicted on CT images with COVID-19. The left column is the original axial CT image and a reformatted coronal image. The middle column the contours of the regions of disease. The right column, regions of disease depicted in red
C. Performance testing
The performance of the deep learning algorithms to segment the lung boundary and main lung vessels were compared with the visually interpreted results of the human expert (D.P.) using the test set (n = 20) of Dataset 1. Likewise, the pneumonic regions labeled in 60 CT scans (20 subjects) of Dataset 2 were used to evaluate the performance of the algorithm to assess the presence and progression of COVID-19. Since the algorithm for elastic lung CT registration has been quantitatively assessed and reported elsewhere [20], only (1) the performance of the deep learning algorithms for lung region segmentation and main lung vessel segmentation, and (2) the performance of the algorithm for pneumonia detection and quantification in COVID-19 subjects were evaluated in this study. The Dice coefficient was used to evaluate the performance of the deep learning algorithms [21]. The Dice coefficient is defined as:
| 1 |
where A is the computerized results and B is the labeled results by the human expert. The overlap between the readers’ delineated pneumonic regions and the computer-detected pneumonic regions was used to evaluate the performance of the computer algorithm. The readers’ outlines of the lung boundary and pneumonic regions were considered the gold standard “truth” in this study. Pneumonic regions detected by the software that did not overlap with radiologist-delineated pneumonic regions were considered false positives (FP). To characterize the detection-localization accuracy of the computer algorithm, we focused on regions > 200 mm3, which are likely to be more clinically relevant. The classic detection-localization characteristics [22] were estimated in terms of the true-positive fraction (TPF), which is the proportion of “true” detected pneumonic regions and the false-positive rate (FPR), which is the average number of FP results per image. TPF and FPR estimates were used for the sensitivity and specificity estimates of the corresponding iROI analysis [22], assuming that a reasonable bound or the number of pneumonic regions for CT scan is 40. The 95% confidence intervals for the estimates were computed using the generalized linear model for clustered binary data (PROC GENMOD, SAS, v.9.4).
Two radiologists (S.K. and Y.G.) independently reviewed and rated randomly presented CT scans from subjects with COVID-19. First, they viewed the original CT scans at T0 and T1 to assess if the disease increased, decreased, or remained the same. Next, they viewed the heatmap the computer software created from the registration of T0 and T1 images. Finally, they subjectively assessed if the heatmap accurately represented disease progression from T0 to T1 on a five 5-point scale: (1) unacceptable, too many errors that affect assessment; (2) poor, obvious errors that may affect assessment; (3) acceptable, minor errors that did not affect assessment; (4) good, minor errors; and (5) excellent, no obvious errors. We assessed the agreement of the two raters using the weighted Kappa coefficients. The statistical analysis (with p values and 95% confidence intervals, CI) was performed using a generalized linear model for a binary outcome, accounting for correlation between the assessment of the images from the same patient (PROC GENMOD, SAS v 9.4, SAS Institute). Inter-rater agreement was evaluated for both binary and multi-category (Likert) quality assessment using a simple weighted kappa statistic (PROC FREQ, SAS v.9.4).
Results
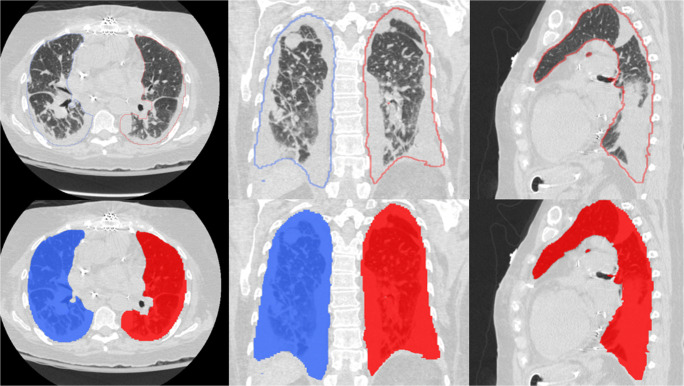
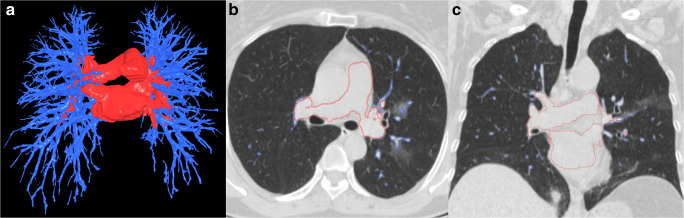
The lung boundaries, main lung vessel, and regions of disease (pneumonia) were reliably segmented with Dice coefficients of 0.95 (CI 0.95–0.96), 0.79 (CI 0.77–0.81), and 0.81 (CI 0.76–0.86), respectively. Although the lungs can be severely affected by COVID-19 infection, the algorithm successfully segmented the lung boundaries (Fig. 2). Furthermore, while our algorithm automatically and robustly segmented the intrapulmonary and extrapulmonary vessels (Fig. 3), a small percentage of very small intrapulmonary vessels were not detected. However, these very small vessels can be filtered out by a size filter or an opening morphological operation. The regions of disease (pneumonia) associated with COVID-19 were automatically and reliably segmented (Figs. 4 and 5). Each CT DICOM file was processed in ~ 6 to 10 min on a typical PC (Intel Core ™ i7-8559 CPU 1.99GHz). Approximately 80% of the processing time was allocated to the deep learning-based segmentation, which was performed on a CPU instead of a GPU to facilitate generalization of the computer software. When GPU (≥ 6 to 8GB memory) is available, it required ~ 2 min to process CT scans.
Fig. 2.
Examples of our algorithm’s automated lung segmentation of the lung boundaries in the presence of severe lung damage
Fig. 3.
Example of intrapulmonary and extrapulmonary vessel segmentation using the U-Net framework. a 3D visualization of the segmentation vessels, and (b, c) the segmentation contours of the vessels overlaid on the original CT images
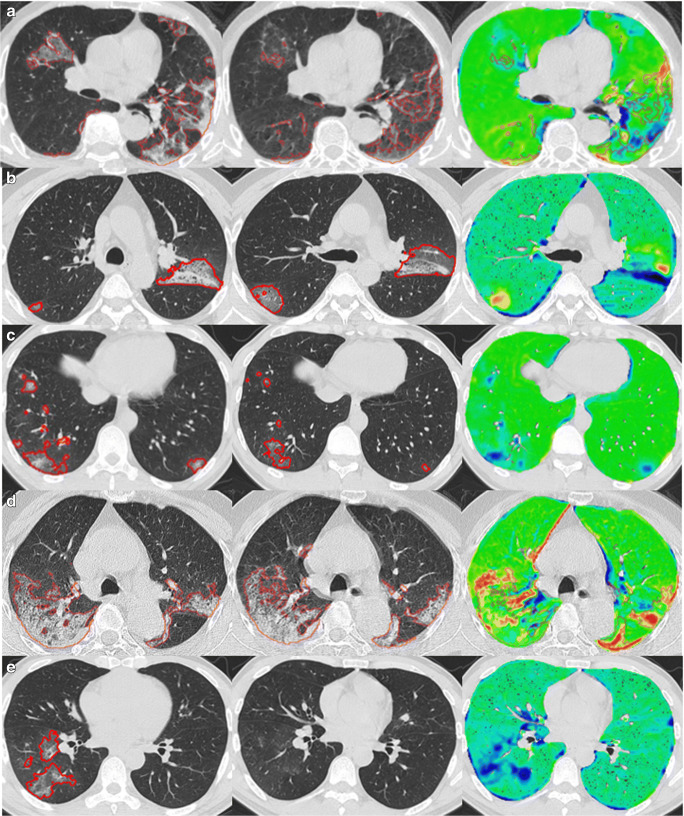
Fig. 5.
Examples demonstrating the visualization of disease progression by subtracting the identified regions of interest on the T0 CT scans from the T1 CT scans. The left panel is the original T0 (baseline) CT scan. The middle panel is the original T1 (follow-up) CT scan. The right panel is the heatmap visualization of disease progression: green regions—limited changes; blue regions—decreased densities from T0 to T1; and yellow and red regions—increased densities from T0 to T1
Two radiologists identified a total of 834 pneumonic regions on 60 CT scans (Dataset 2—test set) from 20 COVID-19 subjects, each > 200 mm3. Individuals had a median of 19 regions (interquartile range (IQR) 10–25). The algorithm detected 95% (796/834) of the regions identified and marked by the radiologists (range, 77 to 100% per scan). There were a total of 257 small false positives at the 95% detection rate or an average of 4.3 per scan (range, 1 to 12 per scan). The algorithm’s sensitivity for detection of the regions marked by the radiologist was 95% (CI 94–97%) with a specificity of 84% (CI 81–86%) for an upper bound for the numbers of non-overlapping pneumonic regions at 40 per CT scan.
The computer software identified a median of 19 separate regions (IQR 10–25) of pneumonia per individual patient on the T0 baseline CT scan. Fifty-five percent (11/20) of the subjects in the Dataset 2 test set demonstrated disease progression and 45% (9/20) demonstrated regression based on the automated assessment.
For both readers, 95% (CI 72 to 99) of the heatmaps were rated at least “acceptable” for representing the disease progression in CT scans of COVID-19 subjects (Tables 2 and 3). There was no significant difference between the two radiologists’ assessments (p = 0.33), with 55% and 60% of the CT scans reported as at least “good” by radiologist 1 and 2, respectively. Radiologists’ ratings on the Likert scale (“unacceptable”/“poor”/“acceptable”/“good”/“excellent”) agreed better than would be expected by chance (κ = 0.37, CI 0.09 to 0.65). Furthermore, 19/20 (95%) of ratings were within one category of each other. While radiologists were blind to the heat maps during their assessment, in a post hoc comparison, the two radiologists generally preferred assessing the progression of disease between two CT scans with the assistance of the heatmaps because of the clear illustration and quick interpretation of the area of disease (Fig. 5) (Table 4).
Table 2.
Dice coefficients for segmentation of the lung, the main lung vessels, and the pneumonia
| Segmentation type | Mean (SD) | Mean–95% CI | Dataset |
|---|---|---|---|
| Lung | 95% (0.8) | (95–96%) | Dataset 1–test set (n = 20) |
| Main lung vessels | 79% (4.7) | (77–81%) | Dataset 1–test set (n = 20) |
| Pneumonia | 81% (13.9) | (76–86%)* | Dataset 2–test set (n = 20) |
*Accounting for the nested data structure (using PROC MIXED, SAS v.9.4)
SD standard deviation, CI confidence interval
Table 3.
Radiologists’ assessment of the heatmap to accurately represent disease progression from T0 to T1
| Radiologist #1 | Radiologist #2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Subject 1 | 〇 | X | ||||||||
| Subject 2 | 〇 | X | ||||||||
| Subject 3 | 〇 | X | ||||||||
| Subject 4 | 〇 | X | ||||||||
| Subject 5 | 〇 | X | ||||||||
| Subject 6 | 〇 | X | ||||||||
| Subject 7 | 〇 | X | ||||||||
| Subject 8 | 〇 | X | ||||||||
| Subject 9 | 〇 | X | ||||||||
| Subject 10 | 〇 | X | ||||||||
| Subject 11 | 〇 | X | ||||||||
| Subject 12 | 〇 | X | ||||||||
| Subject 13 | 〇 | X | ||||||||
| Subject 14 | 〇 | X | ||||||||
| Subject 15 | 〇 | X | ||||||||
| Subject 16 | 〇 | X | ||||||||
| Subject 17 | 〇 | X | ||||||||
| Subject 18 | 〇 | X | ||||||||
| Subject 19 | 〇 | X | ||||||||
| Subject 20 | 〇 | X | ||||||||
1— unacceptable, too many errors that affect assessment; 2—poor, obvious errors that may affect assessment; 3—acceptable, minor errors that did not affect assessment; 4—good, minor errors; and 5—excellent, no obvious errors
Table 4.
Numbers and proportions of cases rated by two radiologists’ for the assessment of the heatmap to accurately represent disease progression from T0 to T1
| Marked at least “acceptable” | Marked as at least “good” | |||||
|---|---|---|---|---|---|---|
| Radiologist | Count | Total cases | Percent | Count | Total cases | Percent |
| 1 | 19 | 20 | 95 | 11 | 20 | 55 |
| 2 | 19 | 20 | 95 | 12 | 20 | 60 |
| Overall | 38 | 40 | 95 | 23 | 40 | 58 |
Discussion
Our computer algorithm automatically and robustly detected and quantified regions of pneumonia associated with COVID-19 as well as disease progression over time. Our unique registration approach was used to innovatively create a heatmap that visually illustrated disease progression. There was a high level of concurrence between human visual determination of progression and that of our automated algorithm. The results of this study on longitudinal CT scans from 20 patients with verified COVID-19 (Table 1) demonstrated the feasibility of the computer software to facilitate the assessment of the presence, severity, and change in the severity of lung abnormalities associated with COVID-19. The computer software could be used to detect and quantify pneumonia depicted on CT images associated with COVID-19 as well as other infectious or inflammatory pulmonary processes. We believe that the computer software can facilitate pneumonia detection, monitor progression, and assess treatment efficacy in both a clinical and research setting with respect to assessment and management of COVID-19.
One innovative aspect of our computer software lies in the integration of the deep learning technology and the computer vision technique to overcome the current limited availability of CT scans from COVID-19 patients for research purposes. Deep learning demonstrated remarkable performance in the detection of lung abnormalities associated with COVID-19, which is challenging for traditional computer vision techniques. For instance, the deep learning approach reliably identified the lung boundary in the presence of severe peripheral consolidation adjacent to the lung pleura (Fig. 2). There have been investigations by other investigators demonstrating the unique strength of the deep learning technology in identifying the complete boundaries of pathological lungs [23, 24]. However, the deep learning approach needs a large dataset with ground truth. It is very difficult to create a labeled dataset for lung vessel segmentation because it is very challenging and extremely tedious to accurately label the vasculature in the lungs. In this study, the intrapulmonary vessels were detected much more easily using traditional computer vision techniques compared with deep learning approaches.
The radiologists reported the heatmaps highlighting the regions of the lung associated with pneumonia were a beneficial addition to visual evaluation of the CT scans to assess disease progression. The radiologists reported that assessing disease progression was better when the novel heatmap was added to CT images compared with visual analysis of CT images alone. In only one case did the heatmap illustrating disease progression perform “poorly” or “unacceptable” as rated by radiologist 1 and 2, respectively, (subject 20, Fig. 5a). In this, case radiologist 2 felt strongly that the presence of the heatmap could possibly lead to a misdiagnosis. While radiologist 1 reported that the heatmap errors could possibly confound diagnosis but had only a small chance of leading to a misdiagnosis. The heatmap error was primarily caused by failure to detect and highlight a large region of the right lung in which the abnormality decreased from T0 to T1. In one case, our algorithm failed to detect the presence of pneumonic regions on the follow-up scans, which nearly resolved. In this case, the heatmap indicated that the presence of the abnormality decreased from T0 to T1. (Fig. 5e). Although the agreement between their Likert ratings (κ = 0.37) was not as high as anticipated, there was no significant difference between the two radiologists when they rated the accuracy of the heatmaps to illustrate disease progression at a rating of at least “acceptable.”
There are several limitations with this study. First, the regions on the CT image identified lung abnormalities associated with the COVID-19 disease may include other types of disease abnormalities that share a similar appearance as pneumonia (e.g., disease of the lung interstitium, non-solid lung nodules, and heart failure). We did not differentiate between abnormalities associated with COVID-19 and the potentially confounding abnormalities associated with other diseases. Second, the presence of comorbidities could affect the segmentation and assessment of COVID-19. However, in our relatively small study population, there were no obvious or reported comorbidities in the subjects’ medical records. Therefore, we cannot comment on how comorbidities may or may not impact our results. Additional procedures are desirable to differentiate the pneumonia regions associated with COVID-19 from other abnormalities. In practice, clinical information or other tests (e.g., RT-PCR) should be considered along with the image findings to exclude the false-positive detections in arriving at the final diagnosis. Notably, all of the subjects in this study were confirmed to have COVID-19. Beyond simply quantifying the presence of a pneumonia associated with COVID-19 or other disease patterns, our computer software can quantify the progression of different regions that do and do not rapidly progress (e.g., days or weeks), which can be critical in assessing both diagnosis and prognosis. For example, in chronic conditions such as idiopathic pulmonary fibrosis, the appearance on CT images will not change significantly between CT scans acquired only days apart, a dramatically different disease course than COVID-19. Third, the algorithm may fail to detect pneumonic regions that are very small and near the vessels or GGOs with very low density. Since these small regions are similar in appearance to ground-glass non-solid pulmonary nodules, traditional nodule detection and segmentation algorithms [25–27] could potentially be leveraged or combined to improve the performance of the computer software. Fourth, when registering two CT scans, the errors of the elastic registration scheme [20] often lead to incorrect alignment near the boundaries of the lungs and the diseased regions (Fig. 5). It is very difficult to completely avoid this issue because of the challenges in registering two CT scans. The two radiologists that evaluated the usefulness of the heatmaps to aid in the assessment of disease progression report that registration error had a limited impact on their visual review and assessment. Finally, only a small number of confirmed COVID-19 cases were available to develop and evaluate the computer software. We recognize that validation on a large dataset and a sophisticated observer study are necessary to assess the computer software and its implication in clinical practice. However, we believe the experimental results demonstrate the feasibility of our approach.
Conclusion
We developed a computer software to automatically detect and quantify regions of pneumonia depicted on CT scans associated with COVID-19. Our results demonstrate the feasibility of the software and its potential as an image-based biomarker to serve as an outcome marker in a clinical trial and ultimately translate to the clinical settings. The study also demonstrates the strength of integrating deep learning and computer vision technologies to overcome the limited availability of a dataset. It should be straightforward to develop a complete computer tool to efficiently label pneumonic regions in a large dataset for developing deep learning software.
Electronic supplementary material
Abbreviations
- AUC
Area under the curve
- BCE
Binary cross-entropy
- CAP
Community-acquired pneumonia
- CI
Confidential interval
- CNN
Convolutional neural network
- COVID-19
Novel coronavirus
- CT
Computed tomography
- FC
Fully connected
- GGO
Ground-glass opacity
- GPU
Graphics processing unit
- IRB
Institutional review boards
- ROC
Receiver operating characteristic
- RT-PCR
Real-time polymerase chain reaction
Funding information
This work is supported by National Institutes of Health (NIH) (Grant No. R01CA237277 and R01HL096613).
Compliance with ethical standards
Guarantor
The scientific guarantors of this publication are Dr. Jiantao Pu and Dr. Chenwang Jin.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
Dr. Andriy Bandos has significant statistical expertise and provided statistical analyses involved in this manuscript.
Informed consent
Written informed consent was waived by the Institutional Review Board.
This study was approved by both the Ethics Committee at the Xian Jiaotong University The First Affiliated Hospital (XJTU1AF2020LSK-012) and the University of Pittsburgh Institutional Review Boards (IRB) (# STUDY20020171).
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiantao Pu, Email: jip13@pitt.edu.
Chenwang Jin, Email: jin1115@mail.xjtu.edu.cn.
References
- 1.Lei J, Li J, Li X, Qi X (2020) CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 10.1148/radiol.2020200236:200236 [DOI] [PMC free article] [PubMed]
- 2.Shi H, Han X, Zheng C (2020) Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China. Radiology. 10.1148/radiol.2020200269:200269 [DOI] [PMC free article] [PubMed]
- 3.Ai T, Yang Z, Hou H et al (2020) Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 10.1148/radiol.2020200642:200642 [DOI] [PMC free article] [PubMed]
- 4.Lee EYP, Ng MY, Khong PL (2020) COVID-19 pneumonia: what has CT taught us? Lancet Infect Dis. 10.1016/S1473-3099(20)30134-1 [DOI] [PMC free article] [PubMed]
- 5.Lin C, Ding Y, Xie B, et al. Asymptomatic novel coronavirus pneumonia patient outside Wuhan: the value of CT images in the course of the disease. Clin Imaging. 2020;63:7–9. doi: 10.1016/j.clinimag.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao W, Zhong Z, Xie X, Yu Q, Liu J (2020) Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. AJR Am J Roentgenol. 10.2214/AJR.20.22976:1-6 [DOI] [PubMed]
- 7.Li Y, Xia L (2020) Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. AJR Am J Roentgenol. 10.2214/AJR.20.22954:1-7 [DOI] [PubMed]
- 8.Zhou S, Wang Y, Zhu T, Xia L (2020) CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. AJR Am J Roentgenol. 10.2214/AJR.20.22975:1-8 [DOI] [PubMed]
- 9.Fang Y, Zhang H, Xie J et al (2020) Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 10.1148/radiol.2020200432:200432 [DOI] [PMC free article] [PubMed]
- 10.Shi H, Han X, Jiang N et al (2020) Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 10.1016/S1473-3099(20)30086-4 [DOI] [PMC free article] [PubMed]
- 11.Hwang EJ, Park S, Jin KN, et al. Development and validation of a deep learning-based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw Open. 2019;2:e191095. doi: 10.1001/jamanetworkopen.2019.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc. 2000;7:593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parveen NR, Sathik MM. Detection of pneumonia in chest X-ray images. J Xray Sci Technol. 2011;19:423–428. doi: 10.3233/XST-2011-0304. [DOI] [PubMed] [Google Scholar]
- 14.Meystre S, Gouripeddi R, Tieder J, Simmons J, Srivastava R, Shah S. Enhancing comparative effectiveness research with automated pediatric pneumonia detection in a multi-institutional clinical repository: a PHIS+ pilot study. J Med Internet Res. 2017;19:e162. doi: 10.2196/jmir.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauczor HU, Heitmann K, Heussel CP, Marwede D, Uthmann T, Thelen M. Automatic detection and quantification of ground-glass opacities on high-resolution CT using multiple neural networks: comparison with a density mask. AJR Am J Roentgenol. 2000;175:1329–1334. doi: 10.2214/ajr.175.5.1751329. [DOI] [PubMed] [Google Scholar]
- 16.Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Springer International Publishing, Cham, pp. 234–241
- 17.van der Heyden B, van de Worp W, van Helvoort A, et al. Automated CT-derived skeletal muscle mass determination in lower hind limbs of mice using a 3D U-Net deep learning network. J Appl Physiol (1985) 2020;128:42–49. doi: 10.1152/japplphysiol.00465.2019. [DOI] [PubMed] [Google Scholar]
- 18.Nemoto T, Futakami N, Yagi M et al (2020) Efficacy evaluation of 2D, 3D U-Net semantic segmentation and atlas-based segmentation of normal lungs excluding the trachea and main bronchi. J Radiat Res. 10.1093/jrr/rrz086 [DOI] [PMC free article] [PubMed]
- 19.Gu S, Meng X, Sciurba FC, et al. Bidirectional elastic image registration using B-spline affine transformation. Comput Med Imaging Graph. 2014;38:306–314. doi: 10.1016/j.compmedimag.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pu J, Fuhrman C, Good WF, Sciurba FC, Gur D. A differential geometric approach to automated segmentation of human airway tree. IEEE Trans Med Imaging. 2011;30:266–278. doi: 10.1109/TMI.2010.2076300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou KH, Warfield SK, Bharatha A, et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol. 2004;11:178–189. doi: 10.1016/S1076-6332(03)00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou K, Liu A, Bandos AI, Ohno-Machado L, Rockette HE (2016) Statistical evaluation of diagnostic performance: topics in ROC analysis. Chapman and Hall/CRC
- 23.Gerard SE, Herrmann J, Kaczka DW, Musch G, Fernandez-Bustamante A, Reinhardt JM. Multi-resolution convolutional neural networks for fully automated segmentation of acutely injured lungs in multiple species. Med Image Anal. 2020;60:101592. doi: 10.1016/j.media.2019.101592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison AP, Xu Z, George K, Lu L, Summers RM, Mollura DJ. Progressive and multi-path holistically nested neural networks for pathological lung segmentation from CT images. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, editors. Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. Cham: Springer International Publishing; 2017. pp. 621–629. [Google Scholar]
- 25.Halder A, Dey D, Sadhu AK (2020) Lung nodule detection from feature engineering to deep learning in thoracic CT images: a comprehensive review. J Digit Imaging. 10.1007/s10278-020-00320-6 [DOI] [PMC free article] [PubMed]
- 26.Pu J, Zheng B, Leader JK, Wang XH, Gur D. An automated CT based lung nodule detection scheme using geometric analysis of signed distance field. Med Phys. 2008;35:3453–3461. doi: 10.1118/1.2948349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pu J, Paik DS, Meng X, Roos JE, Rubin GD. Shape “break-and-repair” strategy and its application to automated medical image segmentation. IEEE Trans Vis Comput Graph. 2011;17:115–124. doi: 10.1109/TVCG.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.