Abstract
Objective: Osteonecrosis of the femoral head (ONFH) is a severe pathological state with multiple etiologies. Steroid hormone metabolism-related genes play an important role in ONFH. The aim of this study was to investigate the relationships between polymorphisms of the drug-metabolizing enzyme gene, cytochrome P450 (CYP450), and the drug transporter gene, ATP-binding cassette subfamily B member 1 (ABCB1), as well as their DNA methylation status with the pathogenesis of steroid-induced ONFH.
Methods: In this case–control study, we evaluated five single nucleotide polymorphisms (SNPs) in two genes in a Han Chinese population, including 79 patients with steroid-induced ONFH and 114 persons who took steroids but did not develop steroid-induced ONFH. SNPs were genotyped by the improved multiplex ligation detection reaction. MethylTarget technology was used to ascertain the methylation status at two CpG islands in the ABCB1 gene for statistical analysis. Finally, interactions between the SNPs and the CpG site's methylation levels were statistically analyzed by methylation quantitative trait locus.
Results: We found that the T allele of the CYP450 rs2242480 locus was associated with steroid-induced ONFH risk reduction (odds ratio [OR] = 0.598, 95% confidence interval [CI]: 0.360–0.992, p = 0.046). In the genetic model analysis, the T allele of the rs2032582 locus in the ABCB1 gene was associated with a reduced risk of steroid-induced ONFH under the dominant model (OR = 0.465, 95% CI: 0.223–0.972, p = 0.042). The CpG sites with significant differences (p < 0.05) in methylation levels between the cases and controls were ABCB1_1_192…ABCB1_2_43. A total of 14 pairs of linear regression tests between SNPs and methylation sites demonstrated statistical significance (p < 0.05).
Conclusions: This study provides evidence for two steroid-induced ONFH susceptibility genes (ABCB1, CYP450) in the Han Chinese population.
Keywords: ABCB1, CYP450, ONFH, single nucleotide polymorphism
Introduction
Osteonecrosis of the femoral head (ONFH) is a complex orthopedic disease. Recent studies have shown that an ONFH attack is associated with decreased fibrinolysis capacity, a tendency toward thrombosis, decreased production of vasoactive substances, and abnormal lipid metabolism (Zalavras et al., 2000; Weinstein et al., 2017). All of these abnormalities disrupt blood flow to the femoral head. The interruption of blood supply to the femoral head leads to apoptosis of bone cells and a change of the internal microstructure of the femoral head, resulting in collapse of the femoral head and eventually joint dysfunction. If patients with ONFH do not receive effective treatment at an early stage, there is a high probability that disabilities will occur. Currently, in China 150,000 to 200,000 incident cases of ONFH are diagnosed annually (Cui et al., 2016). The widespread use of steroid hormones is considered one of the common causes of nontraumatic osteonecrosis. Steroid hormones act on osteoblasts and osteoclasts, reducing the blood supply and strength of the bone, thus leading to femoral head injury (Weinstein, 2012).
Steroid-induced ONFH, however, does not occur in all patients who receive high-dose, long-term steroid therapy. The interactions between genetic susceptibility loci and other risk factors may determine whether the disease will develop and to what particular severity. It has recently been suggested that individual differences in drug sensitivity are due to drug genetic polymorphisms in drug-metabolizing genes. Single nucleotide polymorphisms (SNPs) are increasingly being used as markers of genetic variation because of their high frequency, convenience, and ease of statistical analysis. Possible reasons for individual differences in steroid hormone sensitivity include polymorphisms of steroid hormone receptors, steroid hormone metabolism enzymes, and transporters. Studying the relationship between these SNPs and steroid-induced ONFH will help identify the population at risk of steroid-induced ONFH who use steroid hormones. For these patients, it is necessary to avoid the use of steroid hormones or change the application strategy, and to increase monitoring for steroid-induced ONFH, to provide for early intervention.
Cytochrome P450 (CYP450) is mainly distributed in the endoparasitic reticulum and mitochondrial endometrium. It is a major enzyme involved in lipid-soluble drug metabolism. At present, >12 subfamilies of CYP450 have been identified in the human body. The CYP1, CYP2, and CYP3 subtypes of the enzyme are the ones most closely related to drug metabolism, with obvious individual differences. Brunner-Ziegler et al. (2013) found that patients with CYP2C9 gene mutations in subgroups CYP2C9 * 2 and CYP2C9 * 3 were four times more prone to osteoporosis, and were more sensitive to anticoagulant drugs and had lower bone density in the femoral neck. Phabphal et al. (2013) compared vitamin D and parathyroid hormone levels with bone mineral density in patients, and found that the gene polymorphism of CYP2C9 was related to bone mineral density and that the differences in bone mineral density in the femoral neck were statistically significant. CYP2C19 is an important CYP450 subtype, which mainly exists in liver microsomes. Polymorphisms of this subtype play a major role in drug metabolism in vivo, with significant individual differences. At present, the research on the clinical effects of gene polymorphisms of CYP2C19 has primarily focused on anticoagulation therapy in vivo. Yang et al. (2015) found that CYP2C19 gene polymorphisms do not significantly increase the risk for antiplatelet therapy, but further studies have found that CYP2C19 gene polymorphisms may cause platelet reaggregation and cause the corresponding symptoms. When Rath et al. (2015) detected platelet agglutination status in patients with acute coronary syndrome, they found that the response to anticoagulant drugs of the various CYP2C19*2 gene polymorphisms can affect treatment outcomes. This is of importance for the design of personalized treatment plans. Previous studies have reported that alleles with functional loss of CYP2C19*2 and CYP2C19*3 directly affect drug metabolism of cryptogram and determine the anticoagulant treatment effect of individuals, with significant individual differences (Jeong et al., 2015). Additional studies have found that the ultimate cause of ONFH can be attributed to intravascular coagulation (REF needed). The early use of anticoagulant therapy for ONFH has not been confirmed with good clinical results. The importance of the CYP3A subfamily in statin metabolism has been reported. CYP3A4 and CYP3A5 gene polymorphisms may influence the lipid lowering response and pharmacokinetics characteristics of statins (He et al., 2014; Wei and Zhang, 2015). The CYP3A4 gene is a key enzyme gene in steroid hormone metabolism and plays an important role in steroid-induced ONFH (Guo and Deng, 2019). Studies have suggested that individual differences in steroid-induced ONFH may be linked to genetic predisposition, and that the polymorphisms of CYP450 may directly affect a patient's ability to metabolize these hormones, thus impacting the risk of steroid-induced ONFH (Kaneshiro and Takaoka, 2007).
A variety of synthetic steroid hormones are absorbed and distributed through the ATP-binding cassette subfamily B member 1 (ABCB1), a transporter that has been associated with drug resistance. Thus, the ABCB1 gene is also known as the multidrug-resistant transporter-1 (MDR1) gene. It is located on human chromosome 7q21.12, and encodes a 1280 amino acid protein that is one of the key proteins for drug absorption and transport in cells. There are 48 SNPs in the ABCB1 gene of which 19 are located within exons with different proportions among different ethnic groups (Bodor et al., 2005). The drug transporter P-glycoprotein (P-gp) encoded by the ABCB1 gene is a phosphorescent and glycerogelatin transmembrane transporter, which is a transmembrane ATP-dependent molecular pump that primarily moves active substrates into and out of the cell, thus affecting the efficacy of drugs. Studies have shown that an increase in P-gp activity may be a marker for low risk of steroid-induced ONFH. ABCB1 has become one of the most studied members of the ABC family because it produces multiple drug resistances in tumor cells. Any defect caused by ABCB1 polymorphisms can lead to serious diseases such as breast cancer and Parkinson's disease (Huang et al., 2016; Sharif et al., 2016). Two sites within the ABCB1 gene, C3435T (rs1045642) in exon 26 and G2677T/A (rs2032582) in exon 21, are the focus of current research. The rs1045642 SNP is considered to be functional and has been associated with a variety of diseases. It does not result in a missense mutation; however, it changes the interaction between P-gp and drugs by affecting the time of cotranslation folding, thereby reducing the number and activity of P-gp (Fujii et al., 2012; Sheng et al., 2012). The rs2032582 locus of the gene results in a missense mutation, leading to increased excretion and transport capacity, ultimately reducing the risk of steroid-induced ONFH (Tanabe et al., 2001).
Epigenetic regulation also affects the expression and function of genes related to femoral head necrosis. Currently, most epigenetic studies focus on differences in DNA methylation at CpG islands. Multiple studies have found that DNA methylation may play an important role in regulating the expression and function of ABCB1 in malignant tumor cells (Shi et al., 2011; Onda et al., 2012). However, data on how the methylation status of ABCB1 affects steroid-induced ONFH are limited. Individual differences in the expression and function of ABCB1 in patients may be the result of interactions between genetic and epigenetic variations. Thus, studying the combined effects of genetic and epigenetic changes is important to understand the development of this disease.
Materials and Methods
Study participants
Our study population was restricted to individuals who were ethnically Han Chinese. A total of 79 steroid-induced ONFH patients were enrolled between July 2017 and February 2019 at the Affiliated Hospital of the Institute of Neurology, Anhui University of Chinese Medicine. The patients received standard steroid hormone therapy for treatment of systemic lupus erythematosus, myasthenia graves, multiple sclerosis, acute lymphocyte leukemia, Devic's disease, or organ transplantation. The development of steroid-induced ONFH was confirmed by steroid history, X-ray evaluation, magnetic resonance imaging, and bone scans (Roth et al., 2016). After 3 months of oral steroid hormone (prednisone >30mg/d) treatment, 114 patients in the steroid-resistant group without femoral head necrosis were recruited. All participants provided written informed consent. We also collected basic personal data of patients from all study participants, including name, age, gender, ethnicity, and place of birth.
Genotyping
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI) according to the manufacturer's instructions. SNP genotyping was performed using an improved multiplex ligation detection reaction (iMLDR) technique developed by Genesky Biotechnologies, Inc. (Shanghai, China). A multiplex PCR-ligase detection reaction method was used in the iMLDR. For each SNP, the alleles were distinguished by different fluorescent labels of allele-specific oligonucleotide probe pairs. Different SNPs were further distinguished by different extension lengths at the 3′ end. Two negative controls were included with each experiment: one with double-distilled water as the template and the other with template DNA, but without primers while keeping all other conditions the same. Duplicate tests were performed with all results being consistent. A random sample accounting for ∼5% (n = 20) of the total DNA samples was directly sequenced using Big Dye-terminator technology (version 3.1) run on an ABI 3730XL automated sequencer (Applied Biosystems) to confirm the results of the iMLDR.
CpG islands selection
Two CpG islands located in the proximal promoter of the ABCB1 gene were selected for measurement according to the following criteria: (1) 200 bp minimum length, (2) ≥50% GC content, and (3) ≥0.60 ratio of observed/expected dinucleotides CpG. We did not perform methylation studies of the CYP3A4 gene because no CpG islands were detected in its proximal promoter region.
Methylation detection of the ABCB1 gene
DNA methylation levels were analyzed by MethylTarget™ (Genesky Biotechnologies, Inc.), a NGS-based multiple target CpG methylation analysis method. Specifically, the genomic regions of interest were analyzed and transformed to bisulfite-converted sequences by gene CpG software. PCR primer sets were designed with Methylation Primer software from bisulfate-converted DNA. Genomic DNA (400 ng) was subjected to sodium bisulfite treatment using EZ DNA Methylation™-GOLD Kit (Zymo Research) according to manufacturer's protocols. Multiplex PCRs were performed with optimized primer sets. Libraries from different samples were quantified, normalized, and pooled, followed by sequencing on an Illumina HiSeq according to the manufacturer's protocols. Sequencing was performed using a 2 × 150 bp paired-end mode.
Statistical analyses
SPSS 24.0 statistical packages (SPSS, Chicago, IL) and Microsoft Excel (Chicago, IL) were used to perform the statistical analyses. A p < 0.05 was considered the threshold for whether statistical significance was achieved. A χ2 test was used to check whether the genotypic distribution was consistent with the Hardy–Weinberg equilibrium (HWE) in the control group. The genotypic frequencies in the cases and controls were calculated using χ2 tests. Odds ratio (OR) and 95% confidence interval (CI) were determined using unconditional logistic regression analysis. The genetic models were applied using PLINK software to assess the association of SNPs with the risk of steroid-induced ONFH. Haploview was used to analyze the linkage disequilibrium of different SNPs in the same gene.
Results
The analyses included 79 patients with steroid-induced ONFH (mean age: 50.65 ± 13.95) and 114 patients (mean age: 42.63 ± 19.27) who received steroid therapy but did not develop steroid-induced ONFH. Gender did not differ among the two groups (p > 0.05). Demographic characteristics of the samples are shown in Table 1.
Table 1.
Demographic Characteristics of 79 Patients and 114 Controls
| Characteristics | SONFH (n = 79) (mean ± SD) | Controls (n = 114) (mean ± SD) | t-Value | p-Value |
|---|---|---|---|---|
| Age | 50.65 ± 13.95 | 42.63 ± 19.27 | 3.3305 | 0.001 |
| Gender | Female 45, male 34 | Female 65, male 49 | 0.1273 | 0.8989 |
SONFH, steroid-induced osteonecrosis of the femoral head.
Association of ABCB1 and CYP450 gene polymorphisms with risk of steroid-induced ONFH
SNP information and HWE test results are shown in Table 2. All of the SNPs matched HWE (p > 0.05). The results of the allelic frequencies and genotypic frequencies of the 5 evaluated SNPs for the patients with steroid-induced ONFH and the controls are shown in Table 3. The rs2242480 locus of the gene was significantly associated with steroid-induced ONFH risk (OR = 0.598, 95% CI: 0.360–0.992, p = 0.046). We also evaluated the association of the ABCB1 and CYP450 gene polymorphisms with steroid-induced ONFH under dominant and recessive models (Table 4). Only the ABCB1 rs2032582 T allele was significantly associated with steroid-induced ONFH risk under the dominant model (OR = 0.465, 95% CI: 0.223–0.972, p = 0.042).
Table 2.
Single Nucleotide Polymorphism Information and Hardy–Weinberg Equilibrium Test
| SNP ID | Gene | Location (GRCh37) | Allele | MAF (CHB_1000g) | p-Value of HWE |
|---|---|---|---|---|---|
| rs1045642 | ABCB1 | chr7:87138645 | A/G | 0.658654 | 0.1721 |
| rs1128503 | ABCB1 | chr7:87179601 | G/A | 0.334135 | 0.6399 |
| rs2032582C | ABCB1 | chr7:87160618 | C/A | 0.483173 | 0.3119 |
| rs2032582T | ABCB1 | chr7:87160618 | T/A | 0.141827 | 0.323 |
| rs2242480 | CYP450 | chr7:99361466 | T/C | 0.242788 | 0.5526 |
HWE, Hardy–Weinberg equilibrium; MAF (CHB_1000g), minor allele frequency of Chinese Han population in 1000 g database; SNP, single nucleotide polymorphism.
Table 3.
Analyses of Association of Allele and Genotype Frequency with the Risk of Steroid-Induced Osteonecrosis of the Femoral Head Between Controls and Patients with Steroid-Induced Osteonecrosis of the Femoral Head
| SNP ID | Allele | SONFH | Controls | OR (95% CI) | p-Value | Genotype | SONFH | Controls | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1045642 | A | 65 | 81 | 1.268 (0.836–1.925) | 0.264 | AA | 14 | 18 | 1.243 (0.833–1.853) | 0.443 |
| G | 93 | 147 | AG | 37 | 45 | |||||
| GG | 28 | 51 | ||||||||
| rs1128503 | G | 58 | 80 | 1.095 (0.717–1.672) | 0.675 | GG | 11 | 12 | 1.099 (0.713–1.694) | 0.749 |
| A | 98 | 148 | GA | 36 | 56 | |||||
| AA | 31 | 46 | ||||||||
| rs2032582C | C | 74 | 116 | 0.871 (0.579–1.310) | 0.508 | CC | 15 | 28 | 0.860 (0.561–1.318) | 0.710 |
| A | 82 | 112 | CA | 44 | 60 | |||||
| AA | 19 | 26 | ||||||||
| rs2032582T | T | 16 | 33 | 0.675 (0.357–1.275) | 0.225 | TT | 1 | 0 | 0.647 (0.331–1.263) | 0.085 |
| A | 140 | 195 | TA | 14 | 33 | |||||
| AA | 63 | 81 | ||||||||
| rs2242480 | T | 28 | 61 | 0.598 (0.360–0.992) | 0.046 | TT | 3 | 9 | 0.606 (0.366–1.004) | 0.145 |
| C | 122 | 159 | TC | 22 | 43 | |||||
| CC | 50 | 58 |
CI, confidence interval; OR, odds ratio.
Table 4.
Genetic Model Analyses of Association Between Controls and Patients with Steroid-Induced Osteonecrosis of the Femoral Head
| SNP ID | Recessive model | SONFH | Controls | OR (95% CI) | p-Value | Dominant model | SONFH | Controls | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1045642 | AA | 14 | 18 | 1.149 (0.534–2.471) | 0.8443 | AA+AG | 49 | 59 | 1.599 (0.859–2.977) | 0.139 |
| AG+GG | 65 | 96 | GG | 27 | 49 | |||||
| rs1128503 | GG | 11 | 12 | 1.396 (0.582–3.345) | 0.5013 | AG+GG | 45 | 65 | 0.871 (0.465–1.628) | 0.664 |
| AG+AA | 67 | 102 | AA | 30 | 43 | |||||
| rs2032582C | CC | 15 | 28 | 0.731 (0.361–1.482) | 0.481 | CC+CA | 57 | 83 | 0.879 (0.429–1.799) | 0.724 |
| CA+AA | 63 | 86 | AA | 18 | 25 | |||||
| rs2032582T | TT | 1 | 0 | NA | 0.406 | TT+TA | 14 | 33 | 0.465 (0.223–0.972) | 0.042 |
| TA+AA | 77 | 114 | AA | 61 | 75 | |||||
| rs2242480 | TT | 3 | 9 | 0.467 (0.122–1.788) | 0.366 | TT+TC | 25 | 48 | 0.657 (0.349–1.239) | 0.194 |
| TC+CC | 72 | 101 | CC | 47 | 56 |
Haplotype analysis
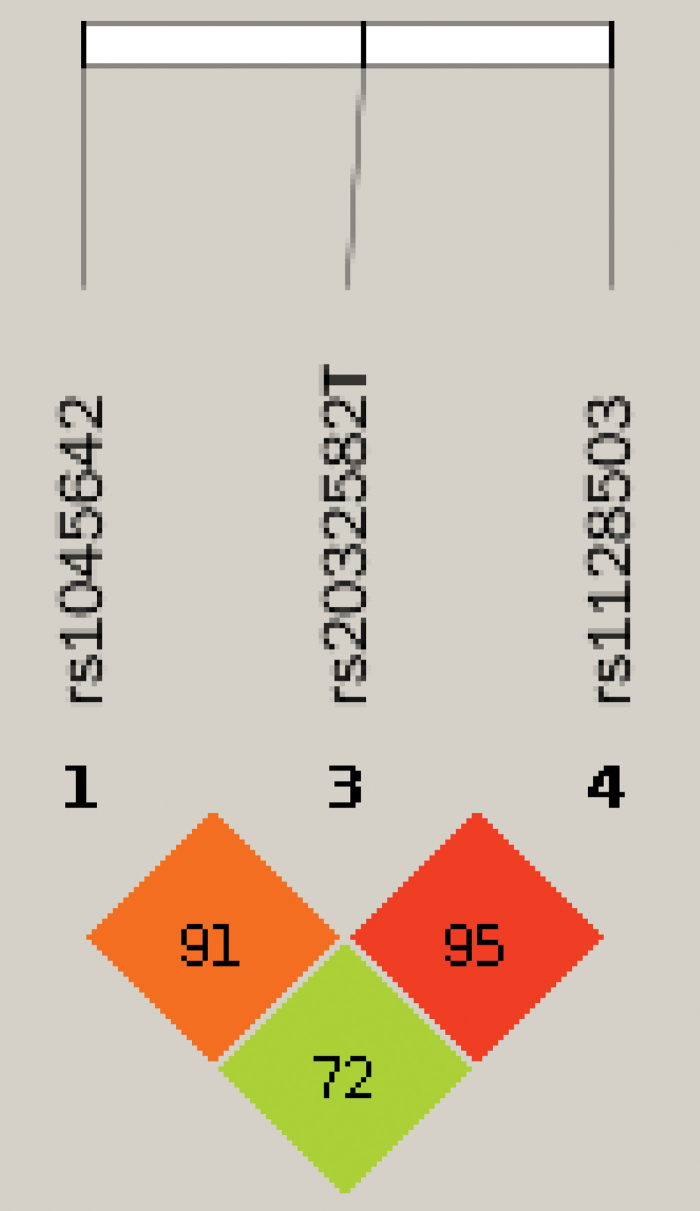
We used Haploview software to analyze the linkage imbalance of the ABCB1 polymorphic sites. In the linkage disequilibrium analyses, the rs2032582 T allele of the ABCB1 gene was found to be strongly linked with the polymorphism at the rs1045642 and rs1128503 loci (Fig. 1). Therefore, haplotype analyses were carried out for these loci to compare the two groups. The results of the association between the ABCB1 haplotypes and steroid-induced ONFH risk are listed in Table 5; however, none showed statistical significance between the case and control groups, suggesting that the haplotype of the ABCB1 gene may be independent of steroid-induced ONFH.
FIG. 1.
Linkage disequilibrium map. Color images are available online.
Table 5.
Haplotype Analysis of Steroid-Induced Osteonecrosis of the Femoral Head and the Controls in the ABCB1 Gene
| Haplotype | SNPs | SONFH | Controls | OR (L95–U95) | p-Value |
|---|---|---|---|---|---|
| GA | rs1045642+rs2032582T | 76 (0.6441) | 114 (0.6628) | — | — |
| AA | rs1045642+rs2032582T | 64 (0.4156) | 81 (0.3553) | 1.26 (0.844–1.886) | 0.257 |
| GT | rs1045642+rs2032582T | 15 (0.1172) | 33 (0.1719) | 0.58 (0.285–1.195) | 0.141 |
| AA | rs2032582T+rs1128503 | 89 (0.7672) | 124 (0.7848) | — | — |
| AG | rs2032582T+rs1128503 | 51 (0.3312) | 71 (0.3114) | 1.13 (0.677–1.899) | 0.633 |
| TG | rs2032582T+rs1128503 | 7 (0.0547) | 9 (0.0511) | 1.07 (0.402–2.841) | 0.895 |
Methylation analysis
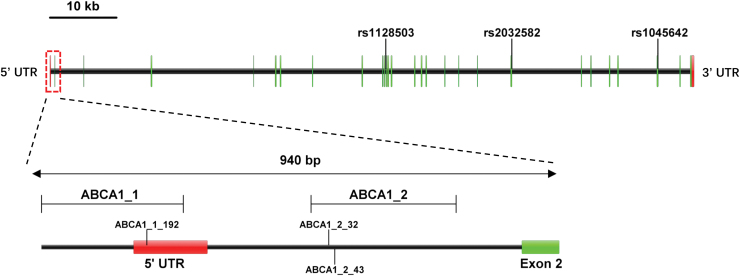
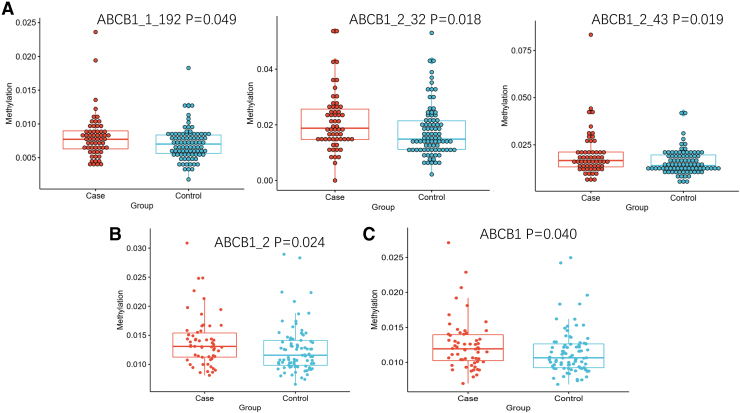
Primer3 software was used to design primers for bisulfite-treated sequences (Table 6 and Fig. 2). We detected a total of 38 CpG sites, and the methylation level for each CpG site was tested for significance. The CpG sites with significant differences in methylation levels were compared between patient groups, with ABCB1_1_192, ABCB1_2_32, and ABCB1_2_43 showing statistically significant differences (p < 0.05). We also determined the mean methylation level of all CpG sites on each amplimer and the mean methylation level of all CpG sites studied as the methylation level of the gene. A statistical analysis of these data shows that the methylation levels of the ABCB1–2 fragment and ABCB1 gene are significantly different between the steroid-induced ONFH group and steroid-resistant group (p = 0.024, p = 0.040, respectively, Table 7 and Fig. 3).
Table 6.
The Position of the Two CpG Regions and Primer Sequences
| Target | Location (GRCh37) | Length | Primer sequences |
|---|---|---|---|
| ABCB1_1 | chr7:87230114-87230372 | 259 | GAAGATATTTYGATTTTAGTGGAAAGATTTA |
| CATTCCTCCTAAAAATTCAACCTATTT | |||
| ABCB1_2 | chr7:87229882-87229619 | 264 | TYGTAGTGGTATTGGATTATGTTGTT |
| CCRTTAATACCCCAACTACTCTAACC |
FIG. 2.
Single nucleotide polymorphism locus and methylation fragment map of ABCB1 gene. Color images are available online.
Table 7.
Differences of Methylation Levels of Sites, Targets, and Gene
| Target | CpG site | SONFH (mean ± SD) | Controls (mean ± SD) | p-Value |
|---|---|---|---|---|
| ABCB1_1 | 192 | 0.81 ± 0.33 | 0.71 ± 0.25 | 0.049 |
| ABCB1_2 | 32 | 2.10 ± 1.05 | 1.77 ± 0.94 | 0.018 |
| ABCB1_2 | 43 | 1.98 ± 1.2 | 1.58 ± 0.65 | 0.019 |
| ABCB1_1 | — | 0.99 ± 0.22 | 0.94 ± 0.19 | 0.063 |
| ABCB1_2 | — | 1.4 ± 0.44 | 1.25 ± 0.41 | 0.024 |
| ABCB1 | — | 1.25 ± 0.36 | 1.15 ± 0.33 | 0.040 |
FIG. 3.
Scatter diagram of methylation difference analysis. (A) The methylation difference analysis of CpG sites. ABCB1_1_192, ABCB1_2_32, and ABCB1_2_43 showing statistically significant differences (p < 0.05). (B) The methylation difference analysis of fragment. The ABCB1–2 fragment is significantly different between the steroid-induced ONFH group and steroid-resistant group (p = 0.024). (C) The methylation difference analysis of gene. The ABCB1 gene is significantly different between the steroid-induced ONFH group and steroid-resistant group (p = 0.040). Color images are available online.
Methylation quantitative trait locus analysis
Finally, the DNA methylation quantitative trait locus (meQTL) method was used to conduct a combined analysis of all SNPs and methylation levels of the CpG sites from all samples. A total of 14 pairs of linear regression tests between SNP and methylation sites were statistically significant (p < 0.05); all results are shown in Table 8.
Table 8.
Methylation Quantitative Trait Locus Analysis Between Single Nucleotide Polymorphisms and CpG Loci
| SNP | Gene | Beta | t-stat | p-value |
|---|---|---|---|---|
| rs1045642 | ABCB1_2|208 | 0.002 | 3.441 | 0.001 |
| rs1045642 | ABCB1_2|27 | 0.003 | 2.694 | 0.008 |
| rs1045642 | ABCB1_2|186 | 0.001 | 2.654 | 0.009 |
| rs1045642 | ABCB1_2|139 | 0.001 | 2.597 | 0.010 |
| rs1045642 | ABCB1_2|43 | 0.002 | 2.561 | 0.011 |
| rs1045642 | ABCB1_2|103 | 0.001 | 2.273 | 0.024 |
| rs1045642 | ABCB1_2|47 | 0.002 | 2.007 | 0.047 |
| rs1045642 | ABCB1_2|206 | 0.001 | 2.007 | 0.047 |
| rs1045642 | ABCB1_2|32 | 0.002 | 1.987 | 0.049 |
| rs2032582C | ABCB1_1|197 | 0.001 | 2.896 | 0.004 |
| rs2032582C | ABCB1_2|208 | 0.001 | 2.262 | 0.025 |
| rs2032582C | ABCB1_1|221 | 0.001 | 2.088 | 0.039 |
| rs2032582C | ABCB1_2|47 | 0.002 | 2.053 | 0.042 |
| rs2032582T | ABCB1_1|197 | −0.001 | −2.369 | 0.020 |
Discussion
Steroid hormones can cause increased blood coagulation and low fibrinolysis, leading to blood vessel thrombosis, bone ischemia, osteonecrosis, and ultimately lead to steroid-induced ONFH. However, the fact that steroid-induced ONFH does not occur in all patients receiving the same steroid hormone therapy means that individual differences in steroid hormone sensitivity due to genetic factors likely play an important role in the development of steroid-induced ONFH. Recently, studies have found genetic polymorphisms in steroid hormone receptors. Drug transport and drug metabolism are also involved in steroid responsiveness in vivo. Prevention of steroid-induced ONFH through understanding of individual risk factors including genetic variation is therefore important clinically.
The CYP450 family is a group of enzymes involved in nearly all lipid-soluble drug redox metabolic processes. Abnormal lipid metabolism is one of the potential causes of nontraumatic ONFH, resulting in vascular occlusion and abnormal blood supply to the femoral head. Although the exact mechanism of the damage to bone circulation caused by high-level and long-term exposure to exogenous steroids remains unclear, CYP3A activity plays a crucial role in the development of steroid-induced bone circulation. Masada et al. (2008) reported that the activity of CYP3A was inversely proportional to the incidence and degree of steroid-induced ONFH. Guo and Deng (2019) found the same result in a meta-analysis. These animal studies suggest that low CYP3A activity in the liver is a risk factor for steroid-induced ONFH. Therefore, the exact location and biological function of SNPs in human CYP3A genes deserve further study. There is evidence of an association between the rs2242480 alleles and clopidogrel response variability in patients with coronary artery disease in Spanish patients (Angiolillo et al., 2006). Danielak et al. (2017) obtained inconsistent results in subsequent studies, suggesting that the rs2242480 locus may not be an important factor affecting the differences in pharmacokinetics and pharmacodynamics of clopidogrel. Based on previous studies, the association between CYP3A4 polymorphisms and steroid-induced ONFH has been demonstrated in the Chinese population. Wang et al. (2016) reported that the CYP3A4 gene rs2242480 polymorphisms were significantly associated with the risk of steroid-induced ONFH. They showed that the incidence of steroid-induced ONFH was significantly increased in CG genotype carriers, while the incidence of steroid-induced ONFH was decreased in TG genotype carriers. In this study, the T allele at CYP450 rs2242480 locus was found to be a protective factor that reduced the risk of steroid-induced ONFH; however, no correlation was found between the rs2242480 genotype and steroid-induced ONFH. Our study provides evidence of an association between the CYP450 rs2242480 polymorphism and steroid-induced ONFH risk. Although we carried out extensive statistical analyses, the ethnicity of the participants was limited to the Han Chinese population. Therefore, further analyses are needed to explain whether the understanding of the genetic and phenotypic characteristics of CYP450 and its interaction with environmental factors is applicable to other ethnicities.
The ABCB1 gene encodes the membrane transport protein P-gp, which is an important determinant of drug uptake, distribution, and elimination. The polymorphisms of ABCB1 have been shown to affect the expression and function of P-gp, and thus determine individual differences in drug resistance (Zhang et al., 2017). Several case–control studies have examined the link between ABCB1 polymorphisms and steroid-induced ONFH. Asano et al. (2003) reported that in 136 Japanese patients who had kidney transplants, 30 cases of steroid-induced ONFH occurred. The TT genotype of the rs1045642 locus of the ABCB1 gene showed a significant decrease. A significant increase in the steroid hormone dose–concentration ratio suggests an increase in p-gp activity in patients with the rs1045642 TT genotype, suggesting that rs1045642 TT genotype reduces the incidence of steroid-induced ONFH by enhancing p-gp pump activity. The rs2032582 polymorphism of the ABCB1 gene also showed a significantly decreased incidence rate of ONFH. Preliminary results indicate that the analysis of SNPs of ABCB1 gene before hormone therapy for related diseases has important significance for the identification of steroid-induced ONFH high-risk individuals. In addition, Yang and Xu (2007) found that these two SNPs reduced the incidence of steroid-induced ONFH. A meta-analysis showed that compared with rs1045642 CC genotype carriers, carriers of the rs1045642 TT genotype had one-third fewer ONFH patients, which further verified that TT genotype may be a protective factor for ONFH (Zhang et al., 2014). Xue et al. (2014) also found the protective effect of T allele (and the TT genotype) of rs1045642 in the Chinese population, yet there was no correlation between rs2032582, rs1128503, and steroid-induced ONFH. In addition, Gong et al. (2013) found no significant association between the rs2032582 polymorphism and steroid-induced ONFH risk in a meta-analysis. A contrary result showed that the rs1045642 polymorphism may be a risk factor for steroid-induced ONFH. The results of a He and Li et al. (2009) study were consistent with those of Gong et al. (2013), indicating that the rs1045642 polymorphism increases the susceptibility to ONFH. The inconsistencies in the above research results may be related to the differences in the research subjects and analytical methods selected for inclusion in each study. Because of the important role of ABCB1 gene in drug metabolism, we focused on the correlation between ABCB1 polymorphisms and steroid-induced ONFH. In this study, no correlation between the rs1045642 polymorphism and steroid-induced ONFH was found, which is different from other research results. We also found no alleles or genotypes of rs2032582C associated with steroid-induced ONFH. However, under the dominant model, the T allele of the rs2032582T locus of the ABCB1 gene was associated with steroid-induced ONFH risk reduction. Few studies have focused on the relationship between rs1128503 gene polymorphisms and steroid-induced ONFH. Therefore, we also analyzed the difference of rs1128503 polymorphisms in steroid-induced ONFH patients. The results of this study were consistent with those of Xue et al. (2014), indicating that the rs1128503 polymorphism was not associated with steroid-induced susceptibility. Different ethnic, demographic, geographical, and other environmental factors may lead to differences in conclusions drawn from the research.
The ABCB1 gene is highly polymorphic; its SNPs and some haplotypes may be related to the occurrence and development of steroid-induced ONFH. However, studies in several countries have shown conflicting results. Therefore, genetic differences cannot fully explain the variation observed among individuals, and many other factors need to be considered including the epigenetic state of the gene. Thus, to obtain more accurate research conclusions, we performed CpG island methylation studies. Our results showed that the CpG island loci ABCB1_1_192, ABCB1_2_32, and ABCB1_2_43were associated with osteonecrosis. The changes of DNA methylation status have potential effects on steroid-induced ONFH prediction and early diagnosis. Fourteen pairs of SNP-methylation sites within the ABCB1 proximal promoter were also found to interact. This suggests that genetic polymorphisms and DNA methylation play a synergistic role in the regulation of ABCB1 expression and may be used to predict steroid-induced ONFH. There are few reports on the interaction of genetic polymorphisms and epigenetic state the genes known to be associated with steroid-induced ONFH. In the future, more studies on epigenome-wide association and genome-wide association need to be carried out to find novel interacting factors to provide information on potential mechanisms of interindividual variation ability, so as to provide more information for further research on precision medicine.
Conclusion
In conclusion, our data suggest that the rs2032582T and rs2242480 SNPs may be associated with the risk of steroid-induced ONFH in the Han Chinese population. Further studies found that the rs2032582T SNP interacts extensively with DNA methylation levels. Therefore, the study of gene polymorphisms of steroid hormone metabolism, transport, and steroid hormone receptor and the synergistic effect of DNA methylation levels play an important role in understanding individual differences in patients' sensitivity to steroid hormones and predicting the possibility of steroid-induced ONFH occurrence.
Acknowledgments
We thank all the participants, hospital staff, and hospitals who provided blood samples for this study.
Ethical Approval
Ethics committee approval was received for this study from the Institute of Neurology of Anhui University of Chinese Medicine.
Author Disclosure Statement
The authors declare that they have no competing interests.
Funding Information
The work was supported by the scientific research fund of Anhui University of Chinese Medicine (no. 2018SYLCZ04).
References
- Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, et al. (2006) Contribution of gene sequence variations of the hepatic cytochrome P450 3A4 enzyme to variability in individual responsiveness to clopidogrel. Arterioscler Thromb Vasc Biol 26:1895–1900 [DOI] [PubMed] [Google Scholar]
- Asano T, Takahashi KA, Fujioka M, et al. (2003) ABCB1 C3435T and G2677T/A polymorphism decreased the risk for steroid-induced osteonecrosis of the femoral head after kidney transplantation. Pharmacogenetics 13:675–682 [DOI] [PubMed] [Google Scholar]
- Bodor M, Kelly EJ, Ho RJ, et al. (2005) Characterization of the human MDR1 gene. AAPS J 7:E1–E5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner-Ziegler S, Giurgea GA, Sunder-Plassmann R, et al. (2013) CYP2C9 genotype and association with bone mineral density: a pilot study. Gene 526:295–298 [DOI] [PubMed] [Google Scholar]
- Cui L, Zhuang Q, Lin J, et al. (2016) Multicentric epidemiologic study on six thousand three hundred and ninety-five cases of femoral head osteonecrosis in China. Int Orthop 40:267–276 [DOI] [PubMed] [Google Scholar]
- Danielak D, Karaźniewicz-Bada M, Wiśniewska K, et al. (2017) Impact of CYP3A4*1G allele on clinical pharmacokinetics and pharmacodynamics of clopidogrel. Eur J Drug Metab Pharmacokinet 42:99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Ota M, Hori H, et al. (2012) Association between the functional polymorphism (C3435T) of the gene encoding P-glycoprotein (ABCB1) and major depressive disorder in the Japanese population. J Psychiatr Res 46:555–559 [DOI] [PubMed] [Google Scholar]
- Gong LL, Fang LH, Wang HY, et al. (2013) Genetic risk factors for glucocorticoid-induced osteonecrosis: a meta-analysis. Steroids 78:401–408 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Deng M (2019) Correlation between steroid-induced osteonecrosis of the femoral head and hepatic CYP3A activity: a systematic review and meta-analysis. J Invest Surg 32:118–126 [DOI] [PubMed] [Google Scholar]
- He BX, Shi L, Qiu J, et al. (2014) The effect of CYP3A4*1G allele on the pharmacokinetics of atorvastatin in Chinese Han patients with coronary heart disease. J Clin Pharmacol 54:462–467 [DOI] [PubMed] [Google Scholar]
- He W, Li K (2009) Incidence of genetic polymorphisms involved in lipid metabolism among Chinese patients with osteonecrosis of the femoral head. Acta Orthop 80:325–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Peng C, Liu Y, et al. (2016) Genetic association of NOS1 exon18, NOS1 exon29, ABCB1 1236C/T, and ABCB1 3435C/T polymorphisms with the risk of Parkinson's disease: a meta-analysis. Medicine (Baltimore) 95:e4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong TD, Kim SM, Kim HJ, et al. (2015) CYP2C19 genotype and early ischemic lesion recurrence in stroke patients treated with clopidogrel. J Stroke Cerebrovasc Dis 24:440–446 [DOI] [PubMed] [Google Scholar]
- Kaneshiro Y, Takaoka K (2007) [Hepatic CYP3A activity in association with development of osteonecrosis of the femoral head]. Clin Calcium 17:902–909 [PubMed] [Google Scholar]
- Masada T, Iwakiri K, Oda Y, et al. (2008) Increased hepatic cytochrome P4503A activity decreases the risk of developing steroid-induced osteonecrosis in a rabbit model. J Orthop Res 26:91–95 [DOI] [PubMed] [Google Scholar]
- Onda K, Suzuki R, Tanaka S, et al. (2012) Decitabine, a DNA methyltransferase inhibitor, reduces P-glycoprotein mRNA and protein expressions and increases drug sensitivity in drug-resistant MOLT4 and Jurkat cell lines. Anticancer Res 32:4439–4444 [PubMed] [Google Scholar]
- Phabphal K, Geater A, Limapichat K, et al. (2013) The association between CYP 2C9 polymorphism and bone health. Seizure 22:766–771 [DOI] [PubMed] [Google Scholar]
- Rath PC, Chidambaram S, Rath P, et al. (2015) A study on the impact of CYP2C19 genotype and platelet reactivity assay on patients undergoing PCI. Indian Heart J 67:114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Beckmann J, Bohndorf K, et al. (2016) S3-Guideline non-traumatic adult femoral head necrosis. Arch Orthop Trauma Surg 136:165–174 [DOI] [PubMed] [Google Scholar]
- Sharif A, Kheirkhah D, Reza SM, et al. (2016) ABCB1-C3435T polymorphism and breast cancer risk: a case-control study and a meta-analysis. J BUON 21:1433–1441 [PubMed] [Google Scholar]
- Sheng X, Zhang L, Tong N, et al. (2012) MDR1 C3435T polymorphism and cancer risk: a meta-analysis based on 39 case-control studies. Mol Biol Rep 39:7237–7249 [DOI] [PubMed] [Google Scholar]
- Shi CJ, Wang F, Ren MF, et al. (2011) Up-regulation of ABCB1/P-glycoprotein by escaping promoter hypermethylation indicates poor prognosis in hematologic malignancy patients with and without bone marrow transplantation. Leuk Res 35:73–79 [DOI] [PubMed] [Google Scholar]
- Tanabe M, Ieiri I, Nagata N, et al. (2001) Expression of P-glycoprotein in human placenta: relation to genetic polymorphism of the multidrug resistance (MDR)-1 gene. J Pharmacol Exp Ther 297:1137–1143 [PubMed] [Google Scholar]
- Wang Y, Li X, Gao Y, et al. (2016) Genetic polymorphisms of CYP3A4 among Chinese patients with steroid-induced osteonecrosis of the femoral head. Medicine (Baltimore) 95:e5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei KK, Zhang LR (2015) Interactions between CYP3A5*3 and POR*28 polymorphisms and lipid lowering response with atorvastatin. Clin Drug Investig 35:583–591 [DOI] [PubMed] [Google Scholar]
- Weinstein RS. (2012) Glucocorticoid-induced osteonecrosis. Endocrine 41:183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein RS, Hogan E, Borrelli MJ, et al. (2017) The pathophysiological sequence of glucocorticoid-induced osteonecrosis of the femoral head in male mice. Endocrinology 158:3817–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhao ZQ, Hong D, et al. (2014) MDR1 gene polymorphisms are associated with glucocorticoid-induced avascular necrosis of the femoral head in a Chinese population. Genet Test Mol Biomarkers 18:196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wang X, Peng L, et al. (2015) CYP2C19 LOF alleles confer no risk for HTPR but higher risk for recurrent ischemic events in clopidogrel treated elderly ACS patients. Int J Cardiol 189:225–227 [DOI] [PubMed] [Google Scholar]
- Yang XY, Xu DH (2007) MDR1(ABCB1) gene polymorphisms associated with steroid-induced osteonecrosis of femoral head in systemic lupus erythematosus. Pharmazie 62:930–932 [PubMed] [Google Scholar]
- Zalavras C, Dailiana Z, Elisaf M, et al. (2000) Potential aetiological factors concerning the development of osteonecrosis of the femoral head. Eur J Clin Invest 30:215–221 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kong X, Wang R, et al. (2014) Genetic association of the P-glycoprotein gene ABCB1 polymorphisms with the risk for steroid-induced osteonecrosis of the femoral head in Chinese population. Mol Biol Rep 41:3135–3146 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xie H, Zhao D, et al. (2017) Association of ABCB1 C3435T polymorphism with the susceptibility to osteonecrosis of the femoral head: a meta-analysis. Medicine (Baltimore) 96:e6049. [DOI] [PMC free article] [PubMed] [Google Scholar]